Abstract
An Arabidopsis (Arabidopsis thaliana) L. Heynh mutant deficient in an isoform of adenylate kinase (ADK; At2g37250) was isolated by reverse genetics. It contains a T-DNA insertion 377 bp downstream of the start point of transcription. The mutant lacks At2g37250 transcripts and has a mild reduction in total cellular ADK activity. Green fluorescent protein-fusion based cellular localization experiments, carried out with the full-length At2g37250, suggested a plastidial localization for this isoform. In keeping with this observation, organelle isolation experiments revealed that the loss in ADK activity was confined to the inner plastid. This plastid stroma ADK gene was found to be expressed tissue constitutively but at much higher levels in illuminated leaves. Phenotypic and biochemical analyses of the mutant revealed that it exhibited higher amino acid biosynthetic activity in the light and was characterized by an enhanced root growth. When the mutant was subjected to either continuous light or continuous dark, growth phenotypes were also observed in the shoots. While the levels of adenylates were not much altered in the leaves, the pattern of change observed in the roots was consistent with the inhibition of an ATP-consuming reaction. Taken together, these data suggest a role for the plastid stromal ADK in the coordination of metabolism and growth, but imply that the exact importance of this isoform is tissue dependent.
Adenylate kinase (ADK; EC 2.7.4.3) catalyzes a reversible transphosphorylation reaction interconverting ADP to ATP and AMP and as such is considered a key enzyme in energy metabolism (Noda, 1973; Pradet and Raymond, 1983). The enzyme is ubiquitous in all living organisms and has been found in different subcellular locations, including the cytosol, mitochondria, and plastids of a wide range of plant species, such as spinach (Spinacia oleracea), wheat (Triticum aestivum), barley (Hordeum vulgare; Birkenhead et al., 1982), tobacco (Nicotiana tabacum; Schlattner et al., 1994), maize (Zea mays; Kleczkowski and Randall, 1991), potato (Solanum tuberosum; Roberts et al., 1997; Regierer et al., 2002), pea (Pisum sativum; Zancani et al., 2001), and rice (Oryza sativa; Kawai and Uchimiya, 1995). In early studies, a wide range of postulates as to the in vivo function of the various isoforms of this enzyme were made on the basis of the correlative data sets. For example, the activities of specific isoforms of ADK appeared to be linked to relative growth rates (Glaser et al., 1975; Goelz and Cronan, 1982), alterations in light regimes (Schiltz et al., 1994), or the maintenance of local variations in nucleotide ratios in order to control the transport of adenylates across membranes (Fricaud et al., 1992; Gellerich, 1992). When taken together, this evidence suggests that changes in isoform expression most probably reflect the energy metabolism of the subcellular compartment that they reside in.
Recently, ADK-encoding genes have been cloned from a wide variety of species (for details, see Regierer et al., 2002). However, only in the cases of maize, rice, and potato have the products of these genes been directly proven to catalyze the ADK reaction (Kawai et al., 1992; Chen et al., 1994; Regierer et al., 2002). To date, very little characterization of the functional roles played by the various isoforms of ADK has been attempted. There are two notable exceptions to this statement, both of which concentrated on the metabolism of the potato tuber. Firstly, the role of the mitochondrial ADK was assessed in isolated tuber mitochondria utilizing a 32P-NMR approach (Roberts et al., 1997). These studies suggested that, immediately following the addition of ADP to isolated tuber mitochondria, ATP synthesis exceeded the rate of oxidative phosphorylation due to a mitochondrial ADK activity that is 2 to 4 times the maximum activity of ATP synthase. They further demonstrated that a pool of sequestered ATP in mitochondria enables ADK and ATP synthase to convert AMP to ATP in the presence of exogenous inorganic phosphate. During this conversion, ADK activity can indirectly influence rates of oxidation of both succinate and NADH via changes in mitochondrial ATP levels. Therefore, ADK activity has the potential both to affect the amount of ADP available for ATP synthesis and to regulate electron transport via the level of ATP. More recently, a reverse genetic approach was taken to assess the role of a plastidial isoform of ADK with respect to metabolism and morphology of the tuber (Regierer et al., 2002). In this previous study, we found that a deficiency of this isoform led to an increase in tuber growth and a concomitant increase in both tuber starch and amino acid content. Furthermore, when assessed within the framework of metabolic control analysis, these results suggest that the plastidial ADK has a very strong negative control coefficient for starch synthesis (Geigenberger et al., 2004), implying that it competes for ATP with starch (and amino acid) biosynthesis under normal physiological conditions.
In this study, we have focused on the characterization of an Arabidopsis (Arabidopsis thaliana) T-DNA knockout of a gene encoding an ADK, at both molecular and physiological levels. Here we confirm the subcellular localization of this enzyme by means of both cellular fractionation studies and by green fluorescent protein (GFP) localization experiments. We additionally carried out an expression analysis at the tissue level and in leaves throughout a diurnal period. Finally, we studied the consequence of this gene mutation on (aerial and root) growth and metabolic regulation under a range of different light regimes. The data will be discussed in the context of current models of amino acid metabolism in plant tissues.
RESULTS
Putative ADKs of Arabidopsis
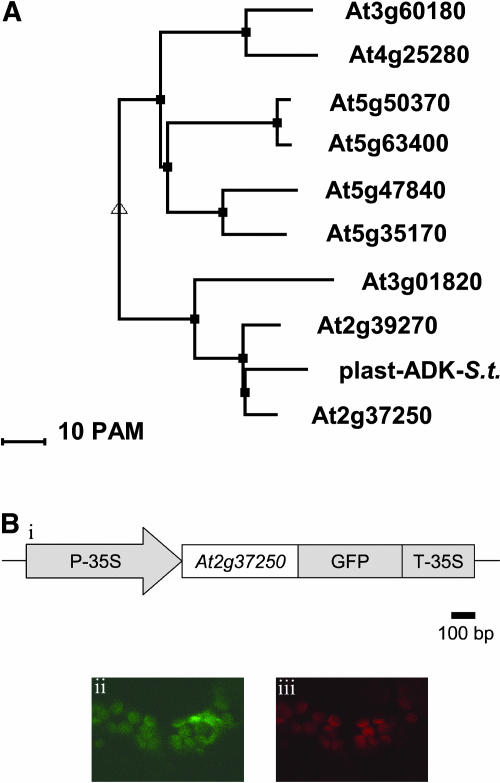
In silico screening of the Arabidopsis genome with the plastidial ADK isoform of potato (Regierer et al., 2002) revealed nine genes of very high sequence homology (e-values under e−20). When analysis was performed at the nucleotide level, these genes formed four phylogenetic clusters (Fig. 1A). The majority of these genes are presently annotated as putative ADKs (or even as unknown proteins; http://mips.gsf.de/proj/thal/db/); their transcription has, however, been verified by microarray experimentation (Yamada et al., 2003). The predicted ADK-encoding gene, At2g37250, fell into the same phylogenetic group as the potato plastidial isoform of ADK− with which it showed 76% identity (Fig. 1A). Despite these similarities, it is important to note that, as yet, none of the Arabidopsis genes have been functionally characterized and that the subcellular localizations presented here were determined merely on the basis of bioinformatic prediction (Emanuelsson et al., 2000; Bendtsen et al., 2004; Small et al., 2004).
Figure 1.
Arabidopsis ADK gene family and subcellular localization of the At2g37250 protein. A, Dendrogram of Arabidopsis sequences homologs to plant ADKs including the plastidial potato Adk (plast-ADK-S.t., accession no. AF411937). Alignments were produced using MULTALIN software. B, Subsection i, GFP fusion construct used for particle bombardment experiments. Detached young leaves of Arabidopsis were transiently transformed with this construct, and after 2 d in darkness the leaves were examined by confocal laser scanning microscopy. Subsection ii, Confocal fluorescence image of a mesophyll cell when excited at 488 nm. Subsection iii, Chlorophyll autofluorescence of the same cells above 520 nm.
In order to confirm the localization of the protein encoded by At2g37250, the complete coding region was fused, at the carboxyl-terminal end, to an enhanced GFP (EGFP) and transiently expressed in leaf cells (Karimi et al., 2002; Fig. 1B). In these cells EGFP fluorescence was detected coincident with chlorophyll autofluorescence (Fig. 1B, ii and iii), suggesting a plastidial location of the protein. When taken together with the structural similarity of this gene to the plastidial isoform of ADK from potato, this subcellular localization provides compelling molecular evidence that this gene is the Arabidopsis ortholog to StpAdK.
Isolation and Genetic Characterization of an Arabidopsis Mutant Harboring a T-DNA Insertion within At2g37250
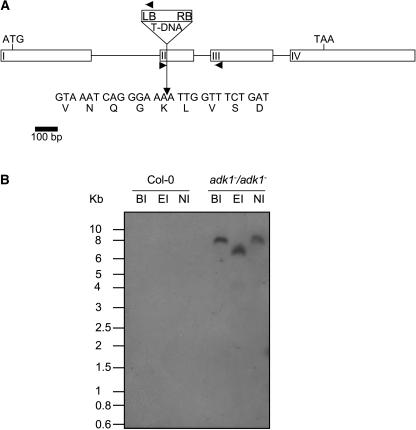
A plant carrying a T-DNA insertion within At2g37250 (from here on named Atpadk1) was identified by PCR screening of the T-DNA insertional-mutant population GABI-Kat (Rosso et al., 2004). A plant homozygous for the mutation was identified by PCR screening of a segregating population. Sequencing of a PCR fragment encompassing the T-DNA/genomic DNA junction revealed that this line carries an insertion within the coding sequence (second exon) of the Atpadk1 gene (Fig. 2A). Segregation analysis of the selectable marker (data not shown) and Southern-blot hybridization (Fig. 2B) indicated that this mutant carries a single T-DNA insertion.
Figure 2.
Isolation and genetic characterization of an Arabidopsis mutant harboring a T-DNA insertion within the At2g37250 gene (Atpadk1). A, The mutant adk1-1, obtained by PCR screening of a T-DNA mutant collection (GABI-Kat), carries an insertion into the second exon of adk1. Arrows on T-DNA and exons II and III denote primer positions for T2 and T3 population screening. LB, Left border; RB, right border. Boxes numbered in Roman numerals denote the exons of the gene. B, Southern blot of genomic DNA from Arabidopsis genotypes digested with the indicated restriction enzymes (EI, EcoRI; BI, BamHI; and NI, NdeI). The membrane was hybridized with a probe spanning 500 bp of the left border of the T-DNA. Numbers on the left indicate molecular masses in kilobases.
Expression Analysis of Atpadk1
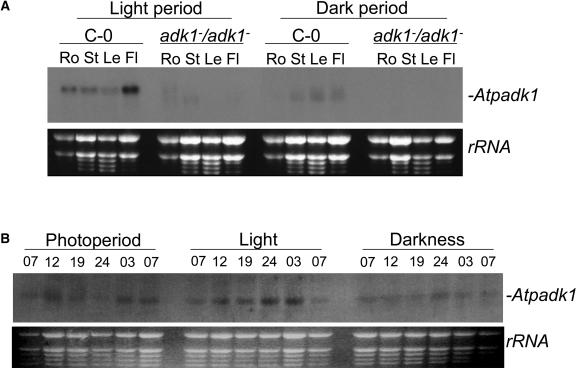
Expression analysis of Atpadk1 in different tissues of wild-type plants showed that the transcript is present in all the organs tested, suggesting a tissue-constitutive expression of the gene (Fig. 3A) with maximal expression observed in floral tissue. The homozygous Atpadk1 mutant, however, completely lacked expression (Fig. 3A). Expression of Atpadk1 was also studied throughout the diurnal cycle in wild-type plants grown for 28 d in a 16-h-light/8-h-dark regime followed by 2 d of growth in which plants were either (1) maintained in the same growth conditions, (2) subjected to continuous light, or (3) subjected to continuous darkness (Fig. 3B). There was a general tendency of increased gene expression in the light, which is consistent with previous observations in rice and potato (Kawai et al., 1992; Regierer et al., 2002), and, consequently, a depressed level of expression during the dark period. However, Atpadk1 appears to be expressed throughout the diurnal cycle (Fig. 3B). On subjection to continuous light, an altered periodicity in Atpadk1 expression was observed with maximal expression coincident with what was the middle of the dark period. In continuous darkness a faint band is observed throughout the time course.
Figure 3.
Expression analysis of Atpadk1 mRNA. A, Northern blot of total RNA extracted from different tissues of wild type (C-0) and homozygous mutant (adk1−/adk1−) in the middle of the light and dark periods. Ro, Root; St, stem; Le, leaf; and Fl, flower. B, Northern blot of leaves RNA from wild-type plants growing under conditions described in “Materials and Methods.” Bottom sections show rRNA as a loading control.
Determination of ADK Activity Levels
We assayed the enzyme activity in both total cell extracts and isolated chloroplasts of the wild type and mutant (Table I). We observed that approximately 80% to 90% of the ADK activity is localized to the plastid in Arabidopsis. This is quantitatively very similar to the distribution observed previously in tobacco cells and leaves (Schlattner et al., 1993). When assayed in the absence of detergent, there was only a minor (and not significant) decrease in both total cellular and plastidial ADK activity of the homozygous mutant. However, in the presence of detergent, the plastidial activity measured in the mutant was 50% of that measured in the wild type.
Table I.
ADK activity in leaves from Col-0 and adk1−/adk1− mutant plants
Arabidopsis leaves from 3-week-old plants were sampled after 12 h in dark, and chloroplasts were isolated as described by Tauberger et al. (2000). ADK activity was measured in both the plastidial fraction and the supernatant with and without Triton (0.01%) following the protocol described by Kleczkowski and Randall (1986). Values are expressed in nmol min−1 mg protein−1 ± se and are means from three replicates. Numbers in bold type denote significant differences determined by the Student's t test (P < 0.05) from Col-0.
| Triton | Col-0 | Atpadk1 | |
|---|---|---|---|
| 0.00% | Total | 2.22 ± 0.05 | 1.84 ± 0.47 |
| Supernatant | 0.22 ± 0.08 | 0.53 ± 0.06 | |
| Plastid | 2.02 ± 0.49 | 1.30 ± 0.68 | |
| 0.01% | Total | 1.53 ± 0.44 | 1.05 ± 0.24 |
| Supernatant | 0.31 ± 0.08 | 0.48 ± 0.01 | |
| Plastid | 1.21 ± 0.09 | 0.56 ± 0.18 |
Phenotypic Characterization of Atpadk1 Mutant Plants
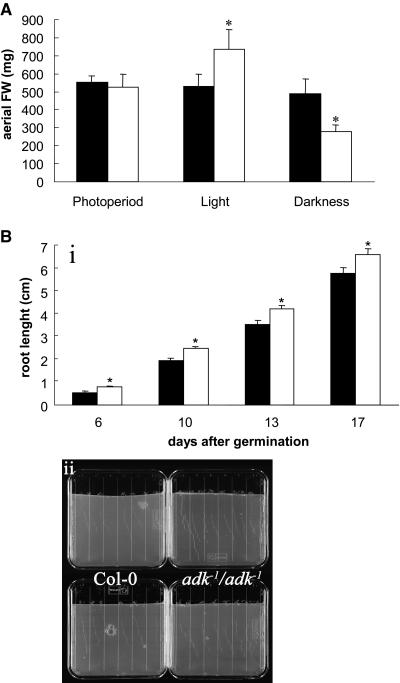
We next grew the mutant in soil under different light regiments (long day, continuous day, and continuous night) and scored morphological parameters of the aerial parts of the plants. Mutant plants growing in a 16-h-light/8-h-dark photoperiod were indistinguishable from those of the control. However, after 2 d in continuous light, the total aerial biomass accumulated by the mutant was 40% higher than the wild type. Intriguingly, a contrasting result was achieved following 2 d in continuous dark (Fig. 4A). In normal photoperiod and continuous light, both the wild type and mutants developed to the reproductive stage synchronically, but in darkness the mutant plants showed a delay of about 2 to 3 d to reach the flowering phase.
Figure 4.
Phenotypic characterization of Arabidopsis adk1− homozygous mutants. A, Fresh weight accumulation in the aerial part of adk1−/adk1− plants. Homozygous mutants were grown under conditions described in “Materials and Methods,” and after 28 d they were transferred to different light regimes for two further days (photoperiod: 16 h light/8 h dark, continuous light and continuous darkness). The entire aerial part of six plants per genotype was detached and weighed. Values are mean ± se of these measurements; asterisk indicates values determined by the t test to be significantly different (P < 0.05) from the control. B, Root length of adk1−/adk1− plants through 17 d after germination. Subsection i, Six to eight seeds per genotype were germinated in vertical plates for 1 d in darkness and then transferred to a 16-h-light/8-h-dark regime. Root lengths were recorded at the indicated times. Values are mean ± se of these measurements; asterisks indicate values determined by the Student's t test to be significantly different (P < 0.05) from the control. Subsection ii, Representative photos of vertical plates 13 d after germination.
In order to analyze root growth, we germinated seeds on vertical agar plates and recorded root length every 3 to 4 d. As shown in Figure 4B, the root lengths of the homozygous mutant plants were between 20% and 40% longer than that those of Col-0.
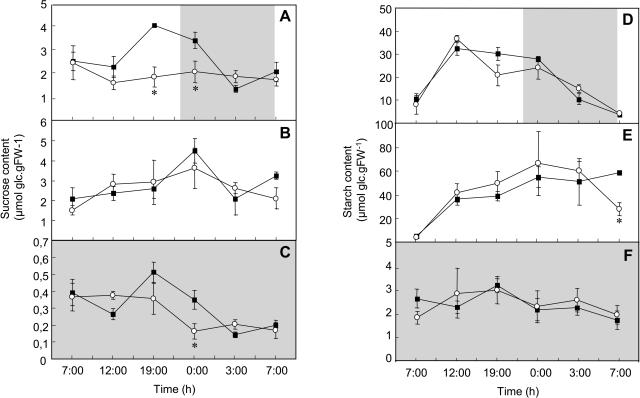
Effect of a Deficiency of Plastidial ADK on Diurnal Carbohydrate Levels
We next evaluated the levels of the major leaf carbohydrates through a diurnal period. In long-day conditions, the Atpadk1 mutant clearly accumulated little or no Suc despite containing similar levels as the wild type at the beginning of the light period (Fig. 5A). There was, however, little difference between the genotypes with respect to starch content (Fig. 5D). Given that the ADK reaction is intimately associated with the level of ATP, we chose to alter the production of this metabolite by subjecting both the wild type and Atpadk1 mutant to continuous light for a period of 2 d before sampling. Although both genotypes displayed differences in the patterns of Suc and starch accumulation (Fig. 5, B and E), there was surprisingly little difference between the genotypes under these conditions (with the exception of a significant decrease in the level of starch in the mutant at the end of the experiment). Similarly, under continuous darkness there was little difference in the levels of Suc (Fig. 5C) or starch (Fig. 5F). The Glc and Fru levels in leaves showed similar patterns when growing the plants under both continuous light and darkness, and no differences between both genotypes were observed (data not shown), except under conditions of continuous darkness when the Glc contents showed a marked increment at 12 am in leaves from the mutant plants (1.26 ± 0.35 and 5.31 ± 0.72 μmol Glc g fresh weight (FW)−1 ± se for Col-0 and Atpadk1, respectively).
Figure 5.
Carbohydrate contents in leaves of adk1−/adk1− Arabidopsis plants. Diurnal changes in Suc (A, B, and C) and starch (D, E, and F) contents in leaves of 2-week-old plants growing under different growth regimes (A and D, photoperiod 16/8; B and E, continuous light; and C and F, continuous darkness). At each time point, samples were taken from mature source leaves, and the data represent the mean ± se of measurements from four to six plants per genotype. Black squares, Col-0; white circles, adk1−/adk1−. Asterisk indicates values determined by the Student's t test to be significantly different (P < 0.05) from the control.
Nucleotide Pool Sizes of Atpadk1 Mutant Plants
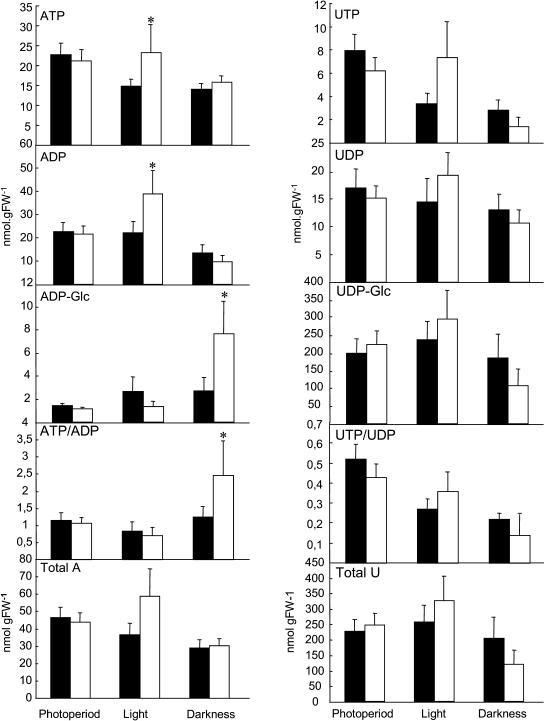
We next determined the levels of nucleotides in plants growing under the various light regimes described above. For this purpose, we determined the levels of ATP, ADP, ADP-Glc, UTP, UDP, and UDP-Glc by HPLC of trichloroacetic acid extracts of leaves from both genotypes as described by Fernie et al. (2001a). The levels of these metabolites were surprisingly indistinguishable between the two genotypes grown under the normal photoperiod (Fig. 6). In contrast, following 2 d in continuous light, the levels of both ADP and ATP were elevated in the mutant with respect to those observed in the wild-type control, whereas in plants that were subjected to continuous dark for 2 d, the ADP-Glc level and the deduced ATP/ADP ratio were elevated in the mutant with respect to the wild type. The levels of the uridinylates were, however, invariant between the genotypes in all light regimes tested.
Figure 6.
Nucleotide levels in leaves of adk1− Arabidopsis plants. Plants were grown as described for Figure 5, and samples were taken 6 h into the photoperiod. Black bars correspond to Col-0 plants and white ones to the homozygous mutant (adk1−/adk1−). Values are means ± se of measurements from four to six plants per genotype. Asterisks indicate those that were determined by the Student's t test to be significantly different (P < 0.05) between genotypes.
For analysis of root material, we grew the two genotypes in hydroponic conditions and collected root samples for nucleotide and amino acid determination. Given that the levels of nucleotides were so low in the roots, it was difficult to detect some of the nucleotides following the HPLC method used above. Therefore, we also used spectrophotometric cycling assays to determine the levels of ATP and ADP (Gibon et al., 2002). The levels of ADP and total adenylates were significantly lower in the mutant while the ATP/ADP ratio was higher, suggesting that the roots had an impaired energy status (Table II). As was observed in the leaves from this mutant, there were no differences in the root uridinylate status (although it should be noted that in this instance we were not able to detect UTP).
Table II.
Nucleotide levels in roots of 3-week-old plants of the Arabidopsis genotypes
Roots were harvested 6 h into the photoperiod from hydroponic cultures as described in “Materials and Methods.” Samples were frozen immediately in liquid N2 and extracted in trichloroacetic acid. Values are means ± se from four to six plants per genotype. Values set in bold type indicate those determined by the Student's t test to be significantly different (P < 0.05) from Col-0.
| Nucleotide | Col-0 | Atpadk1 |
|---|---|---|
| nmol g FW−1 | ||
| ATP | 6.0 ± 1.1 | 3.2 ± 1.2 |
| ADP | 4.4 ± 1.3 | 1.0 ± 0.5 |
| ATP/ADP | 2.4 ± 0.9 | 3.5 ± 0.7 |
| Σ Adenylate | 10.4 ± 2.2 | 4.1 ± 1.5 |
| UDP-Glc | 4.2 ± 1.2 | 3.0 ± 0.6 |
| UDP | 19.0 ± 7.8 | 17.3 ± 3.8 |
Amino Acid Contents of Atpadk1 Mutant Plants
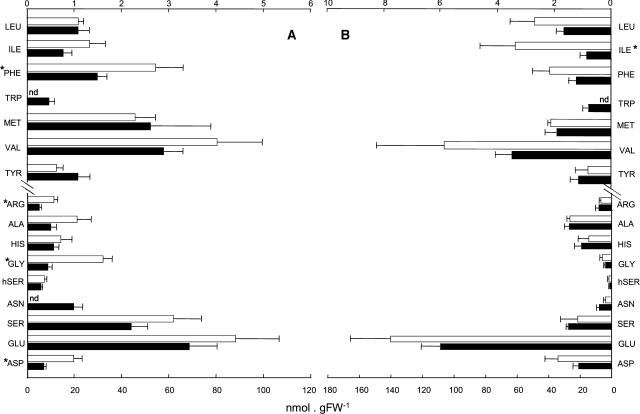
Given that the plastid is the predominant location for the biosynthesis of many amino acids (for example Arg, Met, His, Trp, Lys, Iso, Phe, and Tyr) and that these processes carry a heavy ATP burden (Regierer et al., 2002), we were particularly interested to investigate the amino acid composition in leaves of the Atpadk1 plants. Figure 7 shows the results obtained following determination of the amino acid composition of leaves in the middle of the light (Fig. 7A) and dark (Fig. 7B) periods of the diurnal cycle. The levels of almost all amino acids are marginally higher in the mutant plants (significantly so in the cases of Asp, Gly, Arg, and Phe; Fig. 7A). In sharp contrast, with the exception of elevated Iso contents (Fig. 7B), the amino acid levels are not significantly different between genotypes during the dark period.
Figure 7.
Amino acid levels in leaves of adk1− Arabidopsis plants measured 6 h into either the light (A) or the dark (B) periods. Plants were grown as described for Figure 5. Black bars correspond to Col-0 plants and white ones to homozygous mutant (adk1−/adk1−). Values are means ± se of measurements from four to six plants per genotype. Asterisks indicate those that were determined by the Student's t test to be significantly different between genotypes. Nd, Not detectable.
The amino acid contents of root material from plants grown in hydroponic conditions were also analyzed (Table III). The mutant displayed a tendency of decreased root amino acid content under long-day conditions when assessed on a per gram FW basis; however, this was only statistically significant in the cases of Gly, β-Ala, and γ-aminobutyric acid (and in no instance when analyzed on a per plant basis; see Supplemental Table I, available at www.plantphysiol.org). However, when the plants were subjected to continuous light for 2 d, the levels of amino acids in the roots of the mutant were significantly higher than those of the wild-type control. This was true for all the measured amino acids, with the exception of Gly, Thr, Met, Trp, Phe, Ile, and Lys, and it was particularly pronounced in the cases of Arg, Val, and Leu. Somewhat intriguingly, the pattern of change in the mutant following transfer to continuous light appears to be the opposite of that observed in the wild type. In contrast, the change of both genotypes following transfer to continuous dark was largely conserved, with the root amino acid content generally increasing in both cases. However, the amino acids Gln, Thr, Ala, and Met did not follow this trend since they were either decreased or unaltered in the mutant with respect to plants grown under long-day conditions and as such were present at significantly lower levels than those found in the wild-type roots subjected to continuous darkness.
Table III.
Amino acid content in Arabidopsis roots
Wild type (Col-0) and adk−1/adk−1 plants were grown under various light regimes, and after 2 weeks roots were harvested, extracted, and were quantified by HPLC as described in “Materials and Methods.” Data are means of replicates ± se from six individual plants and presented as nmol g FW−1. Values in bold type denote those determined to be significantly different (P < 0.05) from Col-0 by the Student's t test.
| Amino Acid
|
Photoperiod (16/8)
|
Light
|
Darkness
|
|||
|---|---|---|---|---|---|---|
| Col-0 | adk−1/adk−1 | Col-0 | adk−1/adk−1 | Col-0 | adk−1/adk−1 | |
| Asp | 1,280 ± 210 | 900 ± 100 | 1,000 ± 70 | 2,520 ± 400 | 1,480 ± 210 | 860 ± 200 |
| Glu | 2,190 ± 370 | 1,500 ± 280 | 1,500 ± 150 | 3,100 ± 340 | 2,620 ± 590 | 1,500 ± 280 |
| Asn | 1,200 ± 240 | 790 ± 160 | 1,000 ± 170 | 1,660 ± 150 | 3,050 ± 750 | 2,100 ± 450 |
| Ser | 920 ± 150 | 660 ± 90 | 610 ± 110 | 1,440 ± 200 | 1,480 ± 310 | 1,100 ± 300 |
| Gln | 3,110 ± 610 | 2,200 ± 420 | 2,000 ± 190 | 4,400 ± 680 | 3,130 ± 820 | 1,300 ± 290 |
| Gly | 110 ± 20 | 60 ± 5 | 60 ± 3 | 110 ± 40 | 60 ± 20 | 70 ± 20 |
| h-Ser | 20 ± 6 | 13 ± 4 | 16 ± 3 | 40 ± 3 | 80 ± 7 | 60 ± 10 |
| Thr | 890 ± 180 | 500 ± 90 | 750 ± 72 | 1,940 ± 510 | 1,940 ± 250 | 880 ± 190 |
| His | 100 ± 20 | 60 ± 20 | 70 ± 13 | 150 ± 20 | 390 ± 100 | 260 ± 60 |
| β-Ala | 30 ± 6 | 20 ± 2 | 30 ± 7 | 25 ± 1 | 30 ± 4 | 20 ± 5 |
| Ala | 540 ± 130 | 260 ± 90 | 390 ± 60 | 60 ± 4 | 750 ± 270 | 190 ± 30 |
| Arg | 120 ± 80 | 130 ± 90 | 70 ± 50 | 920 ± 160 | 440 ± 140 | 570 ± 140 |
| GABA | 1,630 ± 220 | 950 ± 160 | 1,350 ± 170 | 2,640 ± 360 | 2,240 ± 440 | 1,650 ± 450 |
| Tyr | 30 ± 4 | 30 ± 10 | 20 ± 1 | 60 ± 5 | 410 ± 100 | 260 ± 60 |
| Val | 180 ± 30 | 140 ± 30 | 120 ± 10 | 340 ± 20 | 1,180 ± 270 | 720 ± 160 |
| Met | 40 ± 8 | 30 ± 4 | 30 ± 2 | 30 ± 7 | 30 ± 3 | 13 ± 3 |
| Trp | 20 ± 2 | 20 ± 3 | 13 ± 2 | 16 ± 3 | 160 ± 50 | 100 ± 20 |
| Phe | 47 ± 8 | 30 ± 4 | 30 ± 2 | 40 ± 9 | 110 ± 30 | 80 ± 10 |
| Ile | 46 ± 7 | 40 ± 10 | 40 ± 3 | 100 ± 10 | 520 ± 120 | 330 ± 70 |
| Leu | 60 ± 7 | 70 ± 20 | 50 ± 4 | 160 ± 10 | 690 ± 160 | 380 ± 90 |
| Lys | 200 ± 50 | 100 ± 40 | 210 ± 50 | 110 ± 14 | 390 ± 90 | 480 ± 160 |
Carbohydrate Contents in Roots and Seeds of Atpadk1 Mutant Plants
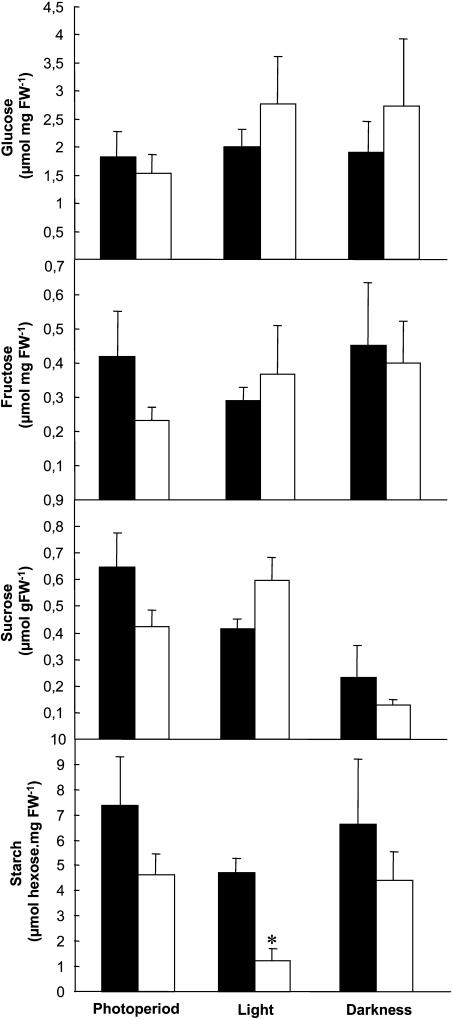
No differences were observed in root carbohydrate content of the two genotypes, with the exception that the starch level in Atpadk1 roots was significantly lower than that of the wild-type control genotype under continuous light conditions (Fig. 8). Given that seeds are the major storage organs in Arabidopsis, we also determined the levels of starch and total protein in mature seeds from six homozygous mutant plants and their corresponding controls. Mature seeds of Col-0 and Atpadk1 contained 26.3 ± 0.8 and 24.4 ± 0.6 μmol of starch (expressed as Glc equivalents per mg dry weight−1 ± se), respectively, while the total protein content of these seeds was 62.7 ± 7.6 and 73.4 ± 8.9 mg (expressed per g dry weight ± se) for Col-0 and Atpadk1 genotypes, respectively.
Figure 8.
Carbohydrate contents in roots of adk1−/adk1− Arabidopsis plants. Plants were grown as described for Figure 5, and samples were taken 6 h into the photoperiod. Black bars correspond to Col-0 plants and the white ones to homozygous mutant (adk1−/adk1−). Values are means ± se of measurements from four to six plants per genotype. Asterisks indicate those that were determined by the Student's t test to be significantly different between genotypes (P < 0.05).
DISCUSSION
Identification of an Arabidopsis T-DNA Insertion Mutant in the Coding Region of At2g37250: A Plastidially Localized ADK
Here we report the novel functional characterization of a gene encoding a plastidial ADK. Evidence in support of the location of this isoform ranges from mere sequence homology to GFP fusion-based cellular-localization studies and the direct determination of subcellular activities of ADK in a verified T-DNA insertion mutant of the gene in question. The subcellular fractionation studies revealed that this mutant was affected only in the activity of this plastidial isoform with no changes apparent in extraplastidial fractions. Several reports have documented the presence of two plastidially localized isoforms of ADK in plants (see Schlattner et al., 1993, 1994), one located in the stroma, the other tightly bound to the envelope of the intermembrane space (Murakami and Strotmann, 1978; Igamberdiev and Kleczkowski, 2003). That the loss of activity on treatment with detergent was similar in the mutant and its corresponding wild type suggests that the gene At2g37250 encodes the nonbound stromal isoform.
Expression analyses revealed that this gene exhibits tissue-constitutive expression and is elevated in the illuminated leaf. These observations are in keeping with those previously made in a range of species. Alterations in the light regime or exogenous supply of Glc have been reported to lead to the induction or suppression of distinct ADK isoform activities of tobacco (Haertle, 1977; Wagner et al., 1983), while the levels of this specific isoform of ADK have been demonstrated to be Suc inducible in rice (Kawai et al., 1992). In addition, this gene was expressed at very high levels in floral tissues, perhaps reflecting the high energy demand in this tissue. However, despite its high level of expression, a deficiency in the plastidial ADK did not impair floral development or pollen viability, as has previously been observed on down-regulation of the TCA cycle (Landschütze et al., 1995; Yiu et al., 2003; Fernie et al., 2004).
Effect of Plastidial ADK Deficiency on Leaf Metabolism and Shoot Growth
Deficiency of the plastidial ADK led to no visible phenotype alteration in long-day growth conditions. However, a significant increase in aerial growth was observed in the mutant when the plants were subjected to 24 h of light. Conversely, plant growth was stunted in the mutant when incubated in continuous darkness. It seems likely that this is a consequence of an altered capacity for ATP production between these conditions with the greatest growth occurring in the presence of the most light. When assessed alongside the expression pattern of this gene, it is intriguing that there is such a large effect on growth in samples grown in continuous darkness since it is much less expressed under these conditions. This result therefore suggests a role for this enzyme in both illuminated and darkened leaves. For this reason, we decided to evaluate the adenylate levels of leaves from plants grown under the various growth conditions. While there was no change in total cellular adenylate status of plants grown in long-day conditions, we observed elevated levels of ATP and ADP in plants kept in continuous light and an elevated ratio of ATP/ADP in plants incubated in continuous darkness. These data, however, are not particularly informative with respect to the phenotypes observed for two reasons. First, they represent the total cellular pool sizes and thus provide no information on the local subcellular concentrations, and second they represent only steady-state levels and therefore do not necessarily provide a true reflection of the energy status of the genotypes. Since it is not technically feasible to measure the subcellular concentrations of adenylates in Arabidopsis plants, we decided to focus our efforts on gauging the energy requirements of the mutant by evaluating the levels of metabolites whose biosynthesis is ATP dependent. Analysis of starch and soluble-sugar levels revealed that these metabolites were largely unaltered with respect to the mutant. This result is at first sight surprising since potato tubers exhibiting a dramatic reduction in the activity of the plastidial ADK were characterized by an approximately 60% increase in starch content (Regierer et al., 2002). However, this difference is most probably due to the different control structure that is apparent in heterotrophic and photosynthetic plant tissues. The supply of ATP to the plastid has a dominant contribution to the control of starch synthesis in the former (Tjaden et al., 1998; Geigenberger et al., 2004), but AGPase or a combination of AGPase, plastidial phospho-Glc isomerase, and plastidial phosphoglucomutase dominate leaf starch synthesis (Neuhaus et al., 1989; Neuhaus and Stitt, 1990; Stitt, 1990). This is most probably best rationalized by the fact that photosynthesis in the chloroplast can produce sufficient ATP to support starch synthesis in the light, whereas in the amyloplast, plastid glycolysis does not suffice as a source of ATP and energy must be imported from the cytosol. In contrast, the levels of almost all the amino acids are marginally increased in the mutant in the light period, whereas these levels were not altered in the dark period. These results, therefore, imply an elevated rate of amino acid biosynthesis in leaves of the mutant during the day. This is particularly interesting since it would be expected during illumination that the ATP produced by photosynthesis would force the ADK reaction in the direction of ADP production. Thus, a deficiency in ADK most probably would lead to an increased availability of ATP for plastidial biosynthesis. In contrast, in the dark, when ATP is no longer produced by photosynthesis, the ADK reaction most likely operates in the other direction, i.e. utilizing ADP to make ATP, and there are no changes in amino acid content. However, it is important to note that these analyses are based on steady-state metabolite analyses and as such do not provide direct evidence of alterations in the rate of synthesis of the various amino acids.
Effect of Plastidial ADK Deficiency on Root Metabolism, Root Growth, and Seed Metabolism
The mutant plants were also characterized by significantly elevated root growth. A similar observation was made on studying the major heterotrophic organs of potato plants exhibiting decreased expression of the homologous gene (Regierer et al., 2002). These potato plants were characterized by up to an 80% increase in total tuber yield as well as significant increases in starch and amino acid content and an increase in total adenlyate concentration. The pattern of change in the adenlyate pools of the Arabidopsis roots were somewhat different, with a significant decrease being observed in the level of ADP and an overall decline in the total adenlyate level when assessed on a per gram FW basis. Such a pattern of change is consistent with our previous speculations that this isoform of ADK is normally operating in the ATP-consuming direction in vivo (Regierer et al., 2002; Geigenberger et al., 2004).
Roots harvested from plants grown under long-day conditions also exhibited little change in amino acid content since, while the mutant contained slightly less amino acids when assessed on a per gram FW basis, the total content of the individual amino acid pools of the roots was unchanged. This result was somewhat surprising to us since the majority of amino acids increased in the potato tuber (Regierer et al., 2002), and several amino acid contents were higher in the mutant leaves. We have previously demonstrated that the potato tuber is capable of de novo biosynthesis of all the proteogenic amino acids (Roessner et al., 2001a, 2001b) and that amino acid biosynthesis in the tuber is regulated by Suc levels. When the amino acid levels of wild-type plants under different growth conditions are scrutinized, it can be seen that those from plants grown in continuous light are lower and those grown in continuous dark are higher than those grown under long-day conditions. It is tempting to speculate that amino acid levels in Arabidopsis root may be under similar control to those of the potato since the changes in the roots are the exact opposite of those observed in leaves following the switch from light to dark. Therefore, on the analysis of the wild-type data alone, it seems exceedingly unlikely that these changes merely reflect changes in the amounts of amino acid transport through either specific (Fischer et al., 2002; Koch et al., 2003) or nonspecific processes. Moreover, the changes in amino acid contents are what would be predicted if Suc repressed transcription of key enzymes of amino acid biosynthesis within the Arabidopsis root.
Looking at the mutant, however, the picture is slightly less clear since it has significantly elevated levels of amino acids when the plants are incubated in continuous light. While it is possible that this is a direct effect of the mutation on root metabolism that is apparent only under high assimilate supply, it would appear likely that these changes in amino acid pool size in the roots most probably reflect changes observed in the leaves. Given that the phloem concentration of amino acids is relatively high in Arabidopsis (see Coruzzi, 2003) and that the characterized amino acid transporters have differing affinities (Fischer et al., 2002; Koch et al., 2003), it is conceivable that changes in leaf amino acid concentration must reach a certain threshold before they are transmitted to the root. When the genotypes are subjected to continuous darkness, the content of some amino acids in roots of the mutant are significantly higher than those documented for the wild type. It suggests that the above hypothesis may be somewhat of an oversimplification. The fact that these roots have altered ATP levels suggests that the availability of ATP may well also play an important role in the regulation of amino acid content within heterotrophic tissues. Given that these two hypotheses appear contradictory suggests that further molecular experiments are required to understand root amino acid biosynthesis in Arabidopsis.
Also differing from the situation observed in potato tubers was the fact that the level of starch was not increased in the Arabidopsis root but, if anything, rather decreased. This decrease was significant in the roots of plants subjected to continuous light (under these conditions there were also minor, nonsignificant increases in soluble sugars, suggesting altered carbon partitioning in these lines). However, it is important to note that Arabidopsis roots contain two orders of magnitude less starch than potato tubers and that starch is far less important in the root than in the potato tuber. The content of starch and protein of the Arabidopsis seed was largely unaltered also. These results therefore suggest that the role of the stromal plastidial ADK is very much tissue dependent, with relatively large changes observed in leaf metabolism and in root growth but little effect on seed metabolism or development.
CONCLUSION
The data presented in this paper reveal that the plastidial ADK At2g37250 is localized in the plastid stroma where it acts to regulate ATP availability for biosynthetic processes. They also confirm early studies in both plants and microbes (Glaser et al., 1975; Goelz and Cronan, 1982; Regierer et al., 2002) that this enzyme plays an important role in the regulation of growth. While the previous studies have concentrated on heterotrophic organisms or tissues, this study revealed that shoot growth can also be, conditionally, affected by the loss of this enzyme. Moreover, these data highlight the diverse roles light can play in regulation of amino acid biosynthesis. While data from the wild-type leaf tissue are in agreement with previous studies suggesting light and Suc-mediated transcriptional regulation of amino acid biosynthesis (Coruzzi, 2003; Roessner-Tunali et al., 2003), the data from the mutant suggest that the rate of amino acid biosynthesis in the leaf also has the potential to be regulated by the rate of ATP production. However, whether this is indicative of ATP signaling (Demidchik et al., 2003) or merely reflects the role of ATP as a cofactor in these biosynthetic pathways, as yet, remains an open question.
MATERIALS AND METHODS
Plant Material and Handling
Arabidopsis (Arabidopsis thaliana) seeds ecotype Colombia and the accession GABI 300-A04 were obtained from the GABI-Kat collection (Max-Planck-Institut für Züchtungsforschung, Cologne, Germany). The seeds were germinated on Murashige and Skoog media (Murashige and Skoog, 1962) containing 2% Suc and were grown in a growth chamber (250 μmol photons m−2 s−1, 22°C) under a 16-h-light/8-h-dark regime before transfer to soil, where (unless otherwise stated) they were grown under the same conditions.
Mutant Population Screening
Screening and selection within the mutant population was done following the GABI-Kat instructions (http://www.mpiz-koeln.mpg.de/GABI-Kat/General_Information/GABI-Kat-sul-selection.html). Approximately 100 seeds of the T3 generation were sterilized and sown on petri dishes containing Murashige and Skoog media supplemented with 11.25 mg/L of Sulfadiazin (4-amino-N-[2-pyrimidinyl] benzene-sulfonamide-Na; Sigma-Aldrich Chemie GmbH, Deisenhofen, Germany). After 24 h in the dark at 4°C, plates were transferred to a long-day regime, and selection was carried out on the criteria of survival until 7 d after germination. The frequency of segregation (1:16) corresponds to that expected following a single insertion event. Resistant lines were subsequently screened for zygosity status by PCR analysis using a pair of primers annealing on the second and third exon of the At2g37250 gene (ADKat5, 5′tcggagattgtaaatcagg3′, and ADKat3, 5′cattaaaacccttgccac3′) and a third primer annealing of the left border of the T-DNA (http://www.mpiz-koeln.mpg.de/GABI-Kat/General_Information/GABI-Kat-pAC161T-DNAmapPr.html; T-DNA, 5′catttggacgtgaatgtag3′). Unless otherwise stated, all biochemical and physiological measurements have been carried out with T4 homozygous plants.
Southern- and Northern-Blot Analysis
Genomic DNA (10 μg) from Col-0 and GABI 300-A04 was isolated and digested with three different restriction enzymes (EcoRI, BamHI, and NdeI) lacking recognition sites on the T-DNA fragment used as a probe. Southern blotting was performed as described by Carrari et al. (2003). The blot was hybridized with an approximately 500-bp fragment, comprising the left border of the T-DNA, using standard conditions (Sambrook et al., 1989) at 60°C. Washes were carried out at low stringency (2× SSC and 1× SSC, 0.1% SDS at 60°C).
Total RNA was isolated using the commercially available Trizol kit (Gibco BRL, Karlsruhe, Germany) according to the manufacturer's suggestions. The RNA (15 μg) was then size fractionated on a 1% agarose MOPS-formaldehyde gel prior to transfer to a nylon membrane filter. Filters were subsequently probed with a 259-bp cDNA fragment amplified by PCR using the gene-specific primers detailed above. This fragment spans 138 and 121 nucleotides of the second and third exons of the gene, respectively. Following PCR amplification, the identity of this fragment was confirmed by sequencing and blasting against the complete Arabidopsis genome. These searches showed that this gene exhibited less than 59% homology to other putative ADK-encoding genes. Hybridization of northern blots was carried out using this fragment as a probe and following standard protocols (Sambrook et al., 1989). Filters were washed once for 20 min at 42°C in 2× SSC, 0.1% SDS; once for 20 min at 42°C in 0.5× SSC, 0.1% SDS; and finally for 20 min at 65°C in 0.1× SSC, 0.1% SDS. After washing, the filters were exposed to x-ray films (Xomat; Eastman-Kodak, Rochester, NY) for 1 to 2 d.
Subcellular Localization Experiments
The ADK gene (At2g37250) was amplified from Arabidopsis Col-0 leaf cDNA by PCR using the following primers 5′caccatggcgagattagtgc3′ and 5′tgctgcgacagactgtttc3′. The PCR fragment was recombined into the entry vector pENTRsd_TOPO (Invitrogen, Carlsbad, CA) making use of the Gateway recombination system. Subsequently, the ADK gene was recombined into the destination vector pK7WGF2 encoding a C-terminal EGFP, driven by cauliflower mosaic virus 35S promoter (Karimi et al., 2002). The plasmid was transiently expressed in leaf cells using PDS-1000 particle delivery system (Bio-Rad Laboratories, Hercules, CA). Fluorescent signals were observed by laser-scanning confocal microscope (Leica DM IRBE microscope, TCS SPII confocal scanner; Leica, Wetzlar, Germany). Images were generated using processing software from Leica.
Analysis of Enzyme Activities
The activity of ADK was measured following the protocol of Kleczkowski and Randall (1986) in leaf material and chloroplast preparations that had been isolated as detailed by Tauberger et al. (2000). Contamination of these preparations by cytosolic marker enzymes did not exceed 10% in either the wild-type or mutant tissue.
Metabolite Analysis
Tissue samples were rapidly frozen in liquid nitrogen. Subsequently the samples were extracted either in ethanol (for determination of carbohydrate and amino acid contents) or in trichloroacetic acid (for the determination of nucleotides) as detailed by Fernie et al. (2001a). The carbohydrate contents were determined spectrophotometrically as described by Fernie et al. (2001b), while the amino acid contents were determined using the HPLC protocol described by Regierer et al. (2002). The levels of nucleotides and nucleosides in leaves were determined using the HPLC protocol defined by Fernie et al. (2001a) while in roots were determined by a cycling assay as described in Gibon et al. (2002). The recoveries of HPLC protocols for nucleotides have been documented previously (see, for example, Geigenberger et al., 1996; Fernie et al., 2001a). Recoveries of ADP and ATP for root material following the cycling assays were determined to be 82% and 88%, respectively.
Statistical Analysis
The t tests have been performed using the algorithm embedded into Microsoft Excel (Microsoft, Seattle). The term significant is used in the text only when the change in question has been confirmed to be significant (P < 0.05) with the t test.
Supplementary Material
Acknowledgments
Discussions and support of Prof. Lothar Willmitzer throughout this work are most gratefully acknowledged. We are also very thankful to Jeannine Mazuch for excellent technical assistance and to Dr. Yves Gibon for advice on cycling assays.
This work was supported by Max-Planck-Gesellschaft (to F.C., A.L., and A.R.F.).
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.056143.
References
- Bendtsen JD, Nielsen H, von Heijne G, Brunak S (2004) Improved prediction of signal peptides: SignalP 3.0. J Mol Biol 340: 783–795 [DOI] [PubMed] [Google Scholar]
- Birkenhead K, Walker D, Foyer C (1982) The intracellular distribution of adenylate kinase in the leaves of spinach, wheat and barley. Planta 156: 171–175 [DOI] [PubMed] [Google Scholar]
- Carrari F, Nunes-Nesi A, Gibon Y, Lytovchenko A, Ehlers-Loureiro M, Fernie AR (2003) Reduced expression of aconitase results in an enhanced rate of photosynthesis and marked shifts in carbon partitioning in illuminated leaves of wild species tomato. Plant Physiol 133: 1322–1335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZT, Fu HI, Liu D, Chang PFL, Narasimhan M, Ferl R, Hasegawa PM, Bressan RA (1994) A nacl-regulated plant gene encoding a brain protein homolog that activates adp ribosyltransferase and inhibits protein-kinase-c. Plant J 6: 729–740 [DOI] [PubMed] [Google Scholar]
- Coruzzi G (2003) Primary N-assimilation into amino acids in Arabidopsis. In CR Somerville, EM Meyerowitz, The Arabidopsis Book. American Society of Plant Biologists, Rockville, MD, doi/10.1199, http://www.aspb.org/publications/arabidopsis/ [DOI] [PMC free article] [PubMed]
- Demidchik V, Nichols C, Oliynyk M, Dark A, Glover BJ, Davies JM (2003) Is ATP a signalling agent in plants? Plant Physiol 133: 456–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, von Heijne G (2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol 300: 1005–1016 [DOI] [PubMed] [Google Scholar]
- Fernie AR, Carrari F, Sweetlove LJ (2004) Respiratory metabolism: glycolysis, the TCA cycle and mitochondrial electron transport. Curr Opin Plant Biol 7: 254–261 [DOI] [PubMed] [Google Scholar]
- Fernie AR, Roessner U, Trethewey RN, Willmitzer L (2001. a) The contribution of plastidial phosphoglucomutase to the control of starch synthesis within the potato tuber. Planta 213: 418–426 [DOI] [PubMed] [Google Scholar]
- Fernie AR, Roscher A, Ratcliffe RG, Kruger NJ (2001. b) Fructose-2,6-bisphosphate activates pyrophosphate: fructose 6-phosphate 1-phosphotransferase and increases triose phosphate to hexose phosphate cycling in heterotrophic cells. Planta 212: 250–263 [DOI] [PubMed] [Google Scholar]
- Fischer WN, Loo DDF, Koch W, Ludewig U, Boorer KJ, Tegeder M, Rentsch D, Wright EM, Frommer WB (2002) Low and high affinity amino acid H+-cotransporters for cellular import of neutral and charged amino acids. Plant J 29: 717–731 [DOI] [PubMed] [Google Scholar]
- Fricaud AC, Walters AJ, Whitehouse DG, Moore AL (1992) The role(s) of adenylate kinase and the adenylate carrier in the regulation of plant mitochondrial respiratory activity. Biochim Biophys Acta 1099: 253–261 [Google Scholar]
- Geigenberger P, Lerchl J, Stitt M, Sonnewald U (1996) Phloem-specific expression of pyrophosphatase inhibits long-distance transport of carbohydrates and amino acids in tobacco plants. Plant Cell Environ 19: 43–55 [Google Scholar]
- Geigenberger P, Stitt M, Fernie AR (2004) Metabolic control analysis and regulation of the conversion of sucrose to starch in growing potato tubers. Plant Cell Environ 27: 655–673 [Google Scholar]
- Gellerich FN (1992) The role of adenylate kinase in dynamic compartmentation of adenine nucleotides in the mitochondrial inner membrane space. FEBS Lett 297: 55–58 [DOI] [PubMed] [Google Scholar]
- Gibon Y, Vigeolas H, Tiessen A, Geigenberger P, Stitt M (2002) Sensitive and high-throughput metabolite assays for inorganic pyrophosphate, ADPGlc, nucleotide phosphates, and glycolytic intermediates based on novel enzymic cycling system. Plant J 30: 221–235 [DOI] [PubMed] [Google Scholar]
- Glaser M, Nulty W, Vagelos PR (1975) Role of adenylate kinase in regulation of macromolecular biosynthesis in a putative mutant of Escherichia coli defective in membrane phospholipid biosynthesis. J Bacteriol 123: 128–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goelz SE, Cronan JE (1982) Adenylate kinase of Escherichia coli evidence for a functional interaction in phospholipid synthesis. Biochemistry 21: 189–195 [DOI] [PubMed] [Google Scholar]
- Haertle U (1977) Untersuchengen zum Einfluß von Licht, Temperatur und Glukose auf des Isoenzymmuster der Adenylatkinase (EC 2.7.4.3) aus Chenopodium rubrum L. PhD thesis. Albrecht Ludwigs Universität, Freiburg, Germany
- Igamberdiev AU, Kleczkowski LA (2003) Membrane potential, adenylate levels and Mg2+ are interconnected via adenylate kinase equilibrium in plant cells. Biochim Biophys Acta 1607: 111–119 [DOI] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A (2002) GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 5: 193–195 [DOI] [PubMed] [Google Scholar]
- Kawai M, Kidou S-I, Kato A, Uchimiya H (1992) Molecular characterization of cDNA encoding for adenylate kinase of rice (Oryza sativa L.). Plant J 2: 845–854 [DOI] [PubMed] [Google Scholar]
- Kawai M, Uchimiya H (1995) Biochemical properties of rice adenylate kinase and subcellular location in plant cells. Plant Mol Biol 27: 943–951 [DOI] [PubMed] [Google Scholar]
- Kleczkowski LA, Randall DD (1986) Maize leaf adenylate kinase: purification and partial characterization. Plant Physiol 81: 1110–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleczkowski LA, Randall DD (1991) Equilibration of adenylates by maize leaf adenylate kinase: effects of magnesium on apparent and true equilibria. J Exp Bot 42: 537–540 [Google Scholar]
- Koch W, Kwart M, Laubner M, Heineke D, Stransky H, Frommer WB, Tegeder M (2003) Reduced amino acid content in transgenic potato tubers due to antisense inhibition of the leaf H+/amino acid symporter StAAP1. Plant J 33: 211–220 [DOI] [PubMed] [Google Scholar]
- Landschütze V, Willmitzer L, Müller-Röber B (1995) Inhibition of flower formation by antisense repression of mitochondrial citrate synthase in transgenic potato plants leads to a specific disintegration of the ovary tissues of the flower. EMBO J 14: 660–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami S, Strotmann H (1978) Adenylate kinase bound to the envelope membranes of spinach chloroplasts. Arch Biochem Biophys 185: 30–38 [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15: 473–497 [Google Scholar]
- Neuhaus HE, Kruckleberg AL, Feil R, Stitt M (1989) Reduced activity of phosphoglucose isomerase in the cytosol and chloroplast of Clarkia xantiana: study of the mechanisms which regulate photosynthate partitioning. Planta 178: 110–112 [DOI] [PubMed] [Google Scholar]
- Neuhaus HE, Stitt M (1990) Control analysis of photosynthate partitioning: impact of reduced activity of ADPglucose pyrophosphorylase or plastid phosphoglucomutase on the fluxes to sucrose and starch in Arabidopsis thaliana (L). Heynh Planta 182: 445–454 [DOI] [PubMed] [Google Scholar]
- Noda LH (1973) Adenylate kinase. In PD Boyer, ed, The Enzymes (Group Transfer Part A), Vol VII. Academic Press, New York, pp 279–305
- Pradet A, Raymond P (1983) Adenine-nucleotide ratios and adenylate energy-charge in energy-metabolism. Annu Rev Plant Physiol Plant Mol Biol 34: 199–224 [Google Scholar]
- Regierer B, Fernie AR, Springer F, Perez-Melis A, Leisse A, Koehl K, Willmitzer L, Geigenberger P, Kossmann J (2002) Starch content and yield increase as a result of altering adenylate pools in transgenic plants. Nat Biotechnol 20: 1256–1260 [DOI] [PubMed] [Google Scholar]
- Roberts JKM, Aubert S, Gout E, Bligny R, Douce R (1997) Cooperation and competition between adenylate kinase, nucleoside diphosphokinase, electron transport and ATP synthase in plant mitochondria studied by P-31-nuclear magnetic resonance. Plant Physiol 113: 191–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessner U, Luedemann A, Brust D, Fiehn O, Linke T, Willmitzer L, Fernie AR (2001. a) Metabolic profiling allows comprehensive phenotyping of genetically or environmentally modified plant systems. Plant Cell 13: 11–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessner U, Willmitzer L, Fernie AR (2001. b) High resolution metabolic phenotyping of genetically and environmentally diverse potato tuber systems: identification of phenocopies. Plant Physiol 127: 749–764 [PMC free article] [PubMed] [Google Scholar]
- Roessner-Tunali U, Urbanczyk-Wochniak E, Czechowski T, Kolbe A, Willmitzer L, Fernie AR (2003) De novo amino acid biosynthesis in potato tubers is regulated by sucrose levels. Plant Physiol 133: 683–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosso MG, Li Y, Strizhov N, Reiss B, Dekker K, Weisshaar B (2004) An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol Biol 53: 247–259 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York
- Schiltz E, Burger S, Grafmuller R, Deppert WR, Haehnel W, Wagner E (1994) Primary structure of maize chloroplast adenylate kinase. Eur J Biochem 222: 949–954 [DOI] [PubMed] [Google Scholar]
- Schlattner U, Wagner E, Greppin H, Bonzon M (1993) Adenylate kinase in tobacco cell cultures: separation and localization of different activities. Plant Physiol Biochem 31: 815–825 [Google Scholar]
- Schlattner U, Wagner E, Greppin H, Bonzon M (1994) Adenylate kinase in tobacco cell cultures: variability and regulation of isoform activity patterns in different cell lines. J Plant Physiol 144: 400–409 [Google Scholar]
- Small I, Peeters N, Legeai F, Lurin C (2004) Predotar: a tool for rapidly screening proteomes for N-terminal targeting sequences. Proteomics 4: 1581–1590 [DOI] [PubMed] [Google Scholar]
- Stitt M (1990) Application of control analysis to sucrose synthesis. In A Cornish-Bowden, ML Cardenas, eds, Control of Metabolic Pathways. Academic Press, London, pp 363–376
- Tauberger E, Fernie AR, Emmermann M, Renz A, Kossmann J, Willmitzer L, Trethewey RN (2000) Antisense inhibition of plastidial phosphoglucomutase provides compelling evidence that potato tuber amyloplasts import carbon from the cytosol in the form of glucose 6-phosphate. Plant J 23: 43–53 [DOI] [PubMed] [Google Scholar]
- Tjaden J, Möhlmann T, Kampfenkel K, Heinrichs G, Neuhaus HE (1998) Altered plastidic ATP/ADP transporter activity influences potato (Solanum tuberosum L.) tuber morphology, yield and composition of tuber starch. Plant J 16: 531–540 [Google Scholar]
- Wagner E, Haertle U, Kossmann I, Frosch S (1983) Metabolic and developmental adaptation of eukaryotic cells as related to endogenous and exogenous control of translocators between subcellular compartments. In W Schwemmler, H Schenk, eds, Endocytobiology II. W. de Gruyter, Berlin, pp 341–351
- Yamada K, Lim J, Dale JM, Chen H, Shinn P, Palm CJ, Southwick AM, Wu HC, Kim C, Nguyen M, et al (2003) Empirical analysis of transcriptional activity in the Arabidopsis genome. Science 302: 842–846 [DOI] [PubMed] [Google Scholar]
- Yiu R, Iketani S, Mikina T, Kubo T (2003) Antisense inhibition of mitochondrial pyruvate dehydrogenase E1α subunit in anther tapetum causes male sterility. Plant J 34: 57–66 [DOI] [PubMed] [Google Scholar]
- Zancani M, Casolo V, Vianello A, Macri F (2001) Involvement of apyrase in the regulation of the adenylate pool by adenylate kinase in plant mitochondria. Plant Sci 161: 927–933 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.