Abstract
Inositol polyphosphates, such as inositol trisphosphate, are pivotal intracellular signaling molecules in eukaryotic cells. In higher plants the mechanism for the regulation of the type and the level of these signaling molecules is poorly understood. In this study we investigate the physiological function of an Arabidopsis (Arabidopsis thaliana) gene encoding inositol polyphosphate kinase (AtIPK2α), which phosphorylates inositol 1,4,5-trisphosphate successively at the D-6 and D-3 positions, and inositol 1,3,4,5-tetrakisphosphate at D-6, resulting in the generation of inositol 1,3,4,5,6-pentakisphosphate. Semiquantitative reverse transcription-PCR and promoter-β-glucuronidase reporter gene analyses showed that AtIPK2α is expressed in various tissues, including roots and root hairs, stem, leaf, pollen grains, pollen tubes, the flower stigma, and siliques. Transgenic Arabidopsis plants expressing the AtIPK2α antisense gene under its own promoter were generated. Analysis of several independent transformants exhibiting strong reduction in AtIPK2α transcript levels showed that both pollen germination and pollen tube growth were enhanced in the antisense lines compared to wild-type plants, especially in the presence of nonoptimal low Ca2+ concentrations in the culture medium. Furthermore, root growth and root hair development were also stimulated in the antisense lines, in the presence of elevated external Ca2+ concentration or upon the addition of EGTA. In addition, seed germination and early seedling growth was stimulated in the antisense lines. These observations suggest a general and important role of AtIPK2α, and hence inositol polyphosphate metabolism, in the regulation of plant growth most likely through the regulation of calcium signaling, consistent with the well-known function of inositol trisphosphate in the mobilization of intracellular calcium stores.
Inositol polyphosphate kinases play important roles in signal transduction by regulating the levels of inositol 1,4,5-trisphosphate [Ins(1,4,5)P3] and inositol 1,3,4,5-tetrakisphosphate [Ins(1,3,4,5)P4; Communi et al., 1995]. Ins(1,4,5)P3 serves as a second messenger responsible for generating transient cytosolic Ca2+ increases (Berridge, 1997), and Ins(1,3,4,5)P4 itself may regulate cytosolic Ca2+ concentration by promoting Ca2+ sequestration (Hill et al., 1988) or by acting together with Ins(1,4,5)P3 to mobilize intracellular Ca2+ stores (Morris et al., 1987).
In higher plants, calcium participates in the regulation of many cellular processes, including cell division, cell expansion, and cytoplasmic streaming (Malho, 1999; Rudd and Franklin-Tong, 2001; Sanders et al., 2002) and is of special importance during the regulation of pollen tube growth and fertilization. Moreover, it is widely reported that tight regulation of cytosolic calcium concentration ([Ca2+]c) is of fundamental importance for polarized growth, including tip growth in pollen tubes, root hairs, and fungal hyphae (Jackson and Heath, 1993; Pierson et al., 1994; Wymer et al., 1997). The steepness of the growing apex-localized gradient of cytosolic Ca2+ correlates with the growth rate of root hairs (Bibikova et al., 1997; Felle and Hepler, 1997; Wymer et al., 1997) and pollen tubes (Pierson et al., 1994). Disruption of the [Ca2+]c gradient results in an arrest of tip growth within minutes (Miller et al., 1992; Pierson et al., 1994). Studies on Lilium pollen tube growth indicate a correlation of growth with cytosolic free Ca2+; the growth rate was high at elevated [Ca2+]c, and low at reduced Ca2+ level (Holdaway-Clarke et al., 1997; Messerli and Robinson, 1997). Recent analysis of the self incompatible response of Papaver rhoeas (Franklin-Tong et al., 2002) showed that extracellular Ca2+ influx along the shank of the pollen tube induced increases in [Ca2+]c, suggesting that [Ca2+]c is involved in the recognition and reorientation of pollen tube growth. All of these studies strongly suggest that [Ca2+]c is required not only for sustaining but also for regulation of tip growth.
Evidence suggests that Ins(1,4,5)P3 regulates pollen tube elongation, e.g. an alteration of Ins(1,4,5)P3 levels modified pollen tube growth (Franklin-Tong et al., 1996; Malho, 1998). Recently, there has been considerable renewed interest in Ins(1,3,4,5)P4, a product of Ins(1,4,5)P3 3-kinase, which phosphorylates Ins(1,4,5)P3 at 3-position of the inositol ring following the identification of an Ins(1,3,4,5)P4 receptor in animals (Cullen, 1998). It is likely therefore that Ins(1,3,4,5)P4 has discrete signaling functions, which may be synergistic with Ins(1,4,5)P3. To date, there is little information on the interrelationship between Ins(1,4,5)P3 and Ins(1,3,4,5)P4 in cytosolic calcium homeostasis of plant cells.
Several cDNAs encoding Ins(1,4,5)P3 3-kinase or Ins(1,4,5)P3 dual specificity 6/3-kinases have been isolated and biochemically characterized, including those from rat (Choi et al., 1990; Thomas et al., 1994), human (Takazawa et al., 1991a, 1991b; Dewaste et al., 2000), chicken (Bertsch et al., 1999), yeast (Saccharomyces cerevisiae; Odom et al., 2000), and Arabidopsis (Arabidopsis thaliana; Stevenson-Paulik et al., 2002; Xia et al., 2003). Mammalian Ins(1,4,5)P3 kinases are generally activated by Ca2+/calmodulin (Sim et al., 1990; Sims and Allbritton, 1998). An Ins(1,4,5)P3 3-kinase from the nematode, Caenorhabditis elegans, has been identified that is insensitive to Ca2+/calmodulin and lacks a consensus calmodulin-binding site (Clandinin et al., 1998). The Arabidopsis inositol polyphosphate kinase, AtIPK2β, also lacks a consensus calmodulin-binding domain and does not interact with calmodulin in a biochemical assay (Xia et al., 2003). The S. cerevisiae inositol polyphosphate kinase 2 (Ipk2) is a dual-specificity kinase, which phosphorylates either Ins(1,4,5)P3 or Ins(1,4,5,6)P4 to produce inositol 1,4,5,6-tetrakisphosphate [Ins(1,4,5,6)P4] or inositol 1,3,4,5,6-pentakisphosphate [Ins(1,3,4,5,6)P5], respectively. The Ipk2 protein was found to be an indispensable component of the ArgR-Mcm1 transcriptional complex in yeast (Odom et al., 2000), which is composed of four proteins (Arg-80p, Arg-81p, Arg-82p, and Mcm1p) that are required for proper control of transcription (Bechet et al., 1970; Messenguy and Dubois, 1993). Arg-82p (also called ArgRIII protein) is identical to Ipk2p. Inositol polyphosphate kinase activity of Arg-82p was independently described by Saiardi et al. (1999, 2000). Arg-82p and Mcm1p are pleiotropic regulators, whereas Arg-80p and Arg-81p serve as specific transcription factors in the metabolism of the amino acid Arg. Disruption of the ARG82/IPK2 gene partially affects cell growth at 30°C, whereas growth is strongly impaired at 37°C. Although the inositol polyphosphate kinase activity of Ipk2p was originally suggested to be necessary for the control of Arg metabolism via the ArgR-Mcm1 complex in vivo (Odom et al., 2000), Dubois et al. (2000) have shown that the kinase activity of Arg-82p/Ipk2p is not required for the regulation of Arg metabolism.
The AtIPK2α and AtIPK2β proteins recently identified in Arabidopsis successively phosphorylate Ins(1,4,5)P3 at the 6- and 3-positions to generate Ins(1,3,4,5,6)P5 without a significant production of Ins(1,3,4,5)P4 (Stevenson-Paulik et al., 2002; Xia et al., 2003). Recombinant glutathione S-transferase AtIPK2α fusion protein exhibited similar apparent Km values for Ins(1,4,5)P3 and Ins(1,3,4,5)P4 of 14.6 and 15.5 μm, respectively (Stevenson-Paulik et al., 2002), but did not phosphorylate Ins(1,4,5)P3 at the 3-position. AtIPK2α and AtIPK2β were both capable of rescuing growth of the yeast ARG82/IPK2 deletion strain. At present, it is not known whether this effect is due to the kinase activity of the plant proteins and/or due to an interaction with yeast ArgR-Mcm1 complex. The two plant proteins, like yeast Ipk2p, also exhibited 5-kinase activity toward Ins(1,3,4,6)P4 and Ins(1,2,3,4,6)P5 (Stevenson-Paulik et al., 2002).
The physiological function of the two Arabidopsis inositol multiphosphate kinases in planta is still unknown. We report here the isolation of a cDNA encoding AtIPK2α from Arabidopsis and confirm the findings of Stevenson-Paulik et al. (2002) on the substrate specificity of this enzyme. AtIPK2α promoter-GUS reporter gene fusions revealed widespread expression of the AtIPK2α gene in Arabidopsis. We used an antisense approach to reduce AtIPK2α transcript levels. Antisense plants exhibited enhanced pollen germination, pollen tube growth, and root elongation under suboptimal Ca2+ concentrations, indicating that AtIKP2α limits growth of the wild type under these conditions. Our results support the hypothesis that AtIKP2α plays a critical role in the regulation of growth probably through the regulation of inositol trisphosphate (IP3)-mediated calcium accumulation in plants.
RESULTS
Isolation of an Arabidopsis cDNA, AtIPK2α, Encoding a Dual-Specificity Inositol Polyphosphate 6/3-Kinase (AtIPK2α)
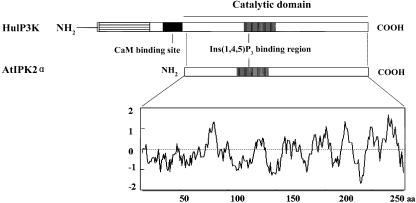
An Arabidopsis expressed sequence tag (EST) encoding a protein that shared high homology with animal Ins(1,4,5)P3 kinase and inositol polyphosphate multikinases was identified by searching the National Center for Biotechnology Information (NCBI) database. Oligonucleotide primers were designed based on the available EST sequence and used to screen a cDNA library established from hypocotyl tissue (see “Materials and Methods”) through a PCR-based strategy. The longest cDNA identified was used for further studies. It had a length of 1,068 bp, encoding a 286-amino acid-long polypeptide of 31.9 kD. During the course of our experiments, Stevenson-Paulik et al. (2002) reported the cloning of a cDNA that encoded AtIPK2α, which was identical to our predicted protein. We therefore adopted the name suggested by Stevenson-Paulik et al. (2002). AtIPK2α shares 73% identical amino acids (84% similarity) with Arabidopsis AtIPK2β (GenBank accession no. AJ404678; Stevenson-Paulik et al., 2002; Xia et al., 2003). It is significantly smaller than human Ins(1,4,5)P3 3-kinase C (683 amino acids, with a molecular mass of 75.2 kD; GenBank accession no. AJ290975). Structural analysis indicated the presence of a catalytic domain in AtIPK2α, similar to that of known Ins(1,4,5)P3 3-kinases; however, a calmodulin-binding site, which typically occurs in the animal enzymes, exists neither in AtIPK2α (Fig. 1) nor in AtIPK2β. We have previously shown that AtIPK2β does not bind calmodulin, consistent with the absence of a predicted calmodulin-binding site (Xia et al., 2003). The AtIPK2α gene is located on Arabidopsis chromosome 5.
Figure 1.
Structural organization of the AtIPK2α protein. Domain organization of AtIPK2α compared with human inositol (1,4,5)P3 kinase (HuIP3K; GenBank accession no. AJ290975). A calmodulin (CaM)-binding site is absent from AtIPK2α. A putative Ins(1,4,5)P3-binding region is localized in the catalytic domain. Hydropathy analysis indicates that AtIPK2α is likely a soluble protein. The cut-off value is calculated based on the characteristics of individual amino acid residues. The detailed description is available at http://au.expasy.org/tools/protscale.html.
Using recombinant AtIPK2α expressed in Escherichia coli, we confirmed the observations of Stevenson-Paulik et al. (2002) that AtIPK2α (like AtIPK2β) is a dual-specificity 6/3-kinase (data not shown).
AtIPK2α Is Highly Expressed in Roots and Pollen Grains
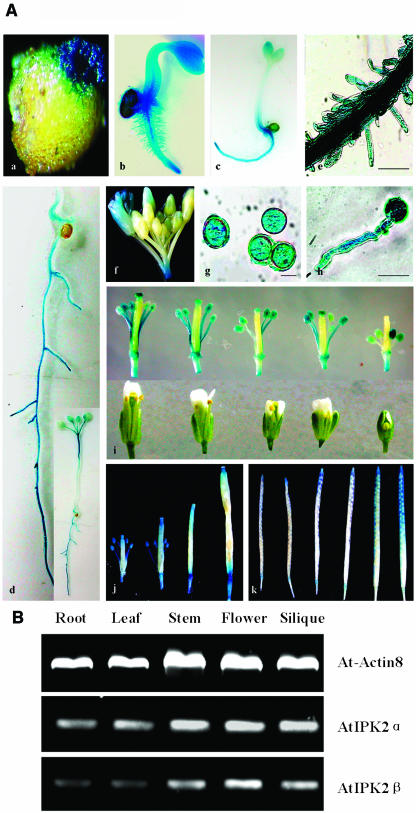
To assess the transcriptional activity of the AtIPK2α gene, a 1.5-kb 5′ upstream region was isolated and fused to the E. coli GUS reporter gene. The promoter-reporter gene construct was transferred into the nuclear genome of Arabidopsis. Ten independent transgenic lines were obtained and GUS expression was analyzed, and all lines showed a very similar pattern of expression. In general, GUS activity was detectable after 2 h of incubation in staining solution. As shown in Figure 2, AtIPK2α was expressed in the emerging seedlings (Fig. 2A, subsection a), in seedlings at different developmental stages (Fig. 2A, subsections b and c), and in roots (Fig. 2A, subsection d), including lateral roots and root hairs (Fig. 2A, subsections d and e). AtIPK2α transcription was also observed in flowers (Fig. 2A, subsections f and i) and was particularly strong in anthers and the stigma (Fig. 2A, subsection i). AtIPK2α transcriptional activity persisted in the stigmatic tissue after fertilization (Fig. 2A, subsection j). AtIPK2α activity was also detected in pollen grains (Fig. 2A, subsection g), in growing pollen tubes (Fig. 2A, subsection h), and in siliques during seed maturation (Fig. 2A, subsection k). Significant GUS activity was also detected in leaves (data not shown).
Figure 2.
Analysis of AtIPK2α expression pattern by promoter-reporter (GUS) gene fusion. A, GUS activity was detected in several tissues. Subsection a, Seeds at the time of germination. Subsections b and c, Young seedlings. Subsection d, Main and lateral roots (the inset shows the whole plantlet). Subsection e, Root hairs. Subsections f and i, Flowers at different developmental stages. Almost no GUS activity was present in the pistil, but strong activity was present in the stigma. Subsection g, Pollen grains. Subsection h, Pollen grain with growing pollen tube. Subsection j, Pistil at different developmental stages (before and after fertilization). Subsection k, Siliques at different developmental stages. Incubation time in GUS staining solution was in all cases 12 to 15 h. Bar = 40 μm. B, Semiquantitative RT-PCR analysis of AtIPK2α and AtIPK2β transcript levels in different Arabidopsis tissues.
Antisense Inhibition of AtIPK2α Results in Superior Pollen Grain Germination and Pollen Tube Growth
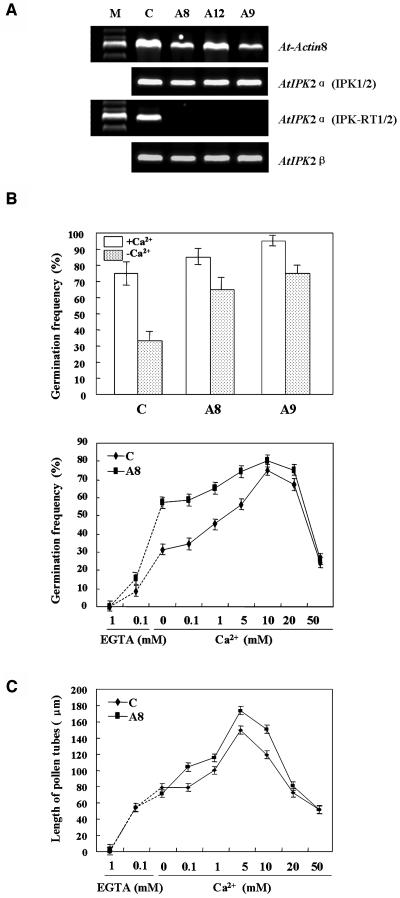
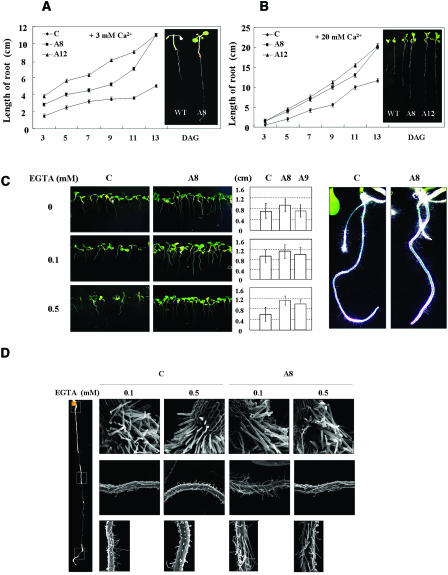
To study the physiological role of AtIPK2α, we used a transgenic approach. The AtIPK2α cDNA was fused in reverse orientation to its own promoter and the resulting antisense construct was then transformed into Arabidopsis using Agrobacterium-mediated transformation. Six independent transgenic lines harboring the AtIPK2α antisense construct were selected. AtIPK2α transcript level was strongly reduced, while the expression of AtIPK2β was not suppressed, as shown by reverse transcription (RT)-PCR (Fig. 3A; AtIPK2α transcript was tested with primers internal, IPK1 and IPK2, and external, IPK-RT1 and IPK-RT2, of the cDNA region used for constructs). Homozygous transgenic plants, obtained after two generations of segregation, were used for analysis. As AtIPK2α was strongly expressed in pollen, we particularly focused our attention on pollen germination and pollen tube growth. Exogenous Ca2+ is critical for pollen grain germination, and pollen grains from control plants exhibited the highest germination frequency (75%) in the presence of exogenous 10 mm Ca2+ (Fig. 3B). Similarly, Fan et al. (2001) observed optimal pollen germination at external 10 mm Ca2+. Germination frequencies were reduced at higher concentrations of Ca2+, i.e. 68% and 25%, at 20 mm and 50 mm Ca2+, respectively. Similarly, at lower Ca2+ concentrations, pollen germination was also reduced (56%, 46%, and 35% at 5 mm, 1 mm, and 0.1 mm added Ca2+, respectively), and only 31% of the pollen grains germinated when no extra Ca2+ was added to the medium. In the presence of the Ca2+-chelating agent EGTA, germination of pollen grains was strongly reduced (8% at 0.1 mm EGTA) or virtually absent (0% at 1 mm EGTA; Figs. 3B and 4, A, C, E, and G). Germination measurements of pollen grains from transgenic plants revealed that reduced AtIPK2α transcript level resulted in an improved germination frequency under identical experimental conditions. As shown in Figure 3B, transgenic pollen also showed a biphasic response to calcium concentration. The germination frequency of transgenic pollen grains was, however, appreciably higher across the range of calcium concentrations: ranging from 58% in the absence of added Ca2+, 59% to 75% at 0.1 to 5 mm Ca2+, and 16% in the presence of 0.1 mm EGTA (Figs. 3B and 4, B, D, F, and H). At high external Ca2+ concentration (50 mm) or in the presence of 1 mm EGTA, pollen grain germination frequencies were similar to that of the control plants.
Figure 3.
Ca2+-dependent pollen germination and growth of pollen tubes are enhanced in AtIPK2α antisense plants. A, Semiquantitative RT-PCR analysis of transgenic Arabidopsis plants. mRNA isolated from 6-d-old seedlings was reverse transcribed into first strand cDNA. Primers IPK1 and IPK2 (internal primers) and IPK-RT1 and IPK-RT2 (external primers, see ‘Materials and Methods’) were used to test for AtIPK2α transcripts, and primers AtIPK2β-1 and AtIPK2β-2 were chosen to determine AtIPK2β transcripts. The following templates were used: C, untransformed plants; A8, A12, and A9, independent transgenic lines. Transcript level of the Arabidopsis actin gene At1g49240 (At-Actin 8) served as an internal control. M, Molecular size marker. B, Germination of pollen grains of control and transgenic lines (A8 and A9) at both with Ca2+ (10 mm) and no Ca2+ conditions (top). Line A8 was selected for further observation of the pollen grains germination at different concentrations of Ca2+ (0.1–50 mm) or in the presence of EGTA (0.1 or 1 mm; bottom). C, Length of pollen tubes at different concentrations of exogenous Ca2+ (0.1–50 mm) or in the presence of EGTA (0.1 or 1 mm EGTA). In B and C, at least 100 pollen grains and pollen tubes were randomly chosen for germination frequency calculation and pollen tube length measurements for each treatment. Data represent the mean ± sd.
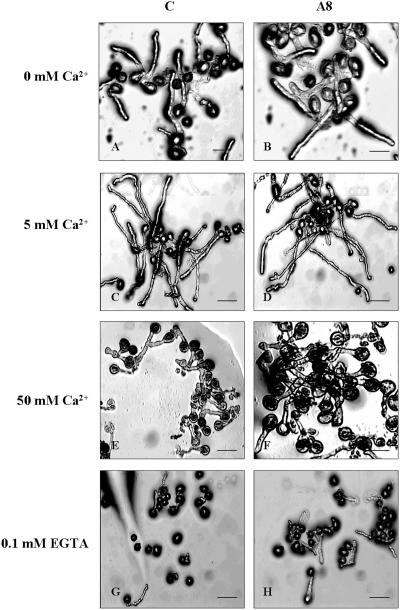
Figure 4.
Visual analysis of pollen grains and pollen tube growth. Pollen sampled from the Arabidopsis wild type (A, C, E, and G) and AtIPK2α antisense line A8 (B, D, F, and H) was germinated in the presence of varying concentrations of supplemented Ca2+ (0, 5, or 50 mm), or in the presence of 0.1 mm EGTA. Pictures were taken 6 h after pollen germination. Bar = 20 μm.
We also analyzed pollen tube lengths. As shown in Figure 3C, pollen tubes from transgenic lines were longer than those from control plants after 6 h of incubation at all concentrations of exogenous Ca2+ tested (see also Fig. 4). Pollen tubes from both transgenic and control plants exhibited maximal growth at an exogenous Ca2+ concentration of 5 mm. At this concentration, pollen tubes from transgenic plants were 16% longer than those from wild-type control plants. Similar data as described above were also obtained with transgenic line A9 (data not shown).
Enhanced Root Growth in AtIPK2α Antisense Lines
Transgenic plants with reduced AtIPK2α transcript level exhibited an improved root growth under different concentrations of exogenous Ca2+ or when EGTA was supplemented. As indicated in Figure 5, root lengths of transgenic plants were elevated when compared with that of the control plants at 3 mm (Fig. 5A) and 20 mm (Fig. 5B) supplemented Ca2+, or at 0.1 or 0.5 mm EGTA (Fig. 5C). Furthermore, scanning electron microscope (SEM) studies showed that the growth of root hairs of control plants was strongly inhibited under conditions of Ca2+ deficiency, whereas root hair growth of transgenic plants was much less compromised (Fig. 5, C and D).
Figure 5.
Ca2+-dependent root and root hair growth is enhanced in AtIPK2α antisense plants. Root growth of Arabidopsis wild-type and AtIPK2α antisense lines (A8 and A12) was tested on Murashige and Skoog medium supplemented with additional 3 mm (A) or 20 mm (B) Ca2+. Note that root growth was enhanced in the antisense lines at both Ca2+ concentrations. DAG, Days after germination. Data indicate means ± sd from 30 determinations (three repeats). In A and B, typical plant phenotypes at 10 DAG are shown on the right. C, Root growth in the presence of EGTA supplemented with Murashige and Skoog medium. Plant photographs on the left show typical growth phenotypes; a quantitative evaluation is shown in the middle part. Columns represent the mean root lengths obtained from 30 determinations (three repeats); error bars indicate ± sd. Data are significantly different (Student's t test; P < 0.05 for 0 and 0.1 mm EGTA; P < 0.01 for 0.5 mm EGTA). Right, Photographic presentation of longer root hairs in the AtIPK2α antisense line A8 at added 0.1 mm EGTA. D, Root hairs formed in the presence of supplemented EGTA at concentrations of 0.1 and 0.5 mm. Images were obtained by SEM. Images were taken from three regions of the root (indicated on the left), i.e. the root-shoot junction zone (top row of figures), the central part of the root (middle row), and the lower part of the root (bottom row). Note, that the AtIPK2α antisense line produced longer root hairs at both concentrations of EGTA.
In addition, the early seedling growth was enhanced in the case of the antisense line both in the absence and presence of external EGTA (data not shown).
AtIPK2α Localizes to the Nucleus and the Plasma Membrane
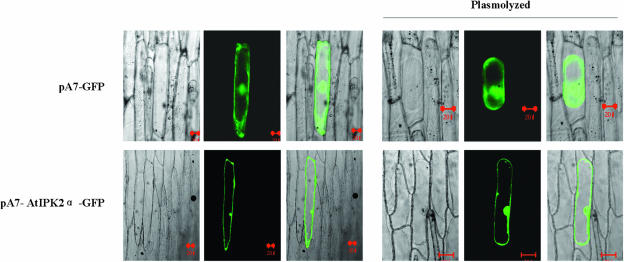
To investigate the localization of AtIPK2α, an AtIPK2α-green fluorescent protein (GFP) fusion protein was transiently expressed in onion (Allium cepa) epidermal cells under the control of the cauliflower mosaic virus 35S promoter. Free GFP was used as a control, which typically accumulates in the cytoplasm and the nucleus (Fig. 6, top). When the AtIPK2α-GFP fusion protein was expressed in onion cells, fluorescence was mostly visible in the nucleus and the plasma membrane (Fig. 6, bottom). However, a transmembrane or membrane-binding domain was not predicted for AtIPK2α when tested with several software tools (data not shown).
Figure 6.
AtIPK2α localizes to both the nucleus and the plasma membrane. Onion epidermal cells were bombarded with plasmids harboring the GFP coding region (pA7-GFP; top, bar = 20 μm), or an AtIPK2α-GFP fusion construct (bottom, bars = 20 or 50 μm in the normal or plasmolyzed cells). GFP accumulated in the cytosolic and nuclear compartments. AtIPK2α-GFP fusion protein accumulated in the nucleus and the plasma membrane. Detection of fluorescence was performed 40 h after bombardment under a confocal laser scanning microscope (wavelength 488 nm). Plasmolysis of onion epidermis cells was induced by 1 m Suc. Right section is the merged image of the ones under normal and UV lights.
Exogenous Ins(1,4,5)P3 Promotes Root Growth
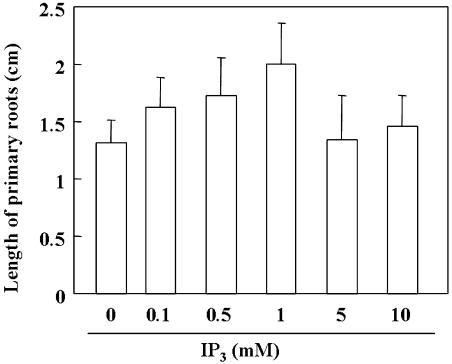
To study whether the accumulated Ins(1,4,5)P3, which is due to the deficiency of AtIPK2α, resulted in the promoted root growth, exogenous Ins(1,4,5)P3 on the root growth were supplemented into the medium and the measurement of primary root growth of 11-d-old Arabidopsis wild-type plants showed that applied Ins(1,4,5)P3 indeed promote root growth. Increased concentrations of Ins(1,4,5)P3 (0.1–1 mm) stimulate root growth and there is an approximately 50% increase at a concentration of 1 mm, while the higher concentrations (5 or 10 mm) didn't have promoting effects (Fig. 7).
Figure 7.
Exogenous Ins(1,4,5)P3 promotes root growth. Growth of primary roots of 11-d-old Arabidopsis wild-type plants was tested on Murashige and Skoog medium supplemented with 0.1, 0.5, 1, 5, or 10 mm Ins(1,4,5)P3. Data indicate means ± sd from 30 determinations (three repeats). Data are significantly different (Student's t test; P < 0.01 for 0 and 0.1 mm IP3; P < 0.01 for 0.5 and 1.0 mm IP3; P < 0.01 for 1 and 5 mm IP3).
DISCUSSION
A number of genes encoding key enzymes of the Arabidopsis phosphoinositide (PI) and inositol polyphosphate pathways have recently been isolated (Mueller-Roeber and Pical, 2002), including phosphatidylinositol synthase (Xue et al., 2000), phosphatidylinositol 4-kinase (Stevenson et al., 1998; Xue et al., 1999), inositol phospholipid kinase (Mikami et al., 1998; Elge et al., 2001), and PI-specific phospholipase C (PI-PLC; e.g. Hirayama et al., 1995; Hartweck et al., 1997). However, few studies have been made of elements that we might expect to operate downstream of the PI pathway in plant cells. It is still not clear from the existing literature whether AtIPK2α operates distal to PI-PLC or in a metabolically more distant or even unrelated pathway. Based on the fact that there are only two IPK genes in the Arabidopsis genome, at least seven PI-PLC genes are known (Hartweck et al., 1997; Mueller-Roeber and Pical, 2002); thus, and not withstanding redundancy of function, simple epistatic analysis of PI-PLC/AtIPK2α interaction is discounted.
Although little is known of the function of discrete protein domains within inositol polyphosphate kinases, it is apparent that Arabidopsis AtIPK2α lacks a calmodulin-binding domain. It is therefore likely that AtIPK2α, like AtIPK2β (Xia et al., 2003), does not bind calmodulin. Thus, both Arabidopsis proteins are likely not regulated in a posttranslational manner by Ca2+/calmodulin.
Promoter-reporter gene studies revealed the strong transcription of AtIPK2α gene in pollen grains, pollen tubes, and root tissues, suggesting a possible role of AtIPK2α in plant fertilization and tip growth. Sequence comparison showed that AtIPK2α shared very high level of identity with AtIPK2β, and comparison of the expression patterns indicated that both are expressed in seedling, leaf, root, and floral organs (stigma and anther). However, there are differences in their expression patterns; in contrast to the weak expression of AtIPK2β in pollen grains and pollen tubes, AtIPK2α was relatively highly transcribed in pollen grain, pollen tube, and roots and leaves (Fig. 2B), implying differential roles for the two genes, which were further suggested by the differential localization of the two proteins. Compared to the nucleus localization of AtIPK2β and its involvement of gene transcription (Xia et al., 2003), localization of AtIPK2α in both plasma and membrane even reveals their different regulatory and functional roles. Both Ins(1,4,5)P3 and calcium served as second messenger molecules and are crucial in root and pollen development. Under AtIPK2α deficiency, the enhanced pollen germination and pollen tube growth, as well as root and root hair growth, especially under nonoptimal Ca2+ concentrations, indeed confirmed the critical roles of AtIPK2α, possibly through regulating calcium signaling, in the developmental processes.
Considering Ins(1,3,4,5)P4 is responsible for Ca2+ sequestration and return of released Ca2+ from calcium stores, deficiency of AtIPK2α may result in the accumulation of Ins(1,4,5)P3, although AtIPK2α does not produce Ins(1,3,4,5)P4 directly. Unbalanced accumulation of Ins(1,4,5)P3 may eventually change the original calcium homeostasis and thus result in an increased level of endogenous Ca2+, which was somehow consistent with and confirmed by the observation that exogenous supplemented Ins(1,4,5)P3 promote root growth (Fig. 7), although it is not conclusive and the detailed functional manner is unclear. Whatever the relationship of AtIPK2α to inositol phosphate metabolism, we provide the direct evidence that AtIPK2α has a developmental function in the regulation of growth phenomena. However, the possibility exists that the physiological function of AtIPK2α is unrelated to the regulation of [Ca2+]c, and that rescue of the temperature sensitive IPK2Δ growth phenotype (and the higher inositol polyphosphate profile of IPK2Δ yeast) by AtIPK2α and AtIPK2β (Stevenson-Paulik et al., 2002; Xia et al., 2003) is not related directly to intracellular calcium. The two plant enzymes may alternatively be involved in the synthesis of InsP6 (also called phytin or phytate), as suggested previously (Stevenson-Paulik et al., 2002).
While a candidate Ins(1,4,5)P3 receptor has not emerged from the yeast genome, indication of important function for higher inositol polyphosphates derived from Ins(1,4,5)P3 has been revealed. Emerging literature (York et al., 1999; Odom et al., 2000; Lemtiri-Chlieh et al., 2003) highlight fundamental roles for yeast and higher plant inositol polyphosphates and InsP6. If AtIPK2α lies in a metabolic pathway to InsP6, then it is possible that the growth phenotypes of AtIPK2α antisense lines relate to the production of higher inositol polyphosphates. Moreover, we should also consider the possibility that the physiological substrate of AtIPK2α is not Ins(1,4,5)P3 or that the function of AtIPK2α lies in transcriptional control unrelated to calcium. Nevertheless, given the importance of calcium to tip growth, it will be worthwhile to establish whether inositol metabolism controlled by AtIPK2α regulates calcium release and further signaling cascade.
MATERIALS AND METHODS
Enzymes and Chemicals
Enzymes used for DNA restriction and modification were obtained from Roche (Mannheim, Germany). DNA primers for PCR and Taq polymerase were from Genecore (Shanghai, China). The Random Labeling kit was obtained from TaKaRa Biotechnology (Dalian, China). d-myo-inositol 1,4,5-tetraphosphate (sodium) was obtained from Echelon (San Jose, CA).
Bacteria and Plants
Escherichia coli XL1-blue cells were used for standard DNA work and for cDNA library amplification. Cells were cultivated at 37°C in Luria-Bertani medium supplemented with appropriate antibiotics using standard methods (Sambrook et al., 1989). Agrobacterium tumefaciens strain GV3101 was used for the transformation of Arabidopsis (Arabidopsis thaliana) L. Heynh. cv C24. Cells were cultivated at 28°C in yeast extract-phosphate medium. Arabidopsis plants were grown in a phytotron with a 16-h-light (22°C; light density approximately 150 μmol photons s−1 m−2)/8-h-dark (18°C) period.
Isolation of the AtIPK2α cDNA
The NCBI EST database (http://www.ncbi.nlm.nih.gov/dbEST/index.html) was screened using human IP3K coding sequence (GenBank accession no. D38169) as a bait, resulting in the identification of a homologous EST from Arabidopsis (accession no. H36803). Primers IPK-1 (5′-AAGATGCAGCTCAAAGTCCC-3′) and IPK-2 (5′-TGACACGGTTGTGGTACCCG-3′) were used to isolate a full-length cDNA from the Arabidopsis cDNA library CD4-16 (Arabidopsis Stock Center, Columbus, OH) employing a PCR-based screening protocol (Alfandari and Darribere, 1994). Plaque-purified phage clones were converted to pBluescript SK derivatives using helper phage ExAssist according to the supplier's (Stratagene, La Jolla, CA) instructions. The clone pAtIPK, which contained the longest cDNA insert, was used for further analyses. DNA sequencing was done by Genecore. Computational analysis was performed with the help of the programs of the Wisconsin Genetics Computer Group (GCG Package, version 10.1). The BLAST search programs provided through the NCBI web interface (http://www.ncbi.nlm.nih.gov/BLAST/) were used for sequence comparisons involving GenBank, EMBL, dbEST, and SwissProt databases. Sequence alignments were performed using the BESTFIT and PILEUP programs.
Isolation of the AtIPK2α 5′ Upstream Regulatory Region and Determination of GUS Activity
A 1.5-kb-long DNA fragment upstream of the translational start codon was amplified by PCR using primers IPK-3 (5′-GCGTCGACGGAACACTCACTTTCCGGTC-3′; added SalI site underlined) and IPK-4 (5′-CGGGATCCCTTGTTTAAATTATGAAACTGAAC-3′; added BamHI site underlined), and genomic DNA from Arabidopsis C24 as template. Amplified DNA products were ligated via the attached SalI/BamHI sites into binary vector pBI101.2 (Jefferson, 1987), leading to plasmid p101-AtIPK-pro (AtIPK2α promoter inserted upstream of the GUS reporter gene). The AtIPK2α promoter in plasmid p101-AtIPK-pro was sequenced to confirm the absence of PCR-induced nucleotide changes. Finally, p101-AtIPK-pro was used to transform Arabidopsis via vacuum infiltration (Bechtold et al., 1993). Transgenic seedlings were identified on sterile agar medium containing kanamycin. GUS activity was determined using 5-bromo-4-chloro-3-indolyl β-d-glucuronic acid as substrate (Jefferson et al., 1987). When required, stained plant tissue was fixed in formaldehyde-acetic acid fixative (distilled water, 95% ethanol, formalin, acetic acid, 80:60:2.4:1.6, v/v/v/v) and analyzed microscopically (Nikon SMZ800; Kanagawa, Japan).
Antisense Inhibition of AtIPK2α Gene Expression
To study the physiological role of AtIPK2α, the full-length cDNA of AtIPK2α was cloned into the vector p101-AtIPK-pro yielding the binary construct, pA-AtIPK. In this plasmid the AtIPK2α coding region was located in reverse orientation under the control of its own promoter. Arabidopsis transformation was performed by vacuum infiltration (Bechtold et al., 1993). Seeds harvested from primary transformants were selected on Murashige and Skoog medium supplemented with 20 mg/L kanamycin, and resistant seedlings were transferred to a phytotron. AtIPK2α transcript levels were determined through coupled RT-PCR. In short, RNA samples extracted from both resistant and control plants were quantitated and used as templates for the synthesis of first-strand cDNA (synthesized with the First-Strand cDNA Synthesis kit from Amersham Biosciences, Shanghai, China). The resulting cDNA pools were used for RT-PCR with either internal primers IPK-1 and IPK-2, or with primers annealing to the 5′ UTR (IPK-RT1, 5′-CTGAAGATTTGTTCTACTACTC-3′), or to the 3′ UTR (IPK-RT2, 5′-TTCTGCTCTTTAGGTAATCAAG-3′), respectively, of the AtIPK2α transcript. Primers from AtIPK2β, AtIPK2β-1 (5′-CGGTTCTTCAAGCCACTTCAGG-3′), and AtIPK2β-2 (5′-CAAGAACAAGCTTCTTCTCAGC-3′) were used to test the transcripts of AtIPK2β in the same pools. The PCR cycling protocol was as follows: 94°C/2 min; 36 × (94°C, 1 min; 60°C, 45 s; 72°C, 1 min); 72°C, 10 min. Primers annealing to Arabidopsis actin transcript (gene locus number At1g49240) were used for positive-control reactions (Act-1, 5′-GCGGTTTTCCCCAGTGTTGTTG-3′; Act-2, 5′-TGCCTGGACCTGCTTCATCATACT-3′). Amplified DNA fragments were separated on 1.5% (w/v) agarose gels and monitored using the GelDoc 2000 system (Bio-Rad, Hercules, CA). Homozygous T3 plants were used for all physiological experiments.
In Vitro Pollen Germination and Measurement of Pollen Tube Growth
Arabidopsis flowers, collected from homozygous transgenic plants 1 to 2 weeks after bolting, were used to analyze pollen phenotypes. Open flowers were dehydrated at room temperature for at least 2 h and pollen grains were obtained by dipping the flowers onto aqueous medium. Pollen grains were germinated on agar medium containing 5 mm MES (pH 5.8, adjusted with Tris), 16.6% (w/v) Suc, 1.5 mm boric acid, 0.8 mm MgSO4, 1 mm KCl, 10 mg/L myo-inositol, 3.65% (w/v) sorbitol, and 1% agar, slightly revised from Fan et al. (2001). CaCl2 was added as required. The frequency of germination of pollen grains and the lengths of pollen tubes were determined after germination at room temperature for 6 h under a microscope (Nikon SMZ800). To examine the affect of extracellular Ca2+ ions on pollen grain germination and pollen tube growth, pollen grains of both control and transgenic plants (two independent lines A8 and A9) were incubated for 6 h at 25°C on medium containing different concentrations of Ca2+ (CaCl2) or EGTA. For each treatment, at least 100 pollen grains and pollen tubes were randomly chosen for germination frequency calculation and pollen tube length measurements. Pollen tubes were observed under a microscope, photographed, and measured with the SMARTSCAPE program (Furi, Shanghai, China). All experiments were repeated three times.
Root Length Measurement and Root Hair Observation
Seeds from homozygous transgenic and Arabidopsis C24 wild-type plants were placed on Murashige and Skoog agar media containing different concentrations of additionally supplied Ca2+ (plus 3 or 20 mm), EGTA (0.1, 0.5, or 1 mm) or Ins(1,4,5)P3 (0.1, 0.5, 1, 5, or 10 mm), and grown in a phytotron for daily measurements of root length and observation of root hairs. For each transgenic line at least 30 seedlings were chosen randomly for measurements. All experiments were repeated three times. For SEM analysis, specimens were dried at critical point, and coated with gold before examination in an S-4160 Field Emission Scanning Electron Microscope (Hitachi, Tokyo Japan) at an accelerating voltage of 10 kV.
Localization of AtIPK2α-GFP Fusion Protein
Localization of AtIPK2 protein was tested by transient expression of an AtIPK2α-GFP fusion protein in onion (Allium cepa) epidermal cells. The AtIPK2α coding region was amplified by PCR using primers IPK-7 (5′-ACGCGTCGACATGCAGCTCAAAGTCCCTG-3′; SalI site underlined, start codon in italics) and IPK-8 (CATGACTAGTGAATCTGCAGACTCATCTG; SpeI site underlined, stop codon substituted). Plasmid pET-AtIPK2αserved as template. The resulting PCR product was cloned into the vector pA7-GFP (kindly provided by Dr. K. Czempinski, University of Potsdam, Germany), yielding plasmid pA7-AtIPK-GFP. In this plasmid expression of the AtIPK2α-GFP fusion transcript was under the control of the Cauliflower Mosaic Virus 35S promoter. The resulting construct was sequenced to confirm an intact in-frame fusion, and to check for the absence of nucleotide errors that may have been introduced by PCR. Plasmid pA7-AtIPK-GFP was used for transient transformation of onion epidermal cells using a particle gun (Bio-Rad). Expression and localization of AtIPK2α-GFP fusion protein in onion cells was observed under a confocal laser scanning microscope (Zeiss LSM 510 META, Wavelength 488 nm). Plasmolysis of onion epidermis cells was induced by 1 m Suc.
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession numbers AJ404678 (AtIPK2β), AJ290975 (HuIP3K), D38169 (human IP3K), and H36803 (Arabidopsis EST).
Acknowledgments
Bernd Mueller-Roeber thanks the Max-Planck Institute of Molecular Plant Physiology, Golm, for providing lab space and access to infrastructure.
This work was supported by the State Key Project of Basic Research (grant no. G1999011604), by the National Natural Sciences Foundation of China (grant no. 30100101), by the Chinese Academy of Sciences, by The British Council (British-German, The Academic Research Collaboration program), by The Royal Society (UK-China Joint Project), and by the Deutsche Forschungsgemeinschaft (SFB 429).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.045427.
References
- Alfandari D, Darribere T (1994) A simple PCR method for screening cDNA libraries. PCR Methods Appl 4: 46–49 [DOI] [PubMed] [Google Scholar]
- Bechet J, Greenson M, Wiame JM (1970) Mutation affecting the repressibility of arginine biosynthetic enzymes in Saccharomyces cerevisiae. Eur J Biochem 12: 31–39 [DOI] [PubMed] [Google Scholar]
- Bechtold N, Ellis J, Pelletier G (1993) In planta Agrobacterium gene transfer by infiltration of adult Arabidopsis thaliana plants. C R Acad Sci Ser III Sci Vie 316: 1194–1199 [Google Scholar]
- Berridge MJ (1997) Elementary and global aspects of calcium signalling. J Physiol 499: 291–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertsch U, Haefs M, Moller M, Deschermeier C, Fanick W, Kitzerow A, Ozaki S, Meyer HE, Mayr GW (1999) A novel A-isoform-like inositol 1,4,5-trisphosphate 3-kinase from chicken erythrocytes exhibits alternative splicing and conservation of intron positions between vertebrates and invertebrates. Gene 228: 61–71 [DOI] [PubMed] [Google Scholar]
- Bibikova TN, Zhigiler A, Gilroy S (1997) Root hair growth in Arabidopsis thaliana is directed by calcium and an endogenous polarity. Planta 203: 495–505 [DOI] [PubMed] [Google Scholar]
- Choi KY, Kim HK, Lee SY, Moon KH, Sim SS, Kim JW, Chung HK, Rhee SG (1990) Molecular cloning and expression of a complementary DNA for inositol 1,4,5-trisphosphate 3-kinase. Science 248: 64–66 [DOI] [PubMed] [Google Scholar]
- Clandinin TR, DeModena JA, Sternberg PW (1998) Inositol trisphosphate mediates a RAS-independent response to LET-23 receptor tyrosine kinase activation in C. elegans. Cell 92: 523–533 [DOI] [PubMed] [Google Scholar]
- Communi D, Vanweyenberg V, Erneux C (1995) Molecular study and regulation of D-myo-inositol 1,4,5-trisphosphate 3-kinase. Cell Signal 7: 643–650 [DOI] [PubMed] [Google Scholar]
- Cullen PJ (1998) Bridging the GAP in inositol 1,3,4,5-tetrakisphosphate signalling. Biochim Biophys Acta 1436: 35–47 [DOI] [PubMed] [Google Scholar]
- Dewaste V, Pouillon V, Shears S, Takazawa K, Erneux C (2000) Cloning and expression of a cDNA encoding human inositol 1,4,5-trisphosphate 3-kinase C. Biochem J 352: 343–351 [PMC free article] [PubMed] [Google Scholar]
- Dubois E, Dewaste V, Erneux C, Messenguy F (2000) Inositol polyphosphate kinase activity of Arg82/ArgRIII is not required for the regulation of the arginine metabolism in yeast. FEBS Lett 486: 300–304 [DOI] [PubMed] [Google Scholar]
- Elge S, Brearley C, Xia H-J, Kehr J, Xue H-W, Mueller-Roeber B (2001) An Arabidopsis inositol phospholipid kinase strongly expressed in procambial cells: synthesis of PtdIns(4,5)P2 and PtdIns(3,4,5)P3 in insect cells by 5-phosphorylation of precursors. Plant J 26: 561–571 [DOI] [PubMed] [Google Scholar]
- Fan LM, Wang YF, Wang H, Wu WH (2001) In vitro Arabidopsis pollen germination and characterization of the inward potassium currents in Arabidopsis pollen grain protoplasts. J Exp Bot 52: 1603–1614 [PubMed] [Google Scholar]
- Felle HH, Hepler PK (1997) The cytosolic Ca2+ concentration gradient of Sinapis alba root hairs as revealed by Ca2+ selective microelectrode tests and fura-dextran ratio imaging. Plant Physiol 114: 39–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin-Tong VE, Drøbak BK, Allan AC, Trewavas AJ (1996) Growth of pollen tubes of Papaver rhoeas is regulated by a slow moving calcium wave propagated by inositol 1,4,5-trisphosphate. Plant Cell 8: 1305–1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin-Tong VE, Holdaway-Clarke T, Straatman KR, Kunkel JG, Hepler PK (2002) Involvement of extracellular calcium influx in the self-incompatibility response of Papaver rhoeas. Plant J 29: 333–345 [DOI] [PubMed] [Google Scholar]
- Hartweck LM, Llewellyn DJ, Dennis ES (1997) The Arabidopsis thaliana genome has multiple divergent forms of phosphoinositol-specific phospholipase C1. Gene 202: 151–156 [DOI] [PubMed] [Google Scholar]
- Hill TD, Dean NM, Boyton AL (1988) Inositol 1,3,4,5-tetrakisphosphate induces Ca2+ sequestration in rat liver cells. Science 242: 1176–1178 [DOI] [PubMed] [Google Scholar]
- Hirayama T, Ohto C, Mizoguchi T, Shinozaki K (1995) A gene encoding a phosphatidylinositol-specific phospholipase C is induced by dehydration and salt stress in Arabidopsis thaliana. Proc Natl Acad Sci USA 92: 3903–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holdaway-Clarke TL, Feijo JA, Hackett GA, Kunkel JG, Hepler PK (1997) Pollen tube growth and the intracellular-cytosolic calcium gradient oscillate in phase while extracellular calcium influx is delayed. Plant Cell 9: 1999–2210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson SL, Heath IB (1993) Roles of calcium ions in hyphal tip growth. Microbiol Rev 57: 367–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA (1987) Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol Biol Report 5: 387–405 [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemtiri-Chlieh F, MacRobbie EA, Webb AA, Manison NF, Brownlee C, Skepper JN, Chen J, Prestwich GD, Brearley CA (2003) Inositol hexakisphosphate mobilizes an endomembrane store of calcium in guard cells. Proc Natl Acad Sci USA 100: 10091–10095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malho R (1998) Role of 1,4,5-inositol trisphosphate-induced Ca2+ release in pollen tube orientation. Sex Plant Reprod 11: 231–235 [Google Scholar]
- Malho R (1999) Coding information in plant cells: the multiple roles of Ca2+ as a second messenger. Plant Biol 1: 487–494 [Google Scholar]
- Messenguy F, Dubois E (1993) Genetic evidence of a role for MCM1 in the regulation of arginine metabolism in Saccharomyces cerevisiae. Mol Cell Biol 13: 2586–2592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messerli M, Robinson KR (1997) Tip localized Ca2+ pulses are coincident with peak pulsatile growth rates in pollen tubes of Lilium longiflorum. J Cell Sci 110: 1269–1278 [DOI] [PubMed] [Google Scholar]
- Mikami K, Katagiri T, Iuchi S, Yamaguchi-Shinozaki K, Shinozaki K (1998) A gene encoding phosphatidylinositol-4-phosphate 5-kinase is induced by water stress and abscisic acid in Arabidopsis thaliana. Plant J 15: 563–568 [DOI] [PubMed] [Google Scholar]
- Miller DD, Callaham DA, Gross DJ, Helper PK (1992) Free Ca2+ gradient in growing pollen tubes of Lilium. J Cell Sci 101: 7–12 [Google Scholar]
- Morris AP, Gallacher DV, Irvine RF, Petersen GH (1987) Synergism of inositol trisphosphate and tetrakisphosphate in activating Ca2+-dependent K+ channels. Nature 330: 653–655 [DOI] [PubMed] [Google Scholar]
- Mueller-Roeber B, Pical C (2002) Inositol phospholipid metabolism in Arabidopsis: characterised and putative isoforms of inositol phospholipid kinase and phosphoinositide-specific phospholipase C. Plant Physiol 130: 22–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odom AR, Stahlberg A, Wente SR, York JD (2000) A role for nuclear inositol 1,4,5-trisphosphate kinase in transcriptional control. Science 287: 2026–2029 [DOI] [PubMed] [Google Scholar]
- Pierson ES, Miller DD, Callaham DA, Shipley MA, Rivers BA, Cresti M, Hepler PK (1994) Pollen tube growth is coupled to the extracellular calcium ion flux and the intracellular calcium gradient: effect of BAPTA-type buffers and hypertonic media. Plant Cell 6: 1815–1828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudd JJ, Franklin-Tong VE (2001) Unravelling response-specificity in Ca2+ signalling pathways in plant cells. New Phytol 151: 7–33 [DOI] [PubMed] [Google Scholar]
- Saiardi A, Caffrey JJ, Snyder SH, Shears SB (2000) Inositol polyphosphate multikinase (ArgRIII) determines nuclear mRNA export in Saccharomyces cerevisiae. FEBS Lett 468: 28–32 [DOI] [PubMed] [Google Scholar]
- Saiardi A, Erdjument-Bromage H, Snowman AM, Tempst P, Snyder SH (1999) Synthesis of diphosphoinositol pentakisphosphate by a newly identified family of higher inositol polyphosphate kinases. Curr Biol 9: 1323–1326 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Sanders D, Pelloux J, Brownlee C, Harper JF (2002) Calcium at the crossroads of signalling. Plant Cell (Suppl) 14: S401–S417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim SS, Kim JW, Rhee SG (1990) Regulation of D-myo-inositol 1,4,5-trisphosphate 3-kinase by cAMP-dependent protein kinase and protein kinase C. J Biol Chem 265: 10367–10372 [PubMed] [Google Scholar]
- Sims CE, Allbritton NL (1998) Metabolism of inositol 1,4,5-trisphosphate and inositol 1,3,4,5-tetrakisphosphate by the oocytes of Xenopus laevis. J Biol Chem 273: 4052–4058 [DOI] [PubMed] [Google Scholar]
- Stevenson JM, Perera IY, Boss WF (1998) A phosphatidylinositol 4-kinase pleckstrin homology domain that binds phosphatidylinositol 4-monophosphate. J Biol Chem 273: 22761–22767 [DOI] [PubMed] [Google Scholar]
- Stevenson-Paulik J, Odom AR, York JD (2002) Molecular and biochemical characterization of two plant inositol polyphosphate 6-/3-/5-kinases. J Biol Chem 277: 42711–42718 [DOI] [PubMed] [Google Scholar]
- Takazawa K, Perret J, DuMont JE, Erneux C (1991. a) Molecular cloning and expression of a human brain inositol 1,4,5-trisphosphate 3-kinase. Biochem Biophys Res Commun 174: 529–535 [DOI] [PubMed] [Google Scholar]
- Takazawa K, Perret J, DuMont JE, Erneux C (1991. b) Molecular cloning and expression of a new putative inositol 1,4,5-trisphosphate 3-kinase isoenzyme. Biochem J 278: 883–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas S, Brake B, Luzio JP, Stanley K, Banting G (1994) Isolation and sequence of a full length cDNA encoding a novel rat inositol 1,4,5-trisphosphate 3-kinase. Biochim Biophys Acta 1220: 219–222 [DOI] [PubMed] [Google Scholar]
- Wymer CL, Bibikova TN, Gilroy S (1997) Cytoplasmic free calcium distribution during the development of root hairs of Arabidopsis thaliana. Plant J 12: 427–439 [DOI] [PubMed] [Google Scholar]
- Xia HJ, Brearley C, Elge S, Kaplan B, Fromm H, Mueller-Roeber B (2003) Arabidopsis inositol polyphosphate 6-/3-kinase is a nuclear protein that complements a yeast mutant lacking a functional ArgR-Mcm1 transcription complex. Plant Cell 15: 449–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue H-W, Hosaka H, Mueller-Roeber B (2000) Cloning of Arabidopsis thaliana phosphatidylinositol synthase and functional expression in the yeast pis mutant. Plant Mol Biol 42: 757–764 [DOI] [PubMed] [Google Scholar]
- Xue H-W, Pical C, Brearley C, Elge S, Mueller-Roeber B (1999) A plant 126-kDa phosphatidylinositol 4-kinase with a novel repeat structure: cloning and functional expression in baculovirus-infected insect cells. J Biol Chem 274: 5738–5745 [DOI] [PubMed] [Google Scholar]
- York JD, Odom AR, Murphy R, Ives EB, Wente SR (1999) A phospholipase C-dependent inositol polyphosphate kinase pathway required for efficient messenger RNA export. Science 285: 96–100 [DOI] [PubMed] [Google Scholar]