Abstract
The trafficking and function of cell surface proteins in eukaryotic cells may require association with detergent-resistant sphingolipid- and sterol-rich membrane domains. The aim of this work was to obtain evidence for lipid domain phenomena in plant membranes. A protocol to prepare Triton X-100 detergent-resistant membranes (DRMs) was developed using Arabidopsis (Arabidopsis thaliana) callus membranes. A comparative proteomics approach using two-dimensional difference gel electrophoresis and liquid chromatography-tandem mass spectrometry revealed that the DRMs were highly enriched in specific proteins. They included eight glycosylphosphatidylinositol-anchored proteins, several plasma membrane (PM) ATPases, multidrug resistance proteins, and proteins of the stomatin/prohibitin/hypersensitive response family, suggesting that the DRMs originated from PM domains. We also identified a plant homolog of flotillin, a major mammalian DRM protein, suggesting a conserved role for this protein in lipid domain phenomena in eukaryotic cells. Lipid analysis by gas chromatography-mass spectrometry showed that the DRMs had a 4-fold higher sterol-to-protein content than the average for Arabidopsis membranes. The DRMs were also 5-fold increased in sphingolipid-to-protein ratio. Our results indicate that the preparation of DRMs can yield a very specific set of membrane proteins and suggest that the PM contains phytosterol and sphingolipid-rich lipid domains with a specialized protein composition. Our results also suggest a conserved role of lipid modification in targeting proteins to both the intracellular and extracellular leaflet of these domains. The proteins associated with these domains provide important new experimental avenues into understanding plant cell polarity and cell surface processes.
Biological membranes consist of a perplexing number of lipids (Edidin, 2003a). The classical model of membranes assumes that these lipids form a homogeneous fluid-like or liquid-disordered (ld) phase, which allows free diffusion of individual molecules and resident proteins (Edidin, 2003b). However, numerous recent studies on model membranes have demonstrated that certain lipids, in particular sphingolipids and cholesterol, may form relatively stable clusters by tight self-association, thus segregating them from surrounding phospholipids (Schroeder et al., 1994; Ahmed et al., 1997; Dietrich et al., 2001; Silvius, 2003). The association of rigid sterol molecules with the long and saturated acyl chains of sphingolipids results in the formation of a more organized, liquid-ordered (lo) phase; lo and ld phases can coexist in the same membrane (Brown and London, 1998; Edidin, 2003b). The lipid raft hypothesis postulates that a sterol- and sphingolipid-rich lo phase is also present in cell membranes and that it forms discrete microdomains or lipid rafts that diffuse in the bulk of the ld phospholipid phase (Simons and Ikonen, 1997; Mayor and Rao, 2004).
There is substantial evidence supporting the existence of plasma membrane (PM) domains in animal cells. Fluorescence resonance energy transfer (Varma and Mayor, 1998; Kenworthy et al., 2000; Sharma et al., 2004), single-particle tracking (Pralle et al., 2000; Dietrich et al., 2002), microscopy (Gaus et al., 2003; Prior et al., 2003), and molecular cross-linking (Friedrichson and Kurzchalia, 1998) have been used to study microdomains. Although estimates of size vary with cell type and method, the emerging consensus is that lipid rafts are small (in the low-nanometer range) but capable of clustering into larger complexes (Harder and Engelhardt, 2004; Mayor and Rao, 2004). A consequence of the tight association of the lipids in microdomains is their reduced solubility in nonionic detergents at low temperatures (Brown and Rose, 1992; London and Brown, 2000; Schuck et al., 2003). Thus, lipid rafts and the proteins associated with them can be separated from nonraft membranes by suitable detergent extraction. The resulting fraction of detergent-resistant membranes (DRMs) is thought to consist of aggregates of the microdomains. This technique has been used extensively to investigate the protein composition of lipid rafts (e.g. Foster et al., 2003; Schuck et al., 2003). The data that have been obtained by analysis of DRMs are largely consistent with those obtained by other biochemical and microscopic approaches (Dietrich et al., 2001; Shogomori and Brown, 2003).
In animal and yeast (Saccharomyces cerevisiae) cells, lipid rafts are believed to function as sorting platforms for proteins destined for the PM (Ikonen, 2001); accordingly, raft formation begins in the endoplasmic reticulum (ER) or Golgi apparatus (Brown and Rose, 1992; Muniz and Riezman, 2000; Helms and Zurzolo, 2004). Even though originally postulated to be responsible for sorting in polarized cells, it is now becoming increasingly accepted that lipid microdomains exist in many different eukaryotic cell types, including cells with no apparent polarity (Bagnat et al., 2001; Ikonen, 2001; Prior et al., 2003). In addition to protein targeting, lipid rafts have been implicated in numerous cell-surface processes, including signal transduction, pathogen entry, secretion, and endocytosis (for review, see Simons and Toomre, 2000; Ikonen, 2001; Harder and Engelhardt, 2004; Mayor and Rao, 2004; Pike, 2004). Specific posttranslational modifications have been reported to confer raft association. Glycosylphosphotidylinositol (GPI) anchoring was the first and best characterized of these signals (Shogomori and Brown, 2003); similarly, N-terminal double acylation and palmitoylation of membrane proteins have also been reported to target proteins to lipid rafts (Morrow et al., 2002; Zacharias et al., 2002; for review, see Resh, 1999).
In plant cells, recent evidence suggests that rafts may play an important role in protein targeting. GPI-anchored proteins (GAPs), which are associated with DRMs in mammalian and yeast cells, are diverse and abundant in Arabidopsis (Arabidposis thaliana; Sherrier et al., 1999; Borner et al., 2002, 2003; Elortza et al., 2003). At least two GAPs, COBRA and SKU5, have a nonuniform (SKU5) or polarized (COBRA) distribution in the PM of Arabidopsis root cells (Schindelman et al., 2001; Sedbrook et al., 2002). Furthermore, the components required for the formation of lipid rafts are present in plants: glucosylceramide, an important component of animal microdomains (Simons and van Meer, 1988; Simons and Ikonen, 1997), is an abundant constituent of plant PMs (Warnecke and Heinz, 2003), and plant membranes contain sterols analogous to cholesterol, including sitosterol, campesterol, and stigmasterol (Clouse, 2002). Finally, the polarized distribution of the putative PM auxin transporter PIN1 is disrupted in the sterol-deficient Arabidopsis mutant orc, demonstrating the potential importance of sterols in plant protein targeting (Willemsen et al., 2003). DRMs can be prepared from tobacco (Nicotiana tabacum) PMs and contain several GAPs, although the identity and enrichment of the GAPs in the DRMs were not determined (Peskan et al., 2000). In general, the specificity of procedures for DRM preparation from plant membranes has not been clearly demonstrated, and also the protein and lipid composition of DRMs remains poorly characterized.
In this study, we developed a protocol to prepare DRMs from Arabidopsis and investigated their composition using immunoblots, proteomics, and lipid analysis. The protein composition of the DRMs indicates that the procedure is highly specific. The results strongly support the hypothesis that Arabidopsis DRMs are predominantly derived from PM sphingolipid- and sterol-rich lipid rafts.
RESULTS
A GPI-Anchored Reporter Protein Is Targeted to the PM
GAPs are often found enriched in DRMs prepared from mammalian and yeast cells. We hypothesized that, in Arabidopsis, GAPs could provide useful biochemical markers for the preparation of DRMs. We therefore generated transgenic Arabidopsis expressing a GPI-anchored reporter protein. The reporter was based on AtAGP4, which was predicted to be GPI anchored (Sherrier et al., 1999), and bacterial phosphinothricin acetyl transferase (PAT), which appears to be free of sorting determinants recognized by the plant secretory machinery (Denecke et al., 1990). A secretory version of PAT with an N-terminal signal peptide was fused to a double-myc tag for immunodetection and the 59 C-terminal amino acids of AtAGP4 containing the predicted GPI-anchoring signal to create the PAT-GPI4 reporter protein (Fig. 1A).
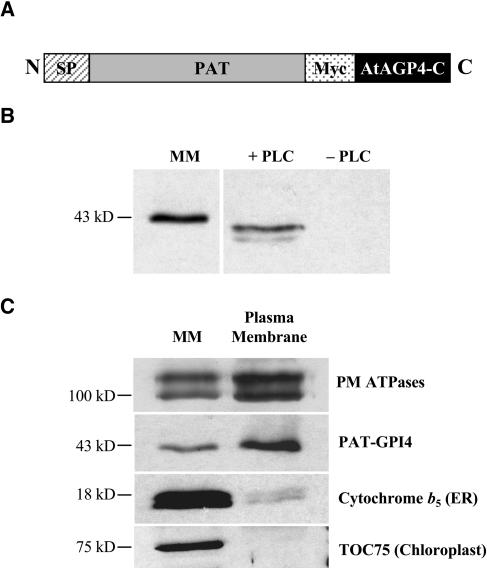
Figure 1.
PAT-GPI4 is a GPI-anchored PM protein. A, Schematic organization of the PAT-GPI4 reporter protein. SP, Signal peptide; Myc, double-myc tag; black bar, 59 C-terminal amino acids of AtAGP4. B, Detection of PAT-GPI4 in MM from callus and sensitivity of PAT-GPI4 to PLC. Aqueous phase proteins of Triton X-114 partitioned MM after PLC treatment (+PLC) or mock PLC treatment (−PLC). Proteins were separated by SDS-PAGE and detected by anti-myc immunoblot. C, PM targeting. The PM-enriched fraction was prepared by dextran-PEG phase partitioning. Proteins were separated by SDS-PAGE and detected by immunoblot. A mixed organelle membrane fraction was included as control. Equal quantities of protein were loaded in each lane.
To investigate GPI anchoring and subcellular localization of PAT-GPI4, callus cultures were generated from roots of transgenic Arabidopsis plants. Mixed organelle membranes (MMs, including most endomembrane organelles, mitochondria, and plastids [Prime et al., 2000]) were prepared by centrifugation in a Suc density gradient. Western blotting with an anti-myc antibody showed that PAT-GPI4 was present in membranes in the callus cells (Fig. 1B). The apparent molecular mass of 43 kD was larger than expected for the reporter construct, perhaps due to dimerization or glycosylation. However, the identity of the protein was subsequently confirmed by mass spectrometry (MS; see below).
We investigated GPI anchoring of the reporter protein using sensitivity to phosphatidylinositol-specific phospholipase C (Pi-PLC; Borner et al., 2003). MM proteins were fractionated by Triton X-114 phase partitioning to separate integral and GPI-anchored proteins in the detergent phase from the peripheral proteins in the aqueous phase. The detergent phase was then incubated with Pi-PLC and subjected again to phase partitioning. As shown in Figure 1B, the PAT-GPI4 protein was released into the aqueous phase by Pi-PLC treatment. Thus, PAT-GPI4 is GPI anchored in the transgenic Arabidopsis callus cells.
Many GAPs are PM localized, and GPI anchoring has been proposed to target proteins to the PM in plants (Sherrier et al., 1999). To investigate the localization of PAT-GPI4 within cells, we prepared a PM fraction by dextran-polyethylene glycol (PEG) partitioning of the MMs and examined these fractions by western blotting. Figure 1C shows the enrichment of PM ATPases in the PM fraction, and depletion of the ER protein cytochrome b5 and the plastid envelope protein TOC75. The distribution of PAT-GPI4 was similar to that of the PM ATPases, indicating that PAT-GPI4 was largely PM localized in the callus cells. This strongly supports the view that GPI anchoring is a PM-targeting signal in plants (Sherrier et al., 1999).
Identification of DRMs in Arabidopsis Cells
We investigated whether PAT-GPI4 was localized in membranes that are resistant to detergent solubilization. In preliminary experiments, we found that high Triton X-100 detergent-to-protein ratios solubilized essentially all the membranes. To find any conditions that could selectively reveal membranes more resistant to detergent extraction, we incubated MMs at 4°C using detergent-to-protein ratios between 2 and 8 (w/w). The membrane extracts were adjusted to 1.8 m Suc and overlaid with 1.6 and 0.15 m Suc steps. After centrifugation, DRMs were seen floating in two bands of slightly different densities above the 1.6 m Suc. Initially, both DRM bands were collected as a single fraction. As a control for the solubilization, MMs were processed in parallel, but without detergent. These floated membranes were called the total membrane (TM) controls.
The protein composition of the membranes extracted with the various detergent-to-protein ratios was analyzed by western blot. Figure 2A shows that the ER protein cytochrome b5 was largely solubilized, particularly at higher detergent-to-protein ratios. In contrast, a substantial fraction of PAT-GPI4 floated with the membranes resistant to detergent. Since the DRMs contained less than 1% of the protein present in the initial MMs, this result indicated that PAT-GPI4 was substantially enriched in the DRMs. The DRMs showed the highest relative enrichment of PAT-GPI4 over cytochrome b5 between a 4-fold and an 8-fold detergent-to-protein ratio. Investigation of the upper (ρ 1.15–1.18 g mL−1) and lower (ρ 1.19–1.2 g mL−1) bands of floating DRMs revealed that the upper DRMs were more highly enriched in PAT-GPI4 (data not shown); therefore, the lower fraction was not investigated further.
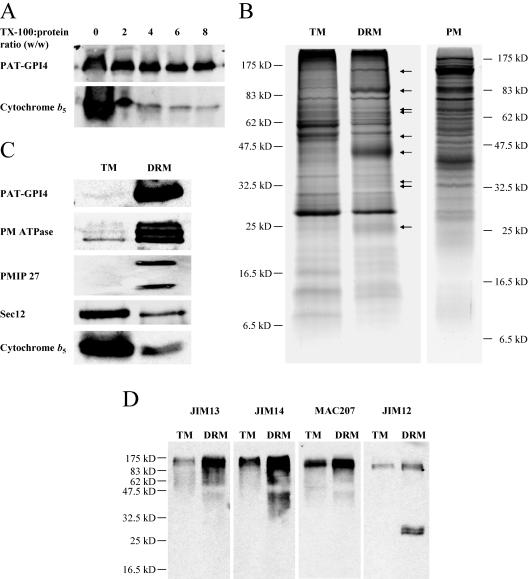
Figure 2.
Analysis of Arabidopsis DRMs by SDS-PAGE and immunoblot reveals a specific protein composition. A, Influence of detergent to protein ratio on DRM composition. DRMs were prepared from callus membranes with detergent ranging from 0:1 to 8:1 (Triton X-100:protein; w/w). Equal proportions of recovered DRMs were separated by SDS-PAGE and proteins were detected by immunoblot. B, Protein composition of Arabidopsis callus TMs and DRMs. Equal quantities of protein were separated by SDS-PAGE and stained with Coomassie Blue. Arrows indicate proteins enriched in the DRMs relative to TMs. Callus PM is shown for comparison. C, TM and DRM proteins analyzed by immunoblot. Equal quantities of protein were loaded in each lane. D, Cell surface glycoproteins in DRMs. Proteins were detected with monoclonal antibodies against arabinogalactan carbohydrates (JIM13, JIM14, and MAC207) or extensin carbohydrates (JIM12).
The difference in solubility of PAT-GPI4 and cytochrome b5 suggested that the Arabidopsis DRMs had a protein composition distinct from TMs. To investigate this further, DRMs were prepared at a 6-fold detergent-to-protein ratio. Similar quantities of proteins from TM and DRM fractions were separated by SDS-PAGE and stained with Coomassie Blue. Certain proteins indeed appeared enriched in the DRMs (Fig. 2B, arrows), whereas many of the major TM proteins were depleted. To investigate in more detail, the two fractions were analyzed by western blotting. Figure 2C shows that, when compared on an equivalent protein basis, PAT-GPI4 showed a very large enrichment in the DRMs. Two other PM proteins, the PM ATPases and a PM intrinsic protein/aquaporin (PMIP), were also substantially enriched in the DRMs. In contrast, two ER proteins, Sec12 and cytochrome b5, were depleted from the DRMs. This comparison indicated that endomembrane proteins show differential solubility in Triton X-100, and furthermore suggested that certain PM proteins are particularly resistant to solubilization.
Arabinogalactan proteins (AGPs) are an abundant and diverse class of proteins, many of which are predicted to be attached to the cell surface by a GPI anchor (Borner et al., 2002, 2003; Schultz et al., 2002). To investigate whether this class of proteins might be localized in DRMs, we used monoclonal antibodies JIM13, JIM14, and MAC207 that recognize specific epitopes in the glycans attached to AGPs. Western blotting revealed an enrichment of these heterogeneously glycosylated proteins in the DRMs (Fig. 2D). In addition, two proteins containing extensin glycosylation recognized by JIM12, of apparent 30-kD molecular mass, were highly enriched in the DRMs. These experiments suggested that certain AGPs and extensin-like proteins are present in DRMs.
Proteomic Analysis of DRM Composition
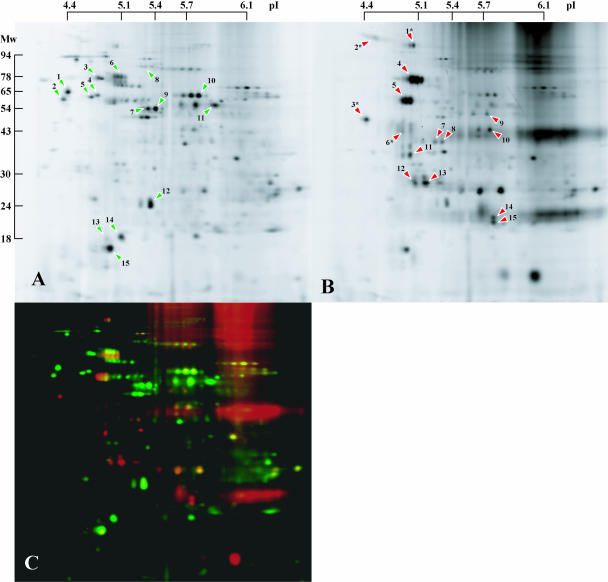
The western-blot analysis indicated that many intracellular proteins were depleted, but not completely solubilized, by detergent treatment. We investigated further the identity of DRM-depleted and DRM-enriched proteins by two-dimensional (2D)-difference gel electrophoresis (DIGE). Equal quantities of control (TM) and DRM membrane proteins were labeled with different CyDyes, pooled, and separated by 2D-PAGE. Although both DRM and TM fractions were complex mixtures of proteins, DIGE allowed a clear distinction between them (Fig. 3). In the DRMs, there was a clear enrichment of many proteins, and most of the TM protein spots were depleted. Identification of the proteins depleted from the DRMs (Fig. 3A, green arrowheads) showed that they were predominantly derived from the ER or mitochondria (Table I). An AtVSR Golgi/prevacuolar compartment-sorting receptor was also solubilized by the detergent (Fig. 3A, spot 8). These findings are consistent with the western analysis, which revealed depletion of intracellular organelle proteins.
Figure 3.
2D-DIGE analysis of Arabidopsis DRMs. TM and DRM fractions were labeled with fluorescent CyDyes, mixed, and separated in single 2D gels. Proteins were detected with a fluorescence scanner. A typical pair of scans is shown. A, TMs. B, DRMs. C, False color overlay of TMs (green) and DRMs (red). Green arrowheads in A indicate Arabidopsis ER, Golgi, and mitochondrial proteins that were depleted from the DRM fraction. Red arrowheads in B indicate proteins specifically enriched in the DRM fraction that were identified by MS/MS. GAPs are marked by an asterisk. Numbers on arrowheads correspond to the identifications listed in Table I.
Table I.
Proteins enriched in or depleted from Arabidopsis DRMs-DIGE analysis
Proteins were separated and analyzed by 2D-DIGE. Proteins enriched in the DRM fraction labeled with red arrowheads in Figure 3B were identified by LC-MS/MS. Proteins depleted from the DRMs labeled with green arrowheads in Figure 3A were identified by comparison of DIGE gels with the 2D database of Arabidopsis organelle proteins (Prime et al., 2000).
| Spot | Protein Family | MIPS Code | Subcellular Localization |
|---|---|---|---|
| Enriched 1 | HIPL1 hedgehog-interacting-protein-like 1 | At1g74790 | PM (GPI anchor) |
| Enriched 2 | GPDL1 glycerophosphodiesterase-like protein 1 | At5g55480 | PM (GPI anchor) |
| Enriched 3 | Unknown protein | At1g29980 | PM (GPI anchor) |
| Enriched 4 | AtVHA-A V1-ATPase subunit | At1g78900 | Endomembranes/PM |
| Enriched 5 | AtVHA-B1 V1-ATPase subunit | At1g76030 | Endomembranes/PM |
| Enriched 5 | AtVHA-B2 V1-ATPase subunit | At4g38510 | Endomembranes/PM |
| Enriched 5 | AtVHA-B3 V1-ATPase subunit | At1g20260 | Endomembranes/PM |
| Enriched 6 | PAT-GPI | –a | PM (GPI anchor) |
| Enriched 7,8 | AtVHA-C V1-ATPase subunit | At1g12840 | Endomembranes/PM |
| Enriched 9 | AtFlot1 (related to flotillin) | At5g25250 | Unknown |
| Enriched 10 | Stomatin like | At4g27585 | Unknown |
| Enriched 11 | Putative quinone reductase | At4g36750 | Unknown |
| Enriched 12 | Putative hypersensitive response protein | At1g69840 | Unknown |
| Enriched 13 | Putative hypersensitive response protein | At5g62740 | Unknown |
| Enriched 14,15 | FQR1 Putative quinone reductase | At5g54500 | Unknown |
| Depleted 1 | Calreticulin | At1g56340 | ER |
| Depleted 2 | Calreticulin | At1g09210 | ER |
| Depleted 3 | Calnexin | At5g61790 | ER |
| Depleted 4 | Protein disulfide isomerase | At1g77510 | ER |
| Depleted 5 | Protein disulfide isomerase | At1g21750 | ER |
| Depleted 6 | BiP | At5g42020 | ER |
| Depleted 7 | HSP60 chaperonin | At3g23990 | Mitochondria |
| Depleted 8 | AtVSR | At2g14740 | Golgi/prevacuole |
| Depleted 9 | ATP synthase β-subunit | At5g08690 | Mitochondria |
| Depleted 10 | Peptidase | At3g02090 | Mitochondria |
| Depleted 11 | ATP synthase α-subunit | At2g07698 | Mitochondria |
| Depleted 12 | Probable ATP synthase 24-kD subunit | At2g21870 | Mitochondria |
| Depleted 13 | Cytochrome b5 | At5g53560 | ER |
| Depleted 14 | ATP synthase δ-subunit | At5g47030 | Mitochondria |
| Depleted 15 | ATP synthase δ-subunit | At3g52300 | Mitochondria |
PAT-GPI was specifically engineered for this study and therefore has no MIPS accession number.
In contrast, many of the proteins in the DRMs were undetectable in the TM fraction, indicating that they were highly enriched by the detergent extraction procedure. We identified 15 DRM-enriched proteins by liquid chromatography coupled with tandem mass spectrometry (LC-MS/MS) of tryptic fragments from excised gel spots (Fig. 3B, red arrowheads; Table I). Consistent with the western analysis, PAT-GPI4 was among these proteins. Three further GAPs were identified (Fig. 3B, spots 1–3), supporting the notion that multiple GAPs are targeted to DRMs. Both the HIPL1 and GPDL1 proteins have previously been confirmed to become GPI anchored, whereas At1g29980 is a predicted GAP (Borner et al., 2003). Five of the proteins were V-type ATPase subunits. In addition, a recently described quinone reductase (FQR1; Laskowski et al., 2002) and a further five previously uncharacterized proteins were identified.
Since many integral membrane proteins are not effectively resolved by 2D-PAGE, we identified DRM proteins by separation with one-dimensional (1D)-PAGE and LC-MS/MS (Fig. 2B; Table II). Proteins were identified both from gel slices containing bands showing enrichment in the DRMs and in the corresponding gel slices from the control TM lane. Proteins with a MASCOT score both above 150 and also at least 2-fold higher from the DRMs than the control TMs were judged to be enriched in the DRMs. This analysis confirmed the DRM enrichment shown by western analysis of PM ATPases and PMIPs, and also of the V-type ATPases and the GPI-anchored GPDL1 revealed by DIGE. Several further DRM-enriched proteins were identified. Two P-glycoproteins (PGP)/multiple drug resistance (MDR) proteins were identified. Five further GAPs, including SKU5 (Sedbrook et al., 2002) and a close homolog, SKS1, were also identified, strengthening the finding that certain GAPs are present in DRMs. Mitochondrial ATPase subunits, ribosomal protein, and β-glucosidase are likely to be present in DRM fractions because they are highly abundant.
Table II.
Membrane proteins enriched in Arabidopsis DRMs-SDS-PAGE analysis
Proteins enriched in the DRM fraction were identified by LC-MS/MS of tryptic peptides from bands excised from a 1D SDS-PAGE gels. Proteins with a MASCOT score both above 150 and also at least 2-fold higher from the DRMs than the control TMs were judged to be enriched in the DRMs. Predicted molecular mass refers to molecular mass prior to any processing.
| Apparent Molecular Mass | Predicted Molecular Mass | Protein Family | MIPS Code | MASCOT Score | Location |
|---|---|---|---|---|---|
| kD | D | ||||
| 125–150 | 139,397 | AtPGP4/AtMDR4 PGP/MDR protein | At2g47000 | 973 | Unknown |
| 138,093 | AtPGP11/AtMDR8 PGP/MDR protein | At1g02520 | 361 | Unknown | |
| 84,511 | GPDL1 glycerophosphodiesterase-like 1 protein | At5g55480 | 197 | PM (GAP) | |
| 90–110 | 104,704 | AtAHA1 PM P3A-type H+ATPase | At2g18960 | 1,061 | PM |
| 104,734 | AtAHA2 PM P3A-type H+ATPase | At4g30190 | 1,147 | PM | |
| 105,650 | AtAHA4 PM P3A-type H+ATPase | At3g47950 | 320 | PM | |
| 104,946 | AtAHA6 PM P3A-type H+ATPase | At2g07560 | 261 | PM | |
| 104,521 | AtAHA8 PM P3A-type H+ATPase | At3g42640 | 235 | PM | |
| 105,598 | AtAHA9 PM P3A-type H+ATPase | At1g80660 | 266 | PM | |
| 105,056 | AtAHA11 PM P3A-type H+ATPase | At5g62670 | 292 | PM | |
| 93,616 | AtVHA-a2 integral V0-ATPase subunit | At2g21410 | 324 | Endomembranes/PM | |
| 93,344 | AtVHA-a3 integral V0-ATPase subunit | At4g39080 | 553 | Endomembranes/PM | |
| 60–80 | 69,111 | AtVHA-A V1-ATPase subunit | At1g78900 | 983 | Endomembranes/PM |
| 66,149 | Jacalin lectin family protein | At3g16460 | 379 | ER/vacuole | |
| 65,767 | SKU5 | At4g12420 | 323 | PM (GAP) | |
| 89,589 | TOC75 chloroplast outer envelope protein | At3g46740 | 253 | Chloroplast | |
| 60,477 | FAD-binding domain-containing protein | At4g20830 | 235 | Unknown | |
| 82,288 | ERD4 early responsive to dehydration stress protein | At1g30360 | 224 | Unknown | |
| 60,025 | β-Glucosidase | At1g66270 | 222 | ER/vacuole | |
| 72,942 | Glycosyl hydrolase family 3 protein | At5g04885 | 212 | PM (GAPa) | |
| 66,120 | SKS-1 | At4g25240 | 163 | PM (GAP) | |
| 50–60 | 54,385 | AtVHA-B2 V1-ATPase subunit | At4g38510 | 767 | Endomembranes/PM |
| 86,221 | ATP synthase α-subunit | At2g07698 | 343 | Mitochondria | |
| 59,805 | ATP synthase β-subunit 1 | At5g08670 | 301 | Mitochondria | |
| 32,797 | Receptor-like protein | At5g41290 | 151 | PM (GAP) | |
| 30–35 | 31,529 | Putative hypersensitive response protein | At3g01290 | 381 | Unknown |
| 31,671 | Putative hypersensitive response protein | At1g69840 | 334 | Unknown | |
| 31,696 | Putative hypersensitive response protein | At5g62740 | 252 | Unknown | |
| 32,138 | Jacalin lectin family protein | At3g16420 | 246 | ER/vacuole | |
| 29,634 | Putative porin | At5g67500 | 219 | Mitochondria/chloroplast | |
| 27,673 | RPS3A 40S ribosomal protein S3b | At2g31610 | 195 | Cytosol | |
| 27,612 | RPS3C 40S ribosomal protein S3b | At5g35530 | 195 | Cytosol | |
| 20,202 | AtLTPL lipid transfer protein-like | At1g27950 | 153 | PM (GAP) | |
| Nonspecific | 30,669 | PIP1A PM intrinsic proteins | At3g61430 | 157 | PM |
| 30,806 | PIP1B PM intrinsic proteins | At2g45960 | 248 | PM | |
| 29,952 | PIP3 PM intrinsic proteins | At4g35100 | 239 | PM |
GPI anchored (G.H.H. Borner, K.S. Lilley, and P. Dupree, unpublished data).
Unable to distinguish between these two proteins from MS data.
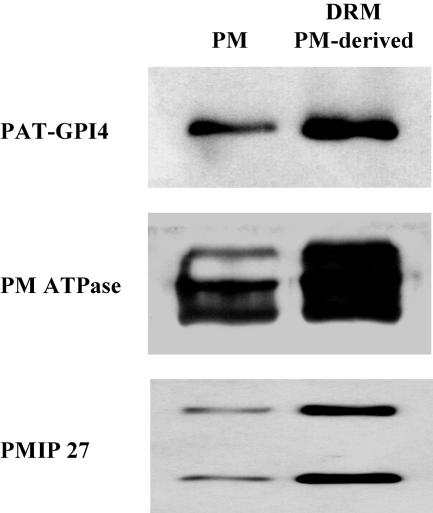
Since the majority of proteins in the DRMs were localized in the PM, we investigated whether DRMs are identical to PMs. When separated on 1D-PAGE, the DRM fraction differed significantly in protein composition from the PM (Fig. 2B). Furthermore, when DRMs were prepared from the PM only, 20% of the extracted PM protein was detergent resistant. Analysis of these DRMs by immunoblot showed enrichment of PAT-GPI4, PM ATPases, and PMIP27 (Fig. 4). Together, these observations suggest that DRMs represent a subset of PM proteins.
Figure 4.
DRMs prepared from PMs are enriched in DRM marker proteins. The PM was prepared from Arabidopsis callus as in Figure 1. DRMs were prepared as in Figure 2, but from the PM. PM and DRM proteins were analyzed by immunoblot. Equal quantities of protein were loaded in each lane.
Lipid Composition of DRMs
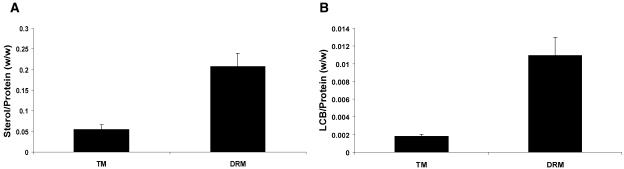
The physical basis for formation of lipid rafts is thought to be the association of sterols and sphingolipids (London and Brown, 2000). DRMs from mammalian cells and from yeast are enriched in these lipid types. If DRMs from Arabidopsis reflect a similar phenomenon, they might be enriched in phytosterols or phytosphingolipids. We investigated first the sterol composition of DRMs and TMs by gas chromatography (GC)-MS of trimethylsilyl derivatives. As shown in Figure 5A, the DRMs contained an approximately 4-fold higher sterol-to-protein ratio than the TMs. The quantity of sterol and the level of enrichment are similar to those seen in some mammalian DRM preparations (Nebl et al., 2002). Since Arabidopsis contains multiple sterols, we investigated whether individual sterols might contribute specifically to the DRMs. Table III shows that, in both fractions, β-sitosterol was the most abundant sterol. There was no evidence for a specific role of an individual sterol in the DRMs.
Figure 5.
Sterol and sphingolipid composition of TMs and DRMs. A, Sterol content. B, Sphingolipid LCB content.
Table III.
Relative sterol compositions of TMs and DRMs
Proportions are calculated as percentage of total sterols detected by GC-MS. Figures are means ± sd (%) of four TM and DRM preparations.
| Sterol | TM Mean ± sd | DRM Mean ± sd |
|---|---|---|
| % | % | |
| β-Sitosterol | 79.4 ± 4.7 | 81.0 ± 2.0 |
| Campesterol | 8.6 ± 1.3 | 10.0 ± 2.1 |
| Unidentified sterol | 12.1 ± 5.2 | 7.6 ± 1.3 |
| Stigmasterol | Trace | 1.2 ± 1.5 |
| Cholesterol | Trace | 0.2 ± 0.4 |
We next investigated any sphingolipid enrichment in the DRMs. Arabidopsis sphingolipids have not been well characterized in comparison to the body of research conducted on yeast and mammalian systems. Sphingolipids are generated by the addition of a polar head group to a ceramide back bone that is composed of a long chain base (LCB), N-acylated to a fatty acid chain. Two complex sphingolipids, cerebrosides, and glycosyl inositol phosphorylceramides (GIPCs) are both present in most plant tissue (Dunn et al., 2004). Depending on the head group, the sphingolipid classes show discrete solubilities in nonaqueous solvents. To better quantify the sphingolipids in the samples, we analyzed their LCBs after direct hydrolysis of total membrane lipids. Figure 5B shows that there was an approximately 5-fold higher LCB-to-protein ratio in the DRMs. Table IV shows the compositional analysis of the individual LCBs present in the total sphingolipids in DRM and TM fractions. Unlike the absence of differential sterol enrichment, this analysis indicated some preferential enrichment of specific LCBs. In particular, the levels of (8Z)-4-hydroxy-8-sphingenine (abbreviated t18:1c) are lower in the DRMs compared with the TMs. Also, the ratio of this LCB compared with its stereoisomer (8E)-4-hydroxy-8-sphingenine (t18:1t) is increased from approximately 3:1 in the TMs to 5:1 in the DRMs. This is consistent with DRMs containing a high proportion of GIPCs, which have a greater 8Z:8E ratio than cerebrosides.
Table IV.
Relative LCB compositions of TMs and DRMs
Proportions are mol%. Figures are means ± sd (%) of four TM and DRM preparations.
| LCB | TM Mean ± sd | DRM Mean ± sd |
|---|---|---|
| % | % | |
| t18:1c | 12.32 ± 0.82 | 5.06 ± 0.61 |
| t18:1t | 35.54 ± 6.17 | 28.10 ± 1.36 |
| t18:0 | 15.12 ± 4.94 | 19.02 ± 6.04 |
| d18:1 | 2.83 ± 0.21 | 2.63 ± 0.46 |
| d18:2 | 12.78 ± 0.92 | 13.14 ± 1.13 |
| d18:0 | 21.41 ± 2.00 | 32.05 ± 6.16 |
DISCUSSION
In this study, the protein and lipid composition of Arabidopsis DRMs was investigated. Previously, plant DRM preparation methods, the specificity of DRM preparation, and the intracellular origin of DRMs were poorly characterized. A protocol for the preparation of sterol- and sphingolipid-enriched DRMs was developed, using as a marker an engineered GAP, PAT-GPI4. Proteomic analysis methods identified over 40 proteins enriched in the DRMs, including 8 GAPs and several other known PM proteins. The DRM fraction was largely depleted of internal organelle proteins. The results clearly indicate that the DRMs are substantially derived from the PM.
The use of detergent resistance is widespread in the mammalian cell biology field to investigate lipid domain phenomena. Although it is relatively simple to prepare membrane fractions apparently resistant to detergent, it is important to ascertain the specificity of the procedure. Abundant proteins that are largely detergent soluble often remain significant contaminants of DRM preparations (Foster et al., 2003). Hence, the presence of a protein in DRMs alone is insufficient evidence for its preferential association with this membrane fraction; a comparative method is required to identify proteins that are specifically enriched in the DRMs. We have achieved this using western and 2D-DIGE analysis of DRMs prepared from Arabidopsis callus mixed organelle membranes. We identified a number of proteins that showed a large and specific enrichment in DRMs, whereas the majority of callus membrane proteins was predominantly solubilized by the detergent extraction. Our DRM extraction protocol can therefore be used to prepare and investigate properties of a specific group of proteins.
The comparative proteomics revealed that the DRMs are likely to be derived substantially from the plant cell PMs. There was a remarkable depletion of mitochondrial and ER proteins (Figs. 2 and 3; Table I). In contrast, many of the enriched proteins are known to reside at the PM (Tables I and II). Several PM ATPases were identified, suggesting this is one of the major DRM proteins. The PM ATPase is also enriched in DRMs from yeast (Bagnat et al., 2000). Second, PMIPs were shown by western analysis and MS to be enriched in the DRMs. A PM aquaporin has also been shown to be present in DRMs from mammalian cells (Palestini et al., 2003). Third, eight cell surface GAPs were enriched in DRMs; GAPs are also abundant constituents of animal and yeast DRMs (Brown and Rose, 1992; Bagnat et al., 2000). Furthermore, the GAP reporter PAT-GPI4 was also PM localized and detergent resistant. Since PAT is a secretory protein and probably devoid of any other targeting signals (Denecke et al., 1990), it is highly likely that PAT-GPI4's PM localization and insolubility are a direct consequence of its GPI anchoring. Also, some arabinogalactosylated proteins are enriched in the DRMs, and these may be attached to the membranes by GPI anchoring (Borner et al., 2002, 2003; Schultz et al., 2002). Collectively, these findings strongly support the hypothesis that plant DRMs are largely derived from the PM.
Several lines of evidence suggest that the Arabidopsis PM is not entirely resistant to Triton X-100 solubilization. First, we compared DRMs and PM by 1D gel electrophoresis and found that the protein composition was significantly different (Fig. 2B). Second, the 2D pattern of DRM proteins (Fig. 3B) was related, but clearly different from that observed for Arabidopsis callus PM proteins shown in Prime et al. (2000). In particular, one of the most prominent markers of the PM identified by Prime et al. (2000), the endomembrane-associated protein (At4g20260), was not detectable in DRM preparations. Thus, this and other PM proteins appear to be detergent soluble. Third, when we prepared DRMs from a PM fraction, only approximately 20% of the PM protein was detergent resistant, indicating substantial solubility of the PM. The DRMs prepared from the PM were also enriched in PAT-GPI4, PM-ATPases, and PMIP 27 relative to the PM fraction (Fig. 4). Similarly, Peskan et al. (2000) and, recently, Mongrand et al. (2004) isolated DRMs from tobacco PM preparations, and analysis of the DRMs by SDS-PAGE showed a different banding pattern compared to whole PM. In conclusion, it appears that, although numerous PM proteins are enriched in DRMs, a large proportion of the Arabidopsis PM is detergent soluble. Thus, the proteins that are enriched in DRMs are most likely a specific subset of PM proteins.
In mammalian cells, flotillin is one of the most frequently used markers for the DRM fraction (Morrow et al., 2002). Here, we have identified a potential Arabidopsis homolog of this protein (At5g25250; Fig. 3B, spot 9; Table I), which we propose to name AtFlot1 (Arabidopsis flotillin 1). The protein shares 50% sequence similarity with human flotillin-1, including a prohibitin-like domain (Morrow et al., 2002) and a conserved Cys (C-35) that is required for palmitoylation and targeting to the PM and DRMs in animals (Morrow et al., 2002). The apparent conservation of this protein and its targeting to DRMs across kingdoms suggests an important function for flotillin in microdomain-related processes. In addition, at least four further proteins with prohibitin-like domains were enriched in the DRMs: three putative hypersensitive response proteins (HIRs; At1g69840, At3g01290, and At5g62740) and a stomatin-like protein (At4g27585). HIRs and stomatins belong to a superfamily of plant proteins, named PID (proliferation, ion, and death), and may be involved in signaling and programmed cell death via modulation of ion channel activity (Nadimpalli et al., 2000). Stomatin is also a known constituent of animal DRMs (Murphy et al., 2004).
DRM-associated quinone reductases (At4g36750 and FQR1) and a putative FAD-linked oxidoreductase (At4g20830) may not have been previously reported in any organism. However, they are likely PM proteins since At4g20830 fractionates with GAPs (Borner et al., 2003; Elortza et al., 2003) and several studies have reported the purification of quinone reductases from plant PMs (Luster and Buckhout, 1989; Serrano et al., 1994; Trost et al., 1997). FQR1 is a primary auxin-response protein that has in vitro quinone-reductase activity (Laskowski et al., 2002). Quinone reductases are thought to function in oxidative stress (Bérczi and Møller, 2000).
In addition to PM ATPases, we identified multiple subunits of the V-type ATPase as proteins highly enriched in the DRMs; V-type ATPases are known constituents of animal DRMs (Foster et al., 2003; Li et al., 2003). Although V-ATPase proton pumps are primarily vacuolar in plants, they have also been described in the PMs of plant and animal cells (Robinson et al., 1996; Prime et al., 2000; Nishi and Forgac, 2002). Similarly, two putative MDRs were identified in the DRMs. In animal cells, these proteins transport small molecules, including sterols, across the PM (Garrigues et al., 2002), and human MDR1 is detergent resistant (Luker et al., 2000). In plants, MDRs have been implicated in vectorial auxin transport (Luschnig, 2002), a process that requires polarized protein targeting.
Finally, it is particularly noteworthy that several of the DRM-enriched proteins, including AtFlot1, all three HIRs, and the putative quinone reductase (At4g36750), have putative acylation motifs near the N terminus. This suggests that these cytosolic proteins may be targeted to PM microdomains by addition of lipid anchors, as is the case in animal cells (Resh, 1999; Morrow et al., 2002; Zacharias et al., 2002).
Sterols and sphingolipids were 4- to 5-fold enriched in DRMs, similar to studies of animal DRMs (Pike, 2004). We found no evidence of the enrichment of specific sterols, although a quantitative change for one particular LCB was observed, and this is indicative of altered sphingolipid composition. The total LCB analysis showed that levels of t18:1c are lower in the DRMs compared with the TMs and that the ratio of t18:1t to t18:1c moved from approximately 3:1 in the TMs to 5:1 in the DRMs. GIPCs are considered to be the most abundant sphingolipid class in Arabidopsis leaf tissue and contain less of the cis form of t18:1 than cerebrosides (Imai et al., 2000; Sperling and Heinz, 2003). Therefore, the reduction in the cis form of t18:1 in the DRM is consistent with a predominance of GIPCs rather than cerebrosides. This is in contrast with the recent work of Mongrand et al. (2004), who reported a cerebroside as the main component of tobacco DRMs. Whether this represents differences between Arabidopsis and tobacco lipid complements is unclear, although it is also possible that the Bligh and Dyer lipid extraction procedure used by Mongrand et al. (2004) underrepresented the levels of GIPCs. In this respect, the approach of analyzing the LCB composition of total sphingolipids may be a more sensitive and quantitative method.
DRMs have been widely equated with lipid rafts in membranes. However, it is important to note that the use of detergents such as Triton X-100 greatly perturbs model membrane organization (Heerklotz, 2002, 2003). Detergents, therefore, cannot be used to probe the fine organization of membrane components and the preexistence of rafts is not proven by the isolation of DRMs. Nevertheless, the detergent resistance of certain proteins probably arises in part from their membrane environment. Thus, the presence in DRMs may be a useful indicator of a protein's presence in specialized membrane domains in the cell (Schuck et al., 2003; Shogomori and Brown, 2003), but correspondence with any microdomains or rafts remains to be demonstrated.
The hypothesis of lipid microdomains or rafts as sorting platforms in plants may help us to understand a variety of recent observations. For example, SKU5 is detergent resistant in root-derived callus cells (Table II), and the nonuniform distribution of the GAP SKU5 in root cells (Sedbrook et al., 2002) could be a consequence of its incorporation into lipid microdomains. COBRA, another GAP, is excluded from the apical cell poles in roots (Schindelman et al., 2001). Similarly, polarized targeting of classical AGPs may arise from GPI anchoring, and it will be important to investigate this possibility. The disturbance of cell polarity and, in particular, polarized auxin transport in sitosterol-deficient Arabidopsis, is evidence that sterols are important for targeting of plant proteins (Willemsen et al., 2003). It will be interesting to investigate the targeting of GAPs in sterol-deficient plants and to investigate whether the DRM composition is affected. It seems very likely that lipid microdomains will emerge as a powerful concept in plant cell biology.
MATERIALS AND METHODS
Plant Culture
All wild-type plants were Arabidopsis (Arabidopsis thaliana) ecotype Columbia (Haughn and Somerville, 1988). Plants were grown hydroponically as described (Wee et al., 1998). Liquid callus cultures were derived from roots and maintained as described (Prime et al., 2000).
Agrobacterium-Mediated Transformation of Arabidopsis
Transformation of Arabidopsis plants was performed by vacuum infiltration as described in Wee et al. (1998).
Molecular Biology
Gene cloning and manipulation were performed according to standard protocols (Sambrook et al., 1990). The PAT-GPI4 construct was based on a gene encoding a secretory version of phosphinothricin acetyl transferase (SP-PAT; Denecke et al., 1990). Sequence encoding a double-myc epitope tag was generated by annealing two partially complementary oligonucleotides with overhanging BglII/XbaI sites. This fragment was cloned into the BglII/XbaI sites of vector pD331, which harbors the SP-PAT construct (pDE331 was a gift of J. Denecke, Leeds, UK). The 3′ end of atagp4 was amplified by PCR with primers trailing XbaI/BamHI sites and joined to the 3′ end of SP-PAT-myc to yield PAT-GPI4. The following oligonucleotides were used: myc forward, CGCAGATCTCCGAACAGAAATTGATTTCGGAGGAAGATTTGATGGAACAAAAATTAATCAGCGAGGAAGACCTCTCTAGACGC; myc reverse, GCGTCTAGAGAGGTCTTCCTCGCTGATTAATTTTTGTTCCATCAAATCTTCCTCCGAAATCAATTTCTGTTCGGAGATCTGCG; atagp4 forward, CGCTCTAGACCTTCTCCTGCTGATGTTCCC; atagp4 reverse, CGCGGATCCTACTTTCCCCATTCCGACAC.
Biochemical Fractionation
Cell membranes (TM) were prepared from callus tissue as described in Borner et al. (2003). Triton X-114 phase partitioning and PLC treatment to determine GPI anchoring were performed as described (Borner et al., 2003). PM fractions were prepared by dextran-PEG partitioning as described in Prime et al. (2000), based on a method by Larsson et al. (1988).
Preparation of DRMs
DRMs were prepared by low-temperature detergent extraction, adapting the protocol described in Fiedler et al. (1993). Membranes were resuspended in cold TNE (25 mm Tris-HCl, 150 mm NaCl, 5 mm EDTA, pH 7.5) containing 4–6:1 (detergent-to-protein, w/w) excess of Triton X-100 (no detergent for control extractions). The final concentration of Triton X-100 was approximately 2%. Extractions were carried out on ice shaking at 100 rpm for 35 min. Extracts were adjusted to 1.8 m Suc/TNE by addition of 3 volumes of cold 2.4 m Suc/TNE. Extracts were overlaid with Suc step gradients 1.6-1.4-1.2-0.15 m (after initial experiments simplified to 1.6-1.4-0.15 m) and centrifuged at 240,000g in a Beckman SW50.1 rotor (Beckman Coulter, Fullerton, CA) for 18 h at 4°C. DRMs were visible as off-white to white bands near the 1.2-1.4 and 1.4-1.6 m interfaces. Control fractions had a gray-green tinge. Fractions of 1 mL (0.5 mL above and 0.5 mL below the center of the bands) were collected to harvest the DRM and control fractions. Membranes were diluted with 4 volumes of cold TNE and pelleted at 100,000g for 2 h in a Beckman 50Ti rotor (Beckman Coulter).
Protein Assays
Protein concentrations were determined using a bicinchoninic-based assay (Pierce Chemical, Rockford, IL). To solubilize membrane proteins, assays were carried out in the presence of 2% to 3% SDS.
Sample Preparation for Gel Electrophoresis
For analysis by 1D SDS-PAGE, membrane pellets were resuspended in standard sample buffer (100 mm Tris-HCl, 20% glycerol, 4% SDS, pH 6.8) and heated to 60°C for 2 min.
For analysis by 2D gel electrophoresis, membrane pellets were resuspended in 5% SDS/TNE and heated to 60°C for 2 min. Proteins were precipitated with 5 volumes of acetone at −20°C for a minimum of 16 h and resuspended in AUT sample buffer (10 mm Tris-HCl, pH 8.5, 7 m urea, 2 m thiourea, 2% ASB14, 0.5% Triton X-100) at room temperature. Samples were labeled with CyDyes Cy3 and Cy5 as described in Borner et al. (2003). All labeling reactions were performed in reciprocal duplicate.
1D SDS-PAGE and Western Analysis
SDS-PAGE and western analysis were performed according to standard protocols (Sambrook et al., 1990). The following antibodies were used: TOC75 (Tranel et al., 1995); PMIP (PMIP27; Barone et al., 1998); cytochrome b5 (gift of J.A. Napier); A14 (c-myc; Santa Cruz Biotechnology, Santa Cruz, CA); PM ATPase (Morsomme et al., 1996); Sec12 (Mogelsvang and Simpson, 1998); JIM12 (Smallwood et al., 1994); JIM13, JIM14, and MAC207 (for review, see Showalter, 2001); goat anti-rabbit, conjugated to horseradish peroxidase (Bio-Rad, Hercules, CA); goat anti-rat, conjugated to horseradish peroxidase (Amersham Biosciences, Buckinghamshire, UK). Enhanced chemiluminescence was used for detection.
2D Gel Electrophoresis and Analysis
After CyDye labeling, samples were mixed with 1 volume of 2d-lysis buffer (Sherrier et al., 1999). Isoelectric focusing in ampholine gel tubes and subsequent SDS-PAGE were carried out according to Sherrier et al. (1999). Gels were scanned using a 2920-2DMasterImager (Amersham Biosciences). Images were exported as TIFF files. False coloration and contrast enhancement of scans were performed within Adobe Photoshop 5.0 LE.
MS
Analysis of proteins by excision of gel slices or spots, trypsinization, and LC-MS/MS was performed as described (Borner et al., 2003) by the Cambridge Centre for Proteomics (CCP; Cambridge, UK). Identifications from 2D gels had MASCOT scores greater than 50, indicating confidence greater than 95%. Peptide identification files are available upon request from the CCP.
Bioinformatics
Sequence alignments were performed with ClustalW (Thompson et al., 1994) at the Network Protein Sequence Analysis (NPS@) server of the Pôle Bio-Informatique Lyonnaise (PBIL): http://npsa-pbil.ibcp.fr/cgi-bin/npsa_automat.pl?page=/NPSA/npsa_clustalw.html (Combet et al., 2000). BLAST searches (Altschul et al., 1990, 1997) against a nonredundant protein database and searches for conserved domains using reverse position-specific (RPS) BLAST (Altschul et al., 1997) were performed at the National Centre for Biotechnology (NCBI) server: http://www.ncbi.nlm.nih.gov/BLAST. BLAST searches of the Arabidopsis protein database were performed at The Arabidopsis Information Resource (TAIR) server: http://www.arabidopsis.org/Blast.
Sterol Analysis
DRM and control membranes were dissolved in methanol:chloroform (2:1, v/v) by ultrasonication in a water bath for 5 min with intermittent vortexing. Volumes of methanol-chloroform were adjusted to yield identical protein concentrations in each sample. To the dissolved samples, dihydrocholesterol was added at a concentration of 16.7 μg mL−1 as internal standard. Samples were vacuum dried and treated with N-methyl-N-trimethylsilyltrifluoroacetamide at 90°C for 15 min. Samples were diluted 1:2.5 in hexane, and 1-μL aliquots were analyzed in duplicate by GC-MS. GC was performed with an Agilent 6890 Series GC system, fitted with a 7683 automatic liquid sampler. The mass spectrometer was a Micromass GCT with a time-of-flight analyzer.
Individual lipids were identified from total ion chromatograms by their mass spectra. The identity of sterols was further confirmed by running authentic standards. Sterols were quantified by peak integration relative to the internal standard. To obtain the relative enrichment of individual sterols in DRM versus control samples, the intensities (i.e. heights) of base peaks of extracted ion chromatograms were compared.
Sphingoid Base Analysis
Sphingolipid analysis of membrane fractions was carried out according to the method of Sperling et al. (1998). Sphingosine (d20:1) was used as an internal standard. LCBs were liberated from DRMs and TMs by alkaline hydrolysis and extracted with chloroform:dioxane:water (6:5:1; v/v/v). The LCB fraction was converted to dinitrophenol derivatives, extracted with chloroform:methanol:water (8:4:3; v/v/v), purified by thin-layer chromatography on silica plates, and analyzed by reversed-phase HPLC using an Agilent 1100 LC system, with MS analysis carried out on a Thermoquest LCQ system with an APCI source.
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes.
Acknowledgments
We thank Zhinong Zhang for the culture of the Arabidopsis callus, Xiaolan Yu for technical assistance, and Jurgen Denecke for the PAT clone. We would also like to thank Julie Howard and Svenja Hester for carrying out MS analyses.
This work was supported by the Biotechnology and Biological Sciences Research Council, by the Biotechnology and Biological Sciences Research Council Investigating Gene Function Initiative GARNet, by a European Community's Framework V Research Training Network Contract (HPRN–CT–2002–00262) Biointeractions (to T.W.), by the Nuffield Foundation, and by the Studienstiftung des Deutschen Volkes (scholarship to G.H.H.B.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.053041.
References
- Ahmed SN, Brown DA, London E (1997) On the origin of sphingolipid/cholesterol-rich detergent-insoluble cell membranes: physiological concentrations of cholesterol and sphingolipid induce formation of a detergent-insoluble, liquid-ordered lipid phase in model membranes. Biochemistry 36: 10944–10953 [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagnat M, Chang A, Simons K (2001) Plasma membrane proton ATPase Pma1p requires raft association for surface delivery in yeast. Mol Biol Cell 12: 4129–4138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagnat M, Keranen S, Shevchenko A, Shevchenko A, Simons K (2000) Lipid rafts function in biosynthetic delivery of proteins to the cell surface in yeast. Proc Natl Acad Sci USA 97: 3254–3259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barone LM, Mu HH, Shih CJ, Kashlan KB, Wasserman BP (1998) Distinct biochemical and topological properties of the 31- and 27-kilodalton plasma membrane intrinsic protein subgroups from red beet. Plant Physiol 118: 315–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bérczi A, Møller IM (2000) Redox enzymes in the plant plasma membrane and their possible roles. Plant Cell Environ 23: 1287–1302 [Google Scholar]
- Borner GHH, Lilley KS, Stevens TJ, Dupree P (2003) Identification of glycosylphosphatidylinositol-anchored proteins in Arabidopsis. A proteomic and genomic analysis. Plant Physiol 132: 568–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borner GHH, Sherrier DJ, Stevens TJ, Arkin IT, Dupree P (2002) Prediction of glycosylphosphatidylinositol-anchored proteins in Arabidopsis. A genomic analysis. Plant Physiol 129: 486–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown DA, London E (1998) Structure and origin of ordered lipid domains in biological membranes. J Membr Biol 164: 103–114 [DOI] [PubMed] [Google Scholar]
- Brown DA, Rose JK (1992) Sorting of GPI-anchored proteins to glycolipid-enriched membrane subdomains during transport to the apical cell surface. Cell 68: 533–544 [DOI] [PubMed] [Google Scholar]
- Clouse SD (2002) Arabidopsis mutants reveal multiple roles for sterols in plant development. Plant Cell 14: 1995–2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Combet C, Blanchet C, Geourjon C, Deléage G (2000) NPS@ Network Protein Sequence Analysis. Trends Biochem Sci 25: 147–150 [DOI] [PubMed] [Google Scholar]
- Denecke J, Botterman J, Deblaere R (1990) Protein secretion in plant cells can occur via a default pathway. Plant Cell 2: 51–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich C, Bagatolli LA, Volovyk ZN, Thompson NL, Levi M, Jacobson K, Gratton E (2001) Lipid rafts reconstituted in model membranes. Biophys J 80: 1417–1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich C, Yang B, Fujiwara T, Kusumi A, Jacobson K (2002) Relationship of lipid rafts to transient confinement zones detected by single particle tracking. Biophys J 82: 274–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn TM, Lynch DV, Michaelson LV, Napier JA (2004) A post-genomic approach to understanding sphingolipid metabolism in Arabidopsis thaliana. Ann Bot (Lond) 93: 483–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edidin M (2003. a) Lipids on the frontier: a century of cell-membrane bilayers. Nat Rev Mol Cell Biol 4: 414–418 [DOI] [PubMed] [Google Scholar]
- Edidin M (2003. b) The state of lipid rafts: from model membranes to cells. Annu Rev Biophys Biomol Struct 32: 257–283 [DOI] [PubMed] [Google Scholar]
- Elortza F, Nuhse TS, Foster LJ, Stensballe A, Peck SC, Jensen ON (2003) Proteomic analysis of glycosylphosphatidylinositol-anchored membrane proteins. Mol Cell Proteomics 2: 1261–1270 [DOI] [PubMed] [Google Scholar]
- Fiedler K, Kobayashi T, Kurzchalia TV, Simons K (1993) Glycosphingolipid-enriched, detergent-insoluble complexes in protein sorting in epithelial cells. Biochemistry 32: 6365–6373 [DOI] [PubMed] [Google Scholar]
- Foster LJ, De Hoog CL, Mann M (2003) Unbiased quantitative proteomics of lipid rafts reveals high specificity for signaling factors. Proc Natl Acad Sci USA 100: 5813–5818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrichson T, Kurzchalia TV (1998) Microdomains of GPI-anchored proteins in living cells revealed by crosslinking. Nature 394: 802–805 [DOI] [PubMed] [Google Scholar]
- Garrigues A, Escargueil AE, Orlowski S (2002) The multidrug transporter, P-glycoprotein, actively mediates cholesterol redistribution in the cell membrane. Proc Natl Acad Sci USA 99: 10347–10352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaus K, Gratton E, Kable EP, Jones AS, Gelissen I, Kritharides L, Jessup W (2003) Visualizing lipid structure and raft domains in living cells with two-photon microscopy. Proc Natl Acad Sci USA 100: 15554–15559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harder T, Engelhardt KR (2004) Membrane domains in lymphocytes: from lipid rafts to protein scaffolds. Traffic 5: 265–275 [DOI] [PubMed] [Google Scholar]
- Haughn GW, Somerville CR (1988) Genetic control of morphogenesis in Arabidopsis. Dev Genet 9: 73–89 [Google Scholar]
- Heerklotz H (2002) Triton promotes domain formation in lipid raft mixtures. Biophys J 83: 2693–2701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heerklotz H, Szadkowska H, Anderson T, Seelig J (2003) The sensitivity of lipid domains to small perturbations demonstrated by the effect of Triton. J Mol Biol 329: 793–799 [DOI] [PubMed] [Google Scholar]
- Helms JB, Zurzolo C (2004) Lipids as targeting signals: lipid rafts and intracellular trafficking. Traffic 5: 247–254 [DOI] [PubMed] [Google Scholar]
- Ikonen E (2001) Roles of lipid rafts in membrane transport. Curr Opin Cell Biol 13: 470–477 [DOI] [PubMed] [Google Scholar]
- Imai H, Morimoto Y, Tamura K (2000) Sphingoid base composition of monoglucosylceramide in Brassicaceae. J Plant Physiol 157: 453–456 [Google Scholar]
- Kenworthy AK, Petranova N, Edidin M (2000) High-resolution FRET microscopy of cholera toxin B-subunit and GPI-anchored proteins in cell plasma membranes. Mol Biol Cell 11: 1645–1655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson C, Widell S, Sommarin M (1988) Inside-out plant plasma-membrane vesicles of high-purity obtained by aqueous 2-phase partitioning. FEBS Lett 229: 289–292 [Google Scholar]
- Laskowski MJ, Dreher KA, Gehring MA, Abel S, Gensler AL, Sussex IM (2002) FQR1, a novel primary auxin-response gene, encodes a flavin mononucleotide-binding quinone reductase. Plant Physiol 128: 578–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Mak A, Richards DP, Naber C, Keller BO, Li L, Shaw AR (2003) Monocyte lipid rafts contain proteins implicated in vesicular trafficking and phagosome formation. Proteomics 3: 536–548 [DOI] [PubMed] [Google Scholar]
- London E, Brown DA (2000) Insolubility of lipids in triton X-100: physical origin and relationship to sphingolipid/cholesterol membrane domains (rafts). Biochim Biophys Acta 1508: 182–195 [DOI] [PubMed] [Google Scholar]
- Luker GD, Pica CM, Kumar AS, Covey DF, Piwnica-Worms D (2000) Effects of cholesterol and enantiomeric cholesterol on P-glycoprotein localization and function in low-density membrane domains. Biochemistry 39: 7651–7661 [DOI] [PubMed] [Google Scholar]
- Luschnig C (2002) Auxin transport: ABC proteins join the club. Trends Plant Sci 7: 329–332 [DOI] [PubMed] [Google Scholar]
- Luster DG, Buckhout TJ (1989) Purification and identification of a plasma membrane associated electron transport protein from maize (Zea mays) roots. Plant Physiol 91: 1014–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayor S, Rao M (2004) Rafts: scale-dependent, active lipid organization at the cell surface. Traffic 5: 231–240 [DOI] [PubMed] [Google Scholar]
- Mogelsvang S, Simpson DJ (1998) Changes in the levels of seven proteins involved in polypeptide folding and transport during endosperm development of two barley genotypes differing in storage protein localisation. Plant Mol Biol 6: 541–552 [DOI] [PubMed] [Google Scholar]
- Mongrand S, Morel J, Laroche J, Claverol S, Carde JP, Hartmann MA, Bonneu M, Simon-Plas F, Lessire R, Bessoule JJ (2004) Lipid rafts in higher plant cells: purification and characterization of Triton X-100-insoluble microdomains from tobacco plasma membrane. J Biol Chem 279: 36277–36286 [DOI] [PubMed] [Google Scholar]
- Morrow IC, Rea S, Martin S, Prior IA, Prohaska R, Hancock JF, James DE, Parton RG (2002) Flotillin-1/reggie-2 traffics to surface raft domains via a novel Golgi-independent pathway. Identification of a novel membrane targeting domain and a role for palmitoylation. J Biol Chem 277: 48834–48841 [DOI] [PubMed] [Google Scholar]
- Morsomme P, de Kerchove d'Exaerde A, De Meester S, Thines D, Goffeau A, Boutry M (1996) Single point mutations in various domains of a plant plasma membrane H(+)-ATPase expressed in Saccharomyces cerevisiae increase H(+)-pumping and permit yeast growth at low pH. EMBO J 15: 5513–5526 [PMC free article] [PubMed] [Google Scholar]
- Muniz M, Riezman H (2000) Intracellular transport of GPI-anchored proteins. EMBO J 19: 10–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy SC, Samuel BU, Harrison T, Speicher KD, Speicher DW, Reid ME, Prohaska R, Low PS, Tanner MJ, Mohandas N, et al (2004) Erythrocyte detergent-resistant membrane proteins: their characterization and selective uptake during malarial infection. Blood 103: 1920–1928 [DOI] [PubMed] [Google Scholar]
- Nadimpalli R, Yalpani N, Johal GS, Simmons CR (2000) Prohibitins, stomatins, and plant disease response genes compose a protein superfamily that controls cell proliferation, ion channel regulation, and death. J Biol Chem 275: 29579–29586 [DOI] [PubMed] [Google Scholar]
- Nebl T, Pestonjamasp KN, Leszyk JD, Crowley JL, Oh SW, Luna EJ (2002) Proteomic analysis of a detergent-resistant membrane skeleton from neutrophil plasma membranes. J Biol Chem 277: 43399–43409 [DOI] [PubMed] [Google Scholar]
- Nishi T, Forgac M (2002) The vacuolar (H+)-ATPases: nature's most versatile proton pumps. Nat Rev Mol Cell Biol 3: 94–103 [DOI] [PubMed] [Google Scholar]
- Palestini P, Calvi C, Conforti E, Daffara R, Botto L, Miserocchi G (2003) Compositional changes in lipid microdomains of air-blood barrier plasma membranes in pulmonary interstitial edema. J Appl Physiol 95: 1446–1452 [DOI] [PubMed] [Google Scholar]
- Peskan T, Westermann M, Oelmuller R (2000) Identification of low-density Triton X-100-insoluble plasma membrane microdomains in higher plants. Eur J Biochem 267: 6989–6995 [DOI] [PubMed] [Google Scholar]
- Pike LJ (2004) Lipid rafts: heterogeneity on the high seas. Biochem J 378: 281–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pralle A, Keller P, Florin EL, Simons K, Horber JK (2000) Sphingolipid-cholesterol rafts diffuse as small entities in the plasma membrane of mammalian cells. J Cell Biol 148: 997–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prime TA, Sherrier DJ, Mahon P, Packman LC, Dupree P (2000) A proteomic analysis of organelles from Arabidopsis thaliana. Electrophoresis 21: 3488–3499 [DOI] [PubMed] [Google Scholar]
- Prior IA, Muncke C, Parton RG, Hancock JF (2003) Direct visualization of Ras proteins in spatially distinct cell surface microdomains. J Cell Biol 160: 165–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resh MD (1999) Fatty acylation of proteins: new insights into membrane targeting of myristoylated and palmitoylated proteins. Biochim Biophys Acta 1451: 1–16 [DOI] [PubMed] [Google Scholar]
- Robinson DG, Haschke HP, Hinz G, Hoh B, Maeshima M, Marty F (1996) Immunological detection of tonoplast polypeptides in the plasma membrane of pea cotyledons. Planta 198: 95–103 [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1990) Molecular Cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Schindelman G, Morikami A, Jung J, Baskin TI, Carpita NC, Derbyshire P, McCann MC, Benfey PN (2001) COBRA encodes a putative GPI-anchored protein, which is polarly localized and necessary for oriented cell expansion in Arabidopsis. Genes Dev 15: 1115–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder R, London E, Brown D (1994) Interactions between saturated acyl chains confer detergent resistance on lipids and glycosylphosphatidylinositol (GPI)-anchored proteins: GPI-anchored proteins in liposomes and cells show similar behavior. Proc Natl Acad Sci USA 91: 12130–12134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuck S, Honsho M, Ekroos K, Shevchenko A, Simons K (2003) Resistance of cell membranes to different detergents. Proc Natl Acad Sci USA 100: 5795–5800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz CJ, Rumsewicz MP, Johnson KL, Jones BJ, Gaspar YM, Bacic A (2002) Using genomic resources to guide research directions. The arabinogalactan protein gene family as a test case. Plant Physiol 129: 1448–1463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedbrook JC, Carroll KL, Hung KF, Masson PH, Somerville CR (2002) The Arabidopsis SKU5 gene encodes an extracellular glycosyl phosphatidylinositol-anchored glycoprotein involved in directional root growth. Plant Cell 14: 1635–1648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano A, Cordoba F, Gonzalez-Reyes JA, Navas P, Villalba JM (1994) Purification and characterization of two distinct NAD(P)H dehydrogenases from onion (Allium cepa L.) root plasma membrane. Plant Physiol 106: 87–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma P, Varma R, Sarasij RC, Ira, Gousset K, Krishnamoorthy G, Rao M, Mayor S (2004) Nanoscale organization of multiple GPI-anchored proteins in living cell membranes. Cell 116: 577–589 [DOI] [PubMed] [Google Scholar]
- Sherrier DJ, Prime TA, Dupree P (1999) Glycosylphosphatidylinositol-anchored cell-surface proteins from Arabidopsis. Electrophoresis 20: 2027–2035 [DOI] [PubMed] [Google Scholar]
- Shogomori H, Brown DA (2003) Use of detergents to study membrane rafts: the good, the bad, and the ugly. Biol Chem 384: 1259–1263 [DOI] [PubMed] [Google Scholar]
- Showalter AE (2001) Arabinogalactan-proteins: structure, expression and function. Cell Mol Life Sci 58: 1399–1417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvius JR (2003) Fluorescence energy transfer reveals microdomain formation at physiological temperatures in lipid mixtures modeling the outer leaflet of the plasma membrane. Biophys J 85: 1034–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons K, Ikonen E (1997) Functional rafts in cell membranes. Nature 387: 569–572 [DOI] [PubMed] [Google Scholar]
- Simons K, Toomre D (2000) Lipid rafts and signal transduction. Nat Rev Mol Cell Biol 1: 31–39 [DOI] [PubMed] [Google Scholar]
- Simons K, van Meer G (1988) Lipid sorting in epithelial cells. Biochemistry 27: 6197–6202 [DOI] [PubMed] [Google Scholar]
- Smallwood M, Beven A, Donovan N, Neill SJ, Peart J, Roberts K, Knox JP (1994) Localization of cell-wall poteins in relation to the developmental anatomy of the carrot root apex. Plant J 5: 237–246 [Google Scholar]
- Sperling P, Heinz E (2003) Plant sphingolipids: structural diversity, biosynthesis, first genes and functions. Biochim Biophys Acta 1632: 1–15 [DOI] [PubMed] [Google Scholar]
- Sperling P, Zahringer U, Heinz E (1998) A sphingolipid desaturase from higher plants. Identification of a new cytochrome b5 fusion protein. J Biol Chem 273: 28590–28596 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTALW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tranel PJ, Froehlich J, Goyal A, Keegstra K (1995) A component of the chloroplastic protein import apparatus is targeted to the outer envelope membrane via a novel pathway. EMBO J 14: 2436–2446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trost P, Foscarini S, Preger V, Bonora P, Vitale L, Pupillo P (1997) Dissecting the diphenylene iodonium-sensitive NAD(P)H:quinone oxidoreductase of zucchini plasma membrane. Plant Physiol 114: 737–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varma R, Mayor S (1998) GPI-anchored proteins are organized in submicron domains at the cell surface. Nature 394: 798–801 [DOI] [PubMed] [Google Scholar]
- Warnecke D, Heinz E (2003) Recently discovered functions of glucosylceramides in plants and fungi. Cell Mol Life Sci 60: 919–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wee EG, Sherrier DJ, Prime TA, Dupree P (1998) Targeting of active sialyltransferase to the plant Golgi apparatus. Plant Cell 10: 1759–1768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willemsen V, Friml J, Grebe M, van den Toorn A, Palme K, Scheres B (2003) Cell polarity and PIN protein positioning in Arabidopsis require sterol methyltransferase1 function. Plant Cell 15: 612–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacharias DA, Violin JD, Newton AC, Tsien RY (2002) Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science 296: 913–916 [DOI] [PubMed] [Google Scholar]