Abstract
Proline transporters (ProTs) mediate transport of the compatible solutes Pro, glycine betaine, and the stress-induced compound γ-aminobutyric acid. A new member of this gene family, AtProT3, was isolated from Arabidopsis (Arabidopsis thaliana), and its properties were compared to AtProT1 and AtProT2. Transient expression of fusions of AtProT and the green fluorescent protein in tobacco (Nicotiana tabacum) protoplasts revealed that all three AtProTs were localized at the plasma membrane. Expression in a yeast (Saccharomyces cerevisiae) mutant demonstrated that the affinity of all three AtProTs was highest for glycine betaine (Km = 0.1–0.3 mm), lower for Pro (Km = 0.4–1 mm), and lowest for γ-aminobutyric acid (Km = 4–5 mm). Relative quantification of the mRNA level using real-time PCR and analyses of transgenic plants expressing the β-glucuronidase (uidA) gene under control of individual AtProT promoters showed that the expression pattern of AtProTs are complementary. AtProT1 expression was found in the phloem or phloem parenchyma cells throughout the whole plant, indicative of a role in long-distance transport of compatible solutes. β-Glucuronidase activity under the control of the AtProT2 promoter was restricted to the epidermis and the cortex cells in roots, whereas in leaves, staining could be demonstrated only after wounding. In contrast, AtProT3 expression was restricted to the above-ground parts of the plant and could be localized to the epidermal cells in leaves. These results showed that, although intracellular localization, substrate specificity, and affinity are very similar, the transporters fulfill different roles in planta.
Environmental stresses such as high salt, low water availability, and low temperature can induce the accumulation of one or several compatible solutes (such as Pro, Gly betaine, polyols, or sugars) in plants and other organisms (Delauney and Verma, 1993; Rhodes and Hanson, 1993; Serrano, 1996). Compatible solutes are involved in either osmotic adjustment or in the protection of cellular structures under stress conditions and can accumulate to high levels in plant organs that undergo dehydration during their maturation, such as pollen or seeds (Chiang and Dandekar, 1995). In general, cellular concentration of compatible solutes can be regulated by increasing biosynthesis, decreasing degradation, and/or modifying rates of uptake or release of these compounds. In plants, research has focused on the metabolism of Pro and Gly betaine and their importance for stress tolerance (Yoshiba et al., 1997; McNeil et al., 1999). However, only limited information on the significance of transport of compatible solutes is available. Physiological experiments indicate that high levels of Pro are present in the phloem sap of alfalfa plants under conditions of low water availability (Girousse et al., 1996). Similarly, Gly betaine was shown to be phloem mobile and is translocated during moderate stress conditions (Mäkelä et al., 1996). These findings support the idea that transport of compatible solutes is important under water stress conditions. However, transport of Pro might also be essential under normal growth conditions. Pro accumulation in developing grapevine berries and maturing pollen occurs independently of changes of the key enzyme(s) of Pro biosynthesis, arguing for a different level of regulation or for import of Pro (Fujita et al., 1998; Stines et al., 1999). Additional indications for Pro transport throughout the plant come from transgenic Arabidopsis (Arabidopsis thaliana) lines with reduced Pro biosynthesis (Nanjo et al., 1999), as the morphological alterations in these lines could be rescued by exogenous Pro application.
In the last few years, genes encoding transporters for the compatible solutes Pro and Gly betaine were isolated from a number of plant species, including Arabidopsis (AtProT1 and AtProT2; Rentsch et al., 1996), tomato (Lycopersicon esculentum; LeProT1–3; Schwacke et al., 1999), rice (Oryza sativa; OsProT; Igarashi et al., 2000), barley (Hordeum vulgare; HvProT; Ueda et al., 2001), orach (Atriplex hortensis; AhProT1; accession no. AAF76897), and mangrove (Avicennia marina; AmT1–3; Waditee et al., 2002). Originally, the transporters were described as specific Pro transporters (ProTs; Rentsch et al., 1996). However, ProTs from tomato and the betaine-accumulating mangrove A. marina were later shown to be general transporters for the compatible solutes Pro and Gly betaine (Schwacke et al., 1999; Waditee et al., 2002). Furthermore, the Arabidopsis ProT2 was shown to mediate transport of Pro and the stress-induced compound γ-aminobutyric acid (GABA), although GABA was recognized with lower affinity (Breitkreuz et al., 1999).
The level of expression of several members of the ProT family is often correlated with conditions of stress or high Pro concentrations. The steady-state mRNA levels of the Arabidopsis ProT2 and the three mangrove ProT homologs (AmT1–3) increased under salt stress conditions, as do the concentrations of Pro and Gly betaine, respectively (Waditee et al., 2002). Furthermore, HvProT expression in barley roots was increased by salt stress treatments (Ueda et al., 2001). In tomato, expression of LeProT1 was shown to be confined to pollen and expression correlated with elevated Pro levels. In contrast, AtProT1 and the rice homolog OsProT did not show any regulation of expression under stress conditions (Rentsch et al., 1996; Igarashi et al., 2000).
Although AtProT1 and AtProT2 were identified and initially characterized several years ago, a detailed comparative analysis of the Arabidopsis ProTs is still missing. In addition, the completion of the sequencing of the Arabidopsis genome revealed the existence of a third, closely related gene. The purpose of this study was to elucidate the function of all three Arabidopsis ProTs. The data presented on the kinetic properties, intracellular localization, and expression patterns indicate that the three Arabidopsis ProTs have distinct roles in the physiology of the plant.
RESULTS
AtProT3 Is a Novel Member of the AProT Gene Family
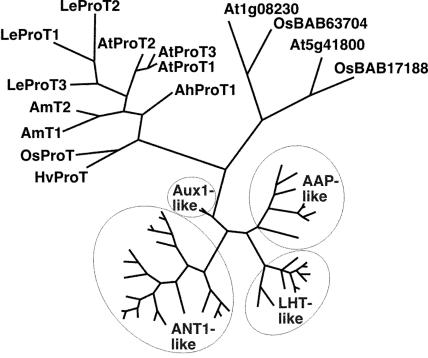
A newly identified ProT homolog, At2g36590 (AtProT3), showed the highest homology to AtProT1 (89.9% identity on the amino acid level) and AtProT2 (83.7%). Phylogenetic analysis revealed that the three Arabidopsis ProTs form a subgroup within the ProT branch of the ATF1 (amino acid transporter family 1) gene family (Fig. 1; Fischer et al., 1998; Wipf et al., 2002). Interestingly, ProT members from a single plant species always cluster together, indicating that duplication of the genes was a relatively recent event. In contrast, in the most closely related group of transporters of the ATF family, the At1g08230-like proteins, rice and Arabidopsis proteins group together. Based on their low sequence identity (approximately 30% on the amino acid level) and their different substrate specificity, we designated At1g08230 and homologs as not belonging to the ProT-like proteins (A. Meyer and D. Rentsch, unpublished data). AtProT3 encodes a protein with a molecular mass of 47.6 kD and 9 to 11 predicted transmembrane domains, which is in agreement with predictions for AtProT1 and AtProT2.
Figure 1.
Phylogenetic relationship between the AtProTs and related proteins. The analysis was performed using the aligned protein sequences of the Arabidopsis ATF gene family that, in addition to the ProTs, contains several groups of amino acid permeases (AAPs, LHTs, ANT-like proteins) as well as potential auxin transporters (AUX1; Fischer et al., 1998; Chen et al., 2001; Wipf et al., 2002). Proteins from other plant species were included for the ProT-like and the At1g08230-like proteins. Maximum parsimony analysis was performed using PAUP 4.0b10, with all DNA characters unweighted and gaps scored as missing characters. The complete alignment was based on 825 amino acids; 573 characters were parsimony informative. AUX1 was used as the outgroup.
The intron-exon structure of AtProT1, AtProT2, and AtProT3 is highly conserved, showing seven exons of comparable length. The main difference was found in the length of the first intron, which is considerably longer in AtProT2 (data not shown). The Arabidopsis genome also contains two partial ProT genes on bacterial artificial chromosome (BAC) clone F19C17 and F5A13 that are highly homologous to AtProT1 and AtProT2, respectively. As the conserved region on BAC F19C17 is relatively short and the ProT homolog on BAC F5A13 contains point mutations leading to a frameshift and an additional stop codon, both homologs do not encode functional Pro transporters. Therefore, in Arabidopsis, only three functional ProTs are present. To discover whether the Arabidopsis ProTs show redundancy of function, the expression patterns and the biochemical properties were compared.
AtProT3 Mediates Growth of a Pro and GABA Transport-Deficient Yeast Mutant under Selective Conditions
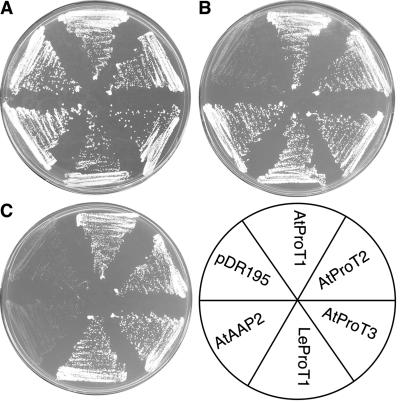
To determine whether AtProT3 differs in its substrate specificity from AtProT1 or AtProT2, the AtProT3 cDNA was isolated by reverse transcription (RT)-PCR. Subsequently, AtProT3 was expressed under the control of the strong PMA1 promoter (vector pDR195) in the yeast (Saccharomyces cerevisiae) strain 22574d (Jauniaux et al., 1987; Rentsch et al., 1995). This strain carries mutations in the general amino acid (gap1), Pro (put4), and the GABA (uga4) permeases and thus is unable to grow on citrulline, Pro, or GABA as the sole nitrogen source. As controls, strain 22574d was transformed with the expression vector pDR195, with pDR195 harboring the cDNAs of AtProT1, AtProT2, LeProT1, or the amino acid permease AtAAP2 (Kwart et al., 1993; Schwacke et al., 1999). Growth under selective conditions showed that, like AtProT1 and AtProT2, AtProT3 was able to mediate growth on Pro and GABA, but not on citrulline (Fig. 2; data not shown). The ability to mediate transport of GABA and Pro, but not of citrulline, distinguishes AtProT3 from the amino acid permeases of the AAP family (Fischer et al., 1998) and also groups AtProT3 to the ProT family with respect to substrate specificity.
Figure 2.
Complementation of a yeast strain (22574d) deficient in the uptake of Pro, GABA, and citrulline. Growth of 22574d cells expressing AtProT1, AtProT2, AtProT3, LeProT1, and AtAAP2 and the strain transformed with the vector pDR195 is shown. Minimal medium was supplemented with ammonium (A), Pro (B), or GABA (C) as the sole nitrogen source.
Kinetic Properties of AtProT1, 2, and 3
To examine whether the AtProTs differ in their kinetic properties, uptake experiments with l-[14C]Pro, [14C]Gly betaine, and [3H]GABA in 22574d cells expressing AtProT1, AtProT2, or AtProT3 were performed. Previous studies described AtProT2 as a transporter with higher affinity for Pro than for GABA (Breitkreuz et al., 1999). However, the affinity of AtProT2 and AtProT1 for Gly betaine, a substrate for LeProT1 from tomato and the ProT homologs from mangrove, as well as the kinetic properties of AtProT3, had not been determined so far (Schwacke et al., 1999; Waditee et al., 2002). Transport assays showed that all three AtProTs were able to mediate uptake of labeled Pro, Gly betaine, and GABA. The affinity of all AtProTs was highest for Gly betaine, intermediate for Pro, and lowest for GABA (Table I). The affinity of AtProT1 for Gly betaine was 2 to 2.5 times the affinity of AtProT2 or AtProT3, and the affinity of AtProT1 and AtProT2 for Pro was 2 times that of AtProT3. In contrast, the Km values for GABA were in a similar range for all three transporters. The maximal transport velocities can only be compared for different substrates of a single transporter because transport rates do not only depend on the intrinsic properties of the transport protein, but also on the number of transporters present at the yeast plasma membrane. In general, for all three transporters, higher affinity correlated with lower capacity, and vice versa. For the ProT homologs AmT1 and AmT2 from mangrove, sodium and potassium ions have been shown to enhance transport activity (Waditee et al., 2002). However, no increased AtProT-mediated transport rates could be measured by the addition of sodium chloride to the assay medium (data not shown).
Table I.
Comparison of the affinities of all three AtProTs for Pro, Gly betaine, and GABA
The uptake of radiolabeled substrates was determined in 22574d cells expressing AtProT1, AtProT2, or AtProT3. The pH of the assay medium was 4.5. Uptake rates were determined at five different substrate concentrations and the apparent affinities and maximal transport rates were determined. Values represent the mean of at least three experiments ± sd.
| Transporter
|
Pro
|
Gly Betaine
|
GABA
|
|||
|---|---|---|---|---|---|---|
| Km | Vmax | Km | Vmax | Km | Vmax | |
| mm | nmol × 10−8 cells × min−1 | mm | nmol × 10−8 cells × min−1 | mm | nmol × 10−8 cells × min−1 | |
| AtProT1 | 0.427 ± 0.017 | 29 ± 1.0 | 0.115 ± 0.009 | 11 ± 0.1 | 4.50 ± 0.26 | 41 ± 4.6 |
| AtProT2 | 0.500 ± 0.005 | 42 ± 1.4 | 0.267 ± 0.018 | 17 ± 0.4 | 4.01 ± 0.64 | 60 ± 4.5 |
| AtProT3 | 0.999 ± 0.036 | 19 ± 2.1 | 0.290 ± 0.027 | 3.6 ± 0.6 | 5.12 ± 0.12 | 34 ± 1.8 |
AtProTs Are Localized at the Plasma Membrane
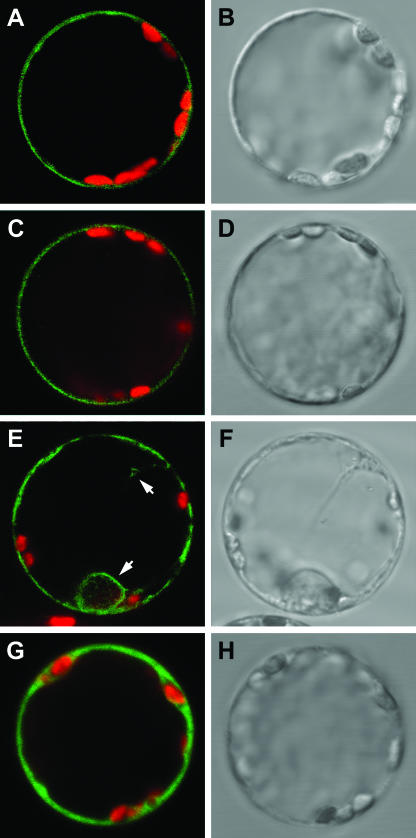
Functional complementation of the yeast Pro/GABA transport mutant by the AtProTs showed that the protein is localized at the yeast plasma membrane. However, their cellular localization in planta has not been demonstrated so far. Therefore, fusion proteins of AtProTs with green fluorescent protein (GFP) were transiently expressed in tobacco (Nicotiana tabacum) protoplasts under the control of the cauliflower mosaic virus 35S promoter (Fig. 3). Fluorescent images obtained by confocal laser-scanning microscopy showed that the signal from the GFP-AtProT1 and GFP-AtProT2 fusion proteins was present as a single fluorescent ring at the periphery of the protoplast, indicative of its localization at the plasma membrane (Fig. 3, A and C). GFP-AtProT3 also localized to the plasma membrane (Fig. 3E) but, in addition, the protoplasts showed some GFP fluorescence on internal membranes. Similar results were obtained for the AtProT-GFP fusions (data not shown). Free GFP localized to the cytosol (Fig. 3G).
Figure 3.
Localization of GFP-AtProT fusion proteins at the plasma membrane of tobacco protoplasts. Confocal laser-scanning microscope images (A, C, E, and G) and corresponding bright-field images (B, D, F, and H) of tobacco protoplasts transiently expressing GFP-AtProT fusion proteins or GFP. A and B, GFP-AtProT1; C and D, GFP-AtProT2; E and F, GFP-AtProT3; G and H, GFP. Arrows in E indicate staining of internal membranes in addition to fluorescence at the plasma membrane. Merged images show GFP fluorescence (green) and chlorophyll fluorescence (red). Diameter of protoplasts is approximately 40 μm.
ProTs Are Differentially Expressed in Arabidopsis
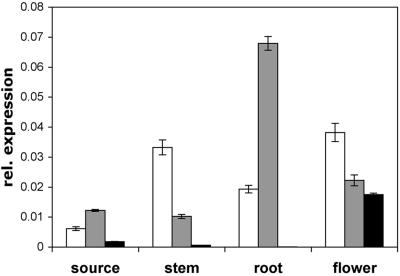
AtProT3 expression in Arabidopsis was extremely weak and hardly detectable by RNA gel-blot analysis. Relative quantification using real-time PCR, with the ubiquitin mRNA as a reference, demonstrated that AtProT3 was expressed in flowers and to very low levels in leaves and stems (Fig. 4). AtProT3 transcripts could not be detected in roots. Quantification of AtProT1 and AtProT2 mRNA levels confirmed results from RNA gel-blot analyses (Rentsch et al., 1996). Expression of AtProT2 in roots was several-fold higher than in other organs, whereas AtProT1 expression was highest in flowers, stems, and roots. In all organs analyzed, the AtProT3 mRNA was the least abundant of all the AtProTs.
Figure 4.
Expression analysis of AtProT1, 2, and 3 in Arabidopsis. Expression was analyzed by relative quantification using real-time PCR, with the ubiquitin mRNA as a reference. RNA from source leaves, stems, roots, and flowers of soil-grown plants was analyzed. Expression of AtProTs is given relative to ubiquitin mRNA levels and is the mean of three different reactions ± sd. Similar results were obtained using independent RNAs as a template. White bars, AtProT1; gray bars, AtProT2; black bars, AtProT3.
To study the tissue specificity of AtProT expression throughout plant development and regulation under stress conditions, 5′ upstream regions of the respective genes were isolated and fused to the Escherichia coli β-glucuronidase (GUS; uidA) gene. Arabidopsis was transformed and 53 (AtProT1-GUS), 30 (AtProT2-GUS), and 68 (AtProT3-GUS) independent transgenic lines were analyzed for reporter gene activity. GUS activity was initially analyzed in leaves, flowers, and siliques of Basta-resistant T1 plants and in Basta-resistant seedlings of the T2 generation. For each construct, more than 90% of the independent lines showed very similar patterns of expression, varying only in the intensity of GUS staining. Three representative lines of each construct were used to determine the expression pattern in more detail, using both tissue culture- and soil-grown plants of the T2 generation.
AtProT1 Is Expressed in the Vascular Tissue
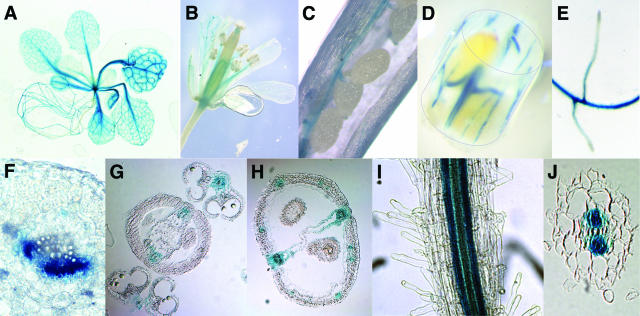
Transgenic plants expressing the uidA gene under the control of the AtProT1 promoter showed strong reporter gene activity in the vascular tissue of leaves, petioles, roots, flowers, siliques, and stems of axenically grown seedlings or mature soil-grown plants (Fig. 5). The expression in the vascular tissue strongly indicates that AtProT1 is important for long-distance transport. In leaves, the vascular tissue of both minor and major veins was stained (Fig. 5A). Cross-sections through leaves showed that GUS activity is confined to the phloem or phloem parenchyma cells (Fig. 5F). In addition, the vascular tissue of sepals, petals, and stamens showed reporter gene activity (Fig. 5, B and G). This staining pattern could also be found in siliques showing strong GUS activity in the vascular strands of the carpels and the funiculi, whereas the seeds themselves did not show expression (Fig. 5, C, D, and H). In roots, staining was not detectable in the root tip and was weak in newly emerging lateral roots (Fig. 5, E and I). Cross-sections of roots showed that expression is restricted to phloem or phloem parenchyma cells (Fig. 5J).
Figure 5.
Expression of the uidA gene under control of the AtProT1 promoter. AtProT1-GUS expression in the vascular tissue of an Arabidopsis seedling (A), flowers (B), siliques (C and D), and roots (E). F, Fresh section through the midvein of a leaf. GUS staining was detected in the phloem or phloem parenchyma cells. Sections of paraffin-embedded X-gluc-stained flowers (G) and siliques (H). In the whole root (I) and root section (J), GUS activity is restricted to the vascular system.
AtProT2-GUS Staining Is Confined to Root Cortex and Epidermal Cells and Induced upon Wounding in Leaves
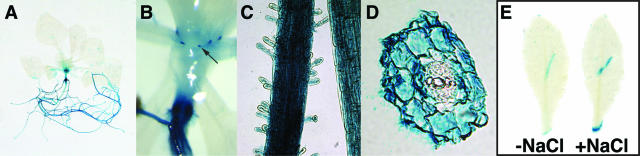
In both seedlings and mature plants, the AtProT2 promoter directed expression in root tissue (Fig. 6A). In addition, stipules of axenically grown plants showed staining (Fig. 6B). Under normal growth conditions, no GUS activity was observed in other tissues. Macroscopically, GUS activity was detected over the entire root, including root hairs, the root tip, and the root cap (Fig. 6C; data not shown). However, cross-sections of roots showed that AtProT2 expression did not overlap with AtProT1 promoter activity because GUS activity in AtProT2-GUS plants was confined to the epidermis and the cortex cells and absent from the stele (Fig. 6D). AtProT2 mRNA levels were previously shown to be elevated under salt stress (Rentsch et al., 1996). We confirmed these findings with independent experiments in this study (data not shown). However, no increase in GUS activity was observed when AtProT2-GUS plants were subjected to different stress treatments such as high salt concentrations, low humidity, or low temperature (data not shown). Also, quantitative GUS assays of salt-treated plants did not reveal significant induction of promoter activity (data not shown). The AtProT2 promoter contains an abscisic acid (ABA)-responsive element (PyACGTGGC), which in other genes has been shown to function as a cis-element in ABA-responsive transcription under water stress (Shinozaki and Yamaguchi-Shinozaki, 1997). However, ABA treatment did not have any effect on AtProT2 promoter activity. Interestingly, AtProT2-GUS activity was induced by mechanical wounding both in detached leaves and in leaves of intact plants, and this effect was increased in the presence of salt (Fig. 6E).
Figure 6.
Histochemical analyses of AtProT2 promoter-GUS fusions. Expression of GUS under control of the AtProT2 promoter in 2-week-old axenically grown Arabidopsis plants (A and B). Arrow indicates staining of stipules (B). C, In roots, staining is visible over the entire root, including root hairs. D, Cross-sections of roots show GUS activity in epidermal and cortex cells. E, Leaves were stained 5 h after mechanical wounding and salt stress treatment (200 mm NaCl).
AtProT3 Promoter Activity Can Be Detected in Epidermal Cells of Leaves
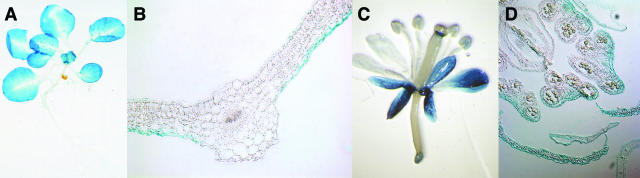
The uidA gene under the control of the AtProT3 promoter showed an expression pattern complementary to AtProT1 and AtProT2. Reporter gene expression under the control of the AtProT3 promoter could be detected in leaves, flowers, and siliques; however, it was absent from roots (Fig. 7, A and C), thus confirming the mRNA quantification experiments. In leaves, staining was strongest in younger leaves (Fig. 7A). Very weakly expressing lines showed staining predominantly in hydathodes and trichomes, whereas in lines with stronger expression (approximately 50% of all lines), GUS staining was detectable in the whole leaf blade but absent around the midvein (Fig. 7A). Cross-sections showed that, in leaves, GUS activity under the control of the AtProT3 promoter was confined to the epidermal cells (Fig. 7B) and was higher in the lower epidermis than in the upper epidermis. In flowers, staining was strongest in sepals and petals (Fig. 7, C and D). In siliques, only the carpels were stained. No expression was detectable in seeds (data not shown).
Figure 7.
Expression of GUS under control of the AtProT3 promoter. A, AtProT3-GUS expression in 2-week-old axenically grown Arabidopsis plants. B, Cross-sections show GUS activity in epidermal cells of leaves. C, In mature soil-grown plants, GUS activity is also present in flowers. D, Cross-section of flowers.
Tissue Specificity of AtProT Promoter Activities Is Conserved in Tobacco
Specificity of promoter activity was maintained in transgenic tobacco lines. GUS activity was detected in the vascular tissue of leaves and roots in AtProT1-GUS lines, in roots of AtProT2-GUS seedlings, and in the leaf blade of AtProT3-GUS transgenics (data not shown).
DISCUSSION
Pro, Gly Betaine, and GABA Are Transported with Different Affinity by the Three AtProTs
The Arabidopsis genome contains a total of three genes and two pseudogenes of the ProT family that show a high degree of amino acid sequence conservation. This raised the question as to whether the individual AtProTs have redundant functions in planta.
Although Arabidopsis accumulates Pro under stress conditions and not Gly betaine, all three AtProTs recognized the latter with highest affinity. The affinities for Gly betaine or Pro varied slightly for individual transporters, whereas only GABA was recognized with the same low affinity by all three AtProTs. Interestingly, LeProT1 also has a higher affinity for Gly betaine (Km = 0.11 mm) than for Pro (Km = 1.9 mm), although, like Arabidopsis, tomato is a Pro accumulator (Schwacke et al., 1999). In contrast, the affinities of the ProT homologs (AmT1 and AmT2) from the Gly betaine-accumulating mangrove A. marina were similar for both Pro and Gly betaine and comparable to the affinity of AtProT1 and AtProT2 for Pro (Waditee et al., 2002). In barley, both Gly betaine and Pro are accumulated in leaves in response to salt stress (Zuniga et al., 1989). However, the barley homolog HvProT is a specific Pro transporter that does not recognize Gly betaine, GABA, or other amino acids (Ueda et al., 2001).
It is interesting to note that transporters recognizing Pro, Gly betaine, and/or GABA have evolved in widely divergent organisms such as yeast, bacteria, and mammals (Grenson, 1992; Kempf and Bremer, 1998; Matskevitch et al., 1999). Their specificity of substrate recognition is quite similar and modified primarily by changes in the relative affinity of one over the other substrate.
All AtProTs Are Localized at the Plasma Membrane
Transporters of compatible solutes are required at several cellular membranes. Long-distance transport of Pro and Gly betaine has been demonstrated and the respective transport systems have to be present at the plasma membrane (Girousse et al., 1996; Mäkelä et al., 1996). Intracellular transport of Pro is also necessary, as Pro is synthesized in the cytosol but, under stress conditions, accumulates both in the cytosol and in the chloroplasts (Büssis and Heineke, 1998). Furthermore, Pro degradation takes place in the mitochondria (Yoshiba et al., 1997). The AtProTs show no obvious targeting signal directing protein import into mitochondria or chloroplasts (ARAMEMNON database; Schwacke et al., 2003). However, targeting to organelles could not be excluded because not all inner envelope proteins depend on a cleavable chloroplast transit peptide (Miras et al., 2002).
So far the subcellular localization has only been determined for one member of the ATF1 amino acid transporter family, AtAAP3, which was shown to be localized in the nuclear membrane, in organelle-like structures, and the plasma membrane, suggesting trafficking or cycling of the transporter (Okumoto et al., 2004). Our results demonstrate that both N- and C-terminal AtProT/GFP fusion proteins are targeted to the plasma membrane when transiently expressed in protoplasts, indicating that, in Arabidopsis, AtProTs mediate the uptake of compatible solutes from the apoplast. More detailed studies are required to determine whether, in addition to its role in uptake of compatible solutes from the apoplast, AtProT3 mediates intracellular transport or whether partial localization of AtProT3/GFP fusion proteins to internal membranes is due to inefficient targeting to the plasma membrane.
Complementary Expression Indicates Different Roles for AtProTs in Arabidopsis
Relative quantification using real-time PCR and analysis of transgenic promoter-GUS lines showed that the expression pattern of AtProTs is complementary and not overlapping. The localization of AtProT1 expression in the vascular tissue is in agreement with in situ localization of AtProT1 mRNA in the phloem tissue of flowers (Rentsch et al., 1996). These data indicate that AtProT1 is involved in long-distance transport of Pro in Arabidopsis and could be responsible for phloem loading, in retrieval of Pro (or Gly betaine) leaking from the phloem, or in xylem-to-phloem transfer along the path. Long-distance transport of Pro has previously been proposed based on correlative comparisons of the expression of genes encoding proteins involved in Pro metabolism along with Pro concentrations in different tissues (Savouré et al., 1995). However, the relative contribution of Pro transport, biosynthesis, degradation, and incorporation into alternative nitrogen sinks (e.g. cell wall proteins) for Pro concentrations in a specific tissue is still largely unknown.
AtProT2 expression, like that of HvProT, was highest in roots where it might be involved in uptake of compatible solutes from the soil or for retrieval of apoplastic amino acids delivered to the roots via the phloem (Schobert and Komor, 1987; Ueda et al., 2001). In maize roots subjected to low water potential, transport was shown to be the main source of Pro in the tip region (Verslues and Sharp, 1999; Raymond and Smirnoff, 2002). AtProT2 would be a good candidate for mediating similar transport processes in Arabidopsis.
RNA gel-blot analyses indicated a role for AtProT2 in stress response, since transcript levels were increased under water stress. However, GUS activity under control of the AtProT2 promoter was not induced by salt, low humidity, or low temperature. This could be explained by the lack of regulatory elements, e.g. present in the long first intron or in the promoter region itself (e.g. Callis et al., 1987). Alternatively, increased steady-state AtProT2 transcript levels under stress conditions might originate from increased mRNA stability rather than from enhanced expression (Gutiérrez et al., 1999). In leaves, AtProT2 expression was induced by mechanical wounding. Mechanical stimulation has been shown to lead to a rapid increase in GABA synthesis, leading to a 20- to 40-fold increase in GABA concentrations (Shelp et al., 1999). AtProT2 might be important for transport of GABA to neighboring cells or for the delivery of Pro to the sites of wounding, where it could serve as a source for nitrogen and reduction equivalents. Pro might also be used as a precursor for the synthesis of (hydroxy) Pro-rich proteins that are abundant in the cell walls and thus would be crucial for increased demand during cell wall reinforcement (Showalter, 1993; Nanjo et al., 1999). Pathogen attack, which also results in mechanical damage, has been shown to induce the Arabidopsis monosaccharide transporter AtSTP4. AtSTP4 was suggested to be important to meet the increased demand for carbohydrates of cells responding to pathogen attack (Truernit et al., 1996). Whether AtProT2 expression is also inducible by pathogens is an interesting aspect that will be tested in further studies.
GUS activity under the control of the AtProT3 promoter, along with mRNA quantification experiments, showed expression only in the aerial part of the plant. In leaves, GUS activity was exclusively localized in the epidermal cells. Differential compartmentation of inorganic ions between the mesophyll and the upper and lower epidermis of leaves has been reported for a number of monocotyledonous and dicotyledonous species (Karley et al., 2000). Also, sugars and amino acids are not evenly distributed in barley leaves, with concentrations being much lower in the epidermis (Dietz et al., 1994; Fricke et al., 1994). Unfortunately, in these very detailed studies, the techniques used for derivatization and detection of amino acids were not suitable for analyzing Pro concentrations. Interestingly, Zuniga et al. (1989) reported that, in water-stressed barley leaves, Pro accumulates preferentially in the epidermal cells and the vascular bundles. The same study also showed that Gly betaine levels in epidermal cells and the vascular bundles were much higher under both stress and normal growth conditions when compared to mesophyll cells or whole leaves. One might speculate that, because of the inevitable water loss by cuticular transpiration, epidermis cells are exposed to a continuous mild water stress that requires higher concentrations of compatible solutes, which might be either synthesized or imported by an AtProT3 homolog.
In summary, our data show that all three AtProTs are plasma membrane-localized transporters that differ only slightly in their substrate preference but fulfill distinct functions in planta due to their complementary and not overlapping expression patterns. Preliminary analyses of T-DNA insertion lines did not reveal any differences in overall growth habit to wild-type plants under standard growth conditions or during salt stress. A comprehensive analysis of mutants deficient in single or multiple AtProTs is necessary to show whether expression of individual AtProTs is differentially regulated in the mutants or whether activity of other amino acid transporters leads to this apparent redundancy. In addition, a more detailed knowledge of cellular Pro concentrations would contribute to an assessment of the physiological role of Pro transport in planta.
MATERIALS AND METHODS
Plant Growth and Transformation
For expression analyses, Arabidopsis thaliana L. ecotype Columbia (Col-0) was grown in a controlled growth chamber (CU-36L5; Percival Scientific, Boone, IA) with a day/night temperature of 22°C/18°C and a 16-h/8-h light/dark regime (100 μmol photons m−2 s−1) in axenic culture on solid AM medium (AM medium is 2.16 g Murashige and Skoog salts, 1% (w/v) Suc supplemented with 0.7% (w/v) BiTek agar [DIFCO Laboratories, Sparks, MD]). Alternatively, plants were grown in soil in a controlled growth chamber (AR-75 L; Percival Scientific) at 22°C/18°C, 65% humidity, and a 16-h/8-h light/dark cycle with uniform illumination of 150 μmol photons m−2 s−1. For transformation and selection of transgenic lines, plants were grown in soil in the greenhouse. Greenhouse temperature was maintained at above 10°C and natural illumination was supplemented to provide a minimum photoperiod of 16 h.
Plants were transformed by Agrobacterium tumefaciens-mediated (GV2260) gene transfer. Bacteria harboring the constructs were suspended in 0.5× Murashige and Skoog medium containing 5% (w/v) Suc, 0.01% (v/v) Silwet L-77, and 44 nm benzylaminopurine and used for vacuum infiltration (Clough and Bent, 1998).
Tobacco (Nicotiana tabacum L. cv SNN) was transformed as described by Köster-Töpfer et al. (1989) and selected on 2 mg L−1 BASTA. Subsequently, the BASTA concentration was increased to 4 mg L−1.
DNA and RNA Work
The open reading frame (ORF) of AtProT3 was isolated by RT-PCR using primers 5′-acaataaccatttggagagg-3′, 5′-aaatccaactaagaataaatacg-3′, and RNA extracted from roots of Arabidopsis Col-0 as a template. The AtProT3 ORF was cloned in the EcoRV site of pSK and verified by sequencing. For yeast (Saccharomyces cerevisiae) complementation assays, the AtProT3 ORF was transferred to pDR195 using XhoI and SacII (Rentsch et al., 1995). AtProT1 and AtProT2 were isolated from pFL61 by cleavage with NotI and inserted into the NotI site of pDR195 (Rentsch et al., 1996).
The promoters of AtProT1 and 3 were amplified by PCR using genomic DNA of Arabidopsis Col-0 as a template (AtProT1, 2,958-bp fragment, 5′-cttgagcttgaacatatgg-3′, 5′-gtttgctaagagactttctc-3′; AtProT3, 2,555-bp fragment, 5′-tgctaaacagagagcatgg-3′, 5′-gacctctccaaatggttattg-3′). The promoters were cloned into the SmaI site of the binary vector pCB308 (Xiang et al., 1999) and sequenced to confirm that no modifications occurred. The promoter fragment of AtProT2 was isolated by inverse PCR on genomic DNA (digested with BglII and religated) of Arabidopsis Col-0 and cloned into the EcoRV site of pSK (2,419-bp fragment, 5′-tggcaaaagacacattcg-3′, 5′-gtttccgacaagaatctg-3′). Sequencing confirmed the accuracy of the fragment and subsequently the promoter was transferred to the SmaI/XbaI site of pCB308.
For translational fusions with GFP, the ORFs of the cDNAs were amplified by PCR and cloned in pUC18-spGFP6 and pUC18-GFP5Tsp for C- and N-terminal fusion proteins (M. Suter-Grotemeyer and D. Rentsch, unpublished data). GFP-AtProT fusions (AtProT1, 5′-gctctagaatgaccgccaccgaag-3′, 5′-ccctcgagttacaaatctgcaaaaacatg-3′; AtProT2, 5′-gctctagaatggatacgagtgaagcaa-3′, 5′-ccctcgagtcaaacatcagcaaaaacatg-3′; AtProT3, 5′-gctctagaatgaactctaagaatcgcat-3′, 5′-ccctcgagttacaaatctgcaaaaacgtg-3′ [ORFs cloned into NheI/SalI site]). AtProT-GFP fusion (AtProT1, 5′-gctctagaatgaccgccaccgaag-3′, 5′-gaagatctagcaaatctgcaaaaacatgga-3′; AtProT2, 5′-gctctagaatggatacgagtgaagcaa-3′, 5′-cggatccttaacatcagcaaaaacatgga-3′; AtProT3, 5′-gctctagaatgaactctaagaatcgcat-3′, 5′-gaagatctagcaaatctgcaaaaacgtgga-3′ [ORFs cloned into SpeI/BglII site]). Sequence identity of all PCR-amplified fragments was verified by sequencing.
RNA was extracted using a method based on phenol extraction (Sambrook et al., 1989). RT was performed using the RetroScript kit (Ambion, Austin, TX), according to the manufacturer's instructions with oligo(dT) primers and 2 μg of total RNA as template.
Relative quantification using real-time PCR was performed on a LightCycler instrument (Roche Diagnostics, Mannheim, Germany). The FastStart DNA Master SYBR green I kit (Roche Diagnostics) was used, according to the manufacturer's instructions, with MgCl2 at a final concentration of 4 mm and 10 pmol of each primer. Primers were designed spanning intron-exon borders to avoid amplification of genomic DNA and allowing specific amplification of the respective AtProT transcripts (AtProT1, 5′-atctctttgcacatatttgcg-3′, 5′-catagcttttgcatagcattc-3′; AtProT2, 5′-ccggaaatacaggccacg-3′, 5′-gtcggacttgcaaaaatatgt-3′; AtProT3, 5′-ttaccgatgtttgcggttgt-3′, 5′-gacttgcaaaaatgtgcaac-3′). Ubiquitin was used as a reference gene (5′-gaatccaccctccacttggtc-3′, 5′-cgtctttcccgttagggtttt-3′).
Yeast Growth, Transformation, and Selection
Yeast 22574d (MATα ura3-1, gap1-1, put4-1, uga4-1; Jauniaux et al., 1987) was transformed according to Dohmen et al. (1991), and transformants were selected on nitrogen-free medium supplemented with 20 mg mL−1 of Glc and 1 mg mL−1 of Pro or GABA. For nonselective conditions, the cells were grown on synthetic dextrose minimal medium (Adams et al., 1998).
Transport Assays
For standard uptake studies, yeast cells were grown to logarithmic phase in synthetic dextrose minimal medium. Cells were harvested at an OD600nm of 0.5, washed twice in water, and resuspended in buffer A (0.6 m sorbitol and 50 mm potassium phosphate, pH 4.5) to a final OD600nm of 5.5. Prior to the uptake measurements, the cells were supplemented with 100 mm Glc and incubated for 5 min at 30°C. To start the reaction, 100 μL of this cell suspension were added to 100 μL of the same buffer containing 9.25 kBq labeled L-[14C]Pro, [3H]GABA (γ-aminobutyric acid; Amersham, Braunschweig, Germany), or [14C]Gly betaine (Biotrend, Cologne, Germany), and appropriate amounts of the respective unlabeled amino acids. The transport activity of the yeast mutant 22574d transformed with the empty vector pDR195 was subtracted as background from the observed rates. Samples were removed after 20, 60, 120, and 240 s, transferred to 4 mL of ice-cold buffer A, filtered on glass fiber filters, and washed with 8 mL of buffer A. The uptake of carbon-14 or tritium was determined by liquid scintillation spectrometry. Transport measurements were repeated independently and represent the mean of at least three experiments.
Transient Expression in Protoplasts
Tobacco protoplasts were isolated and transformed as described (Di Sansebastiano et al., 1998). Images were obtained after 16 to 28 h with a confocal laser-scanning microscope (DMR; Leica Microsystems, Wetzlar, Germany) using the TCS 4D operating system (Leica). GFP was detected with the filter set for fluorescein isothiocyanate, whereas chlorophyll epifluorescence was detected with the filter set for trimethylrhodamine isothiocyanate. The stored digital images were pseudocolored as red or green images using Photoshop 7.0 (Adobe Systems, Mountain View, CA) in correspondence to the real red or green colors.
Staining for β-Glucuronidase Activity
For histochemical localization of β-glucuronidase activity, plant material was infiltrated with 2 mm X-gluc in staining buffer (100 mm sodium phosphate, pH 7.0, 10 mm EDTA, 3 mm K4[Fe2(CN)6], 0.5 mm K3[Fe2(CN)6], 0.1% (v/v) Triton X-100). Staining was performed overnight at 37°C. For thin sections, plant material was embedded in paraffin (Peel-a-way 60–62°C; Polysiences, Warrington, PA) after staining according to the protocol of Sessions et al. (1999). Embedded material was sectioned to 12 μm on a Reichert-Jung 2040 Autocut (Pacific Southwest Lab Equipment, Vista, CA), and images were taken using a digital camera (Axiocam; Zeiss, Jena, Germany).
Acknowledgments
We are grateful to Ricardo Flückiger for help with the protoplast transformation, to Christa Köhler for tobacco transformation, and to Tanja Sikler and Christopher Ball for taking care of the plants. We wish to thank Wolf B. Frommer for helpful suggestions during the project and V.R. Franceschi for critical reading of the manuscript.
This work was supported by the Swiss National Foundation SNF (grant no. 31–64918.01) and by the Deutsche Forschungsgemeinschaft (grant no. SFB466). T.W. was a recipient of a scholarship of the Studienstiftung des Deutschen Volkes.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.055079.
References
- Adams A, Gottschling DE, Kaiser C, Stearns T (1998) Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Breitkreuz KE, Shelp BJ, Fischer WN, Schwacke R, Rentsch D (1999) Identification and characterization of GABA, proline and quaternary ammonium compound transporters from Arabidopsis thaliana. FEBS Lett 450: 280–284 [DOI] [PubMed] [Google Scholar]
- Büssis D, Heineke D (1998) Acclimation of potato plants to polyethylene glycol-induced water deficit. II. Contents and subcellular distribution of organic solutes. J Exp Bot 49: 1361–1370 [Google Scholar]
- Callis J, Fromm M, Walbot V (1987) Introns increase gene expression in cultured maize cells. Genes Dev 1: 1183–1200 [DOI] [PubMed] [Google Scholar]
- Chen LS, Ortiz-Lopez A, Jung A, Bush DR (2001) ANT1, an aromatic and neutral amino acid transporter in Arabidopsis. Plant Physiol 125: 1813–1820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang HH, Dandekar AM (1995) Regulation of proline accumulation in Arabidopsis thaliana (L.) Heynh during development and in response to desiccation. Plant Cell Environ 18: 1280–1290 [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Delauney AJ, Verma DPS (1993) Proline biosynthesis and osmoregulation in plants. Plant J 4: 215–223 [Google Scholar]
- Dietz KJ, Hollenbach B, Hellwege E (1994) The epidermis of barley leaves is a dynamic intermediary storage compartment of carbohydrates, amino acids and nitrate. Physiol Plant 92: 31–36 [Google Scholar]
- Di Sansebastiano G-P, Paris N, Marc-Martin S, Neuhaus J-M (1998) Specific accumulation of GFP in a non-acidic vacuolar compartment via a C-terminal propeptide-mediated sorting pathway. Plant J 15: 449–457 [DOI] [PubMed] [Google Scholar]
- Dohmen RJ, Strasser AWM, Honer CB, Hollenberg CP (1991) An efficient transformation procedure enabling long-term storage of competent cells of various yeast genera. Yeast 7: 691–692 [DOI] [PubMed] [Google Scholar]
- Fischer WN, Andre B, Rentsch D, Krolkiewicz S, Tegeder M, Breitkreuz K, Frommer WB (1998) Amino acid transport in plants. Trends Plant Sci 3: 188–195 [Google Scholar]
- Fricke W, Leigh RA, Tomos AD (1994) Concentrations of inorganic and organic solutes in extracts from individual epidermal, mesophyll and bundle sheath cells of barley leaves. Planta 192: 310–316 [Google Scholar]
- Fujita T, Maggio A, Garcia-Rios M, Bressan RA, Csonka LN (1998) Comparative analysis of the regulation of expression and structures of two evolutionarily divergent genes for Delta1-pyrroline-5-carboxylate synthetase from tomato. Plant Physiol 118: 661–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girousse C, Bournoville R, Bonnemain JL (1996) Water deficit-induced changes in concentrations in proline and some other amino acids in the phloem sap of alfalfa. Plant Physiol 111: 109–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grenson M (1992) Amino acid transporters in yeast: structure, function and regulation. In JJLLM De Pont, ed, Molecular Aspects of Transport Proteins. Elsevier Science Publishers B.V., Amsterdam, pp 219–245
- Gutiérrez RA, MacIntosh GC, Green PJ (1999) Current perspectives on mRNA stability in plants: multiple levels and mechanisms of control. Trends Plant Sci 4: 429–438 [DOI] [PubMed] [Google Scholar]
- Igarashi Y, Yoshiba Y, Takeshita T, Nomura S, Otomo J, Yamaguchi-Shinozaki K, Shinozaki K (2000) Molecular cloning and characterization of a cDNA encoding proline transporter in rice. Plant Cell Physiol 41: 750–756 [DOI] [PubMed] [Google Scholar]
- Jauniaux JC, Vandenbol M, Vissers S, Broman K, Grenson M (1987) Nitrogen catabolite regulation of proline permease in Saccharomyces cerevisiae: cloning of the Put4 gene and study of Put4 RNA levels in wild-type and mutant strains. Eur J Biochem 164: 601–606 [DOI] [PubMed] [Google Scholar]
- Karley AJ, Leigh RA, Sanders D (2000) Where do all the ions go? The cellular basis of differential ion accumulation in leaf cells. Trends Plant Sci 5: 465–470 [DOI] [PubMed] [Google Scholar]
- Kempf B, Bremer E (1998) Uptake and synthesis of compatible solutes as microbial stress responses to high-osmolality environments. Arch Microbiol 170: 319–330 [DOI] [PubMed] [Google Scholar]
- Köster-Töpfer M, Frommer WB, Rocha-Sosa M, Rosahl S, Schell J, Willmitzer L (1989) A class II patatin promoter is under developmental control in both transgenic potato and tobacco plants. Mol Gen Genet 219: 390–396 [DOI] [PubMed] [Google Scholar]
- Kwart M, Hirner B, Hummel S, Frommer WB (1993) Differential expression of two related amino acid transporters with differing substrate specificity in Arabidopsis thaliana. Plant J 4: 993–1002 [DOI] [PubMed] [Google Scholar]
- Mäkelä P, Peltonen Sainio P, Jokinen K, Pehu E, Setala H, Hinkkanen R, Somersalo S (1996) Uptake and translocation of foliar-applied glycinebetaine in crop plants. Plant Sci 121: 221–230 [Google Scholar]
- Matskevitch I, Wagner CA, Stegen C, Broer S, Noll B, Risler T, Kwon HM, Handler JS, Waldegger S, Busch AE, et al (1999) Functional characterization of the betaine/gamma-aminobutyric acid transporter BGT-1 expressed in Xenopus oocytes. J Biol Chem 274: 16709–16716 [DOI] [PubMed] [Google Scholar]
- McNeil SD, Nuccio ML, Hanson AD (1999) Betaines and related osmoprotectants. Targets for metabolic engineering of stress resistance. Plant Physiol 120: 945–949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miras S, Salvi D, Ferro M, Grunwald D, Garin J, Joyard J, Rolland N (2002) Non-canonical transit peptide for import into the chloroplast. J Biol Chem 277: 47770–47778 [DOI] [PubMed] [Google Scholar]
- Nanjo T, Kobayashi M, Yoshiba Y, Sanada Y, Wada K, Tsukaya H, Kakubari Y, Yamaguchi-Shinozaki K, Shinozaki K (1999) Biological functions of proline in morphogenesis and osmotolerance revealed in antisense transgenic Arabidopsis thaliana. Plant J 18: 185–193 [DOI] [PubMed] [Google Scholar]
- Okumoto S, Koch W, Tegeder M, Fischer WN, Biehl A, Leister D, Stierhof YD, Frommer WB (2004) Root phloem-specific expression of the plasma membrane amino acid co-transporter AAP3. J Exp Bot 55: 2155–2168 [DOI] [PubMed] [Google Scholar]
- Raymond MJ, Smirnoff N (2002) Proline metabolism and transport in maize seedlings at low water potential. Ann Bot (Lond) 89: 813–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rentsch D, Hirner B, Schmelzer E, Frommer WB (1996) Salt stress-induced proline transporters and salt stress-repressed broad specificity amino acid permeases identified by suppression of a yeast amino acid permease-targeting mutant. Plant Cell 8: 1437–1446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rentsch D, Laloi M, Rouhara I, Schmelzer E, Delrot S, Frommer WB (1995) NTR1 encodes a high affinity oligopeptide transporter in Arabidopsis. FEBS Lett 370: 264–268 [DOI] [PubMed] [Google Scholar]
- Rhodes D, Hanson AD (1993) Quaternary ammonium and tertiary sulfonium compounds in higher plants. Annu Rev Plant Physiol Plant Mol Biol 44: 357–384 [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Savouré A, Jaoua S, Hua X-J, Ardiles W, Van Montague M, Verbruggen N (1995) Isolation, characterization and chromosomal localization of a gene encoding the delta1-pyrroline-5-carboxylate-synthetase in Arabidopsis thaliana. FEBS Lett 372: 13–19 [DOI] [PubMed] [Google Scholar]
- Schobert C, Komor E (1987) Amino acid uptake by Ricinus communis roots: characterization and physiological significance. Plant Cell Environ 10: 493–500 [Google Scholar]
- Schwacke R, Grallath S, Breitkreuz KE, Stransky E, Stransky H, Frommer WB, Rentsch D (1999) LeProT1, a transporter for proline, glycine betaine, and gamma-amino butyric acid in tomato pollen. Plant Cell 11: 377–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwacke R, Schneider A, van der Graaff E, Fischer K, Catoni E, Desimone M, Frommer WB, Flügge UI, Kunze R (2003) ARAMEMNON, a novel database for Arabidopsis integral membrane proteins. Plant Physiol 131: 16–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano R (1996) Salt tolerance in plants and microorganisms: toxicity targets and defense responses. Int Rev Cytol 165: 1–52 [DOI] [PubMed] [Google Scholar]
- Sessions A, Weigel D, Yanofsky MF (1999) The Arabidopsis thaliana MERISTEM LAYER 1 promoter specifies epidermal expression in meristems and young primordia. Plant J 20: 259–263 [DOI] [PubMed] [Google Scholar]
- Shelp BJ, Bown AW, McLean MD (1999) Metabolism and functions of gamma-aminobutyric acid. Trends Plant Sci 4: 446–452 [DOI] [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K (1997) Gene expression and signal transduction in water-stress response. Plant Physiol 115: 327–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Showalter AM (1993) Structure and function of plant cell wall proteins. Plant Cell 5: 9–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stines AP, Naylor DJ, Hoj PB, van Heeswijck R (1999) Proline accumulation in developing grapevine fruit occurs independently of changes in the levels of delta(1)-pyrroline-5-carboxylate synthetase mRNA or protein. Plant Physiol 120: 923–931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truernit E, Schmid J, Epple P, Illig J, Sauer N (1996) The sink-specific and stress-regulated Arabidopsis STP4 gene: enhanced expression of a gene encoding a monosaccharide transporter by wounding, elicitors, and pathogen challenge. Plant Cell 8: 2169–2182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda A, Shi WM, Sanmiya K, Shono M, Takabe T (2001) Functional analysis of salt-inducible proline transporter of barley roots. Plant Cell Physiol 42: 1282–1289 [DOI] [PubMed] [Google Scholar]
- Verslues PE, Sharp RE (1999) Proline accumulation in maize (Zea mays L.) primary roots at low water potentials. II. Metabolic source of increased proline deposition in the elongation zone. Plant Physiol 119: 1349–1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waditee R, Hibino T, Tanaka Y, Nakamura T, Incharoensakdi A, Hayakawa S, Suzuki S, Futsuhara Y, Kawamitsu Y, Takabe T, et al (2002) Functional characterization of betaine/proline transporters in betaine-accumulating mangrove. J Biol Chem 277: 18373–18382 [DOI] [PubMed] [Google Scholar]
- Wipf D, Ludewig U, Tegeder M, Rentsch D, Koch W, Frommer WB (2002) Conservation of amino acid transporters in fungi, plants and animals. Trends Biochem Sci 27: 139–147 [DOI] [PubMed] [Google Scholar]
- Xiang CB, Han P, Lutziger I, Wang K, Oliver DJ (1999) A mini binary vector series for plant transformation. Plant Mol Biol 40: 711–717 [DOI] [PubMed] [Google Scholar]
- Yoshiba Y, Kiyosue T, Nakashima K, Yamaguchi-Shinozaki K, Shinozaki K (1997) Regulation of levels of proline as an osmolyte in plants under water stress. Plant Cell Physiol 38: 1095–1102 [DOI] [PubMed] [Google Scholar]
- Zuniga GE, Argandona VH, Corcuera LJ (1989) Distribution of glycine betaine and proline in water stressed and unstressed barley leaves. Phytochemistry 28: 419–420 [Google Scholar]