Abstract
Photoperiod is the primary environmental factor affecting flowering time in rapid-cycling accessions of Arabidopsis (Arabidopsis thaliana). Winter-annual Arabidopsis, in contrast, have both a photoperiod and a vernalization requirement for rapid flowering. In winter annuals, high levels of the floral inhibitor FLC (FLOWERING LOCUS C) suppress flowering prior to vernalization. FLC acts to delay flowering, in part, by suppressing expression of the floral promoter SOC1 (SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1). Vernalization leads to a permanent epigenetic suppression of FLC. To investigate how winter-annual accessions integrate signals from the photoperiod and vernalization pathways, we have examined activation-tagged alleles of FT and the FT homolog, TSF (TWIN SISTER OF FT), in a winter-annual background. Activation of FT or TSF strongly suppresses the FLC-mediated late-flowering phenotype of winter annuals; however, FT and TSF overexpression does not affect FLC mRNA levels. Rather, FT and TSF bypass the block to flowering created by FLC by activating SOC1 expression. We have also found that FLC acts as a dosage-dependent inhibitor of FT expression. Thus, the integration of flowering signals from the photoperiod and vernalization pathways occurs, at least in part, through the regulation of FT, TSF, and SOC1.
Flowering time in most plant species is regulated by a combination of endogenous controls and environmental cues (Boss et al., 2004). This combination of signals helps to ensure that flowering takes place at the proper point in the plant's development, as well as at a favorable time of year, thereby maximizing the chances of successful reproduction. Two of the most common environmental factors affecting flowering time are day length (photoperiod) and temperature. In rapid-cycling strains of Arabidopsis (Arabidopsis thaliana), photoperiod is the primary environmental signal regulating flowering time; plants flower more rapidly under inductive long days (LD) than short days (SD). The B-box zinc-finger-containing transcription factor CONSTANS (CO) plays a critical role in the regulation of flowering time in response to photoperiod (Putterill et al., 1995). co mutants flower late in LD, but flowering is unaffected in SD (Koornneef et al., 1991). Thus, CO acts to promote flowering under LD. Recently, the molecular mechanism for the LD-specific promotion of flowering by CO has been elucidated. CO expression is regulated at both the RNA and protein levels. CO protein level is regulated via the photoreceptors PHYTOCHROME A and CRYPTOCHROME 2 and accumulates only during the light (Valverde et al., 2004). CO transcription, in turn, is circadian regulated with expression peaking late in the day (Suarez-Lopez et al., 2001). This peak in mRNA levels takes place at the end of the day in LD but during the night in SD. Thus, only in LD is CO transcription coincident with the light required for protein accumulation.
In contrast to rapid-cycling strains of Arabidopsis whose flowering is regulated primarily by photoperiod, flowering in many naturally occurring accessions is also promoted by vernalization. Vernalization is the promotion of flowering by a prolonged exposure to cold temperatures (Chouard, 1960), such as would be experienced during winter in temperate climates. These accessions are delayed in flowering unless vernalized and thus behave as winter annuals. The vernalization-responsive block to flowering is caused by the interaction of two genes, FLC (FLOWERING LOCUS C) and FRI (FRIGIDA). FLC encodes a MADS-domain-containing transcription factor that acts as a repressor of flowering (Michaels and Amasino, 1999; Sheldon et al., 1999), and FRI encodes a protein of unknown biochemical function that is required for FLC to be expressed to high levels (Johanson et al., 2000). Thus, both genes are required to block flowering, and loss-of-function mutations in either gene results in early flowering (in the absence of FRI, FLC is not expressed and in the absence of the floral repressor FLC, there is no effect of FRI [Michaels and Amasino, 1999, 2001]). It is interesting to note that rapid-cycling accessions of Arabidopsis are naturally occurring mutants in FRI or FLC (Johanson et al., 2000; Corre et al., 2002; Loudet et al., 2002; Gazzani et al., 2003; Michaels et al., 2003). Vernalization promotes flowering by causing a permanent epigenetic repression of FLC (Michaels and Amasino, 1999; Sheldon et al., 1999). Recent studies have shown that changes in chromatin structure via histone modifications at the FLC locus play a critical role in the repression of FLC by vernalization (Bastow et al., 2004; Sung and Amasino, 2004).
In winter-annual Arabidopsis, the decision to flower is influenced by environmental information from the photoperiod and vernalization pathways. The molecular confluence of these pathways takes place at the level of two floral integrators, FT (Kardailsky et al., 1999; Kobayashi et al., 1999) and SOC1 (SUPPRESSOR OF OVEREXPRESSION OF CONSTANS)/ AGAMOUS-LIKE 20 (Borner et al., 2000; Lee et al., 2000; Samach et al., 2000). Both FT and SOC1 act as strong floral promoters; overexpression of either gene leads to a dramatic early-flowering phenotype. The expression of FT appears to be controlled primarily by photoperiod; CO promotes flowering by activating FT expression (Kardailsky et al., 1999; Kobayashi et al., 1999). SOC1 expression, in contrast, is strongly regulated by vernalization (Lee et al., 2000). In the absence of vernalization, SOC1 is repressed by FLC. Following cold treatment, however, FLC expression is suppressed and SOC1 is expressed at high levels. Although FT and SOC1 are most strongly regulated by photoperiod and vernalization, respectively, crosstalk between pathways does occur. CO overexpression increases SOC1 levels and elevated levels of FLC expression decrease FT levels (Samach et al., 2000). To further characterize the integration of flowering signals, we examined activation-tagged mutants of FT and the FT homolog, TSF (TWIN SISTER OF FT; Kardailsky et al., 1999; Kobayashi et al., 1999) in a winter-annual background.
RESULTS
Identification of Activation Alleles of FT and TSF
Two T-DNA-mutagenized populations were generated in either wild-type Wassilewskija (Ws) or a late-flowering vernalization-responsive line containing FRI-SF2 in the Columbia (Col) background (FRI-Col) (Lee et al., 1994). Plants were transformed with the activation-tagging vector pSKI015 (Weigel et al., 2000), which carries 4 copies of the 35S cauliflower mosaic virus enhancer element. Several early-flowering plants were isolated from the T1 generations, suggesting that they contained dominant mutations due to gene activation. To determine the site of T-DNA integration in these mutants, genomic DNA flanking the site of T-DNA insertion was isolated using Thermal Asymmetric Interlaced PCR (Liu et al., 1995) and sequenced.
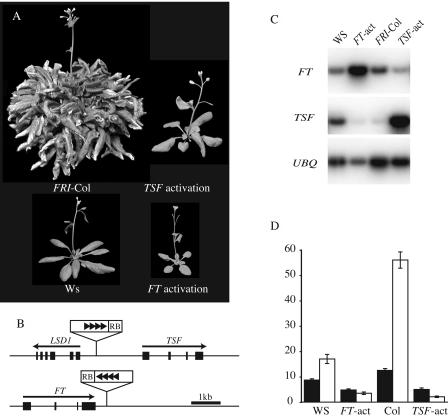
An early-flowering mutant from the Ws population was found to contain a T-DNA insertion 0.4 kb downstream of the 3′ end of the floral promoter FT, suggesting that the early-flowering phenotype of this mutant is due to the activation of FT (Fig. 1, A and B). This model is supported by the fact that the position and orientation of the T-DNA relative to FT is nearly identical to that in a previously described activation-tagged FT mutant and the two mutants exhibit similar early-flowering phenotypes (Kardailsky et al., 1999). A second mutant from the FRI-Col population contained a T-DNA 1.6 kb upstream of the translational start site of TSF (Fig. 1, A and B). Like FT, TSF overexpression also causes early flowering, suggesting that the early-flowering phenotype of this mutant is due to an activation of TSF (Kobayashi et al., 1999). Indeed, reverse transcription (RT)-coupled PCR showed that FT and TSF levels are elevated in these mutants (Fig. 1C). Interestingly, TSF mRNA levels are reduced in the FT activation mutant; similarly, FT levels are reduced in the TSF activation, indicating that negative feedback may play a role in the regulation of FT and TSF.
Figure 1.
Activation-tagged alleles of TSF and FT. A, The early-flowering phenotype of the TSF activation mutant in the FRI-Col background and of the FT activation mutant in the Ws background. Plants were grown under LD. B, Location and orientation of the four 35S enhancer elements (black triangles) and T-DNA right border (RB) relative to TSF and FT. C, RT-PCR analysis of FT and TSF expression in wild type and activation-tagged mutants. RNA was extracted from 7-d-old seedlings. D, Flowering time of FT and TSF activation mutants in fri-null backgrounds (FT-act in the Ws background and TSF-act in the Col background). Black and white bars represent plants grown in LD and SD, respectively. Flowering time is expressed as the number of rosette leaves formed by the primary shoot apical meristem prior to the initiation of flowering. Error bars indicate 1 sd.
FT and TSF Activation Mutants Flower More Rapidly under Short Days
To determine the effects of FT and TSF activation on flowering time in a non-FRI-containing background, the TSF activation-tagged line was crossed to Col and plants were identified in the F2 generation that were homozygous for the T-DNA and the Col allele of fri (which is a naturally occurring null). Genotypes were verified using PCR-based markers.
Previous studies have shown that overexpression of FT or TSF driven by the constitutive 35S promoter results in a strong early-flowering phenotype and the formation of terminal flowers (Kardailsky et al., 1999; Kobayashi et al., 1999). Consistent with these overexpression studies, the FT and TSF activation-tagged mutants flowered much earlier than the corresponding wild types in both LD and SD (Fig. 1D), and terminal flowers were observed in a small fraction of the plants (data not shown). An interesting distinction between the activation-tagged mutants and previously described FT overexpression lines is that the activation-tagged mutants flowered with fewer leaves in SD than in LD. This phenotype was not restricted to the fri-null background; in all genetic situations tested, the FT and TSF activation alleles caused earlier flowering in SD than in LD (see below).
FT and TSF Activation Alleles Strongly Suppress the Late-Flowering Phenotype of FRI and FLC
The photoperiod and vernalization pathways monitor the two primary environmental factors controlling flowering time in Arabidopsis. A major component of the photoperiodic regulation of flowering is the regulation of FT levels by CO, whereas the regulation of flowering by vernalization is achieved largely through the regulation of FLC. To investigate the interaction between these two pathways, the effects of FT and TSF activation alleles were evaluated in a FRI-containing background.
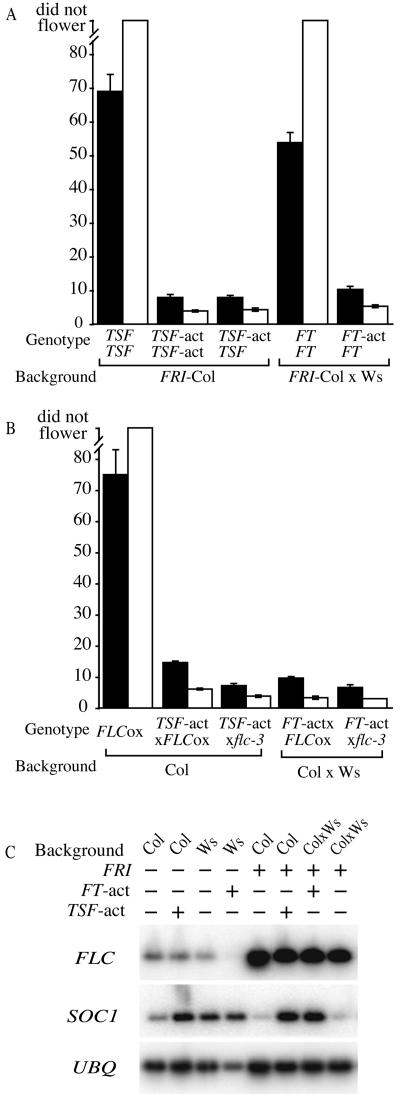
In a FRI-containing background, TSF activation completely suppressed the delayed flowering effect of FRI in LD (Fig. 2A). Moreover, the early-flowering phenotype exceeded that of a fri null; the TSF activation in the FRI background flowered with approximately five fewer leaves than Col, a fri null (Col data shown in Fig. 1D). The effect of TSF activation under SD was even more dramatic. The TSF activation mutant in the FRI background flowered after forming approximately 4 leaves, whereas FRI-Col did not flower even after forming >100 leaves (Fig. 2A). As in LD, the TSF activation in the FRI background flowered much earlier than Col, which flowered with approximately 55 leaves in SD (Fig. 1D). Because the early-flowering phenotype of TSF activation is much stronger than a loss of FRI activity, the activation of TSF clearly does more to promote flowering than to simply counteract the effects of FRI and FLC. To determine the dosage dependence of the TSF-activation allele, the TSF-activation mutant was crossed to the FRI-Col. The flowering time of the F1 plants was indistinguishable from the homozygous TSF-activation mutant (Fig. 2A), indicating that the TSF-activation allele behaves dominantly.
Figure 2.
Activation alleles of TSF and FT suppress the late-flowering phenotype of FRI and FLC overexpression. Black and white bars represent plants grown in LD and SD, respectively. Flowering time is expressed as the number of rosette leaves formed by the primary shoot apical meristem prior to the initiation of flowering. Error bars indicate 1 sd. A, Flowering time of TSF and FT activation alleles in a FRI-containing background. B, Effect of TSF and FT activation on flowering time in a 35S∷FLC background (FLCox). C, RT-PCR analysis of the effect of TSF and FT activation alleles on the expression of FLC and SOC1. UBIQUITIN (UBQ) was included as a control for loading.
To determine the effect of FT activation in a FRI-containing background, the FT-activation mutant and Ws as a control were crossed to FRI-Col and the resulting F1 plants were grown under LD and SD (FRI is completely dominant). Similar to the results with the TSF-activation allele, the FT-activation allele also strongly suppressed the effects of FRI (Fig. 2A). Under LD, the F1 plants from the FT-activation/FRI-Col cross flowered similarly to wild-type Ws (Fig. 1D), which is a fri-null. Under SD, the early-flowering phenotype was even stronger, with the F1 plants flowering with 12 fewer leaves than Ws (Figs. 1D and 2A).
FT and TSF Activation Suppresses the Late-Flowering Phenotype of FLC Overexpression
FT- and TSF-activation alleles strongly suppress the late-flowering phenotype caused by FRI and FLC. One possible model to explain this early-flowering phenotype is that FT and TSF activation may directly suppress FLC expression. Alternatively, the activation of FT and TSF may bypass the block to flowering created by FRI and FLC. To differentiate between these models, the activation-tagged alleles of FT and TSF were crossed to a line containing FLC under control of the constitutive 35S promoter (Odell et al., 1985) in an flc-3 mutant background. The flowering time of F1 plants resulting from crosses between the activation-tagged mutants and 35S∷FLC were similar to that of crosses between the mutants and FRI-Col (Fig. 2, A and B). Given that FT and TSF activation alleles cause a similar early-flowering phenotype in FRI-containing or FLC-overexpression backgrounds, FT and TSF are not likely to affect FLC expression, but rather the elevated expression of FT or TSF appears to bypass the block to flowering created by FLC. Indeed, FT or TSF overexpression does not affect FLC mRNA levels (see below).
FT and TSF Act As Positive Regulators of SOC1, a Downstream Target of FLC
To gain molecular insight into the strong suppression of the late-flowering phenotype of FRI or 35S∷FLC by activation alleles of FT or TSF, we investigated the effects of FT and TSF activation on FLC and SOC1 expression. SOC1 is a floral promoter that is negatively regulated by FLC, and SOC1 overexpression has been shown to suppress the late-flowering phenotype of FRI-containing lines (Lee et al., 2000). Thus, a possible model for the suppression of FLC-mediated late flowering by FT and TSF activation is that SOC1 is up-regulated by FT and TSF. To test this model, RT-PCR analysis was performed using total RNA isolated from 7-d-old seedlings of FRI-containing lines with or without FT or TSF activation alleles (Fig. 2C). All lines showed high levels of FLC expression regardless of the presence or absence of FT- or TSF-activation alleles. SOC1 expression, however, was up-regulated by FT or TSF activation. Thus, it appears that FT and TSF suppress the late-flowering phenotype of lines containing high levels of FLC, at least in part, through the up-regulation of SOC1.
Histochemical Analysis of Gene Expression
FLC and SOC1 are expressed at the highest levels in shoot and root apicies (Lee et al., 2000; Michaels and Amasino, 2000; Samach et al., 2000; Hepworth et al., 2002), whereas FT is most strongly expressed in the vasculature of the leaves (Takada and Goto, 2003). Because of these differences in spatial expression patterns, β-glucuronidase (GUS) fusions were used to further investigate the interactions between FLC, SOC1, FT, and TSF. Because FRI, FLC, and the FT and TSF activation alleles all behave dominantly, gene expression patterns were determined in F1 plants resulting from crosses to lines containing SOC1∷GUS, FLC∷GUS, and FT∷GUS.
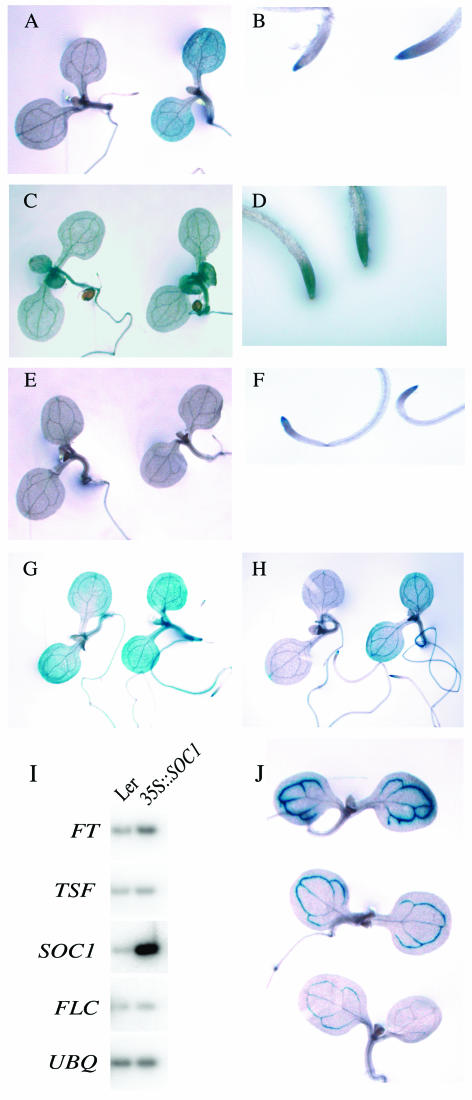
Consistent with RNA expression data, the SOC1∷GUS fusion is negatively regulated by FRI and FLC (Fig. 3A). In the absence of FRI, SOC1∷GUS is expressed broadly in seedlings with the highest staining in the shoot and root tips. When the SOC1∷GUS line is crossed to FRI-Col, GUS staining is strongly reduced in the shoot apex and the cotyledons (Fig. 3A). SOC1∷GUS expression in the root tip, however, is relatively unaffected by FRI (Fig. 3B). This result is surprising because FLC is expressed at high levels in both the root tip and shoot tip in a FRI background (Fig. 3, C and D). One possible explanation for this result is that, in the root tip, FLC is not expressed in the necessary cell types or is not expressed to sufficient level to suppress SOC1. Another possibility is that FLC requires the presence of other factors to suppress SOC1 and that one or more of these factors are not expressed in roots. To differentiate between these models, SOC1∷GUS expression was also determined in a 35S∷FLC background (Fig. 3, E and F). The pattern of SOC1∷GUS expression in the FLC overexpression background was similar to that in a FRI-containing background; expression was suppressed in the shoot, but was unaffected in the root tip. Thus, it seems that FLC expression alone is insufficient to suppress SOC1 expression in roots, suggesting that other factors, which are present in the shoot, may be lacking in roots.
Figure 3.
Histochemical analysis of the interactions between FLC, SOC1, TSF, and FT. To minimize variation in GUS staining, plants that appear in the same section were grown, fixed, and stained in parallel. A and B, Effect of FRI on SOC1∷GUS expression. F1 seedlings and root tips resulting from the cross of SOC1∷GUS to FRI-Col (left) or Ws (right). C and D, FLC∷GUS expression in seedlings and root tips in a FRI-containing background. E and F, Comparison of the effects of FRI and 35S∷FLC on SOC1∷GUS expression. F1 seedlings and root tips resulting from the cross of SOC1∷GUS to FRI-Col (left) or 35S∷FLC (right). G, Effect of FT activation on SOC1∷GUS expression. F1 seedlings resulting from the cross of SOC1∷GUS to Ws (left) and the FT activation mutant (right). Plants are in a fri-null background. H, Effect of TSF activation on SOC1∷GUS expression. F1 seedlings resulting from the cross of SOC1∷GUS to FRI-Col (left) and to the TSF activation mutant (right). Plants are in a FRI-containing background. I, Effect of SOC1 overexpression. RT-PCR analysis of FT, TSF, SOC1, and FLC expression in wild-type and 35S∷SOC1 lines. UBIQUITIN (UBQ) was included as a control for loading. Plants are in the Landsberg erecta (Ler) background. J, Inhibition of FT expression by FRI and FLC. F1 seedlings resulting from the cross of FT∷GUS in a Col background to Col (top), FRI-Col (middle), or 35S∷FLC (bottom).
SOC1∷GUS expression is up-regulated by both the FT and TSF activation alleles in the presence or absence of FRI (Fig. 3, G and H). Comparison of crosses of SOC1∷GUS to wild type and the FT activation mutant revealed that SOC1∷GUS expression is increased throughout the plant by FT activation (Fig. 3G). Likewise, TSF activation increased SOC1∷GUS expression in a FRI background (Fig. 3H). Although both the FT and TSF activation alleles increased SOC1∷GUS expression, it is interesting to note that the expression patterns of SOC1∷GUS differ slightly. In the FT-activation background, SOC1∷GUS expression is relatively uniform throughout the plant, whereas in the TSF-activation background expression is highest in the vasculature of the cotyledons. One possible explanation for the concentration of SOC1∷GUS expression in the vasculature is that the TSF-activation allele may be expressed to highest levels in the vasculature, similar to FT∷GUS. (In some cases activation alleles maintain aspects of endogenous gene regulation [Weigel et al., 2000].)
We also investigated whether there might be reciprocal regulation of FT or TSF by SOC1. FT and TSF expression were examined in wild type and a 35S∷SOC1 line by RT-PCR. TSF expression was similar in both backgrounds (Fig. 3I); thus, SOC1 does not appear to regulate TSF. FT expression was slightly higher in the 35S∷SOC1 line, suggesting that SOC1 may regulate FT. It should be noted, however, that the extreme early-flowering phenotype of the 35S∷SOC1 line (approximately 2 leaves) makes it difficult to ensure that wild type and 35S∷SOC1 are at the same developmental stage (i.e. the 35S∷SOC1 seedlings may have initiated flowering, while wild type remained vegetative). Thus, the increase in FT expression caused by 35S∷SOC1 may reflect a difference in developmental stage of the plants rather than a direct effect of SOC1 overexpression.
The FT∷GUS line was crossed to both Col and FRI-Col to determine the effect of FRI and FLC on FT∷GUS expression (Fig. 3J). When crossed to Col, strong FT∷GUS expression was observed in the vasculature of the cotyledons. The level of FT∷GUS expression was reduced in the FRI-containing line, which has higher levels of FLC. Thus, FLC negatively regulates FT expression. The FT∷GUS line was also crossed to 35S∷FLC to determine the effect of increased FLC levels in the cotyledons. This line showed an even greater reduction in FT∷GUS expression. Thus, FLC acts as a dosage-dependent suppressor of FT.
FT and TSF Have Overlapping Roles in the Promotion of Flowering
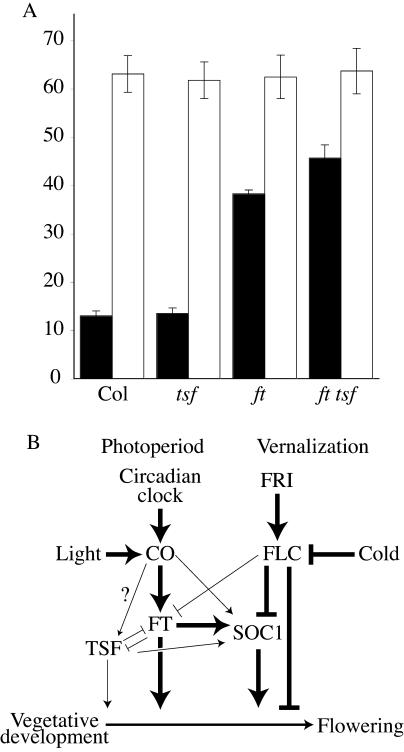
The similar phenotypes of activation or overexpression of FT and TSF suggest that they have similar functions in the regulation of flowering. Loss-of-function mutations in ft were identified on the basis of their late-flowering phenotype in LD. To determine if tsf loss-of-function mutations affect flowering, we obtained a line containing a T-DNA insertion in TSF from the SALK collection (Alonso et al., 2003). Homozygous tsf-mutant plants were identified using PCR-based markers and grown in both LD and SD. Under both conditions, the tsf mutant flowered similarly to wild type (Fig. 4A). A possible explanation for the lack of a flowering-time phenotype in the tsf mutant is that FT and TSF have overlapping or partially redundant roles in the promotion of flowering and that FT activity can compensate for the lack of TSF. Therefore, we investigated the effect of the tsf mutant in an ft-mutant background by crossing ft and tsf and isolating the ft tsf double mutant in the F2 generation. Genotypes were verified using PCR-based markers. In LD, the ft tsf double mutant formed approximately 12 more leaves than the ft single mutant before flowering. Thus, FT and TSF have overlapping roles in the promotion of flowering, with FT playing the dominant role.
Figure 4.
Interactions between flowering time genes. A, Flowering phenotype of tsf loss-of-function mutations in wild-type and ft-mutant backgrounds. Black and white bars represent plants grown in LD and SD, respectively. Flowering time is expressed as the number of rosette leaves formed by the primary shoot apical meristem prior to the initiation of flowering. Error bars indicate 1 sd. B, A model for the regulatory relationships between flowering genes. Line thickness is intended as a speculative measure of the strength of promotion or inhibition.
DISCUSSION
Much is now known of the molecular mechanisms by which the photoperiod and vernalization pathways regulate flowering time in Arabidopsis (e.g. Boss et al., 2004). Less is known, however, about how signals from these two pathways are integrated to control the transition to flowering. Rapid-cycling strains of Arabidopsis do not have a vernalization requirement for early flowering; thus, photoperiod is the major environmental factor regulating flowering time in these accessions. Flowering in winter-annual accessions, in contrast, is controlled by both photoperiod and vernalization, and therefore winter-annual Arabidopsis is an attractive system in which to study the integration of the photoperiod and vernalization pathways.
In the course of screening for early-flowering mutants in fri-mutant and FRI-containing backgrounds, we identified activation-tagged alleles of FT and TSF. The function of FT in the regulation of flowering time is most closely associated with the promotion of flowering in response to inductive photoperiods. ft mutants flower relatively normally in SD, but are late flowering in LD (Koornneef et al., 1991). Subsequent studies have shown that FT expression is positively regulated by CO (Kardailsky et al., 1999; Kobayashi et al., 1999), which is regulated by light and the circadian clock (Suarez-Lopez et al., 2001; Valverde et al., 2004). The activation mutants have elevated steady-state RNA levels of FT and TSF and exhibit a strong early-flowering phenotype in fri-null backgrounds consistent with previous overexpression studies (Kardailsky et al., 1999; Kobayashi et al., 1999). Interestingly, the activation-tagged alleles of FT and TSF also strongly suppressed the late-flowering phenotype caused by FRI and FLC in a winter-annual strain. These winter-annual strains containing activation alleles of FT and TSF were used to investigate the integration of flowering signals between the photoperiod and vernalization pathways.
The late-flowering vernalization-responsive phenotype of winter-annual Arabidopsis is caused by the up-regulation of FLC by FRI (Michaels and Amasino, 1999; Sheldon et al., 1999). Although the activation alleles of FT and TSF effectively suppress the late-flowering phenotype of winter annuals, FLC expression remains high. Expression of SOC1, a promoter of flowering that is negatively regulated by FLC, however, is up-regulated by FT and TSF activation. Thus, the regulation SOC1 by both FLC and FT represents a mechanism for the integration of signals from the photoperiod and vernalization pathways.
It should be noted, however, that previous work suggests that positive regulation of SOC1 by the photoperiod pathway is not solely accomplished through FT and TSF. Studies have shown that CO and FLC antagonistically regulate SOC1 via separate promoter elements (Hepworth et al., 2002). In those experiments, FLC was shown to bind directly to SOC1 promoter sequences in gel shift assays. CO was not shown to bind SOC1 promoter sequences, but other experiments using a translational fusion of CO to the ligand-binding domain of the glucocorticoid receptor in cycloheximide-treated plants indicate the CO can up-regulate SOC1 in the absence of the translation of new proteins (Samach et al., 2000), suggesting that CO is a direct regulator of SOC1. In total, these experiments indicate that CO may positively regulate SOC1 through both indirect mechanisms (i.e. CO promotes FT expression, which in turn promotes SOC1 expression) and direct mechanisms (e.g. CO can bind to the SOC1 promoter as part of a complex).
Experiments using a SOC1∷GUS fusion also demonstrated the regulation of SOC1 by FT and TSF and revealed an interesting aspect of the regulation of SOC1 by FLC. Despite the fact that both FLC and SOC1 are expressed in the shoot and root tips, FLC is only effective in suppressing SOC1 in the shoot. In lines containing FRI or 35S∷FLC, SOC1∷GUS expression is suppressed in the shoot but is unchanged in the root, suggesting that additional shoot-specific factors may be required for the suppression of SOC1 by FLC. It is interesting to note that differences in the expression requirements for root and shoot expression of FLC have also been observed; in a pie1 mutant background, FLC expression is suppressed in the shoot but is not affected in roots (Noh and Amasino, 2003).
An interesting phenotype of the FT and TSF activation mutants is that, in all genetic backgrounds tested (fri-null, FRI-containing, and 35S∷FLC backgrounds), plants flowered after forming fewer leaves in SD than LD. The reason for earlier flowering in SD is not clear. One possibility is that the slower overall growth in SD provides additional time for FT and TSF to act. This SD-plant phenotype was not observed in plants containing 35S∷FT or 35S∷TSF constructs, which flowered with a similar number of leaves in LD or SD (Kardailsky et al., 1999; Kobayashi et al., 1999). These overexpression lines, however, flower after forming four or fewer leaves in LD or SD. (In our laboratory, transgenic plants containing 35S∷FT in a FRI-containing background flowered with approximately three leaves in either photoperiod.) Thus, the transition to flowering may occur so rapidly in these lines as to obscure any effect of photoperiod.
In summary, the results of these and previous experiments show that the integration of flowering signals from the photoperiod and vernalization pathways occurs, at least in part, through the regulation of FT, TSF, and SOC1 (Fig. 4B). CO and FLC are regulated by the photoperiod and vernalization pathways, respectively. Both pathways, however, regulate the floral integrators FT, TSF, and SOC1; FT is positively regulated by CO and negatively regulated by FLC, whereas SOC1 is negatively regulated by FLC and positively regulated by CO, FT, and TSF. In species such as Arabidopsis that have a quantitative response to both photoperiod and vernalization (i.e. plants will eventually flower even in the absence of inductive photoperiods or vernalization), it is possible that the levels of these integrators may provide a composite picture of the favorableness of the environment for flowering.
MATERIALS AND METHODS
Plant Material and Mutagenesis
FRI-SF2 in the Col background (Lee et al., 1994) and flc-3 (Michaels and Amasino, 1999) have been described previously. ft-1 introgressed into the Col background (Kardailsky et al., 1999) was kindly provided by D. Weigel (Max Planck Institute for Developmental Biology, Tubingen, Germany). The tsf mutant was obtained from the Salk collection (SALK 087522; Alonso et al., 2003). T-DNA and fast-neutron mutagenized populations have been described previously (Michaels and Amasino, 1999).
Growth Conditions
All plants were grown under 120 μE m−2 s−1 of cool-white fluorescent light at 22°C. LD conditions consisted of 16 h of light followed by 8 h of darkness; SD consisted of 8 h of light followed by 16 h of darkness. Plants used for RNA analysis were grown under constant light for 7 d prior to tissue harvest.
Gene Expression Analysis
For RT-PCR analysis, RNA isolation, RT, and PCR were performed as described previously (Michaels et al., 2004). Primers used for the detection of SOC1, UBIQUITIN, and FLC have been described previously (Michaels et al., 2004). For the detection of FT (5′-ACCTCAGGAACTTCTATACTTTGG-3′ and 5′-TACTATAGGCATCATCACCGTTCG-3′) and TSF (5′-ATGTCTTTAAGTCGTAGAGATCCTCTTGTGGT-3′ and 5′-CTACGTTCTTCTTCCCCCACAGCCATTC-3′), the indicated primers were used.
GUS Constructs
FT∷GUS (Takada and Goto, 2003) and SOC1∷GUS (Hepworth et al., 2002) constructs have been described previously. The FLC∷GUS fusion was created by inserting the GUS gene into an NheI site located in the sixth exon of a 16-kb genomic clone containing 5.4 kb upstream of the FLC start codon and 5 kb downstream of the stop codon.
Distribution of Materials
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes, subject to the requisite permission from any third part owners of all or parts of the material. Obtaining any permissions will be the responsibility of the requestor.
Acknowledgments
We thank Yannick Jacob for critical reading of this paper and Ann Marie DeGruccio for assistance in plant culture.
This work was supported by the College of Agricultural and Life Sciences and the Graduate School of the University of Wisconsin and by the National Science Foundation (grant nos. 0133663 and 0209786).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.052811.
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Bastow R, Mylne JS, Lister C, Lippman Z, Martienssen RA, Dean C (2004) Vernalization requires epigenetic silencing of FLC by histone methylation. Nature 427: 164–167 [DOI] [PubMed] [Google Scholar]
- Borner R, Kampmann G, Chandler J, Gleissner R, Wisman E, Apel K, Melzer S (2000) A MADS domain gene involved in the transition to flowering in Arabidopsis. Plant J 24: 591–599 [DOI] [PubMed] [Google Scholar]
- Boss PK, Bastow RM, Mylne JS, Dean C (2004) Multiple pathways in the decision to flower: enabling, promoting, and resetting. Plant Cell (Suppl) 16: S18–S31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chouard P (1960) Vernalization and its relations to dormancy. Annu Rev Plant Physiol 11: 191–238 [Google Scholar]
- Corre VL, Roux F, Reboud X (2002) DNA polymorphism at the FRIGIDA gene in Arabidopsis thaliana: extensive nonsynonymous variation is consistent with local selection for flowering time. Mol Biol Evol 19: 1261–1271 [DOI] [PubMed] [Google Scholar]
- Gazzani S, Gendall AR, Lister C, Dean C (2003) Analysis of the molecular basis of flowering time variation in Arabidopsis accessions. Plant Physiol 132: 1107–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepworth SR, Valverde F, Ravenscroft D, Mouradov A, Coupland G (2002) Antagonistic regulation of flowering-time gene SOC1 by CONSTANS and FLC via separate promoter motifs. EMBO J 21: 4327–4337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johanson U, West J, Lister C, Michaels S, Amasino R, Dean C (2000) Molecular analysis of FRIGIDA, a major determinant of natural variation in Arabidopsis flowering time. Science 290: 344–347 [DOI] [PubMed] [Google Scholar]
- Kardailsky I, Shukla VK, Ahn JH, Dagenais N, Christensen SK, Nguyen JT, Chory J, Harrison MJ, Weigel D (1999) Activation tagging of the floral inducer FT. Science 286: 1962–1965 [DOI] [PubMed] [Google Scholar]
- Kobayashi Y, Kaya H, Goto K, Iwabuchi M, Araki T (1999) A pair of related genes with antagonistic roles in mediating flowering signals. Science 286: 1960–1962 [DOI] [PubMed] [Google Scholar]
- Koornneef M, Hanhart CJ, van der Veen JH (1991) A genetic and physiological analysis of late-flowering mutants in Arabidopsis thaliana. Mol Gen Genet 229: 57–66 [DOI] [PubMed] [Google Scholar]
- Lee H, Suh SS, Park E, Cho E, Ahn JH, Kim SG, Lee JS, Kwon YM, Lee I (2000) The AGAMOUS-LIKE 20 MADS domain protein integrates floral inductive pathways in Arabidopsis. Genes Dev 14: 2366–2376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I, Michaels SD, Masshardt AS, Amasino RM (1994) The late-flowering phenotype of FRIGIDA and LUMINIDEPENDENS is suppressed in the Landsberg erecta strain of Arabidopsis. Plant J 6: 903–909 [Google Scholar]
- Liu YG, Mitsukawa N, Oosumi T, Whittier RF (1995) Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J 8: 457–463 [DOI] [PubMed] [Google Scholar]
- Loudet O, Chaillou S, Camilleri C, Bouchez D, Daniel-Vedele F (2002) Bay-0 x Shahdara recombinant inbred line population: a powerful tool for the genetic dissection of complex traits in Arabidopsis. Theor Appl Genet 104: 1173–1184 [DOI] [PubMed] [Google Scholar]
- Michaels S, Amasino R (1999) FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11: 949–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels S, Amasino R (2000) Memories of winter: vernalization and the competence to flower. Plant Cell Environ 23: 1145–1154 [Google Scholar]
- Michaels SD, Amasino RM (2001) Loss of FLOWERING LOCUS C activity eliminates the late-flowering phenotype of FRIGIDA and autonomous-pathway mutations but not responsiveness to vernalization. Plant Cell 13: 935–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels SD, Bezerra IC, Amasino RM (2004) FRIGIDA-related genes are required for the winter-annual habit in Arabidopsis. Proc Natl Acad Sci USA 101: 3281–3285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels SD, He Y, Scortecci KC, Amasino RM (2003) Attenuation of FLOWERING LOCUS C activity as a mechanism for the evolution of summer-annual flowering behavior in Arabidopsis. Proc Natl Acad Sci USA 100: 10102–10107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noh YS, Amasino RM (2003) PIE1, an ISWI family gene, is required for FLC activation and floral repression in Arabidopsis. Plant Cell 15: 1671–1682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odell JT, Nagy F, Chua NH (1985) Identification of DNA sequences required for activity of the cauliflower mosaic virus 35S promoter. Nature 313: 810–812 [DOI] [PubMed] [Google Scholar]
- Putterill J, Robson F, Lee K, Simon R, Coupland G (1995) The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 80: 847–857 [DOI] [PubMed] [Google Scholar]
- Samach A, Onouchi H, Gold SE, Ditta GS, Schwarz-Sommer Z, Yanofsky MF, Coupland G (2000) Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 288: 1613–1616 [DOI] [PubMed] [Google Scholar]
- Sheldon CC, Burn JE, Perez PP, Metzger J, Edwards JA, Peacock WJ, Dennis ES (1999) The FLF MADS box gene. A repressor of flowering in Arabidopsis regulated by vernalization and methylation. Plant Cell 11: 445–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez-Lopez P, Wheatley K, Robson F, Onouchi H, Valverde F, Coupland G (2001) CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature 410: 1116–1120 [DOI] [PubMed] [Google Scholar]
- Sung S, Amasino RM (2004) Vernalization in Arabidopsis thaliana is mediated by the PHD finger protein VIN3. Nature 427: 159–164 [DOI] [PubMed] [Google Scholar]
- Takada S, Goto K (2003) TERMINAL FLOWER2, an Arabidopsis homolog of HETEROCHROMATIN PROTEIN1, counteracts the activation of FLOWERING LOCUS T by CONSTANS in the vascular tissues of leaves to regulate flowering time. Plant Cell 15: 2856–2865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valverde F, Mouradov A, Soppe W, Ravenscroft D, Samach A, Coupland G (2004) Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science 303: 1003–1006 [DOI] [PubMed] [Google Scholar]
- Weigel D, Ahn JH, Blazquez MA, Borevitz JO, Christensen SK, Fankhauser C, Ferrandiz C, Kardailsky I, Malancharuvil EJ, Neff MM, et al (2000) Activation tagging in Arabidopsis. Plant Physiol 122: 1003–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]