Abstract
Among the chromosome fragility-associated human syndromes that present cancer predisposition, Fanconi anemia (FA) is unique due to its large genetic heterogeneity. To date, mutations in 21 genes have been associated with an FA or an FA-like clinical and cellular phenotype, whose hallmarks are bone marrow failure, predisposition to acute myeloid leukemia and a cellular and chromosomal hypersensitivity to DNA crosslinking agents exposure. The goal of this review is to trace the history of the identification of FA genes, a history that started in the eighties and is not yet over, as indicated by the cloning of a twenty-first FA gene in 2016.
Keywords: DNA repair, leukemia, Fanconi anemia, chromosomal abnormalities
Introduction
Fanconi anemia (FA) is a rare human genetic syndrome associated with bone marrow failure (BMF), myelodysplasia (MDS) and a predisposition to acute myeloid leukemia (AML) and head and neck cancer. FA was described in 1927 by the Swiss pediatrician Giuseppe Fanconi, who reported a family with three affected siblings exhibiting anemia and developmental defects (Lobitz and Velleuer, 2006).
The clinical phenotype of FA patients is extremely heterogeneous. Beyond their hematological problems, which constitute the major hallmark of the disease, approximately 70% of these patients present developmental abnormalities, including abnormal radius, absent or supernumerary thumbs, microcephaly, microphthalmia, slow growth rate, café-au-lait spots, skin hyper- and hypo-pigmentation, kidney and urogenital defects, and hypoplasia of the testes. The estimated frequency of the syndrome is 1 in 250,000 - 350,000 live births, with a carrier frequency of approximately 1 in 200 (Rosenberg et al., 2011; Fanconi Anemia Research Fund Inc, 2014).
During the seventies, several groups around the world contributed to the definition of the two major cellular characteristics of the pathology: its particular chromosome fragility and its hypersensitivity to DNA interstrand crosslink (ICL)-inducing agents (Fujiwara and Tatsumi, 1975; Latt et al., 1975; Fornace et al., 1979; Novotna et al., 1979; Ishida and Buchwald, 1982). Indeed, FA cells appear exquisitely sensitive at both the cellular (survival) and chromosomal levels to the exposure to chemicals such as mitomycin C (MMC), diepoxybutane, cis-platinum and photoactivated psoralens. Since it is difficult to distinguish FA patients from individuals suffering from other inherited or idiopathic BMF syndromes on their clinical characteristics alone, the diagnosis of FA is based on the chromosomal response to ICL-inducing agents. Indeed, cytogeneticists score both the basal and induced frequency of chromosome aberrations as well as their subtypes, i.e., tri- and quadriradials, whose presence is quite specific for FA cells (Pinto et al., 2009; Fanconi Anemia Research Fund Inc, 2014).
Based on both the chromosome fragility and the hypersensitivity to exposure to DNA damaging agents, it was quickly suspected that the proteins whose loss of function caused FA must be involved in the DNA damage response and, more specifically, in a DNA repair mechanism. Indeed, although alternative functions associated with each individual or subgroup of FANC proteins exist (Joenje et al., 1981; Rosselli et al., 1994; Fagerlie et al., 2001; Pang et al., 2001; Briot et al., 2008; Pagano et al., 2003, 2012; Zanier et al., 2004; Myers et al., 2011; Justo et al., 2014; Parodi et al., 2015; Sumpter et al., 2016), the well-established “canonical” function of the proteins is to work along a “linear” pathway that addresses replication stresses, assuring the transmission of a stable genome from one cell to the daughters and acting both during DNA replication to cope with stalled replication forks and in G2 and M phases to resolve underreplicated regions before cell division (Ceccaldi et al., 2016; Lopez-Martinez et al., 2016; Michl et al., 2016). How the other, noncanonical functions of the FANC proteins that are involved in cytokine production/response, inflammation (Rosselli et al., 1994; Fagerlie et al., 2001; Pang et al., 2001; Zanier et al., 2004; Briot et al., 2008), mitophagy Sumpter et al., 2016), and oxygen free radical metabolism (Joenje et al., 1981; Pagano et al., 2003; Pagano et al., 2012) as well as the subtle defects in immunity (Myers et al., 2011; Justo et al., 2014; Nguyen et al., 2014; Parodi et al., 2015) impact the clinical and cellular phenotypes of the patients remains a challenge for the future understanding of the pathology.
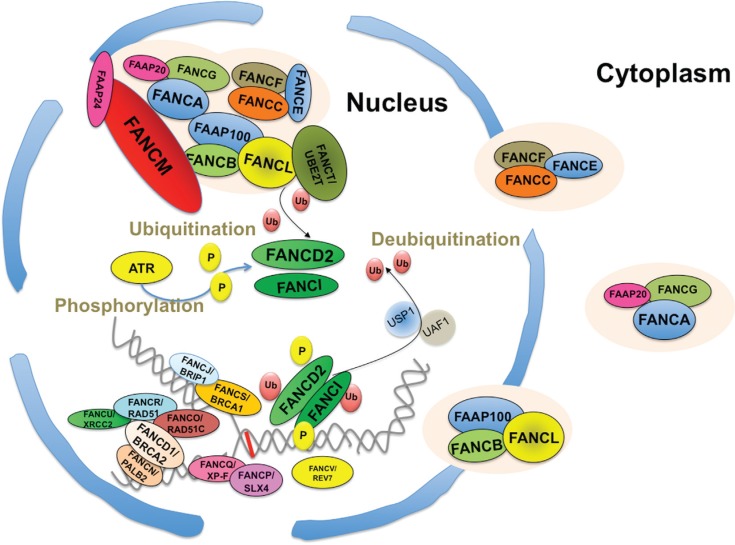
The 21 currently identified FANC and FANC-like genes (Table 1) encode proteins assembled into three biochemically and functionally defined groups (Wang, 2007). The first group contains several FA and FA-associated proteins (some named FAAPs) that coimmunoprecipitate in the same supramolecular complex. This group is formally called “FANCcore complex” and exhibits the E3 ubiquitin ligase activity responsible for the monoubiquitination of two downstream FANC proteins, FANCD2 and FANCI, which constitute group II. The third group consists of proteins directly involved in DNA metabolism, including structure-specific endonucleases (XP-F and SLX4) and several proteins involved in homologous recombination. Therefore, cells that are defective in genes coding for proteins from group III have normal levels of FANCD2 and FANCI monoubiquitination. The majority of patients (not less than 85%) harbor mutations in genes encoding proteins of the first group. Figure 1 schematically demonstrates the subcellular localization and the assembling pattern of the FANC proteins inside the nucleus in the presence of DNA damage and replication stress, where some of them form subnuclear foci, which can be observed by immunofluorescence microscopic analysis. Briefly, the strongest evidence in the literature supports the presence of three main FANCcore subcomplexes in the cytoplasm and/or in the nucleus, representing triads of proteins - FANCA, FANCG and FAAP20; FANCC, FANCE and FANCF; and FANCB, FANCL and FAAP100. In the nucleus, they assemble into a unique complex on the “cargo” FANCM, which is also associated with the FAAP24 and MHF 1/2 proteins as well as the Bloom-Associated Proteins (BLAPs) (Meetei et al., 2003b; Guo et al., 2009), which are prevented from sliding onto DNA by lesions or a stalled fork. Following an interaction with UBE2T, the complex locally monoubiquitinates FANCD2 and FANCI, a process that requires ATM- and/or ATR- and CHK1-mediated phosphorylation events on both the FANCcore complex proteins and FANCD2 and FANCI. The FANCcore complex monoubiquitinates FANCD2 and FANCI on lysine 561 and 523, respectively. Together, monoubiquitinated FANCD2 and FANCI represent the central link between the upstream FANC proteins (group I or the FANCcore complex) devoid of direct “DNA repair function” and the downstream proteins of group III that are involved in DNA metabolism. Indeed, following their monoubiquitination, FANCD2 and FANCI assemble in chromatin-associated foci, where they colocalize with several FANC and non-FANC proteins directly involved in homologous recombination. The USP1:UAF1 dimer promotes the deubiquitination of both FANCD2 and FANCI, a step necessary to optimally complete the process of DNA repair and for the rescue of stalled replication forks. Several recent reviews have summarized how and when the FANC pathway assumes its role of the guardian of genome integrity (Wang, 2007; Bogliolo and Surralles, 2015; Ceccaldi et al., 2016; Lopez-Martinez et al., 2016; Michl et al., 2016;).
Table 1. The 21 currently identified FANC and FANC-like genes.
| Complementation group | Extimated frequency | Gene name | Gene alias | Chromosomal position | Protein M.W. (kDa) | Core component | Cloning date | Bona fide FANC gene |
|---|---|---|---|---|---|---|---|---|
| A | 60-70 | FANCA | 16q24.3 | 162,7 | Yes | 1996 | Yes | |
| B | rare | FANCB | FAAP95 | Xp22.2 | 97,7 | Yes | 2004 | Yes |
| C | 10-15 | FANCC | 9q22.3 | 63,4 | Yes | 1992 | Yes | |
| D1 | 1-5 | FANCD1 | BRCA2 | 13q12.3 | 384,2 | 2002 | ||
| D2 | 1-5 | FANCD2 | 3p25.3 | 164,1 | 2001 | Yes | ||
| E | rare | FANCE | 6p21.3 | 58,7 | Yes | 2000 | Yes | |
| F | rare | FANCF | 11p15 | 42,2 | Yes | 2000 | Yes | |
| G | 10-15 | FANCG | XRCC9 | 9p13.3 | 68,5 | Yes | 1998 | Yes |
| I | rare | FANCI | 15q26.1 | 149,3 | Yes | 2007 | Yes | |
| J | rare | FANCJ | BACH1; BRIP1 | 17q22.2 | 140,9 | 2005 | ? | |
| L | rare | FANCL | 2p16.1 | 42,9 | Yes | 2003 | Yes | |
| M | rare | FANCM | 14q21.2 | 232,2 | Yes | 2005 | ||
| N | rare | FANCN | PALB2 | 16p12.12 | 131,3 | 2007 | ||
| O | rare | FANCO | RAD51C | 17q22 | 42,2 | 2010 | ||
| P | rare | FANCP | SLX4 | 16p13.3 | 200 | 2011 | ||
| Q | rare | FANCQ | ERCC4; XPF | 16p13.12 | 104,5 | 2013 | ||
| R | rare | FANCR | RAD51 | 15q15.1 | 37 | 2015 | ||
| S | rare | FANCS | BRCA1 | 17q21 | 207,7 | 2015 | ||
| T | rare | FANCT | UBE2T | 1q32.1 | 22,5 | 2015 | Yes | |
| U | rare | FANCU | XRCC2 | 7q36.1 | 31,9 | 2016 | ||
| V | rare | FANCV | REV7 | 1p36.22 | 24,3 | 2016 | Yes |
Figure 1. Schematic representation of the subcellular distribution of the FANC proteins, their association and their relocalization in nuclear foci at stalled replication forks. In unstressed conditions, three subcomplexes are present in the nucleus and/or the cytosol: FANCA, FANCG and FAAP20; FANCC, FANCE and FANCF; and FANCB, FANCL and FAAP100. In the presence of DNA damage (the red line represents an interstrand crosslink) that leads to stalled replication forks, all the FANC proteins shuttle into the nucleus to form the FANCcore complex to monoubiquitinate FANCD2 and FANCI, which in turn assemble to subnuclear foci, where they colocalize with several other proteins involved in homologous recombination, including other FANC and FANC-like representatives. The USP1:UAF1 dimer deubiquitinates both FANCD2 and FANCI.
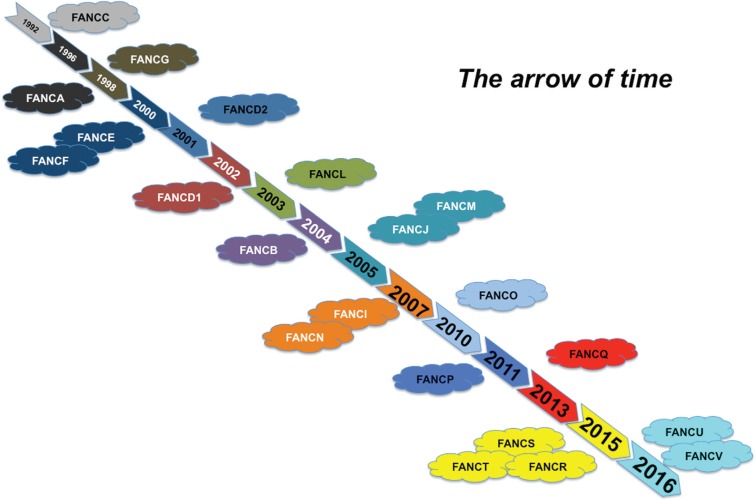
Here, we sought to retrace the story of the identification of the FANC genes (Table 1 and Figure 2). This story exemplifies the evolution of genetics and of molecular biology techniques during the last three decades. Indeed, FA is paradigmatic for several aspects of the human genetics field.
Figure 2. Milestones in the FANC pathway research: a timeline indicating the steps in the discovery of the FANC-BRCA network from the first gene discovered in 1992 to the present.
It is important to note that in recent years, the criteria to be considered as a “bona fide FA gene” have become more stringent and are now based on the clinical phenotype. Indeed, whereas the loss of function of all the identified genes leads to the primary FA cellular characteristics, including the ICL hypersensitivity and chromosome fragility, the clinical traits of some patients fail to reach the canonical features of the FA syndrome, namely BMF and the MDS. The genes mutated in those patients are now excluded from the “bona fide FA gene” group and are considered as “FA-like genes” (Bofliolo and Surralles, 2015). Nevertheless, the number of bona fide FANC and FANC-like genes continues to grow and it is unlikely to stop anytime soon.
History of the identification of FANC genes
One disease, many genes
The existence of genetic heterogeneity in FA was demonstrated at the beginning of the eighties by the pioneering work of the groups of Manuel Buchwald and Karl Sperling, which used a cell fusion approach (Zakrzewski and Sperling, 1980; Duckworth-Rysiecki et al., 1985). In particular, Duckworth-Risiecky and collaborators reported the existence of two FA complementation groups: A and non-A. The complementation analysis was based on the rescue of the cellular and chromosomal sensitivity to MMC exposure in the hybrid cells compared to the two cell lines fused to obtain the hybrid (Duckworth-Rysiecki et al., 1985).
The nineties
Seven years later, in 1992, the Buchwald group published two seminal manuscripts, with the first recognizing that the non-A group was heterogeneous, assembling three complementation groups: B, C and D (Strathdee et al., 1992a). The second manuscript reported the cloning of the first FANC gene, FANCC (Strathdee et al., 1992b). To identify the mutated gene, Strathdee and collaborators followed a functional complementation strategy. They transfected an EBV-based cDNA expression library into the HSC-536 B-lymphoblastoid cell line (previously assigned to the FA-C complementation group) and isolated three overlapping cDNA sequences able to independently complement the huge cellular and chromosomal hypersensitivity to MMC of the transfected cells. The proband carried a mutation of maternal origin that changed leucine 544 to a proline (L544P), a modification predicted to disrupt an alpha helical secondary structure of the protein (Strathdee et al., 1992b). Initially elusive, the mutation affecting the allele of paternal origin, a deletion of 327 bp resulting in the removal of exons 1 and 2, was identified some years later (Parker et al., 1998).
A new complementation group was added to the list in 1995. By analyzing 13 unrelated FA patients, Joenje et al. (1995) identified a fifth FA complementation group, named FA-E. The following year, two groups cloned FANCA, the most frequently mutated FANC gene, using two alternative strategies. One group identified the gene by the same functional complementation approach as used by the Buchwald group (Lo Ten Foe et al., 1996). Alternatively a consortium of several laboratories working to identify breast cancer susceptibility genes adopted the chromosome walking strategy (Fanconi anaemia/Breast Cancer, 1996) after the localization of a putative FANCA gene in the q24.3 region of the chromosome 16 (Pronk et al., 1995).
In 1997, the number of complementation groups established by the original cell fusion approach grew again, totaling 8: A to G (Joenje et al., 1997). In 1998, de Winter et al. (1998) used functional complementation to isolate a cDNA able to rescue the MMC hypersensitivity of a standard FA-G cell line. The identified sequence, renamed FANCG, was similar to that of a gene cloned one year before on the basis of its capability to complement the MMC hypersensitivity of a CHO UV40 mutant clone called XRCC9 (Liu et al., 1997).
The 2000s
The third millennium opened with the cloning of two new FANC genes. Still using the functional complementation cloning approach, de Winter and collaborators identified the genes whose loss of function was associated with the FA complementation groups E and F, i.e., FANCE, (de Winter et al., 2000a), previously mapped on chromosome 6 (Waisfisz et al., 1999), and FANCF (de Winter et al., 2000b).
In 2001, Timmers and collaborators reported that the cell lines originally categorized in the FA complementation group D (FA-D) could be separated into two groups, named D1 and D2, and they identified the gene mutated in the D2 group, FANCD2, by positional cloning and chromosome transfer, owing to the previous identification of the gene on chromosome 3p (Whitney et al., 1995; Hejna et al., 2000; Timmers et al., 2001).
The next year, D'Andrea and colleagues decided to test the hypothesis that the inactivation of BRCA1 and BRCA2, the most famous and frequently mutated genes in familial predisposition to breast cancer and whose loss of function results in a cellular phenotype similar to that described for FA, could also be involved in FA. A systematic sequencing of BRCA1 and BRCA2 was performed in several FA cell lines that belonged to complementation groups without an assigned gene. This “target gene” approach allowed the identification of biallelic variations in BRCA2 in the FA-D1 standard cell line HSC-62. The variants were successively validated as inactivating mutations (Howlett et al., 2002), assigning BRCA2 to the list of FANC genes as FANCD1. Howlett and collaborators also identified some variants of BRCA1 by examining the HSC230 cell line, the FA-B standard cell line. However, these variants failed to be confirmed as bona fide inactivating mutations.
In 2003, a big step forward in the genetics of FA was achieved thanks to the work of Meetei et al. (2003b) in the Weidong Wang laboratory. They purified a BLM-associated supramolecular complex containing two salt-concentration-dependent separable groups of proteins as follows: the BLM-associated proteins (BLMAPs) and Fanconi anemia-associated polypeptides (FAAPs). Mass spectrometry analysis of the isolated FAAPs identified some of the known FANC proteins (FANCA, FANCC, FANCE, FANCF, and FANCG), and the unknown components were identified as FAAP43, 90/95, 100, and 250/300 on the basis of their molecular mass. The biochemical approach of the Wang group allowed the cloning of new FANC genes by a “protein to gene” walking route. Indeed, after the identification of the amino acid composition of the FAAPs, the authors were able to match this to the sequence of each corresponding gene, to look for mutations in cells from FA patients and/or to determine the gene function(s) by analyzing the phenotypic consequences of the engineered inactivation of these genes in model cells or mice. Moreover, the work of Meetei and collaborators also provided the first indication that at least some of the FANC proteins work together inside a molecular complex.
The same year, Meetei et al. (2003a) identified FAAP43 as the PHD finger protein 9 coding gene, or PHF9. It was known that the inactivation of the mouse homolog of PHF9, Pog (for proliferation of germ cells), resulted in infertility and the MMC hypersensitivity of bone marrow cells, the two more consistent features presented by the already obtained FA mouse models. To definitively validate that PHF9 belonged to the FANC gene family, Meetei and coworkers identified inactivating PHF9 mutations in a cell line, EUFA868, isolated from a patient not previously assigned to a complementation group. The EUFA868 cell line used to clone PHF9 was assigned to FA-L and PHF9 was also named FANCL (Meetei et al., 2003a). PHF9/FANCL encodes the ubiquitin E3 ligase of the FANCcore complex, which mediates the FANCD2 and FANCI monoubiquitination (Meetei et al., 2004b). A second FA patient bearing a mutation in FANCL was identified six years later (Ali et al., 2009).
In 2004, Meetei et al. (2004a) identified the coding sequence of FAAP95 as being similar to the one named FLJ34064, a sequence localized on the X chromosome. Mutations in FLJ34064 were found in several FA cell lines, including the HSC-230 cell line, the standard for FA complementation group B. Thus, FAAP95 was renamed FANCB. FANCB is the only known FANC gene localized on the X chromosome. Consequently, its inactivation affects only males. FANCB is silenced via the methylation of its promoter on the X chromosome that undergoes inactivation during embryogenesis. Since the X-inactivation is stochastic, i.e., it affects either the paternal or the maternal X chromosome randomly, it is expected that the expression of a gene subjected to inactivation will exhibit mosaicism. In the case of FANCB+/- female carriers, the large majority of the lymphocytes and fibroblasts of the body express the WT gene, suggesting that the cells that express the mutated gene are counterselected, probably due to their growth difficulties, and are rapidly lost (Meetei et al., 2004a).
In 2005, Meetei et al. (2005) identified FAAP250 as KIAA1596, a human protein with sequence similarities to DNA repair proteins, including the yeast MPH1 and the human ERCC4/XP-F. siRNA-mediated depletion of KIAA1596 in cellular models affected FANCD2 monoubiquitination and increased MMC sensitivity, arguing for the assignment of FAAP250 to the FANC gene family. Mutations were then identified in an FA patient who was not assigned to a known complementation group. The gene was called FANCM, and the new complementation group FA-M. Nevertheless, the attempts to complement the MMC hypersensitivity of cell lines derived from the patient by transfection of the wild-type FANCM cDNA failed. Surprisingly, it was demonstrated that he also carried biallelic mutations in FANCA. Therefore, even if the loss of function of FANCM by targeted mutagenesis in mice or by siRNA-mediated depletion in human cells results in an FA-like cellular phenotype and in spite of its interaction with several other FANC proteins, the lack of patients with the major features of FA and only the loss of FANCM function impedes the assignment of the protein to the group of the “bona fide” FANC genes (Meetei et al., 2005; Singh et al., 2009).
In the same year, 2005, three groups identified the gene mutated in FA-J cells and named it FANCJ (Levitus et al., 2005; Levran et al., 2005; Litman et al., 2005). Following unsuccessful attempts to identify the mutated gene by a complementation cloning strategy, Levitus et al. (2005) opted for a positional cloning strategy and identified in eight FA-J cell lines several pathogenic mutations in the gene encoding the DEAH-box DNA helicase and binding partner of BRCA1 BRIP1/BACH1 (BRCA1-Immunoprecipitated Protein 1/BRCA1-associated C-terminal helicase-1), which was previously cloned by Cantor et al. (2001). Using a genome-wide scan, Levran et al. (2005) identified a homozygous region on chromosome 17q23 in which there were two interesting candidate genes: RAD51C and BRIP1. Inactivating mutations were found only in BRIP1/BACH1. Finally, Litman et al. (2005) identified BRCA1/BACH1 mutations in two families that were associated with an early onset of breast cancer and found the same recurrent nonsense mutation, the R798X mutation in exon 17, in both the breast cancer and FA-J families. This mutation affected the helicase domain of the protein, and since it was identified in people of different ethnic origin, it likely represents a hot spot of mutation or an inactivating event that remains compatible with survival (Levitus et al., 2005). Thus, BRIP1/BACH1 is also called FANCJ.
In 2007, following alternative approaches, three independent groups cloned the 13th FANC gene, FANCI, which is the paralog of FANCD2 (Dorsman et al., 2007; Meijer, 2007; Sims et al., 2007; Smogorzewska et al., 2007). FANCI was identified by a linkage analysis approach (Dorsman et al., 2007), by a bioinformatical screening for FANCD2 homologs (Sims et al., 2007) and by a proteomic search for ATM and ATR targets (Smogorzewska et al., 2007). It was described as the gene mutated in cells belonging to FA complementation group I.
Also in 2007, the 14th FANC gene was cloned and assigned to FA-N, a previously unrecognized FA complementation group (Reid et al., 2007; Xia et al., 2007). The identified gene, FANCN, was known to encode PALB2, isolated by immunoprecipitation one year prior as the Partner and Localizer of BRCA2 (Xia et al., 2006). Reid et al. (2007) followed a candidate gene approach, sequencing 82 FA patients with unelucidated genetic causes and identified mutations inactivating PALB2 in 7 individuals belonging to independent families. By Western blot analysis, Xia and collaborators noticed the lack of a full-length PALB2 in an unassigned FA cell line. Subsequent DNA sequencing allowed the identification of the inactivating mutations in PALB2 (Xia et al., 2007).
In 2010, aiming to identify the gene responsible for the pathology in a Pakistani family with FA by a genome-wide mapping approach, Vaz et al. (2010) identified a homozygous mutation in the RAD51C gene. Successively, in vitro functional studies showed that the identified mutation resulted in the loss of RAD51 focus formation in response to DNA damage, a defect that could be rescued by the ectopic expression of wild-type RAD51C. On this basis, the authors proposed to assign the acronym FANCO to RAD51C. RAD51C is also recognized as a gene associated with breast and ovarian cancer predisposition (Somyajit et al., 2012).
In 2011, several groups focused their work on SLX4, a gene previously identified in yeast and flies as well as in humans and involved in the cellular response to DNA ICLs (Mullen et al., 2001; Wu et al., 2004; Lee et al., 2005; Fekairi et al., 2009; Munoz et al., 2009; Svendsen et al., 2009). Kim et al. (2011) and Stoepker et al. (2011) decided to sequence the SLX4 gene in several patients with an FA-like phenotype, who until that time had not been assigned to any of the sixteen known complementation FA groups. They successfully identified some patients with mutations in the SLX4 coding sequence. On the other hand, Crossan et al. (2011) found that Slx4-null mice recapitulated the features of FA. Thus, SXL4 was also named FANCP. It codes for a structure-specific endonuclease that can be found in a complex with XP-F/ERCC1 and MUS81/EME1, proteins involved in protecting the genome during S and M phases.
Surprisingly, in 2013, inactivating mutations in ERCC4/XP-F, whose loss of function was previously associated with the skin cancer predisposition syndrome Xeroderma pigmentosum complementation group F, were also identified to be associated with an FA-like phenotype by whole-exome and Sanger sequencing of the DNA of unclassified FA individuals (Bogliolo et al., 2013). ERCC4/XP-F was thus renamed FANCQ. Analyses of the consequences of the identified mutations clearly demonstrated that compared to the NER-associated mutations, these mutations altered different regions, affecting an alternative function of the protein (Bogliolo et al., 2013). The association of XP-F mutations with an FA phenotype was successfully validated by an independent analysis (Kashiyama et al., 2013). Therefore, genetic defects in the structure-specific endonuclease XP-F/ERCC1 can result in xeroderma pigmentosum, Cockayne syndrome, Fanconi anemia, XFE progeria and cerebro-oculo-facio-skeletal syndrome (Manandhar et al., 2015).
Three important articles were published in 2015. Using whole-genome sequencing, Ameziane et al. (2015) identified a heterozygous dominant negative de novo mutation in the RAD51-encoding gene in an atypical FA patient and suggested adding the acronym FANCR to the major homologous recombination player known to play a role in both the resistance (if overexpressed) or sensitivity (when mutated or underexpressed) of cancer cells to radio- and chemotherapies and whose haploinsufficiency is involved in the congenital mirror movement neurological disorder (Depienne et al., 2012).
The second 2015 article was published by Sawyer et al. (2015). They described a patient with a complex FA-like phenotype carrying hereditary biallelic mutations in BRCA1. Indeed, the patient was identified in 2013 (Domchek et al., 2013), but the clinical phenotype of the patient was ascertained definitively only two years later. Therefore, thirteen years after the discovery that the gene for FA-D1 is BRCA2, BRCA1 has likewise obtained the acronym FANCS.
Likewise, Hira et al. (2015) described one individual harboring the classical cellular features and symptoms of FA and bearing biallelic mutations in the gene coding for an E2 ubiquitin-conjugating enzyme, UBE2T. UBE2T was originally identified by Zhang et al. (2000) and recognized as the principal ubiquitin E2 ligase of the FANCcore complex by Machida et al. (2006). UBE2T was, indeed, renamed FANCT.
In 2016, XRCC2 was identified as FANCU (Park et al., 2016). However, the patient who carried the XRCC2 mutations failed to show bone marrow failure. XRCC2 belongs to a group of RAD51 paralogs, which includes RAD51B, C and D. Therefore, with XRCC2/FANCU, the list of genes of which inactivating mutations could be associated with an FA-like phenotype and with breast cancer predisposition and/or homologous recombination now includes BRCA2, BRCA1, PALB2, BRIP1/BACH1, RAD51 and RAD51C.
Also in 2016, the 21st and last (but probably not for long) FA or FA-like gene was cloned (Bluteau et al., 2016). These authors identified a child with severe BMF harboring biallelic inactivating mutations in the gene encoding the translesion DNA synthesis (TLS) protein subunit REV7 (also known as MAD2L2), which was named FANCV. FANCV plays a central role in the bypass of the unhooked ICL downstream FANCD2/FANCI, allowing the progression of the process that leads to the HR-mediated rescue of the DNA replication impeded by the stall and collapse of an ongoing replication fork at the DNA lesion.
What next?
While 21 FANC genes have been identified and although the alphabet is near its end, the story is probably far from being finished. Obviously, patients bearing mutations in new genes are expected to be extremely rare. However, just looking at FANCM and the FANCcore complex partners, no fewer than five genes could claim the title of “FANC or FANC-like gene”: FAAP20, FAAP24 and FAAP100, MHF1 and MHF2. The depletion or deletion of these proteins results in an FA-like cellular phenotype and mouse mutants, when derived, present a phenotype similar to that of the majority of the FANC-KO mouse models. However, because patients with these mutations are rare, the formal attribution of a FANC acronym to previous genes is currently impossible. Loss-of-function mutations of USP1, the FANCD2/FANCI deubiquitinase, result in an FA-like phenotype in a mouse model and in human cells. Indeed, to be unable to monoubiquitinate FANCD2 or to have FANCD2 constitutively monoubiquitinated represents a similarly poor fate for a cell. However, again, no patient bearing USP1 mutations and presenting the FA clinical symptoms have yet been identified. Moreover, the loss of function of the other components of the structure-specific endonuclease heterodimers MUS81-EME1 (and possibly also EME2), XPF-ERCC1 and SLX4-SLX1 could also be associated with patients with an FA-like phenotype, although, for the moment, their potential mutations are associated with either lethal or extremely strong clinical phenotypes that probably preclude the possibility of their assignment to FA. Additionally, mutations in several other known HR-associated proteins could also result in an FA or an FA-like phenotype in some rare families.
In conclusion, the story of the identificatioin of the FANC genes allows to appreciate the evolution of the genetic and molecular techniques to identify disease-associated genes and to better define their lonks with the pathological traits. Also, considering the divergent clinical phenotypes associated with the loss of function of the gene products involved in the resistance to DNA crosslinking agents, it seems important to stress again that not all the ICL-repair proteins can nowadays claim to be members of the FANC gene group, even if they are involved in the FANC pathway. In the future, the upper part of the FANC pathway, consisting of the FANCcore complex-encoding genes FANCD2/FANCI and some of the proteins of the third group (FANCQ, FANCV), will probably be considered separately from the bottom part, whose associated gene products are involved in homologous recombination biochemistry and in breast and ovarian cancer predisposition, for which biallelic germinal inactivation results in strong clinical phenotypes.
Acknowledgments
A. Gueiderikh is a recipient of a PhD fellowship sponsored by the Fondation Philanthropia at Gustave Roussy. F. Rosselli was the recipient of a “Chaire Franco-brésilienne dans l'état de Sao Paulo”, sponsored by the USP and the French Consulate of Sao Paulo.
Footnotes
Associate Editor: Carlos F. M. Menck
References
- Ali AM, Kirby M, Jansen M, Lach FP, Schulte J, Singh TR, Batish SD, Auerbach AD, Williams DA, Meetei AR. Identification and characterization of mutations in FANCL gene: A second case of Fanconi anemia belonging to FA-L complementation group. Hum Mutat. 2009;30:E761–E770. doi: 10.1002/humu.21032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ameziane N, May P, Haitjema A, van de Vrugt HJ, van Rossum-Fikkert SE, Ristic D, Williams GJ, Balk J, Rockx D, Li H, et al. A novel Fanconi anaemia subtype associated with a dominant-negative mutation in RAD51. Nat Commun. 2015;6:8829–8829. doi: 10.1038/ncomms9829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bluteau D, Masliah-Planchon J, Clairmont C, Rousseau A, Ceccaldi R, Dubois d'Enghien C, Bluteau O, Cuccuini W, Gachet S, et al. Biallelic inactivation of REV7 is associated with Fanconi anemia. J Clin Invest. 2016;126:3580–3584. doi: 10.1172/JCI88010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogliolo M, Schuster B, Stoepker C, Derkunt B, Su Y, Raams A, Trujillo JP, Minguillon J, Ramirez MJ, Pujol R, et al. Mutations in ERCC4, encoding the DNA-repair endonuclease XPF, cause Fanconi anemia. Am J Hum Genet. 2013;92:800–806. doi: 10.1016/j.ajhg.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogliolo M, Surralles J. Fanconi anemia: A model disease for studies on human genetics and advanced therapeutics. Curr Opin Genet Dev. 2015;33:32–40. doi: 10.1016/j.gde.2015.07.002. [DOI] [PubMed] [Google Scholar]
- Briot D, Mace-Aime G, Subra F, Rosselli F. Aberrant activation of stress-response pathways leads to TNF-alpha oversecretion in Fanconi anemia. Blood. 2008;111:1913–1923. doi: 10.1182/blood-2007-07-099218. [DOI] [PubMed] [Google Scholar]
- Cantor SB, Bell DW, Ganesan S, Kass EM, Drapkin R, Grossman S, Wahrer DC, Sgroi DC, Lane WS, Haber DA, et al. BACH1, a novel helicase-like protein, interacts directly with BRCA1 and contributes to its DNA repair function. Cell. 2001;105:149–160. doi: 10.1016/s0092-8674(01)00304-x. [DOI] [PubMed] [Google Scholar]
- Ceccaldi R, Sarangi P, D'Andrea AD. The Fanconi anaemia pathway: New players and new functions. Nat Rev Mol Cell Biol. 2016;17:337–349. doi: 10.1038/nrm.2016.48. [DOI] [PubMed] [Google Scholar]
- Crossan GP, van der Weyden L, Rosado IV, Langevin F, Gaillard PH, McIntyre RE, Sanger Mouse Genetics Project. Gallagher F, Kettunen MI, Lewis DY, et al. Disruption of mouse Slx4, a regulator of structure-specific nucleases, phenocopies Fanconi anemia. Nat Genet. 2011;43:147–152. doi: 10.1038/ng.752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Winter JP, Waisfisz Q, Rooimans MA, van Berkel CG, Bosnoyan-Collins L, Alon N, Carreau M, Bender O, Demuth I, Schindler D, et al. The Fanconi anaemia group G gene FANCG is identical with XRCC9. Nat Genet. 1998;20:281–283. doi: 10.1038/3093. [DOI] [PubMed] [Google Scholar]
- de Winter JP, Leveille F, van Berkel CG, Rooimans MA, van Der Weel L, Steltenpool J, Demuth I, Morgan NV, Alon N, Bosnoyan-Collins L, et al. Isolation of a cDNA representing the Fanconi anemia complementation group E gene. Am J Hum Genet. 2000a;67:1306–1308. doi: 10.1016/s0002-9297(07)62959-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Winter JP, Rooimans MA, van Der Weel L, van Berkel CG, Alon N, Bosnoyan-Collins L, de Groot J, Zhi Y, Waisfisz Q, Pronk JC, et al. The Fanconi anaemia gene FANCF encodes a novel protein with homology to ROM. Nat Genet. 2000b;24:15–16. doi: 10.1038/71626. [DOI] [PubMed] [Google Scholar]
- Depienne C, Bouteiller D, Meneret A, Billot S, Groppa S, Klebe S, Charbonnier-Beaupel F, Corvol JC, Saraiva JP, Brueggemann N, et al. RAD51 haploinsufficiency causes congenital mirror movements in humans. Am J Hum Genet. 2012;90:301–307. doi: 10.1016/j.ajhg.2011.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domchek SM, Tang J, Stopfer J, Lilli DR, Hamel N, Tischkowitz M, Monteiro AN, Messick TE, Powers J, Yonker A, et al. Biallelic deleterious BRCA1 mutations in a woman with early-onset ovarian cancer. Cancer Discov. 2013;3:399–405. doi: 10.1158/2159-8290.CD-12-0421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsman JC, Levitus M, Rockx D, Rooimans MA, Oostra AB, Haitjema A, Bakker ST, Steltenpool J, Schuler D, Mohan S, et al. Identification of the Fanconi anemia complementation group I gene, FANCI. Cell Oncol. 2007;29:211–218. doi: 10.1155/2007/151968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duckworth-Rysiecki G, Cornish K, Clarke CA, Buchwald M. Identification of two complementation groups in Fanconi anemia. Somatic Cell Mol Genet. 1985;11:35–41. doi: 10.1007/BF01534732. [DOI] [PubMed] [Google Scholar]
- Fagerlie S, Lensch MW, Pang Q, Bagby GC., Jr The Fanconi anemia group C gene product: Signaling functions in hematopoietic cells. Exp Hematol. 2001;29:1371–1381. doi: 10.1016/s0301-472x(01)00755-x. [DOI] [PubMed] [Google Scholar]
- Fanconi Anaemia Research Fund Inc and Breast Cancer Positional cloning of the Fanconi anaemia group A gene. Nat Genet. 1996;14:324–328. doi: 10.1038/ng1196-324. [DOI] [PubMed] [Google Scholar]
- Fanconi Anemia Research Fund Inc . Fanconi anemia: Guidelines for Diagnosis and Management. 4th edition. Fanconi Anemia Research Fund Inc; Eugene: 2014. 429 p [Google Scholar]
- Fekairi S, Scaglione S, Chahwan C, Taylor ER, Tissier A, Coulon S, Dong MQ, Ruse C, Yates JR, 3rd, Russell P, et al. Human SLX4 is a Holliday junction resolvase subunit that binds multiple DNA repair/recombination endonucleases. Cell. 2009;138:78–89. doi: 10.1016/j.cell.2009.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornace AJ, Jr., Little JB, Weichselbaum RR. DNA repair in a Fanconi's anemia fibroblast cell strain. Biochim Biophys Acta. 1979;561:99–109. doi: 10.1016/0005-2787(79)90494-5. [DOI] [PubMed] [Google Scholar]
- Fujiwara Y, Tatsumi M. Repair of mitomycin C damage to DNA in mammalian cells and its impairment in Fanconi's anemia cells. Biochem Biophys Res Commun. 1975;66:592–598. doi: 10.1016/0006-291x(75)90551-3. [DOI] [PubMed] [Google Scholar]
- Guo R, Xu D, Wang W. Identification and analysis of new proteins involved in the DNA damage response network of Fanconi anemia and Bloom syndrome. Methods. 2009;48:72–79. doi: 10.1016/j.ymeth.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hejna JA, Timmers CD, Reifsteck C, Bruun DA, Lucas LW, Jakobs PM, Toth-Fejel S, Unsworth N, Clemens SL, Garcia DK, et al. Localization of the Fanconi anemia complementation group D gene to a 200-kb region on chromosome 3p25.3. Am J Hum Genet. 2000;66:1540–1551. doi: 10.1086/302896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hira A, Yoshida K, Sato K, Okuno Y, Shiraishi Y, Chiba K, Tanaka H, Miyano S, Shimamoto A, Tahara H, et al. Mutations in the gene encoding the E2 conjugating enzyme UBE2T cause Fanconi anemia. Am J Hum Genet. 2015;96:1001–1007. doi: 10.1016/j.ajhg.2015.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howlett NG, Taniguchi T, Olson S, Cox B, Waisfisz Q, De Die-Smulders C, Persky N, Grompe M, Joenje H, Pals G, et al. Biallelic inactivation of BRCA2 in Fanconi anemia. Science. 2002;297:606–609. doi: 10.1126/science.1073834. [DOI] [PubMed] [Google Scholar]
- Ishida R, Buchwald M. Susceptibility of Fanconi's anemia lymphoblasts to DNA-cross-linking and alkylating agents. Cancer Res. 1982;42:4000–4006. [PubMed] [Google Scholar]
- Joenje H, Arwert F, Eriksson AW, de Koning H, Oostra AB. Oxygen-dependence of chromosomal aberrations in Fanconi's anaemia. Nature. 1981;290:142–143. doi: 10.1038/290142a0. [DOI] [PubMed] [Google Scholar]
- Joenje H, Lo Ten Foe JR, Oostra AB, van Berkel CG, Rooimans MA, Schroeder-Kurth T, Wegner RD, Gille JJ, Buchwald M, Arwert F. Classification of Fanconi anemia patients by complementation analysis: Evidence for a fifth genetic subtype. Blood. 1995;86:2156–2160. [PubMed] [Google Scholar]
- Joenje H, Oostra AB, Wijker M, di Summa FM, van Berkel CG, Rooimans MA, Ebell W, van Weel M, Pronk JC, Buchwald M, et al. Evidence for at least eight Fanconi anemia genes. Am J Hum Genet. 1997;61:940–944. doi: 10.1086/514881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Justo GA, Bitencourt MA, Pasquini R, Castelo-Branco MT, Almeida-Oliveira A, Diamond HR, Rumjanek VM. Immune status of Fanconi anemia patients: Decrease in T CD8 and CD56dim CD16+ NK lymphocytes. Ann Hematol. 2014;93:761–767. doi: 10.1007/s00277-013-1953-4. [DOI] [PubMed] [Google Scholar]
- Kashiyama K, Nakazawa Y, Pilz DT, Guo C, Shimada M, Sasaki K, Fawcett H, Wing JF, Lewin SO, Carr L, et al. Malfunction of nuclease ERCC1-XPF results in diverse clinical manifestations and causes Cockayne syndrome, xeroderma pigmentosum, and Fanconi anemia. Am J Hum Genet. 2013;92:807–819. doi: 10.1016/j.ajhg.2013.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Lach FP, Desetty R, Hanenberg H, Auerbach AD, Smogorzewska A. Mutations of the SLX4 gene in Fanconi anemia. Nat Genet. 2011;43:142–146. doi: 10.1038/ng.750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latt SA, Stetten G, Juergens LA, Buchanan GR, Gerald PS. Induction by alkylating agents of sister chromatid exchanges and chromatid breaks in Fanconi's anemia. Proc Natl Acad Sci U S A. 1975;72:4066–4070. doi: 10.1073/pnas.72.10.4066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W, St Onge RP, Proctor M, Flaherty P, Jordan MI, Arkin AP, Davis RW, Nislow C, Giaever G. Genome-wide requirements for resistance to functionally distinct DNA-damaging agents. PLoS Genet. 2005;1:e24. doi: 10.1371/journal.pgen.0010024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitus M, Waisfisz Q, Godthelp BC, de Vries Y, Hussain S, Wiegant WW, Elghalbzouri-Maghrani E, Steltenpool J, Rooimans MA, Pals G, et al. The DNA helicase BRIP1 is defective in Fanconi anemia complementation group J. Nat Genet. 2005;37:934–935. doi: 10.1038/ng1625. [DOI] [PubMed] [Google Scholar]
- Levran O, Attwooll C, Henry RT, Milton KL, Neveling K, Rio P, Batish SD, Kalb R, Velleuer E, Barral S, et al. The BRCA1-interacting helicase BRIP1 is deficient in Fanconi anemia. Nat Genet. 2005;37:931–933. doi: 10.1038/ng1624. [DOI] [PubMed] [Google Scholar]
- Litman R, Peng M, Jin Z, Zhang F, Zhang J, Powell S, Andreassen PR, Cantor SB. BACH1 is critical for homologous recombination and appears to be the Fanconi anemia gene product FANCJ. Cancer Cell. 2005;8:255–265. doi: 10.1016/j.ccr.2005.08.004. [DOI] [PubMed] [Google Scholar]
- Liu N, Lamerdin JE, Tucker JD, Zhou ZQ, Walter CA, Albala JS, Busch DB, Thompson LH. The human XRCC9 gene corrects chromosomal instability and mutagen sensitivities in CHO UV40 cells. Proc Natl Acad Sci U S A. 1997;94:9232–9237. doi: 10.1073/pnas.94.17.9232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo Ten Foe JR, Rooimans MA, Bosnoyan-Collins L, Alon N, Wijker M, Parker L, Lightfoot J, Carreau M, Callen DF, Savoia A, et al. Expression cloning of a cDNA for the major Fanconi anaemia gene, FAA. Nat Genet. 1996;14:320–323. doi: 10.1038/ng1196-320. [DOI] [PubMed] [Google Scholar]
- Lobitz S, Velleuer E. Guido Fanconi (1892-1979): A jack of all trades. Nat Rev Cancer. 2006;6:893–898. doi: 10.1038/nrc2009. [DOI] [PubMed] [Google Scholar]
- Lopez-Martinez D, Liang CC, Cohn MA. Cellular response to DNA interstrand crosslinks: The Fanconi anemia pathway. Cell Mol Life Sci. 2016;73:3097–3114. doi: 10.1007/s00018-016-2218-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machida YJ, Machida Y, Chen Y, Gurtan AM, Kupfer GM, D'Andrea AD, Dutta A. UBE2T is the E2 in the Fanconi anemia pathway and undergoes negative autoregulation. Mol Cell. 2006;23:589–596. doi: 10.1016/j.molcel.2006.06.024. [DOI] [PubMed] [Google Scholar]
- Manandhar M, Boulware KS, Wood RD. The ERCC1 and ERCC4 (XPF) genes and gene products. Gene. 2015;569:153–161. doi: 10.1016/j.gene.2015.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meetei AR, de Winter JP, Medhurst AL, Wallisch M, Waisfisz Q, van de Vrugt HJ, Oostra AB, Yan Z, Ling C, Bishop CE, et al. A novel ubiquitin ligase is deficient in Fanconi anemia. Nat Genet. 2003a;35:165–170. doi: 10.1038/ng1241. [DOI] [PubMed] [Google Scholar]
- Meetei AR, Sechi S, Wallisch M, Yang D, Young MK, Joenje H, Hoatlin ME, Wang W. A multiprotein nuclear complex connects Fanconi anemia and Bloom syndrome. Mol Cell Biol. 2003b;23:3417–3426. doi: 10.1128/MCB.23.10.3417-3426.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meetei AR, Levitus M, Xue Y, Medhurst AL, Zwaan M, Ling C, Rooimans MA, Bier P, Hoatlin M, Pals G, et al. X-linked inheritance of Fanconi anemia complementation group B. Nat Genet. 2004a;36:1219–1224. doi: 10.1038/ng1458. [DOI] [PubMed] [Google Scholar]
- Meetei AR, Yan Z, Wang W. FANCL replaces BRCA1 as the likely ubiquitin ligase responsible for FANCD2 monoubiquitination. Cell Cycle. 2004b;3:179–181. [PubMed] [Google Scholar]
- Meetei AR, Medhurst AL, Ling C, Xue Y, Singh TR, Bier P, Steltenpool J, Stone S, Dokal I, Mathew CG, et al. A human ortholog of archaeal DNA repair protein Hef is defective in Fanconi anemia complementation group M. Nat Genet. 2005;37:958–963. doi: 10.1038/ng1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer GA. The 13th Fanconi anemia gene identified: FANCI - Importance of the ‘Fanconi anemia pathway’ for cellular oncology. Cell Oncol. 2007;29:181–182. doi: 10.1155/2007/871608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michl J, Zimmer J, Tarsounas M. Interplay between Fanconi anemia and homologous recombination pathways in genome integrity. EMBO J. 2016;35:909–923. doi: 10.15252/embj.201693860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen JR, Kaliraman V, Ibrahim SS, Brill SJ. Requirement for three novel protein complexes in the absence of the Sgs1 DNA helicase in Saccharomyces cerevisiae. Genetics. 2001;157:103–118. doi: 10.1093/genetics/157.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz IM, Hain K, Declais AC, Gardiner M, Toh GW, Sanchez-Pulido L, Heuckmann JM, Toth R, Macartney T, Eppink B, et al. Coordination of structure-specific nucleases by human SLX4/BTBD12 is required for DNA repair. Mol Cell. 2009;35:116–127. doi: 10.1016/j.molcel.2009.06.020. [DOI] [PubMed] [Google Scholar]
- Myers KC, Bleesing JJ, Davies SM, Zhang X, Martin LJ, Mueller R, Harris RE, Filipovich AH, Kovacic MB, Wells SI, et al. Impaired immune function in children with Fanconi anaemia. Br J Haematol. 2011;154:234–240. doi: 10.1111/j.1365-2141.2011.08721.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen TV, Riou L, Aoufouchi S, Rosselli F. Fanca deficiency reduces A/T transitions in somatic hypermutation and alters class switch recombination junctions in mouse B cells. J Exp Med. 2014;211:1011–1018. doi: 10.1084/jem.20131637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novotna B, Goetz P, Surkova NI. Effects of alkylating agents on lymphocytes from controls and from patients with Fanconi's anemia. Studies of sister chromatid exchanges, chromosome aberrations, and kinetics of cell division. Hum Genet. 1979;49:41–50. doi: 10.1007/BF00277685. [DOI] [PubMed] [Google Scholar]
- Pagano G, Manini P, Bagchi D. Oxidative stress-related mechanisms are associated with xenobiotics exerting excess toxicity to Fanconi anemia cells. Environ Health Perspect. 2003;111:1699–1703. doi: 10.1289/ehp.6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagano G, Talamanca AA, Castello G, Pallardo FV, Zatterale A, Degan P. Oxidative stress in Fanconi anaemia: From cells and molecules towards prospects in clinical management. Biol Chem. 2012;393:11–21. doi: 10.1515/BC-2011-227. [DOI] [PubMed] [Google Scholar]
- Pang Q, Christianson TA, Keeble W, Diaz J, Faulkner GR, Reifsteck C, Olson S, Bagby GC. The Fanconi anemia complementation group C gene product: Structural evidence of multifunctionality. Blood. 2001;98:1392–1401. doi: 10.1182/blood.v98.5.1392. [DOI] [PubMed] [Google Scholar]
- Park JY, Virts EL, Jankowska A, Wiek C, Othman M, Chakraborty SC, Vance GH, Alkuraya FS, Hanenberg H, Andreassen PR. Complementation of hypersensitivity to DNA interstrand crosslinking agents demonstrates that XRCC2 is a Fanconi anaemia gene. J Med Genet. 2016;53:672–680. doi: 10.1136/jmedgenet-2016-103847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker L, dos Santos C, Buchwald M. The delta327 mutation in the Fanconi anemia group C gene generates a novel transcript lacking the first two coding exons. Hum Mutat. 1998;1998(Suppl 1):S275–S277. doi: 10.1002/humu.1380110187. [DOI] [PubMed] [Google Scholar]
- Parodi A, Kalli F, Svahn J, Stroppiana G, De Rocco D, Terranova P, Dufour C, Fenoglio D, Cappelli E. Impaired immune response to Candida albicans in cells from Fanconi anemia patients. Cytokine. 2015;73:203–207. doi: 10.1016/j.cyto.2015.02.016. [DOI] [PubMed] [Google Scholar]
- Pinto FO, Leblanc T, Chamousset D, Le Roux G, Brethon B, Cassinat B, Larghero J, de Villartay JP, Stoppa-Lyonnet D, Baruchel A, et al. Diagnosis of Fanconi anemia in patients with bone marrow failure. Haematologica. 2009;94:487–495. doi: 10.3324/haematol.13592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pronk JC, Gibson RA, Savoia A, Wijker M, Morgan NV, Melchionda S, Ford D, Temtamy S, Ortega JJ, Jansen S, et al. Localisation of the Fanconi anaemia complementation group A gene to chromosome 16q24.3. Nat Genet. 1995;11:338–340. doi: 10.1038/ng1195-338. [DOI] [PubMed] [Google Scholar]
- Reid S, Schindler D, Hanenberg H, Barker K, Hanks S, Kalb R, Neveling K, Kelly P, Seal S, Freund M, et al. Biallelic mutations in PALB2 cause Fanconi anemia subtype FA-N and predispose to childhood cancer. Nat Genet. 2007;39:162–164. doi: 10.1038/ng1947. [DOI] [PubMed] [Google Scholar]
- Rosenberg PS, Tamary H, Alter BP. How high are carrier frequencies of rare recessive syndromes? Contemporary estimates for Fanconi Anemia in the United States and Israel. Am J Med Genet A. 2011;155A:1877–1883. doi: 10.1002/ajmg.a.34087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosselli F, Sanceau J, Gluckman E, Wietzerbin J, Moustacchi E. Abnormal lymphokine production: A novel feature of the genetic disease Fanconi anemia. II. In vitro and in vivo spontaneous overproduction of tumor necrosis factor alpha. Blood. 1994;83:1216–1225. [PubMed] [Google Scholar]
- Sawyer SL, Tian L, Kahkonen M, Schwartzentruber J, Kircher M, University of Washington Centre for Mendelian Genomics. Consortium FC, Majewski J, Dyment DA, Innes AM, et al. Biallelic mutations in BRCA1 cause a new Fanconi anemia subtype. Cancer Discovery. 2015;5:135–142. doi: 10.1158/2159-8290.CD-14-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims AE, Spiteri E, Sims RJ, 3rd, Arita AG, Lach FP, Landers T, Wurm M, Freund M, Neveling K, Hanenberg H, et al. FANCI is a second monoubiquitinated member of the Fanconi anemia pathway. Nat Struct Mol Biol. 2007;14:564–567. doi: 10.1038/nsmb1252. [DOI] [PubMed] [Google Scholar]
- Singh TR, Bakker ST, Agarwal S, Jansen M, Grassman E, Godthelp BC, Ali AM, Du CH, Rooimans MA, Fan Q, et al. Impaired FANCD2 monoubiquitination and hypersensitivity to camptothecin uniquely characterize Fanconi anemia complementation group M. Blood. 2009;114:174–180. doi: 10.1182/blood-2009-02-207811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smogorzewska A, Matsuoka S, Vinciguerra P, McDonald ER, 3rd, Hurov KE, Luo J, Ballif BA, Gygi SP, Hofmann K, D'Andrea AD, et al. Identification of the FANCI protein, a monoubiquitinated FANCD2 paralog required for DNA repair. Cell. 2007;129:289–301. doi: 10.1016/j.cell.2007.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somyajit K, Subramanya S, Nagaraju G. Distinct roles of FANCO/RAD51C protein in DNA damage signaling and repair: Implications for Fanconi anemia and breast cancer susceptibility. J Biol Chem. 2012;287:3366–3380. doi: 10.1074/jbc.M111.311241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoepker C, Hain K, Schuster B, Hilhorst-Hofstee Y, Rooimans MA, Steltenpool J, Oostra AB, Eirich K, Korthof ET, Nieuwint AW, et al. SLX4, a coordinator of structure-specific endonucleases, is mutated in a new Fanconi anemia subtype. Nat Genet. 2011;43:138–141. doi: 10.1038/ng.751. [DOI] [PubMed] [Google Scholar]
- Strathdee CA, Duncan AM, Buchwald M. Evidence for at least four Fanconi anaemia genes including FACC on chromosome 9. Nat Genet. 1992a;1:196–198. doi: 10.1038/ng0692-196. [DOI] [PubMed] [Google Scholar]
- Strathdee CA, Gavish H, Shannon WR, Buchwald M. Cloning of cDNAs for Fanconi's anaemia by functional complementation. Nature. 1992b;356:763–767. doi: 10.1038/356763a0. [DOI] [PubMed] [Google Scholar]
- Sumpter R, Jr., Sirasanagandla S, Fernandez AF, Wei Y, Dong X, Franco L, Zou Z, Marchal C, Lee MY, Clapp DW, et al. Fanconi anemia proteins function in mitophagy and immunity. Cell. 2016;165:867–881. doi: 10.1016/j.cell.2016.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svendsen JM, Smogorzewska A, Sowa ME, O'Connell BC, Gygi SP, Elledge SJ, Harper JW. Mammalian BTBD12/SLX4 assembles a Holliday junction resolvase and is required for DNA repair. Cell. 2009;138:63–77. doi: 10.1016/j.cell.2009.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmers C, Taniguchi T, Hejna J, Reifsteck C, Lucas L, Bruun D, Thayer M, Cox B, Olson S, D'Andrea AD, et al. Positional cloning of a novel Fanconi anemia gene, FANCD2. Mol Cell. 2001;7:241–248. doi: 10.1016/s1097-2765(01)00172-1. [DOI] [PubMed] [Google Scholar]
- Vaz F, Hanenberg H, Schuster B, Barker K, Wiek C, Erven V, Neveling K, Endt D, Kesterton I, Autore F, et al. Mutation of the RAD51C gene in a Fanconi anemia-like disorder. Nat Genet. 2010;42:406–409. doi: 10.1038/ng.570. [DOI] [PubMed] [Google Scholar]
- Waisfisz Q, Saar K, Morgan NV, Altay C, Leegwater PA, de Winter JP, Komatsu K, Evans GR, Wegner RD, Reis A, et al. The Fanconi anemia group E gene, FANCE, maps to chromosome 6p. Am J Hum Genet. 1999;64:1400–1405. doi: 10.1086/302385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W. Emergence of a DNA-damage response network consisting of Fanconi anaemia and BRCA proteins. Nat Rev Genet. 2007;8:735–748. doi: 10.1038/nrg2159. [DOI] [PubMed] [Google Scholar]
- Whitney M, Thayer M, Reifsteck C, Olson S, Smith L, Jakobs PM, Leach R, Naylor S, Joenje H, Grompe M. Microcell mediated chromosome transfer maps the Fanconi anaemia group D gene to chromosome 3p. Nat Genet. 1995;11:341–343. doi: 10.1038/ng1195-341. [DOI] [PubMed] [Google Scholar]
- Wu HI, Brown JA, Dorie MJ, Lazzeroni L, Brown JM. Genome-wide identification of genes conferring resistance to the anticancer agents cisplatin, oxaliplatin, and mitomycin C. Cancer Res. 2004;64:3940–3848. doi: 10.1158/0008-5472.CAN-03-3113. [DOI] [PubMed] [Google Scholar]
- Xia B, Sheng Q, Nakanishi K, Ohashi A, Wu J, Christ N, Liu X, Jasin M, Couch FJ, Livingston DM. Control of BRCA2 cellular and clinical functions by a nuclear partner, PALB2. Mol Cell. 2006;22:719–729. doi: 10.1016/j.molcel.2006.05.022. [DOI] [PubMed] [Google Scholar]
- Xia B, Dorsman JC, Ameziane N, de Vries Y, Rooimans MA, Sheng Q, Pals G, Errami A, Gluckman E, Llera J, et al. Fanconi anemia is associated with a defect in the BRCA2 partner PALB2. Nat Genet. 2007;39:159–161. doi: 10.1038/ng1942. [DOI] [PubMed] [Google Scholar]
- Zakrzewski S, Sperling K. Genetic heterogeneity of Fanconi's anemia demonstrated by somatic cell hybrids. Hum Genet. 1980;56:81–84. doi: 10.1007/BF00281573. [DOI] [PubMed] [Google Scholar]
- Zanier R, Briot D, Dugas du Villard JA, Sarasin A, Rosselli F. Fanconi anemia C gene product regulates expression of genes involved in differentiation and inflammation. Oncogene. 2004;23:5004–5013. doi: 10.1038/sj.onc.1207677. [DOI] [PubMed] [Google Scholar]
- Zhang QH, Ye M, Wu XY, Ren SX, Zhao M, Zhao CJ, Fu G, Shen Y, Fan HY, Lu G, et al. Cloning and functional analysis of cDNAs with open reading frames for 300 previously undefined genes expressed in CD34+ hematopoietic stem/progenitor cells. Genome Res. 2000;10:1546–1560. doi: 10.1101/gr.140200. [DOI] [PMC free article] [PubMed] [Google Scholar]