Abstract
The present study aimed to evaluate the impact of small interfering RNA (siRNA) targeting of the survivin gene in human tumor cells and the effect of decreased survivin expression on the proliferation and apoptosis of tumor cells. Human tumor cell lines (MSA-MB-231, SGC-7901, HeLa, A549, SK-OV-3 and Raji, PC-3) were cultured in vitro and divided into three groups: survivin siRNA-treated, scrambled negative control siRNA-treated and an untreated control group. The level of survivin mRNA and protein expression was subsequently determined by reverse transcription-quantitative polymerase chain reaction and western blot analysis, respectively. Cell proliferation was also examined by an MTT assay following transfection and the apoptotic rate of cells was detected by Hoechst and Annexin V/propidium iodide staining. It was observed that relative to the control group, expression of survivin mRNA and protein in the survivin siRNA-treated group was significantly downregulated. Furthermore, siRNA targeting of survivin lead to the inhibition of tumor cell proliferation, as well as an increase in their apoptotic rate in vitro. These data suggest that survivin may be a potential tumor biomarker and a novel target for the treatment of cancer.
Keywords: small interfering RNA, survivin, tumor cells, expression, proliferation, apoptosis
Introduction
Identifying effective therapy for the treatment of malignant tumors is a global challenge, and while the development of medical technology has led to an increase in cancer therapies, a treatment strategy of adequate efficacy has not yet been determined. Strategies employing a combination of surgery, radiotherapy, chemotherapy and immunotherapy, as well as clinical-based rehabilitation, have improved the quality of life for cancer patients and increased the 5-year-survival rate (1,2). Combination therapies integrate the research achievements from various specializations, making the treatment more diversified. Combination therapy may exert the advantages and avoid the drawbacks of individual treatments, allowing for the achievement of maximum therapeutic effect. However, due to the presentation of serious complications, side effects (including pain, infection, cardiovascular and gastrointestinal side effects and depression) (3), tumor recurrence and metastasis following therapy, studies investigating alternative therapeutics are warranted. Optimal cancer therapies require the capacity to specifically inhibit tumor cells, while having minimal adverse effects on normal cells. Therefore, the identification of a molecular target that specifically inhibits tumor cell growth is warranted.
Survivin is a member of the inhibitor of apoptosis family of proteins, which collectively has roles in the inhibition of caspase activation and regulation of mitosis. Survivin protein is expressed at high levels in the majority of human tumors and fetal tissue, though is absent in terminally differentiated cells (4). In tumor tissue, survivin promotes the anti-apoptotic activity of tumor cells, while also contributing to tumor angiogenesis and resistance to chemoradiotherapy (5–7). Specifically, survivin has been found to promote the formation and development of tumors by the following mechanisms: i) interaction with cell division protein kinase 4 leading to acceleration in the S-phase of the cell cycle (8); ii) increasing the expression of human angiopoietin 1, thus promoting the formation of capillary networks and proliferation of endothelial cells (9); iii) overexpression in the G2/M phase of the cell cycle in a cycle-regulated manner and association with microtubules of the mitotic spindle, thus inducing aberrant mitosis (10); and iv) suppressing the release of mitochondrial cytochrome C, thus inhibiting apoptosis and promoting tumor progression (11).
As a commonly upregulated gene in tumor cells, survivin is considered to be a potential cancer biomarker and a novel target for cancer treatment (12). A number of studies have documented the overexpression of survivin in different human tumor types, including leukemia, brain tumors, liver, breast, gastric, lung, colon, prostate, lymph, oesophageal, pancreatic, skin, cervical and ovarian cancer (13). Therefore, studies are aiming to identify novel strategies that target the expression and function of survivin for the treatment of cancer.
RNA interference (RNAi) is an evolution-conserved cell defense mechanism mediated by small interfering RNA (siRNA) (14). Target genes are knocked down by siRNA inhibition of the corresponding mRNA, resulting in suppressed gene expression (15). Compared with conventional antisense strategies, RNAi has demonstrated greater efficacy in the silencing of specific genes by allowing low level target gene expression and subsequent protein function, thus resulting in fewer adverse effects, such as pain, nausea and vomiting, tiredness and sleeping problems, occurring in patients (12). However, rapid degradation of siRNA within the extracellular environment and an inability to traverse cellular membranes means that the delivery of siRNA to its intracellular targets remains a challenge (16).
In the present study, liposomes were used for the intracellular delivery of siRNA against survivin. siRNA liposomal vehicles were successfully constructed and transfected into cell lines (MSA-MB-231, SGC-7901, HeLa, A549, SK-OV-3 and Raji, PC-3) of a number of common human tumor types (breast cancer, gastric carcinoma, cervical carcinoma, lung carcinoma, ovarian carcinoma, lymphoma, prostate carcinoma, respectively). Changes in the level of survivin protein expression and inhibition of tumor cell proliferation and apoptosis were then determined by reverse transcription-quantitative polymerase chain reaction (RT-qPCR), western blot analysis, the MTT method, Hoechst 33258 staining and Annexin V/PI staining in vitro, in order to determine the efficacy of survivin as a potential gene target in the treatment of cancer.
Materials and methods
Cell lines and culture
Tumor cell lines MDA-MB-231 (human breast carcinoma), SGC-7901 (human gastric carcinoma), HeLa (human cervical carcinoma) and A549 (human lung carcinoma) were obtained from Biomics Biotechnologies Co., Ltd. (Nantong, China). SK-OV-3 (human ovarian carcinoma), Raji (human lymphoma) and PC-3 (human prostate carcinoma) cell lines were purchased from the Cell Banks of Type Culture Collection at the Chinese Academy of Sciences (Beijing, China). MDA-MB-231, SGC-7901 and Raji cells were maintained in RPMI 1640 (Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS; Hyclone; GE Healthcare, Logan, UT, USA), and HeLa and A549 cells were maintained in Dulbecco's modified Eagle medium (DMEM; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS. SK-OV-3 cells were maintained in McCoy's 5A modified medium (Life Technologies; Thermo Fisher Scientific, Inc.) and PC-3 were maintained in DMEM/ nutrient mixture F12 (Thermo Fisher Scientific, Inc.) supplemented with 10% FBS. All cell lines were cultured at 37°C in 5% CO2 for 1–3 days.
siRNA constructs
siRNAs were synthesized using the following sequences: Survivin, forward 5′-GCAUCUCUACAUUCAAGAA-3′ and reverse 5′-UUCUUGAAUGUAGAGAUGC-3′; a scrambled sequence (si-NC), forward 5′-UUCUCCGAACGUGUCACGU-3′ and reverse 5′-ACGUGACACGUUCGGAGAA-3′. si-NC was used as the negative control and was non-homologous to all known human DNA sequences.
Preparation of siRNA-liposomes
siRNA was incorporated into liposomes for intracellular delivery. Disaturated phosphatidylcholine (Avanti Polar Lipids, Alablaster, AL, USA), cholesterol, dioctadecyldimethylammonium chloride and N-palmitoyl-sphingosine-1-succinyl (also known as methoxypolyethylene glycol 2000; Sigma-Aldrich; Merck Millipore, Darmstadt, Germany) were mixed in 100% ethanol at a molar ratio of 25:45:25:2.5 to produce pre-formed vesicles (PFV). siRNA (in a 30% ethanol solution) and PFV's (in a 30% ethanol solution) were warmed to 35–40°C for 10 min, respectively. Subsequently, siRNA was added slowly to the PFVs while the solution was stirred continuously to form siRNA-liposomes. Following the encapsulation, the solution was concentrated by centrifugal filtration (Merck Millipore) and sterilized by filtration using a 0.22-mm Supor membrane filter (Gelman Sciences, Ann Arbor, MI, USA), and the final products were produced. The average diameters of siRNA-survivin-liposomes and siRNA-NC-liposomes were 70.7±29.077 and 64.9±26.128 nm, respectively.
In vitro transfection
Before transfection, the cancer cells at the growth stage were harvested and a cell suspension was prepared (1×105 cells/ml) with the use of DMEM-H medium (Life Technologies; Thermo Fisher Scientific, Inc.). Following this, 1 ml cell suspension was added in each well. The wells were divided into three groups: Survivin siRNA group (survivin siRNA nanoliposome), NC group (NC siRNA nanoliposome) and normal group (without transfection). The cells were plated and grown to 70–80% confluence prior to transfection. Survivin and NC siRNA-liposomes were mixed with Opti-MEM (Life Technologies; Thermo Fisher Scientific, Inc.) and left to stand for 5 min at room temperature. Subsequently, the survivin and NC siRNA-liposomes were mixed with an equal volume of cell suspension and cultured at 37°C in 5% CO2. After 4–6 h, the media were changed to fresh media for growth, as described, and cells were incubated for an additional 48 h at 37°C. A total of 24 h after transfection, cells were fixed with 4% paraformaldehyde for 30 min and washed with phosphate buffered saline (PBS) three times, 5 min each time, and stained with Hoechst 33258 (Life Technologies; Thermo Fisher Scientific, Inc.) for 10–20 min. Transfection efficiency was observed using a fluorescence microscope (magnification, ×100; Nikon Corp., Tokyo, Japan).
RNA extraction and RT-qPCR
Total RNA was extracted from the transfected cell lines with RISO™ RNA Isolation Reagent (Biomics Biotechnologies Co., Ltd.), according to the manufacturer's instructions. cDNA was synthesized with a SensiMix™ SYBR-Green One-Step kit (Quantace; Bioline Reagents, Ltd., London, UK) according to the manufacturer's instructions. qPCR was performed using an ABI PRISM real-time PCR system (Applied Biosystems; Thermo Fisher Scientific, Inc.). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an endogenous control. PCR primers were as follows: survivin, forward 5′-ACGACCCCATAGAGGAACAT-3′ and reverse, 5′-TCCGCAGTTTCCTCAAATTC-3′; GAP DH, forward 5′-GAAGGTGAAGGTCGGAGTC-3′ and reverse 5′-GAAGATGGTGATGGGATTTC-3′. The RT reaction was performed at 42°C for 30 min. qPCR was then performed under the following conditions: The initial PCR incubation was 95°C for 10 min and the amplification loop (95°C for 20 sec, 58°C for 30 sec and 72°C for 30 sec) was repeated 45 times. Following the reaction, the amplification and melting curves were analyzed. Relative quantification was performed using the comparative 2-ΔΔCq method (17), using GAPDH as a reference gene. All analyses were performed in triplicate.
Western blot analysis
The expression of survivin protein was determined by western blotting. Cells were washed with PBS and placed on ice for 4 min following a 3–5 min incubation in a boiling water bath, then centrifuged at 13,500 × g at 4°C for 15 min. Following centrifugation, the protein pellet was resuspended and the resulting cell protein extract samples were analyzed by 8% SDS-PAGE (15 µl/lane), then transferred to polyvinylidene fluoride membrane filters (EMD Millipore, Billerica, MA, USA). Membranes were incubated at room temperature for 2 h in 5% non-fat dry milk, and rabbit polyclonal anti-survivin antibody (1:500; ab469; Abcam, Cambridge, UK) and mouse anti-β-actin monoclonal antibody (1:400; BM0005; Wuhan Boster Biological Technology, Ltd., Wuhan, China) were added and incubated overnight at 4°C. Following three washes with Tris-buffered saline with Tween, goat anti-rabbit and anti-mouse secondary antibodies (both 1:500; BA1054 and BA1051, respectively; both Wuhan Boster Biological Technology, Ltd.) were added and incubated at room temperature for 2 h. The gray-scale values of the blots were quantified with the computer-based system ImageJ 1.441 (National Institutes of Health, Bethesda, MA, USA). The results were shown as mean values from three replications.
Cell proliferation assay
Growth of the cancer cell lines was evaluated using an MTT assay. The assay was performed at 0, 24, 48, 72 and 96 h post-transfection. Cells were plated in medium supplemented with serum into 96-well plates at a density of 1×104 cells/ml (100 µl). Absorbance was determined following incubation with MTT solution at 37°C without light for 4 h. The supernatant was discarded and 150 µl DMSO was added. Optical density (OD) values were measured at a wavelength of 570 nm with a microplate reader, with normalization to a blank control (dimethyl sulfoxide). The inhibition rate was calculated relative to the OD value in the normal control group.
Hoechst staining
Apoptotic cells were detected by Hoechst 33258 staining. Cells in survivin siRNA, NC and normal groups were fixed with 4% paraformaldehyde for 10 min at room temperature and washed three times with PBS, followed by staining with Hoechst 33258 (Thermo Fisher Scientific, Inc.) for 10–20 min. Apoptotic features were observed under a fluorescence microscope.
Annexin V/propidium iodide (PI) staining
To quantify apoptosis, cells were double-stained with an Annexin V-Fluorescein isothiocyanate Apoptosis Detection kit (Beyotime Institute of Biotechnology, Haimen, China) and PI, according to the manufacturer's protocol. The percentages of cells in different cell cycle phases were then analyzed by FACS Calibur flow cytofluorometry (BD Biosciences, San Jose, CA USA) for the presence of viable (Annexin V− and PI−), early apoptotic (Annexin V+ and PI−) and late apoptotic (Annexin V+ and PI+) cells, as described by a previous method (18).
Statistical analysis
Data are presented as the mean ± standard deviation. Results were statistically analyzed using SPSS 10.0 software (SPSS, Inc., Chicago, IL, USA). A t-test was performed to determine statistical differences between the different groups and P<0.05 was considered to indicate a statistically significant difference.
Results
siRNA transfection suppresses survivin gene expression in human cancer cell lines
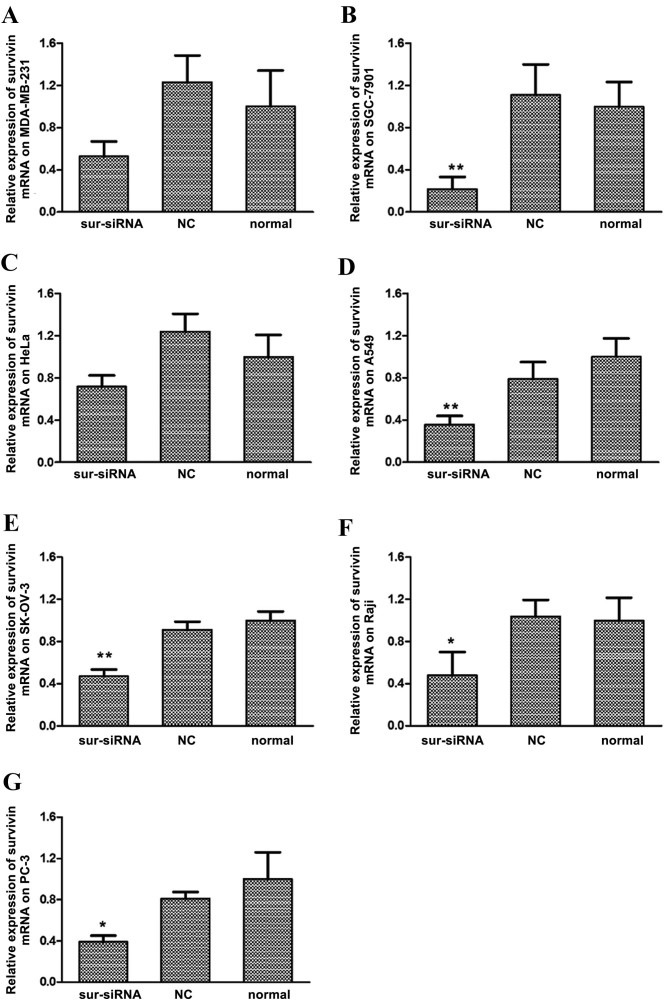
Expression of survivin mRNA in the MDA-MB-231, SGC-7901, HeLa, A549, SK-OV-3, Raji and PC-3 cancer cell lines was suppressed following siRNA transfection, as indicated by results of RT-qPCR (Fig. 1A-G, respectively). Among them, cell line SGC-7901 exhibited the greatest sensitivity to survivin siRNA, whereby expression of survivin mRNA significantly decreased to 22% relative to the normal control group (P<0.01, Fig. 1B). Expression of survivin mRNA also significantly decreased compared with the control group in the A549 (36%; P<0.01, Fig. 1D), SK-OV-3 (47%, P<0.01, Fig. 1E) Raji (45%; P<0.05, Fig. 1F) and PC-3 (39%; P<0.05; Fig. 1G) cell lines. Marked decreases in survivin mRNA were observed in MDA-MB-231 (50%, Fig. 1A) and HeLa (65%, Fig. 1C) cells, relative to their respective untreated (normal) control groups. For all cell lines, differences in survivin mRNA levels between the si-NC and normal control groups were not significant.
Figure 1.
Relative levels of survivin mRNA expression in human cancer cell lines following transfection with survivin siRNA. Levels of survivin mRNA expression were determined by reverse transcription-quantitative polymerase chain reaction following survivin siRNA-transfection of the human cancer cells lines: (A) MDA-MB-231, (B) SGC-7901, (C) HeLa, (D) A549, (E) SK-OV-3, (F) Raji and (G) PC-3. *P<0.05 and **P<0.01 vs. normal control group of each cell line, respectively. siRNA, small interfering RNA; NC, scrambled negative control siRNA; normal, untreated negative control.
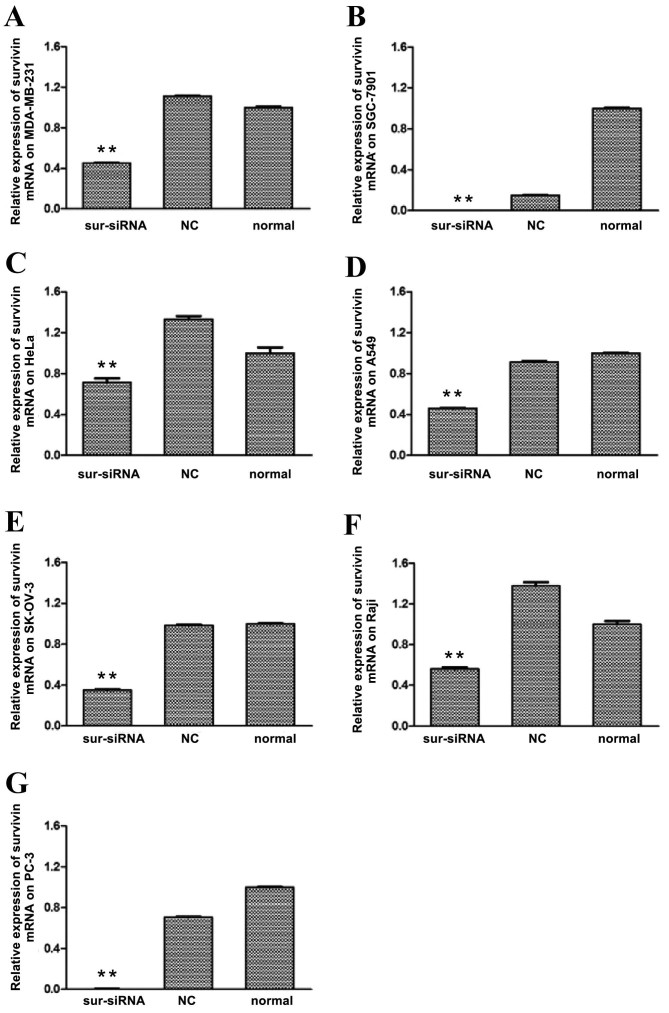
The level of survivin protein expression in each cell line following survivin siRNA transfection was subsequently determined by western blot analysis. As depicted in Fig. 2, levels of survivin protein in the survivin siRNA groups of all cell lines were significantly (P<0.01) downregulated relative to that in the control groups, particularly for SGC-7901 and PC-3, where the expression was almost completely inhibited. The survivin protein expression levels of the remaining cancer cells were reduced to 40–70% of the level of the normal control group. For all cell lines, differences in survivin protein levels between the si-NC and normal control groups were not significant.
Figure 2.
Relative levels of survivin protein expression in human cancer cell lines following transfection with survivin siRNA. Levels of survivin protein expression were determined by western blot analysis following survivin siRNA-transfection of the human cancer cells lines: (A) MDA-MB-231, (B) SGC-7901, (C) HeLa, (D) A549, (E) SK-OV-3, (F) Raji and (G) PC-3. **P<0.01 vs. normal control group. siRNA, small interfering RNA; NC, scrambled negative control siRNA; normal, untreated negative control.
Survivin siRNA inhibits cancer cell proliferation
The inhibitory effects of survivin siRNA in each cell line were analyzed by an MTT assay at 24, 48, 72 and 96 h post-transfection. Inhibition rates are listed in Table I. The data indicates that proliferation of the cancer cell lines was reduced following survivin knockdown by siRNA. At 96 h post-transfection, significant inhibition of cell proliferation was detected in all cell lines relative to the si-NC group (P<0.05).
Table I.
Inhibition rate of survivin siRNA on cancer cell lines.
| Inhibition rate (mean ± standard deviation, %) | |||||
|---|---|---|---|---|---|
| Cell lines | Groups | 24 h | 48 h | 72 h | 96 h |
| MDA-MB-231 | siRNA | 19.2±4.8a | 17.5±1.4a | 16.6±3.7a | 9.9±2.2a |
| si-NC | 9.1±5.1 | 4.1±2.2 | 4.6±1.9 | −1.5±3.7 | |
| SGC-7901 | siRNA | 9.0±0.8 | 15.5±5.6 | 5.3±1.8 | 6.8±4.8a |
| si-NC | 5.2±3.5 | 8.3±5.0 | 1.1±4.1 | 1.1±3.7 | |
| HeLa | siRNA | 7.1±2.2 | 18.6±2.2 | 25.5±4.6a | 46.3±1.8a |
| si-NC | 8.6±3.6 | 18.3±3.4 | 5.8±4.2 | 33.2±4.3 | |
| A549 | siRNA | 4.0±1.0 | 1.4±2.2 | 25.4±2.5a | 29.4±1.2a |
| si-NC | 2.8±2.4 | 2.3±2.1 | 2.9±3.7 | 3.3±3.1 | |
| SK-OV-3 | siRNA | 1.2±2.1 | 2.8±4.0 | −0.9±4.3 | 7.0±0.9a |
| si-NC | −1.7±3.3 | 3.0±3.2 | −2.4±3.8 | 0.4±1.8 | |
| Raji | siRNA | 2.0±0.4 | 3.0±0.02 | 1.7±5.0 | 10.8±4.0a |
| si-NC | 1.4±0.3 | 0.5±0.7 | 0.5±3.1 | 7.4±7.0 | |
| PC-3 | siRNA | 1.2±1.8 | 0.4±1.5 | 4.7±2.1 | 5.5±2.7a |
| si-NC | 0.2±2.5 | 1.0±0.9 | 1.4±1.9 | −0.5±4.1 | |
P<0.05 vs. si-NC control group. siRNA, small interfering RNA; si-NC, scrambled negative control siRNA.
Survivin knockdown induces apoptosis of cancer cell lines
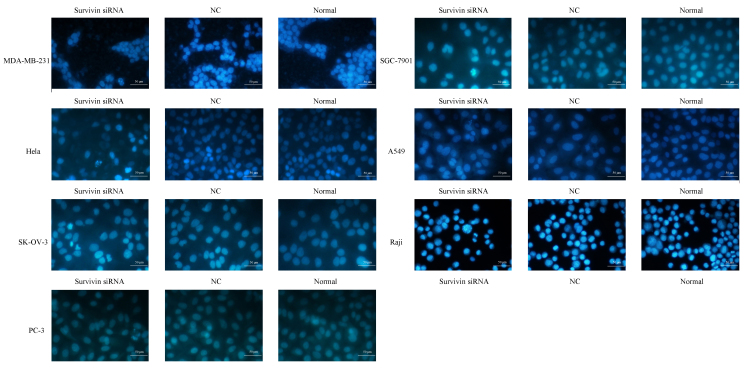
Hoechst 33258 staining was performed in order to evaluate the levels of cell apoptosis following survivin mRNA knockdown (Fig. 3). Cell nuclei were invariably stained blue by Hoechst 33258, however the nuclei morphology and extent of staining were distinct between normal and apoptotic cells. Normal cells with intact nuclei appeared light blue in color, while apoptotic cells with condensed or fragmented nuclei exhibited bright blue, fragmented nuclei of variable sizes. As depicted in Fig. 3, the nuclei of cancer cell lines transfected with survivin siRNA exhibited granular bright blue fluorescence, reflecting the typical morphological characteristics of apoptosis. These indicators of apoptosis were not observed in the si-NC and normal control groups.
Figure 3.
Morphological changes of apoptotic cells following siRNA knockdown of survivin in cancer cell lines (MDA-MB-231, SGC-7901, HeLa, A549, SK-OV-3, Raji and PC-3). Apoptosis of cells was detected by Hoechst 33258 staining and visualized using a fluorescence microscope (magnification, ×400). Survivin siRNA transfectants exhibited higher rates of apoptosis relative to the NC and normal control groups. siRNA, small interfering RNA; NC, scrambled negative control siRNA; normal, untreated negative control.
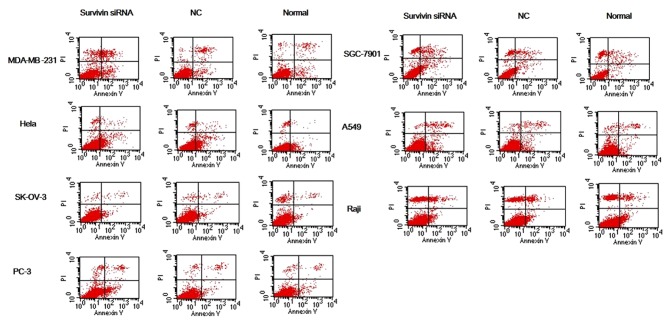
Cells in each group were subsequently double-stained with Annexin V/PI and analyzed with flow cytometry (Fig. 4). The apoptosis rate of MDA-MB-231 and PC-3 cells was significantly increased in the survivin siRNA-transfected groups compared with the apoptosis rate in the si-NC and normal control groups (all P<0.05; Table II). The apoptosis rate was significantly increased in the SGC-7901, HeLa and A549 survivin siRNA-transfected groups compared with the apoptosis rate in the normal control group (all P<0.05; Table II); however, the differences for SK-OV-3 and Raji were not significant.
Figure 4.
Apoptosis of cancer cell lines transfected with survivin siRNA was detected by Annexin V/PI double staining. Following siRNA knockdown of survivin in cancer cell lines (MDA-MB-231, SGC-7901, HeLa, A549, SK-OV-3, Raji and PC-3), levels of cell apoptosis were evaluated by Annexin V/PI double staining followed by flow cytofluorometry analysis. Early and late stage apoptosis (Annexin V+/PI− and Annexin V+/PI+, respectively) in the survivin siRNA groups were increased to some extent relative to the si-NC and normal control groups. siRNA, small interfering RNA; NC, scrambled negative control siRNA; normal, untreated negative control.
Table II.
Effect of survivin siRNA on the apoptosis rate of different cancer cells detected by flow cytometry after Annexin-V/propidium iodide staining.
| Apoptosis rate (mean ± standard deviation, %) | |||
|---|---|---|---|
| Cell lines | Survivin siRNA group | NC group | Normal group |
| MDA-MB-231 | 17.21±2.13a,b | 4.29±1.47 | 3.27±1.32 |
| SGC-7901 | 19.45±2.58c | 13.77±2.71 | 12.72±2.83 |
| HeLa | 5.37±1.04c | 4.45±1.01 | 2.62±0.91 |
| A549 | 19.89±3.18c | 15.29±2.86 | 13.38±2.15 |
| SK-OV-3 | 4.87±0.84 | 4.66±0.84 | 4.53±0.81 |
| Raji | 8.98±1.03 | 8.65±1.02 | 7.89±0.92 |
| PC-3 | 7.38±1.56b,d | 3.77±1.19 | 3.72±0.89 |
Values are presented as the mean ± standard deviation for three separate experiments performed in triplicate.
P<0.01
P<0.05 vs. normal group
P<0.01
P<0.05 vs. NC group. NC, scrambled negative control siRNA; si, small interfering.
Discussion
Malignant tumors are characterized by uncontrolled and accelerated proliferation of cancer cells and are associated with high morbidity and mortality. In 2012, 14.1 million new cancer cases occurred globally, and cancer resulted 8.2 million deaths (19,20). In the treatment of cancer, combined therapy (21), which includes surgery, chemotherapy, radiotherapy and biological therapy, is generally adopted. Of these, biological therapy is considered to have the greatest efficacy at each stage of tumor development, as well as the least side effects (22). The occurrence and development of tumors is in part due to synergistic and antagonistic interactions between multiple genes, including oncogenes, tumor suppressor genes, apoptosis inhibiting genes and reverse transcriptases. Therefore, along with uncontrolled proliferation of tumor cells, tumorigenesis is also associated with restricted tumor cell apoptosis (23,24).
Survivin is an intracellular protein that belongs to the inhibitor of apoptosis family (5). The protein is considered to be a potential target of cancer therapy, due to its observed overexpression in the majority of malignancies. Therefore, the potential mechanisms of survivin regarding its inhibition of apoptosis have been evaluated (25). As survivin is considered to serve a key regulatory role in cancer progression, methods that suppress survivin expression may promote tumor cell apoptosis and thus have anticancer effects.
RNAi is a post-transcriptional gene silencing mechanism that uses siRNA, and may be used to analyze cancer biology (26). In the present study, RNAi was used to determine the potential role of survivin in human cancer. Specific siRNA against survivin was transfected into a number of human cancer cell lines and its effect on cell proliferation and apoptosis was assessed using RT-qPCR, western blot analysis, MTT assay and Hoechst and Annexin V/PI staining. Using RT-qPCR and western blotting, it was determined that the expression of survivin was suppressed following transfection with survivin siRNA, In addition, data from the MTT assay indicated that proliferation of cancer cells lines was reduced. Subsequent staining assays also demonstrated that siRNA knockdown of survivin led to higher rates of apoptosis in cancer cells. Collectively, these data suggest that survivin may serve a key role in cancer progression.
siRNA technology may be a potential therapeutic method of treating tumors, however the difficulties in delivering siRNA into target cells has limited its use (27). Thus, studies investigating effective delivery systems for siRNA are warranted. A previous study documented the intracellular delivery of siRNA by lipid-based agents and carriers, including atelocollagen, protamine-antibody fusion protein and polyethyleneimine (28). Prior to initiation of the present study, a cationic liposome was optimized and used as an siRNA transfer vehicle. This liposome was chosen due to its ability to efficiently traverse the cell membrane, relatively high stability and biological compatibility.
The distinct distribution of survivin in cancer tissues makes it a suitable target for RNAi-based antitumor therapy, and downregulation of survivin by RNAi has been documented in gastric cancer (29), rectal cancer (30), bladder cancer (31) and lung cancer cells (14). The results of these studies indicated that RNAi-targeting successfully inactivated the anti-apoptotic function of survivin and inhibited tumor growth. Furthermore, expression of survivin in tissues and organs is considered to be an indicator of precancerosis (32), while also being associated with tumor invasion and metastasis (33), sensitivity to chemotherapy (34) and degree of lymphatic metastasis (35). Therefore, survivin is a potential biomarker for the early diagnosis, treatment and prognosis of cancer.
In conclusion, the present study achieved downregulation of survivin in a number of common human cancer cell lines by RNAi. It was observed that siRNA knockdown of survivin inhibited the growth of tumor cells and induced their apoptosis, demonstrating that survivin is a potential gene target in cancer therapy. However, due to differences between in vitro experiments and clinical therapy, the present study was unable to simulate an in vivo environment completely, which made the obtained results biased to some degree. Thus, further studies in animal models are required in the future.
Acknowledgements
The present study was supported by the National High Technology Research and Development Program of China (grant no. 2012AA020804).
References
- 1.Sydorak RM, Applebaum H. Soft Tissue Tumors. In: Mattei P, editor. Fundamentals of Pediatric Surgery. Springer-Verlag; New York, NY: 2011. pp. 755–760. [DOI] [Google Scholar]
- 2.Li W. The new concept of cancer supportive treatment. Electronic Journal of Metabolism and Nutrition of Cancer. 2014;1 [Google Scholar]
- 3.Horiot JC. At last-progress in the assessment of the adverse effects of cancer treatments. Lancet Oncol. 2007;8:568–570. doi: 10.1016/S1470-2045(07)70183-3. [DOI] [PubMed] [Google Scholar]
- 4.Barrett PT, Petrides K, Eysenck SB, Eysenck HJ. The Eysenck personality questionnaire: An examination of the factorial similarity of PE, N and L across 34 countries. Personality Individual Differences. 1998;25:805–819. doi: 10.1016/S0191-8869(98)00026-9. [DOI] [Google Scholar]
- 5.Ambrosini G, Adida C, Altieri DC. A novel anti-apoptosis gene, survivin, expressed in cancer and lymphoma. Nat Med. 1997;3:917–921. doi: 10.1038/nm0897-917. [DOI] [PubMed] [Google Scholar]
- 6.Zaffaroni N, Daidone MG. Survivin expression and resistance to anticancer treatments: Perspectives for new therapeutic interventions. Drug Resistance Updates. 2002;5:65–72. doi: 10.1016/S1368-7646(02)00049-3. [DOI] [PubMed] [Google Scholar]
- 7.Kong LJ, Zhao J, Fan WW. Survivin and Tumor Angiogenesis. Zhong Liu Xue Za Zhi. 2009;15:950–952. (In Chinese) [Google Scholar]
- 8.Suzuki A, Hayashida M, Ito T, Kawano H, Nakano T, Miura M, Akahane K, Shiraki K. Survivin initiates cell cycle entry by the competitive interaction with Cdk4/p16 (INK4a) and Cdk2/cyclin E complex activation. Oncogene. 2000;19:3225–3234. doi: 10.1038/sj.onc.1203665. [DOI] [PubMed] [Google Scholar]
- 9.Mesri M, Morales-Ruiz M, Ackermann EJ, Bennett CF, Pober JS, Sessa WC, Altieri DC. Suppression of vascular endothelial growth factor-mediated endothelial cell protection by survivin targeting. Am J Pathol. 2001;158:1757–1765. doi: 10.1016/S0002-9440(10)64131-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li F, Ambrosini G, Chu EY, Plescia J, Tognin S, Marchisio PC, Altieri DC. Control of apoptosis and mitotic spindle checkpoint by survivin. Nature. 1998;396:580–584. doi: 10.1038/25141. [DOI] [PubMed] [Google Scholar]
- 11.Kayaselcuk F, Nursal T, Polat A, Noyan T, Yildirim S, Tarim A, Seydaoglu G. Expresion of survivin, Bcl-2, P53 and bax in breast carcioma and ductal intraepithelial neoplasia (DIN 1a) J Exp Clin Cancer Res. 2004;23:105–112. [PubMed] [Google Scholar]
- 12.Ryan BM, O'Donovan N, Duffy MJ. Survivin: A new target for anti-cancer therapy. Cancer Treat Rev. 2009;35:553–562. doi: 10.1016/j.ctrv.2009.05.003. [DOI] [PubMed] [Google Scholar]
- 13.Altieri DC. Survivin, versatile modulation of cell division and apoptosis in cancer. Oncogene. 2003;22:8581–8589. doi: 10.1038/sj.onc.1207113. [DOI] [PubMed] [Google Scholar]
- 14.Chen XQ, Yang S, Li ZY, Lu HS, Kang MQ, Lin TY. Effects and mechanism of downregulation of survivin expression by RNA interference on proliferation and apoptosis of lung cancer cells. Mol Med Rep. 2012;5:917–922. doi: 10.3892/mmr.2012.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baker B, Eldrup A, Manoharan M, et al., inventors. US patent 10/700,884. RNA-induced silencing complexes; RNA interference; gene expression inhibition. 2003 2005 Oct 4; issued February 17.
- 16.Aliabadi H Montazeri, Landry B, Mahdipoor P, Uludag H. Induction of apoptosis by survivin silencing through siRNA delivery in a human breast cancer cell line. Mol Pharm. 2011;8:1821–1830. doi: 10.1021/mp200176v. [DOI] [PubMed] [Google Scholar]
- 17.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 18.Mirandola P, Sponzilli I, Gobbi G, Marmiroli S, Rinaldi L, Binazzi R, Piccari GG, Ramazzotti G, Gaboardi GC, Cocco L, Vitale M. Anticancer agents sensitize osteosarcoma cells to TNF-related apoptosis-inducing ligand downmodulating IAP family proteins. Int J Oncol. 2006;28:127–133. [PubMed] [Google Scholar]
- 19.Stewart B, Wild C, editors. World cancer report 2014. International Agency for Research on Cancer; Lyon: 2014. [PubMed] [Google Scholar]
- 20.Organization WH. ES Fact sheet 310: Top causes of death. Centro De Prensa. 2014 [Google Scholar]
- 21.Arnold LD, Ji QS, Buck E, Haley JD, Mulvihill MJ, inventors. U.S. Patent 8,575,164. Combination cancer therapy. 2013
- 22.Yang Y, Huang S. Discuss the strengths and weaknesses between tumour biotherapy and chemoradiotherapy. Medical Information. 2011;24:3503–3504. [Google Scholar]
- 23.Hengartner MO. The biochemistry of apoptosis. Nature. 2000;407:770–776. doi: 10.1038/35037710. [DOI] [PubMed] [Google Scholar]
- 24.Reed JC. Dysregulation of apoptosis in cancer. J Clin Oncol. 1999;17:2941–2953. doi: 10.1200/JCO.1999.17.9.2941. [DOI] [PubMed] [Google Scholar]
- 25.Chiou SK, Jones MK, Tarnawski AS. Survivin-an anti-apoptosis protein: Its biological roles and implications for cancer and beyond. Med Sci Monit. 2003;9:PI25–PI29. [PubMed] [Google Scholar]
- 26.Merritt WM, Bar-Eli M, Sood AK. The dicey role of Dicer: Implications for RNAi therapy. Cancer Res. 2010;70:2571–2574. doi: 10.1158/0008-5472.CAN-09-2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Semple SC, Akinc A, Chen J, Sandhu AP, Mui BL, Cho CK, Sah DW, Stebbing D, Crosley EJ, Yaworski E, et al. Rational design of cationic lipids for siRNA delivery. Nat Biotechnol. 2010;28:172–176. doi: 10.1038/nbt.1602. [DOI] [PubMed] [Google Scholar]
- 28.Medarova Z, Pham W, Farrar C, Petkova V, Moore A. In vivo imaging of siRNA delivery and silencing in tumors. Nat Med. 2007;13:372–377. doi: 10.1038/nm1486. [DOI] [PubMed] [Google Scholar]
- 29.Wenying Z, Zhaoning J, Zhimin Y, Dongyun C, Lili S. Survivin siRNA inhibits gastric cancer in nude mice. Cell Biochem Biophys. 2012;62:337–341. doi: 10.1007/s12013-011-9315-0. [DOI] [PubMed] [Google Scholar]
- 30.Rödel F, Hoffmann J, Distel L, Herrmann M, Noisternig T, Papadopoulos T, Sauer R, Rödel C. Survivin as a radioresistance factor, and prognostic and therapeutic target for radiotherapy in rectal cancer. Cancer Res. 2005;65:4881–4887. doi: 10.1158/0008-5472.CAN-04-3028. [DOI] [PubMed] [Google Scholar]
- 31.Ning S, Fuessel S, Kotzsch M, Kraemer K, Kappler M, Schmidt U, Taubert H, Wirth MP, Meye A. siRNA-mediated down-regulation of survivin inhibits bladder cancer cell growth. Int J Oncol. 2004;25:1065–1071. [PubMed] [Google Scholar]
- 32.Endoh A, Asanuma K, Moriai R, Yamada M, Koyanagi Y, Sato T, Yagihasi A, Nakamura M, Kobayashi D, Watanabe N. Expression of survivin mRNA in CD34-positive cells. Clin Chim Acta. 2001;306:149–151. doi: 10.1016/S0009-8981(01)00416-8. [DOI] [PubMed] [Google Scholar]
- 33.Lee GH, Joo YE, Koh YS, Chung IJ, Park YK, Lee JH, Kim HS, Choi SK, Rew JS, Park CS, Kim SJ. Expression of survivin in gastric cancer and its relationship with tumor angiogenesis. Eur J Gastroenterol Hepatol. 2006;18:957–963. doi: 10.1097/01.meg.0000230086.83792.56. [DOI] [PubMed] [Google Scholar]
- 34.Smith SD, Wheeler MA, Plescia J, Colberg JW, Weiss RM, Altieri DC. Urine detection of survivin and diagnosis of bladder cancer. JAMA. 2001;285:324–328. doi: 10.1001/jama.285.3.324. [DOI] [PubMed] [Google Scholar]
- 35.Yao X, Liu F, Qi X, Wu B, Yin HL, Ma HH, Shi QL, Zhou XJ, Li JS. Expression of survivin in human gastric adenocarcinomas: Correlation with proliferation and apoptosis. Zhonghua Wai Ke Za Zhi. 2004;42:145–148. (In Chinese) [PubMed] [Google Scholar]