Abstract
We have performed a comparative analysis of the insertion sites of engineered Arabidopsis (Arabidopsis thaliana) insertional mutagenesis vectors that are based on the maize (Zea mays) transposable elements and Agrobacterium T-DNA. The transposon-based agents show marked preference for high GC content, whereas the T-DNA-based agents show preference for low GC content regions. The transposon-based agents show a bias toward insertions near the translation start codons of genes, while the T-DNAs show a predilection for the putative transcriptional regulatory regions of genes. The transposon-based agents also have higher insertion site densities in exons than do the T-DNA insertions. These observations show that the transposon-based and T-DNA-based mutagenesis techniques could complement one another well, and neither alone is sufficient to achieve the goal of saturation mutagenesis in Arabidopsis. These results also suggest that transposon-based mutagenesis techniques may prove the most effective for obtaining gene disruptions and for generating gene traps, while T-DNA-based agents may be more effective for activation tagging and enhancer trapping. From the patterns of insertion site distributions, we have identified a set of nucleotide sequence motifs that are overrepresented at the transposon insertion sites. These motifs may play a role in the transposon insertion site preferences. These results could help biologists to study the mechanisms of insertions of the insertional mutagenesis agents and to design better strategies for genome-wide insertional mutagenesis.
Insertional mutagenesis techniques are key resources for studying the gene functions of Arabidopsis (Arabidopsis thaliana). These techniques use either maize (Zea mays) transposable elements (Fedoroff, 1989) or Agrobacterium tumefaciens T-DNA (Koncz et al., 1992; Azpiroz-Leehan and Feldmann, 1997) as mutagens. The maize transposable elements transpose through an excision-integration mechanism in which the element excises from the donor site and then inserts into a new target site. Two commonly used maize transposable elements are Dissociation (Ds) and defective Suppressor-mutator (Spm; dSpm). Insertion of Ds transposons is mediated by the Arabidopsis activator/Ds system (Sundaresan et al., 1995), in which Arabidopsis lines carrying a gene trap or enhancer trap Ds element on a T-DNA vector are crossed to lines expressing the transposase on another T-DNA vector. Stable unlinked insertion events are recovered and enriched by a combination of positive selection for kanamycin resistance on the Ds element and negative selection against both T-DNAs. The dSpm transposon element is produced by the Arabidopsis Enhancer/Spm system (Tissier et al., 1999), which uses a similar selection scheme.
T-DNA insertional mutagenesis techniques use a portion of the tumor-inducing plasmid from A. tumefaciens that in nature induces crown galls by transferring T-DNA into the nucleus of plant cells. Upon infection, the T-DNA is transferred into host cells and inserts into the nuclear genome (Gordon, 1998). Though the mechanism of T-DNA integration into the plant nuclear genome is not completely understood, it is generally considered to involve illegitimate recombination (Tinland, 1996). Unlike the transposable elements, which will often excise after integration into the genome, the T-DNA integration is stable for several generations. GABI-Kat T-DNA (Li et al., 2003; Rosso et al., 2003) and FLAGdb/FST T-DNA (Balzergue et al., 2001; Samson et al., 2002) are two major publicly available T-DNA agents in Arabidopsis.
Previous studies (Rubin and Spradling, 1982; Jones et al., 1990; Bancroft and Dean, 1993; Thomas et al., 1994; James et al., 1995; Smith et al., 1996; Azpiroz-Leehan and Feldmann, 1997; Craig, 1997; Machida et al., 1997; Parinov et al., 1999; Liao et al., 2000; Szabados et al., 2002) suggest that while T-DNA-based mutagenesis agents insert randomly into the genome, transposon agents have marked preferences for particular regions. Such information is important for large insertional mutagenesis projects since it is frequently asked how many insertions are required to recover at least one knockout mutation in every Arabidopsis gene. In many transgenesis experiments, it would be useful to exploit elements that have strong target site preferences to ensure that each transgene will have the same genomic context, ideally in the same target site. A better understanding of insertion site preferences might allow the design of modified elements targeted to particular sites of one's choice. Therefore, we have undertaken a study of the insertion site preferences for insertional mutagenesis agents in Arabidopsis.
RESULTS
We have developed Arabidopsis thaliana Insertion Database (ATIDB) to store information about insertional mutagenesis lines and to analyze the distributions of their insertion sites (Pan et al., 2003). From ATIDB, we extracted the insertion sites of Ds and dSpm transposons and two subsets of T-DNAs that have well-estimated insertion site annotations (Table I). We then analyzed the insertion distributions of the various insertional mutagenesis agents in the regions of the genome that differed in GC content and in the relative positions of protein-coding genes. Using pattern discovery techniques (Bailey and Elkan, 1994; Brazma et al., 1998; Helden et al., 2000), we searched for motifs at the insertion sites of each insertional mutagenesis agent to determine whether specific sequence motifs act as insertion targets.
Table I.
Insertion datasets
| Insertion Type | Number of Lines Available in ATIDB | Number of Insertion Sites Used for Analysis | Data Source |
|---|---|---|---|
| Ds transposon | 6,729 | 6,318 | Cold Spring Harbor Laboratory |
| dSpm transposon | 14,156 | 6,358 | John Innes Centre, UK |
| GABI-Kat T-DNA | 8,450 | 7,131 | Max Planck Institute for Plant Breeding Research, Germany |
| FLAGdb T-DNA | 14,095 | 11,762 | FLAGdb/FST, France |
Transposon-Based Agents Have Marked Preference for High GC Content whereas T-DNA-Based Agents Show Preference for Low GC Content
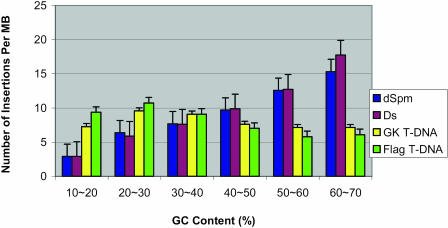
We stratified the Arabidopsis genome by GC content as described in “Materials and Methods” and calculated the insertion frequency of each of the four insertional mutagenesis agents. As there are few extremely GC-poor (0%–10%) or GC-rich (70%–100%) regions, we removed these regions from our analysis. As shown in Figure 1, the insertion frequencies of Ds and dSpm transposons significantly increase with increasing GC content, from approximately 3 insertions/Mb in 10% to 20% GC regions to 17 insertions/Mb in 60% to 70% GC regions. In contrast, the insertion frequency of T-DNAs shows a preference for low GC content regions, especially the 20% to 30% GC region. The window size change from 20 to 50 bp and the starting position variation of the window had no noticeable effect on these measurements.
Figure 1.
Insertion frequency by GC content. Insertion events have been normalized to 1,000 insertions of each insertional mutagenesis agent to allow for comparison between the agents.
Transposon Insertions Preferentially Occur at the 5′ Ends of Coding Regions while T-DNA-Based Agents Favor Upstream Regions
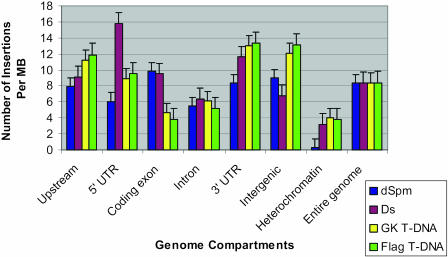
Using gene annotation data from The Institute for Genomic Research (TIGR; Wartman et al., 2003), we calculated the insertion frequency of each of the agents within the upstream regions of protein-coding genes, 5′ untranslated regions (5′ UTRs), coding exons, introns, 3′ untranslated regions (3′ UTRs), and intergenic and heterochromatic regions. As a baseline for comparison, we used insertional frequencies across the entire genome. The result is shown in Figure 2. For insertions within genes and their immediate environs, we found that T-DNA insertions favor the immediate upstream region, 5′ UTRs, and 3′ UTRs. In contrast, Ds and dSpm transposon insertions were relatively evenly distributed among different compartments with the exception of an apparently strong preference of Ds insertions for the 5′ and 3′ UTRs and a very low frequency of dSpm insertions in heterochromatin. Ds and dSpm transposon insertions have higher insertion site densities in exons than in introns, whereas the T-DNA insertions have higher insertion site densities in introns than in exons. Both transposon and T-DNA insertions have lower insertion site densities in heterochromatin than in euchromatin. In heterochromatin, the T-DNAs have more insertions than do the transposable elements. Interestingly, the insertion site densities of T-DNAs in exons are almost as low as those in the heterochromatin. Because insertion lines are selected for expression of the vector, insertions are more likely to be selected if they occur in transcriptionally active euchromatin. The relative increase in heterochromatic insertion frequency in T-DNA versus the transposon insertions may have to do with the fact that T-DNA lines often have multiple copies, allowing for selection of lines that have insertions in both euchromatin and heterochromatin. Transposon lines are mainly single copy, and heterochromatic insertions could be less likely to be selected.
Figure 2.
Distribution of 1,000 insertions of each insertional mutagenesis agent within genome structure. Insertion events have been normalized to 1,000 insertions of each agent to allow for comparison between the agents.
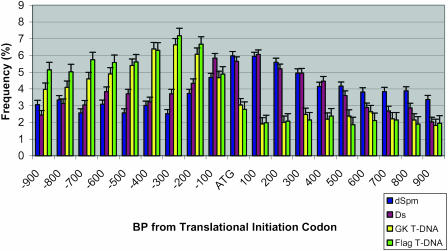
To determine whether there is an insertion site bias relative to the coding regions, we took the region 900 bp upstream of a translation start codon and divided it equally into 9 subregions. We also took the region 1,000 bp downstream of the start codon and divided it equally into 10 subregions. Then, we calculated the positional distribution of insertion sites relating to the start codon (Fig. 3). There is a striking preference of the T-DNA to insert toward the upstream region starting from approximately 100 bp upstream of the start codon. In contrast, more transposon insertions are located downstream of the translation start sites, especially in the region of 200 bp downstream of ATG.
Figure 3.
Distribution of different insertions relating to translation initiation codon.
To determine whether the insertion preferences of transposon and T-DNA insertions to different genome regions are reflections of differing preferences for GC-rich regions, we calculated GC content of 20-bp genomic sequences with centers at the insertion sites of each insertional mutagenesis agent in different regions, in parallel with sequences randomly taken from each region. As shown in Table II, GC content of transposon insertion sites is higher than that of T-DNA insertion sites in almost every region. This confirms that transposon insertions prefer high GC content sites, whereas T-DNA insertions favor low GC content regions. The coding exons have high GC content, which may lead to a higher frequency of transposon insertion sites than that of T-DNA insertion sites in this region. The transposon Ds insertion sites have high GC content in 5′ UTRs, which is probably a reason causing the preference of these insertions to this compartment. The other genome regions do not show that GC contents at different insertion sites are significantly different from the control.
Table II.
GC content (%) of 20-bp genomic sequences with centers at the insertion sites of each insertional mutagenesis agent in different genome structures and of 20-bp genomic sequences randomly taken from each region
| Genome Structure | dSpm Transposon | Ds Transposon | GABI-Kat T-DNA | FLAGdb T-DNA | Control |
|---|---|---|---|---|---|
| Upstream | 36.89 | 37.17 | 33.78 | 33.07 | 35.56 |
| 5′ UTR | 38.05 | 39.76 | 36.83 | 36.38 | 36.92 |
| Coding Exon | 46.50 | 46.20 | 43.66 | 43.62 | 43.20 |
| Intron | 32.90 | 33.15 | 32.76 | 32.61 | 32.40 |
| 3′ UTR | 33.28 | 34.35 | 31.52 | 33.29 | 34.96 |
| Intergenic | 35.63 | 35.02 | 33.31 | 31.92 | 35.87 |
Transposon Insertions Recognize a Set of Sequence Motifs
As described in “Materials and Methods,” we used the Multiple EM for Motif Elicitation (MEME) algorithm to search for motifs that are overrepresented at insertion sites using randomly selected subsets of the insertion sites as training sets. Motifs that were identified by MEME in two independent training sets were pursued further. In this manner, we identified three full invariant candidate motifs and six partial motifs with one or more ambiguous bases (Table III) to study further. We found that motif 11 contains motif 1, both motifs 21 and 22 contain motif 2, and motifs 31, 32, and 33 all contain motif 3. To assess the significance of these candidate motifs, we calculated their occurrence frequencies in 20-bp genomic sequences centered at the insertion sites of the corresponding insertional mutagenesis agent. As a control, we used 6 sets of 500 20-bp sequences randomly selected from the entire genome. As shown in Table III, nine candidate motifs are overrepresented at Ds insertion sites. Motifs 1, 2, 11, 21, 22, 31, and 33 are also overrepresented at dSpm insertion sites (P value < 0.05 or 0.01).
Table III.
Occurrence frequencies (%) of the candidate motifs between 20-bp genomic sequences centered at insertion sites and the same length genomic sequences retrieved randomly from Arabidopsis genome
A single digital motif code represents a full motif while a double digital motif code represents a partial motif. If the first motif code number of a partial motif is the same as a motif code number of a full motif, the motif is a partial motif of the full motif. The symbols * and ** indicate P values less than 0.05 and 0.01, respectively. [AT] and [TA] represent A or T, [TC] and [CT] represent C or T, [GAT] represents G, A, or T, and so on. These representations are also applied in Tables IV and V.
| Motif Code | Consensus Sequence | dSpm | Ds | GABI-Kat T-DNA | FLAGdb T-DNA | Control |
|---|---|---|---|---|---|---|
| 1 | CCTCCTC | GGAGGAG | 0.43* | 0.53* | 0.20 | 0.17 | 0.30 |
| 11 | CC[TA]CC[AT]C | G[TA]GG[AT]GG | 1.23* | 1.27* | 0.57 | 0.57 | 0.77 |
| 2 | GGTGGTG | CCACCAC | 0.67** | 0.47** | 0.23 | 0.13 | 0.20 |
| 21 | G[GAT]TGGTG | C[CTA]ACCAC | 1.30** | 1.47** | 0.67 | 0.67 | 0.57 |
| 22 | C[AT][TC]CA[CT]C | G[GA]TG[AG][TA]G | 2.70* | 3.87** | 1.93 | 1.73 | 2.10 |
| 3 | TCTTCT | AGAAGA | 2.83 | 3.47* | 2.70 | 2.67 | 2.93 |
| 31 | [GA]GA[AG]GA | [CT]CT[TC]CT | 6.47* | 7.40** | 5.57 | 5.73 | 5.63 |
| 32 | [GA]AGAAG[AG] | [CT]TCTTC[TC] | 2.73 | 3.23* | 2.60 | 2.63 | 2.80 |
| 33 | TC[TC]TCT[TC]C | AG[AG]AGA[AG]G | 1.13* | 1.13* | 0.77 | 0.77 | 0.83 |
None of the above motifs occur more frequently near GABI-Kat T-DNA and FLAGdb T-DNA insertion sites than over the entire genome (the control), suggesting that T-DNA insertion site preference may be independent of small sequence motifs.
Motifs May Play a Role in Insertion Site Preference of Transposable Elements
To distinguish whether the identified sequence motifs play a role in insertion site preference of transposable elements independently of GC content or position relative to protein coding genes, we stratified the data set by calculating the occurrence frequency of each motif in 20-bp genomic sequences centered at either Ds or dSpm insertion sites in the region of 60% to 70% GC content and the region 200 bp downstream of the translation start site, respectively (Table IV). As a control, we used 6 sets of 500 20-bp sequences selected at random from either testing region of Arabidopsis genome, through which we wished to control for any possible associations between the identified motifs and nucleotide sequences common in either testing region. All of the identified motifs are also overrepresented at the 3′ ends of Ds flanking sequences and the 5′ ends of dSpm flanking sequences (Table V).
Table IV.
Frequencies (%) of occurrence of motifs in 20-bp genomic sequences centered at the insertion sites of transposable elements in specific regions
*, **, and *** indicate 0.05, 0.01, and 0.001 significance of levels, respectively.
| Motif Code | Consensus Sequence | Insertion Type | Region of 60%–70% GC Content | Region of 200 bp Downstream of ATG | Over Entire Genome |
|---|---|---|---|---|---|
| 11 | CC[TA]CC[AT]C | G[TA]GG[AT]GG | dSpm | 10.10*** | 2.70** | 1.23* |
| Ds | 5.60* | 2.10* | 1.27* | ||
| Control | 5.20 | 1.80 | 0.77 | ||
| 21 | G[GAT]TGGTG | C[CTA]ACCAC | dSpm | 3.90** | 2.50** | 1.30** |
| Ds | 3.90* | 3.10** | 1.47** | ||
| Control | 3.13 | 1.00 | 0.57 | ||
| 22 | C[AT][TC]CA[CT]C | G[GA]TG[AG][TA]G | dSpm | 8.10** | 6.50** | 2.70* |
| Ds | 3.90 | 6.30** | 3.87** | ||
| Control | 4.90 | 3.97 | 2.10 | ||
| 31 | [GA]GA[AG]GA | [CT]CT[TC]CT | dSpm | 9.50* | 11.10* | 6.47* |
| Ds | 11.40** | 12.00* | 7.40** | ||
| Control | 7.93 | 10.40 | 5.63 | ||
| 32 | [GA]AGAAG[AG] | [CT]TCTTC[TC] | dSpm | 2.00 | 5.80 | 2.73 |
| Ds | 2.50** | 6.20* | 3.23* | ||
| Control | 1.23 | 4.40 | 2.80 | ||
| 33 | TC[TC]TCT[TC]C | AG[AG]AGA[AG]G | dSpm | 2.50** | 2.40* | 1.13* |
| Ds | 1.10* | 2.90** | 1.13* | ||
| Control | 0.73 | 2.07 | 0.83 |
Table V.
Frequencies (%) of the partial motifs occur in 20-bp genomic sequences of both ends of flanking sequences of transposon Ds and dSpm elements
| Motif Code | Consensus Sequence | Flanking Sequence Position | Frequencies in Ds Sequence | Frequencies in dSpm Sequence |
|---|---|---|---|---|
| 11 | CC[TA]CC[AT]C | G[TA]GG[AT]GG | 5′ end | 0.6 | 1.3 |
| 3′ end | 0.7 | 0.4 | ||
| 21 | G[GAT]TGGTG | C[CTA]ACCAC | 5′ end | 0.6 | 0.9 |
| 3′ end | 0.7 | 0.4 | ||
| 22 | C[AT][TC]CA[CT]C | G[GA]TG[AG][TA]G | 5′ end | 1.6 | 3.8 |
| 3′ end | 3.7 | 1.4 | ||
| 31 | [GA]GA[AG]GA | [CT]CT[TC]CT | 5′ end | 3.5 | 5.8 |
| 3′ end | 10.0 | 2.9 | ||
| 32 | [GA]AGAAG[AG] | [CT]TCTTC[TC] | 5′ end | 1.6 | 2.7 |
| 3′ end | 7.1 | 1.3 | ||
| 33 | TC[TC]TCT[TC]C | AG[AG]AGA[AG]G | 5′ end | 0.7 | 0.7 |
| 3′ end | 2.6 | 0.5 |
Because a full motif is included in one or more partial motifs (Table III), we need only to consider the occurrence frequencies of the partial motifs in a region. As shown in Table IV, motifs 11, 21, 22, 31, and 33 at dSpm transposon insertion sites and motifs 11, 21, 31, 32, and 33 at Ds transposon insertion sites are overrepresented in the region of 60% to 70% GC content. The summed total occurrence frequencies of the overrepresented motifs at Ds and dSpm insertion sites are approximately 34% and 25%, respectively, compared to the control with 22% and 19%. This demonstrates that the insertion site preference to the GC-rich area is correlated with the distribution of these motifs. Interestingly, motifs 11, 21, 22, 31, and 33 at dSpm transposon insertion sites and all of the 6 partial motifs at Ds transposon insertion sites are also overrepresented in the region 200 bp downstream from the translation start codon. The summed frequencies of the overrepresented motifs at dSpm and Ds insertion sites are approximately 25% and 33%, respectively, compared to the control with 19% and 24%. This suggests that downstream insertion preference of the transposon-based agents may in part be due to the distribution of these motifs.
DISCUSSION
In this study, we used large data sets from several insertional mutagenesis agents to analyze insertion site distributions and thereby provided powerful representation of the site integration process and a good comparison among different insertional mutagenesis agents in Arabidopsis. We found that both transposon and T-DNA agents have insertion site preferences but that these preferences are distinctly different.
Our results suggest that both Ds and dSpm transposon-based insertion mutagenesis in Arabidopsis prefer sites with high GC content. This result has not previously been reported in plants and may shed light on the mechanism of insertion of these vectors. Alternatively, it may be that insertions in high GC content regions are preferentially selected for the GC content of the flanking region enhances expression of the vector's selectable antibiotic resistance marker. Interestingly, our results are similar to the results of Liao et al. (2000), who reported that P transposable element insertion sites in Drosophila melanogaster prefer regions of high GC content.
In contrast to the results with transposon-based agents, we found that T-DNA-based agents prefer low GC content regions. This is consistent with the finding of Brunaud et al. (2002) who showed that FLAGdb/FST T-DNA insertions have a preference for AT-rich regions. However, a possible confounding factor in this analysis is that in the two T-DNA-based agents studied, a similar adaptor-ligation PCR walking method was used to recover the plant genomic DNA flanking the T-DNA insertion sites. Since an early step of this protocol is restriction enzyme digestion of genomic DNA, one might argue that the restriction sites might have some preferences for GC content regions and particular genome compartments and could influence the perceived distribution of T-DNA insertions. However, this seems not the case because different restriction enzymes with different GC contents were used in the two populations (Balzergue et al., 2001; Rosso et al., 2003) but resulted in similar patterns of insertion site distributions. The dSpm transposons were also amplified using a restriction enzyme digestion step that had no effect to GC content analysis.
Transposon-based insertional mutagenesis agents preferentially occur at the 5′ ends of gene coding regions, while T-DNA insertions favor the regions immediately upstream of the start codon, 3′ UTRs, and intergenic regions. Our findings are supported by a previous study (Parinov et al., 1999) that showed that the preferential insertion of Ds transposable elements is at the 5′ ends of coding regions. Also consistent with our findings are reports (Tinland, 1996; Sessions et al., 2002; Alonso et al., 2003) that T-DNA has a bias for insertion into transcribed regions. Our study further suggests that transposon-based insertional mutagenesis agents have a higher rate in exons than in introns, while T-DNA-based agents reverse the relationship. This may be explained by the preference of transposons for GC-rich regions and the preference of T-DNAs for AT-rich areas because exons are more GC rich than introns and transcribed regions.
While we cannot determine whether some of these biases are the result of primary insertion-site preference at the time of vector insertion or are due to subsequent selection of mutated lines, in practical terms these observations show that these two kinds of mutagenesis agents could complement one another well, and neither alone is sufficient to achieve the goal of saturation mutagenesis in Arabidopsis. These observations also suggest that transposons may be more effective for gene disruptions and gene traps, while T-DNAs are more effective for activation tagging and enhancer trapping. The latter has been proven to be practically useful in Arabidopsis due to extensive functional redundancy in its genome, while knockout mutants often result in no obvious phenotype.
Another interesting result of our study is that transposon-based insertional mutagenesis agents in Arabidopsis may recognize a small set of sequence motifs. Craig (1997) found that in several systems, the key determinant in target selection is a direct interaction between the transposase and the target DNA. In some cases, the transposase interacts preferentially with particular sequences; in other cases, the transposase prefers a particular DNA structure, most notably bent DNA. Based on the patterns of insertion site distributions within different GC content regions and gene compartments found above, we have identified nine sequence motifs associated with dSpm and Ds transposon insertions. These motifs may have complementary tendency in the sequences at an end of either Ds and dSpm transposon elements, because these motifs are also overrepresented at the 3′ ends of Ds and the 5′ ends of dSpm flanking sequences. In addition, these sequence motifs might provide unique geometric solutions for optimal transposase-target interaction. Some of these motifs occur more frequently in the regions of high GC content and the 5′ ends of genes and may account to some extent for the preference of transposon insertions to these regions. However, the total frequency of occurrence of our identified motifs is only 25% to 34% in regions of dSpm or Ds insertions. The reason may be that some motifs do not occur exactly at insertion sites but at some distance that is larger than the window of 10 bp that we chose for our training sets. Kuromori et al. (2004) found that the nucleotide contents of perfect, 8-bp target-site-duplication sequences statistically showed a complementary tendency in the 8-bp duplicated sequence of Ds transposons, suggesting that there are also base preferences at each position of the target-site-duplication sequences. There may also be other factors besides sequence motifs that determine transposon insertion sites. For example, in D. melanogaster, the P-element transposition mechanism recognizes a structural feature, a 14-bp palindromic pattern at insertion sites (Liao et al., 2000). Sleeping Beauty, the most active Tc1/mariner-type transposable element in vertebrates, prefers a palindromic AT-repeat, in which the central TA is the canonical target site (Vigdal et al., 2002). However, we did not find any palindromic patterns at Ds and dSpm transposon insertion sites in Arabidopsis, implying that the mechanism of interaction of the Ds and dSpm transposases with the genomic sequence follows a less rigid set of rules than the vertebrate and arthropod transposon systems. We noticed that several sequence motifs are preferred by both transposon Ds and dSpm insertions. This suggests that common host binding sequences might be involved in the insertion integration process. Further efforts will be needed to understand the relationship between insertion site preference and associated sequence motifs in Arabidopsis.
In contrast to our findings in the transposon-based insertional mutagenesis agents, we were unable to identify sequence motifs associated with T-DNA insertion using the motif discovery algorithm described in “Materials and Methods.” However, Brunaud et al. (2002) found that there are micro-similarities (base preferences and consensus sequences) in T-rich sequences between the host genome and the T-DNA border at the insertion site, which may be instrumental in determining the selectivity of T-DNA for the insertion site. The difference between their analysis and ours is that they excluded insertion sites with filler DNA (intermediate sequences between T-DNA and plant DNA) in the training data, and we did not. This may be the main reason we were not able to detect any sequence motifs in T-DNA insertion data. In other words, the motifs Brunaud et al. (2002) observed may be detectable only under more specific conditions.
MATERIALS AND METHODS
Insertion Sites
We annotated the insertion sites for four major insertional mutagenesis agents in Arabidopsis (Arabidopsis thaliana): the maize (Zea mays) transposable elements Ds and dSpm and the Agrobacterium tumefaciens T-DNAs from GABI-Kat and FLAGdb/FST and then stored them in ATIDB at http://gremlin6.zool.iastate.edu/. The annotation of Ds and dSpm transposon insertion sites was described by Pan et al. (2003). The FLAGdb/FST T-DNA flanking sequences were produced from a single PCR amplification (Balzergue et al., 2001), and therefore, their precise insertion sites can also be obtained using the same annotation method as the transposon insertion sites were obtained from. For GABI-Kat T-DNAs, deducing the precise insertion sites from flanking sequence is not always straightforward due to frequently observed filler DNA and microsimilarities, as well as different sequencing strategy employed in these two T-DNA agents. If these T-DNA flanking sequences were obtained directly from one or more PCR products without isolation, two or more overlapping reads could exist in the same sequence and obscure the precise insertion site. We downloaded all of the flanking sequences at http://www.mpiz-koeln.mpg.de/GABI-Kat/expert_download/ and picked up those with T-DNA locating just before the start of each sequence. After blasting, we selected the insertion sites of the T-DNA flanking sequences that had BLAST query start position less than or equal to 20 bp.
We retrieved all insertion sites of the four insertional mutagenesis agents from ATIDB at http://gremlin6.zool.iastate.edu/cgi-perl/insertion_tab?Line_type=X (where X = GT:ET for Ds, X = SM for dSpm transposons, X = FTN for FLAGdb and X = GTN for GABI-Kat T-DNAs). After removal of duplicated insertion sites, there are 6,318 Ds and 6,258 dSpm transposon and 7,131 and 11,762 GABI-Kat and FLAGdb T-DNA insertion sites that were used in further analysis (Table I).
Genome Data
Release 4 of the Arabidopsis genome and its annotation data were downloaded from the FTP site of the TIGR Arabidopsis genome annotation database (Wartman et al., 2003). We used these data in the following calculations and analyses.
Calculation of Insertion Distributions in Different GC Content Regions
We partitioned the genome into deciles according to GC content across nonoverlapping 20, 30, 40, and 50-bp windows, respectively, and calculated the insertion frequencies in each compartment by dividing the number of insertion sites in each partition by the total number of basepairs in the partition.
Calculation of Insertion Distributions in Different Genome Compartments
We calculated the numbers of insertion sites in upstream regions, 5′ UTRs, coding exons, introns, 3′ UTRs, and intergenic regions in the entire genome of Arabidopsis. The number of insertion sites per megabase pair in each compartment was obtained by dividing the number of insertion sites in that compartment by the total size of the compartment. We defined the upstream region as 900 bp upstream of the translation start codon. If a 5′ UTR was annotated, we truncated the upstream region after the transcriptional start site. The 3′ UTR includes the untranslated exons and introns at the 3′ end of a gene. The intergenic region is the region between two nearby genes. The heterochromatic region used for this analysis is at the location from 1,591,342 to 2,001,638 bp on chromosome 4, which is within the interval that includes bacterial artificial chromosomes from T5L23 to T27D20 as described in the previous publication (Cold Spring Harbor Laboratory, 2000).
Motif Discovery
We used MEME (Bailey and Elkan, 1994) as a motif search tool to find candidate motifs at the sites of transposon and T-DNA insertions. For each of the 4 insertional mutagenesis agents, we generated training sets of 20-bp and 50-bp genomic sequences centered at the insertion sites in 4 specific regions, respectively. These regions include those with 50% to 60% and 60% to 70% GC content and those of 200 bp downstream of the translation start codon and of 100 bp upstream of the start codon to 300 bp. Each set was divided into two subsets. One of the two subsets was used as a training sequence set for MEME. We selected the reverse complement case, assumed 5 motifs and a motif length from 6 to 20 bp. The program generated a set of motifs. We then chose the motifs with a large number of insertion sites, low E values calculated by MEME, and simple patterns as draft candidate motifs. After that, we used the second subset to train MEME using the same parameters that we used earlier. If an output motif of MEME had the same or very similar pattern to a candidate identified during the first round, the motif was selected as a candidate motif.
To verify the candidate motifs, we used the Regulatory Sequence Analysis Tools-DNA pattern match program (Helden et al., 2000) to determine whether the motifs occur more frequently around the insertion sites than in the genome overall as described in the sequence pattern discovery algorithm (Brazma et al., 1998). As a negative control, 6 sets of 500 20-bp sequences were sampled randomly from the Arabidopsis genome. These were compared to another 6 sets of 500 20-bp sequences, centered at the insertion sites of an insertional mutagenesis agent, independent of the MEME training sets, and selected evenly from the Arabidopsis genome. For the control and the test set, we searched for the candidate motif consensus sequences using the pattern match program. We tested the significance of the differences between frequencies of motif occurrences, at least once in each 20-bp sequence, in the control and test sets using statistical test for inferences on the difference between means in independent samples (Freund and Wilson, 1993). We used a similar methodology to test for the occurrences of motif consensus sequences in specific genome compartments and at both ends of transposon flanking sequences.
Acknowledgments
We thank Robert Martienssen's group at Cold Spring Harbor Laboratory, Mike Bevan's group at John Innes Centre, UK, Bernd Weisshaar's group at the Max Planck Institute for Plant Breeding Research, Germany, the FLAGdb/FST, France, and the Institute of Molecular Agrobiology, Singapore for allowing us to use their insertional mutagenesis data. We are grateful to Peter D'Eustachio for reading this manuscript. We also thank Michael Zhang, Jonathan Clarke, and Hong Liu for helpful comments on the data analysis and thank two anonymous referees for valuable suggestions.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.053215.
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Azpiroz-Leehan R, Feldmann KA (1997) T-DNA insertion mutagenesis in Arabidopsis: going back and forth. Trends Genet 13: 152–156 [DOI] [PubMed] [Google Scholar]
- Bailey TL, Elkan C (1994) Fitting a mixture model by expectation maximization to discover motifs in biopolymers. In Proceedings of the Second International Conference on Intelligent Systems for Molecular Biology. AAAI Press, Menlo Park, CA, pp 28–36 [PubMed]
- Balzergue S, Dubreucq B, Chauvin S, Le-Clainche I, Le Boulaire F, de Rose R, Samson F, Biaudet V, Lecharney A, Cruaud C, et al (2001) Improved PCR-walking for large-scale isolation of plant T-DNA borders. Biotechniques 30: 496–504 [DOI] [PubMed] [Google Scholar]
- Bancroft I, Dean C (1993) Transposition pattern of the maize element Ds in Arabidopsis thaliana. Genetics 134: 1221–1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brazma A, Jonassen I, Vilo J, Ukkonen E (1998) Predicting gene regulatory elements in silico on a genomic scale. Genome Res 8: 1202–1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunaud V, Balzergue S, Dubreucq B, Aubourg S, Samson F, Chauvin S, Bechtold N, Cruaud C, DeRose R, Pelletier G, et al (2002) T-DNA integration into the Arabidopsis genome depends on sequences of pre-insertion sites. EMBO Rep 3: 1152–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig NL (1997) Target site selection in transposition. Annu Rev Biochem 66: 437–474 [DOI] [PubMed] [Google Scholar]
- Fedoroff N (1989) Maize transposable elements. In M Howe, D Ber, eds, Mobile DNA. American Society for Microbiology, Washington, pp 375–411
- Freund RJ, Wilson WJ (1993) Statistical Methods. Academic Press, San Diego
- Gordon MP (1998) Discovery of the T-DNA of Agrobacterium tumefaciens. In S Kung, S Yang, eds, Discoveries in Plant Biology, Vol 1. World Scientific, Singapore, pp 111–115
- Helden JV, André B, Collado-Vides J (2000) A web site for the computational analysis of yeast regulatory sequences. Yeast 16: 177–187 [DOI] [PubMed] [Google Scholar]
- James DW Jr, Lim E, Keller J, Plooy I, Ralston E, Dooner HK (1995) Directed tagging of the Arabidopsis FATTY ACID ELONGATION1 (FAE1) gene with the maize transposon Activator. Plant Cell 7: 309–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JDG, Carland FC, Lim E, Ralston E, Dooner HK (1990) Preferential transposition of the maize element Activator to linked chromosomal locations in tobacco. Plant Cell 2: 701–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koncz C, Nemeth K, Redei GP, Schell J (1992) T-DNA insertional mutagenesis in Arabidopsis. Plant Mol Biol 20: 963–976 [DOI] [PubMed] [Google Scholar]
- Kuromori T, Hirayama T, Kiyosue Y, Takabe H, Mizukado S, Sakurai T, Akiyama K, Kamiya A, Ito T, Takuya T, et al (2004) A collection of 11800 single-copy Ds transposon insertion lines in Arabidopsis. Plant J 37: 897–905 [DOI] [PubMed] [Google Scholar]
- Li Y, Rosso MG, Strizhov N, Viehoever P, Weisshaar B (2003) GABI-Kat SimpleSearch: a flanking sequence tags (FST) database for the identification of T-DNA insertion mutants in Arabidopsis thaliana. Bioinformatics 19: 1441–1442 [DOI] [PubMed] [Google Scholar]
- Liao GC, Rehm EJ, Rubin GM (2000) Insertion site preferences of the P transposable element in Drosophila melanogaster. Proc Natl Acad Sci USA 97: 3347–3351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machida C, Onouchi H, Koizumi J, Hamada S, Semiarti E, Torikai S, Machida Y (1997) Characterization of the transposition pattern of the Ac element in Arabidopsis thaliana using endonuclease I-Scel. Proc Natl Acad Sci USA 94: 8675–8680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X, Liu H, Clarke J, Jones J, Bevan M, Stein L (2003) ATIDB: Arabidopsis Thaliana insertion database. Nucleic Acids Res 31: 1245–1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parinov S, Sevugan M, Ye D, Yang W-C, Kumaran M, Sundaresan V (1999) Analysis of flanking sequences from Dissociation insertion lines: a database for reverse genetics in Arabidopsis. Plant Cell 11: 2263–2270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosso MG, Li Y, Strizhov N, Reiss B, Dekker K, Weisshaar B (2003) An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol Biol 53: 247–259 [DOI] [PubMed] [Google Scholar]
- Rubin GM, Spradling AC (1982) Genetic transformation of Drosophila with transposable element vectors. Science 218: 348–353 [DOI] [PubMed] [Google Scholar]
- Samson F, Brunaud V, Balzergue S, Dubreucq B, Lepiniec L, Pelletier G, Caboche M, Lecharny A (2002) FLAGdb/FST: a database of mapped flanking insertion sites (FSTs) of Arabidopsis thaliana T-DNA transformants. Nucleic Acids Res 30: 94–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessions A, Burke E, Presting G, Aux G, McElver J, Patton D, Dietrich B, Ho P, Bacwaden J, Ko C, et al (2002) The high-throughput Arabidopsis reverse genetics system. Plant Cell 14: 2985–2994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D, Yanai Y, Liu Y-G, Ishiguro S, Okada K, Shibata D, Whittier RF, Fedoroff NV (1996) Characterization and mapping of Ds-GUS-T-DNA lines for targeted insertional mutagenesis. Plant J 10: 721–732 [DOI] [PubMed] [Google Scholar]
- Sundaresan V, Springer P, Volpe T, Haward S, Jones JD, Dean C, Ma H, Martienssen R (1995) Patterns of gene action in plant development revealed by enhancer trap and gene trap transposable elements. Genes Dev 9: 1797–1810 [DOI] [PubMed] [Google Scholar]
- Szabados L, Kovacs I, Oberschall A, Abraham E, Kerekes I, Zsigmond L, Nagy R, Alvarado M, Krasovskaja I, Gal M, et al (2002) Distribution of 1000 sequenced T-DNA tags in the Arabidopsis genome. Plant J 32: 233–242 [DOI] [PubMed] [Google Scholar]
- The Cold Spring Harbor Laboratory, Washington University Genome Sequencing Center, and PE Biosystems Arabidopsis Sequencing Consortium (2000) The complete sequence of a heterochromatic island from a higher eukaryote. Cell 100: 377–386 [PubMed] [Google Scholar]
- Thomas CM, Jones DA, English JJ, Carroll BJ, Bennetzen JL, Harrison K, Burbidge A, Bishop GJ, Jones JD (1994) Analysis of the chromosomal distribution of transposon-carrying T-DNAs in tomato using the inverse polymerase chain reaction. Mol Gen Genet 242: 573–585 [DOI] [PubMed] [Google Scholar]
- Tinland B (1996) The integration of T-DNA into plant genomes. Trends Plant Sci 1: 178–183 [Google Scholar]
- Tissier AF, Marillonnet S, Klimyuk V, Patel K, Torres MA, Murphy G, Jones JDG (1999) Multiple independent defective Suppressor-mutator transposon insertions in Arabidopsis: a tool for functional genomics. Plant Cell 11: 1841–1852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigdal TJ, Kaufman CD, Izsvak Z, Voytas DF, Ivics Z (2002) Common physical properties of DNA affecting target site selection of sleeping beauty and other Tc1/mariner transposable elements. J Mol Biol 323: 441–452 [DOI] [PubMed] [Google Scholar]
- Wartman JR, Haas BJ, Hannick LI, Smith RK Jr, Maiti R, Ronning CM, Chan AP, Yu C, Ayele M, Whitelaw CA, et al (2003) Annotation of the Arabidopsis genome. Plant Physiol 132: 461–468 [DOI] [PMC free article] [PubMed] [Google Scholar]