Abstract
Ser acetyltransferase (SATase; EC 2.3.1.30) catalyzes the formation of O-acetyl-Ser from l-Ser and acetyl-CoA, leading to synthesis of Cys. According to its position at the decisive junction of the pathways of sulfur assimilation and amino acid metabolism, SATases are subject to regulatory mechanisms to control the flux of Cys synthesis. In Arabidopsis (Arabidopsis thaliana) there are five genes encoding SATase-like proteins. Two isoforms, Serat3;1 and Serat3;2, were characterized with respect to their enzymatic properties, feedback inhibition by l-Cys, and subcellular localization. Functional identity of Serat3;1 and Serat3;2 was established by complementation of a SATase-deficient mutant of Escherichia coli. Cytosolic localization of Serat3;1 and Serat3;2 was confirmed by using fusion construct with the green fluorescent protein. Recombinant Serat3;1 was not inhibited by l-Cys, while Serat3;2 was a strongly feedback-inhibited isoform. Quantification of expression patterns indicated that Serat2;1 is the dominant form expressed in most tissues examined, followed by Serat1;1 and Serat2;2. Although Serat3;1 and Serat3;2 were expressed weakly in most tissues, Serat3;2 expression was significantly induced under sulfur deficiency and cadmium stress as well as during generative developmental stages, implying that Serat3;1 and Serat3;2 have specific roles when plants are subjected to distinct conditions. Transgenic Arabidopsis plants expressing the green fluorescent protein under the control of the five promoters indicated that, in all Serat genes, the expression was predominantly localized in the vascular system, notably in the phloem. These results demonstrate that Arabidopsis employs a complex array of compartment-specific SATase isoforms with distinct enzymatic properties and expression patterns to ensure the provision of Cys in response to developmental and environmental changes.
Sulfur is an essential macronutrient in the plant life cycle. After reduction, inorganic sulfate is integrated in organic compounds via Cys biosynthesis. This pathway plays a central role in sulfur assimilation, and it involves several enzymatic reactions (Leustek and Saito, 1999). Ser acetyltransferase (SATase; EC 2.3.1.30), which catalyzes the formation of O-acetyl-l-Ser (OAS) from l-Ser and acetyl-CoA, links the Ser metabolism to Cys biosynthesis (Leustek and Saito, 1999; Saito, 2004). Subsequently, Cys is formed by the condensation of sulfide and OAS, catalyzed by Cys synthase (OAS [thiol] lyase; EC 4.2.99.8). This final step of Cys biosynthesis exists in the three major compartments of plant cells: cytosol, chloroplasts, and mitochondria. SATase (Smith, 1972; Ascano and Nicholas, 1977; Brunold and Suter, 1982; Ruffet et al., 1995) and Cys synthase (Brunold and Suter, 1982; Lunn et al., 1990; Droux et al., 1992; Rolland et al., 1992; Yamaguchi and Masada, 1995; Kuske et al., 1996) have been demonstrated in these three compartments from several plants.
SATase plays a regulatory role in whole sulfur assimilation through the production of OAS, which is a positive key metabolite in the Cys biosynthetic pathway (Saito, 2000). Regarding regulation of SATase, two major mechanisms are proposed: allosteric feedback inhibition of SATase activity by l-Cys and modulation of SATase activity through reversible formation of a protein complex with Cys synthase (Leustek et al., 2000; Saito, 2000; Hell and Hillebrand, 2001).
The presence of feedback regulation in SATase isoforms differs with plant species and subcellular compartments. Feedback inhibition by Cys has been reported with the watermelon (Citrullus vulgaris) cytosolic SATase (Saito et al., 1995), the Arabidopsis (Arabidopsis thaliana) cytosolic isoform (Howarth et al., 1997; Noji et al., 1998), the cytosolic isoform ASAT5 from Chinese chive (Allium tuberosum; Urano et al., 2000), the plastidic isoform from spinach (Spinacea oleracea; Noji et al., 2001), the pea (Pisum sativum) plastidic isoform (Droux, 2003), and the soybean (Glycine max) cytosolic isoform (Chronis and Krishnan, 2004). In contrast, insensitivity to Cys inhibition has been reported with the plastidic/cytosolic and mitochondrial SATases from Arabidopsis (Noji et al., 1998) and cytosolic and mitochondrial isoforms from pea (Droux, 2003). The transcriptional regulation of mRNA levels in response to sulfur nutrition was not notable in watermelon and Arabidopsis (Saito et al., 1995; Takahashi, et al., 1997). These findings suggest that the feedback inhibition is an important regulatory mechanism for OAS levels. However, only a few cDNAs of SATase have been investigated for their subcellular localization and feedback inhibition by l-Cys.
In the Arabidopsis genome, five genes putatively encode SATase (Hell et al., 2002). Until now, of these five SATase isoforms, only three have been well characterized with respect to their biochemical characterization and subcellular localization (Noji et al., 1998). The fourth isoform Serat3;1 was partially characterized recently (Howarth et al., 2003). In this study, we present the completion of molecular cloning for the SATase gene family and a comprehensive biochemical characterization of two family members Serat3;1 and Serat3;2, including their subcellular localization. Furthermore, the expression analysis of the entire SATase-like gene family revealed differential expression patterns and intensities during plant development and under stress conditions.
RESULTS
Phylogenic Relation and Clustering of SATase-Like Gene Family
The investigation of Arabidopsis Genome Initiative reveals the presence of five putative SATase-like genes. Each isoform localizes to one of the five chromosomes in the genome (Hell et al., 2002). cDNAs of four SATase isoforms, SAT-c (Howarth et al., 1997; Noji et al., 1998), SAT-p (Murillo et al., 1995; Ruffet et al., 1995; Noji et al., 1998), SAT-m (Bogdanova et al., 1995; Hell and Bogdanova, 1995; Roberts and Wray, 1996; Noji et al., 1998), and SAT-106 (Howarth et al., 2003), were cloned. The fifth gene (At4g35640) is the only family member that has not been verified functionally by complementation using a SAT-deficient cysE mutant of E. coli.
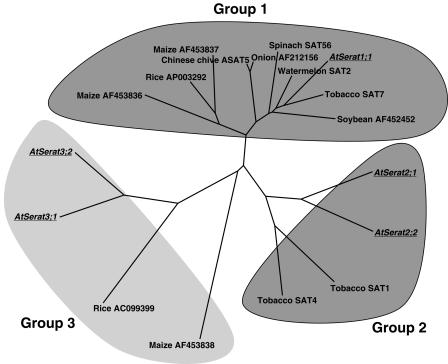
Until now, no systematic nomenclature for SATase-like proteins in plants has been proposed. In particular, Arabidopsis SATase genes are referred to with several different names that would potentially lead to unnecessary confusion. The phylogenetic tree was calculated using the amino acid sequences of 18 members of the plant SATase-like gene family available in the databases (Fig. 1). SATase-like proteins are divided into at least three subfamilies assigned as groups 1, 2, and 3. We have designated the Arabidopsis SATase-like gene family gene as AtSerat (Arabidopsis thaliana serine acetyltransferase-like protein) with three subfamilies (Serat1∼3) according to the grouping of the phylogenic tree (Table I). The described five Arabidopsis isoforms within this nomenclature are: Serat1;1 (At5g56760, SAT-c), Serat2;1 (At1g55920, SAT-p), Serat2;2 (At3g13110, SAT-m), Serat3;1 (At2g17640, SAT-106), and Serat3;2 (At4g35640; Fig. 1).
Figure 1.
Molecular phylogenic tree of the amino acid sequences of SATase-like proteins. The tree was constructed based on the alignment of the full-length sequences using ClustalW program. Arabidopsis Serat isoforms are underlined. Gene designation and accession numbers are in .
Table I.
Gene designation of SATases from higher plants
| Group | Plant Species | Gene Designation | Gen Bank Accession No. | Description | Reference |
|---|---|---|---|---|---|
| 1 | Arabidopsis thaliana | AtSerat1;1 | U30298 | cDNA encoding a SATase isoform named SAT52; SAT-c; and SAT5. At5g56760 | Howarth et al. (1997); Noji et al. (1998); This paper |
| Nicotiana tabacum | AJ414053 | cDNA encoding a SATase isoform named SAT7. | Wirtz and Hell (2003) | ||
| Citrullis vulgaris | D49535, L34076 | cDNA encoding a SATase isoform named SAT2. | Saito et al. (1995) | ||
| Spinacea oleracea | D88529 | cDNA encoding a SATase isoform named SAT56. | Noji et al. (2001) | ||
| Glycine max | AF452452 | cDNA encoding a cytosolic SATase isoform. Accession AF452452. | Chronis and Krishnan (2004) | ||
| Allium tuberosum | AB040502, AB040503 | cDNA encoding a SATase isoform named ASAT5. | Urano et al. (2000) | ||
| Allium cepa | AF212156 | Pither-Joyce, M.D., and McCallum, J.A., 1999, submitted in EMBL accession AF212156. | |||
| Zea mays | AF453837 | Bolchi, A., and Petrucco, S., 2001, submitted in EMBL accession AF453837. | |||
| Z. mays | AF453836 | Petrucco, S., and Bolchi, A., 2001, submitted in EMBL accession AF453836. | |||
| Oryza sativa | AP003292 | Sasaki, T., et al., 2002, cited in EMBL accession AP003292. | Sasaki et al. (2002) | ||
| 2 | A. thaliana | AtSerat2;1 | L42212, Z34888 | cDNA encoding a SATase isoform named SAT5; SAT-B; SAT-p; and SAT1. At1g55920 | Murillo et al. (1995); Ruffet et al. (1995); Noji et al. (1998); This paper; |
| A. thaliana | AtSerat2;2 | X82888, U22964 | cDNA encoding a SATase isoform named SAT1-6; SAT-A; Sat-1; SAT-m; and SAT3. At3g13110 | Bogdanova et al. (1995); Hell and Bogdanova (1995); Roberts and Wray (1996); Noji et al. (1998); This paper | |
| N. tabacum | AJ414051 | cDNA encoding a SATase isoform named SAT1. | Wirtz and Hell (2003) | ||
| N. tabacum | AJ414052 | cDNA encoding a SATase isoform named SAT4. | Wirtz and Hell (2003) | ||
| 3 | A. thaliana | AtSerat3;1 | AF112303 | cDNA encoding a SATase isoform named SAT106. At2g17640 | Howarth et al. (2003); This paper |
| A. thaliana | AtSerat3;2 | AF331847 | Yamada, K., et al. 2003, submitted in EMBL accession AF331847, cDNA named SAT4. At4g35640 | This paper | |
| O. sativa | AC099399 | Wing, R.A., et al., 2002, submitted in EMBL accession AC099399. | |||
| Z. mays | AF453838 | Petrucco, S., and Bolchi, A., 2001, submitted in EMBL accession AF453838. |
cDNA Cloning of a New Member Serat3;2
The previously uncharacterized SATase-like cDNA-encoding Serat3;2 isoform was isolated based on the information of the Arabidopsis genome sequence using 3′-,5′-RACE PCR. In the database, two distinct sequences for Serat3;2 were found, CAB80280 (Howarth et al., 2003) and AF331847. The first 203 deduced amino acids in the sequence were conserved and only differed in the C-terminal region. For the deduced sequence of CAB80280 the last four amino acids were HGES, whereas in the AF331847 sequence the last four amino acids were ERRH. To clarify this discrepancy, several independent clones were sequenced and the correct C terminus agreed with the deduced sequence of AF331847. Sequence analysis revealed an open reading frame of 1,068 nucleotides, encoding for 355 amino acid residues. The first ATG triplet, which is 54 nucleotides away from the 5′-end of Serat3;2, was assigned as the translational start point because the sequence around the Met codon (TTTGGTCATGGC) matches the consensus sequence for the start codon of plant genes (Lütcke et al., 1987). A 3′-untranslated region of 192 nucleotides downstream of the translation stop codon is present in the cDNA sequence.
The deduced amino acid sequence of Serat3;2 was aligned with other SATases from Arabidopsis. The homology with the other SATases is high throughout the central region. The phylogenetic tree (Fig. 1) indicates that Serat3;2 is closely related to Serat3;1, and both are separated from other SATases forming a unique group different from those characterized previously.
Functional Complementation of an E. coli Mutant with Serat3;1 and Serat3;2
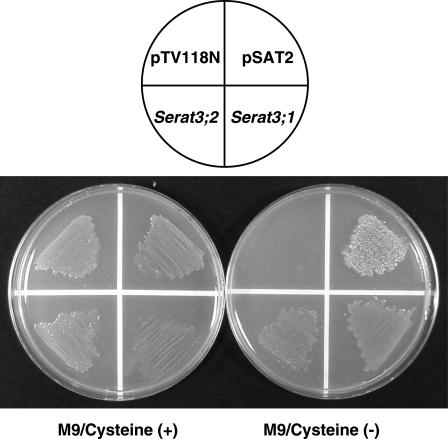
The functional identity of the isolated cDNA, Serat3;2, together with Serat3;1 was confirmed by successful complementation with an E. coli Cys-auxotrophic mutant lacking an endogenous SATase activity. Bacterial expression vectors, pSerat3;1 and pSerat3;2, carrying the coding regions of Serat3;1 and Serat3;2 under the transcriptional control of a lacZ promoter of pTV118N were constructed. E. coli JM39/5 transformed with these vectors was able to grow on M9 minimal medium without Cys, in a similar manner as pSAT2 expressing watermelon SATase (Saito et al., 1995), whereas E. coli transformed with an empty vector, pTV118N, could not grow without supplementation of Cys (Fig. 2). This indicates the authenticity of Serat3;1 and Serat3;2 encoding the functional SATase.
Figure 2.
Functional complementation of E. coli Cys-auxotroph by expression of Serat3;1 and Serat3;2. Genetic complementation of Cys− E. coli JM39/5 by transformation with expression plasmids, pSerat3;1 and pSerat3;2, carrying Serat3;1 and Serat3;2 cDNAs from Arabidopsis. Transformed bacteria were spread on M9 minimal agar plates with 0.5 mm l-Cys (left plate) or without l-Cys (right plate). pSAT2-carrying SATase gene of Chinese chive (Saito et al., 1995) was used as a positive control. pTV118N is the vector background as a negative control.
Catalytic and Regulatory Properties of Serat3;1 and Serat3;2
Using purified recombinant proteins, the catalytic and regulatory properties of Serat3;1 and Serat3;2 were investigated. The cDNAs were inserted in frame downstream from the malE gene of E. coli, which encodes a maltose-binding protein. With this system, the purified proteins of Serat3;1 and Serat3;2 were obtained by the cleavage of the maltose-binding protein with factor Xa. The recombinant proteins were visualized on SDS-PAGE gel as the expected 35-kD (Serat3;1) and 38-kD (Serat3;2) recombinant proteins in the soluble fraction. The Km values for Serat3;1 and Serat3;2 were determined by OAS formation and detected by HPLC. The Km values of these two SATases for l-Ser and acetyl-CoA were much higher than those of the three previously reported isoforms (Noji et al., 1998; Table II), indicating low catalytic efficiencies of Serat3;1 and Serat3;2 for both substrates.
Table II.
Kinetic constants of recombinant Arabidopsis Serat proteins
| Isoforms
|
Feedback Inhibition by l-Cys
|
Subcellular Localization
|
Km (mm)
|
Inhibition (Ki) by l-Cys (μm)
|
Reference
|
||
|---|---|---|---|---|---|---|---|
| L-Ser | Acetyl-CoA | L-Ser | Acetyl-CoA | ||||
| Serat1;1 | Sensitive | Cytosol | 2.71 | 0.28 | 10.8 (Noncompetitive) | 7.4 (Competitive) | Noji et al. (1998) |
| Serat2;1 | Insensitive | Chloroplast/cytosol | 1.64 | 0.16 | – | – | Noji et al. (1998) |
| Serat2;2 | Insensitive | Mitochondrion | 1.68 | 0.02 | – | – | Noji et al. (1998) |
| Serat3;1 | Insensitive | Cytosol | 121.4 | 24.5 | – | – | This paper |
| Serat3;2 | Sensitive | Cytosol | 39.5 | 45.1 | 17.3 (Noncompetitive) | 2.5 (Competitive) | This paper |
–, No inhibition by l-cysteine.
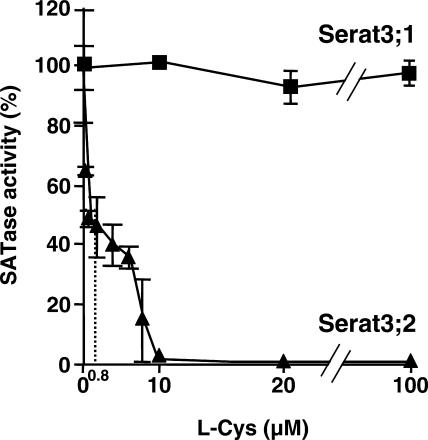
The feedback inhibition by l-Cys on SATase reaction was analyzed using purified recombinant proteins. The activity of Serat3;2 was inhibited by l-Cys in a non-competitive manner to l-Ser and in a competitive manner to acetyl-CoA, with Ki values of 17.3 μm and 2.5 μm, respectively. The concentration for 50% inhibition (IC50) was 0.8 μm, being similar to Serat1;1 (1.8 μm; Noji et al., 1998), spinach SATase (7.6 μm; Noji et al., 2001), and watermelon SATase (2.9 μm; Saito et al., 1995). In contrast to Serat3;2, Serat3;1 was insensitive to feedback inhibition by l-Cys (Fig. 3).
Figure 3.
Feedback inhibition of recombinant Arabidopsis Serat3;1 and Serat3;2 by l-Cys. Assay was carried out as described in “Materials and Methods.” IC50 for Serat3;2 is determined as 0.8 μm. Data are the means of triplicate analyses ± sd.
Cytosolic Localization of Serat3;1 and Serat3;2
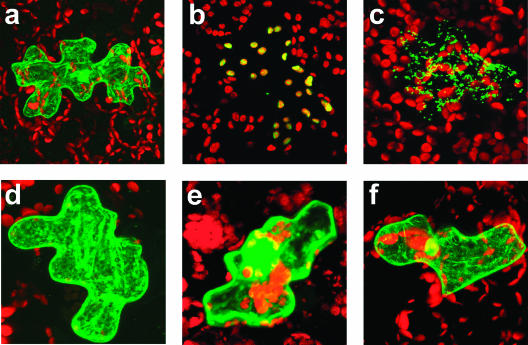
The subcellular localization of Serat3;1 and Serat3;2 was determined in Arabidopsis cells using fusion proteins of green fluorescent protein (GFP) with the N-terminal peptides of these proteins. The N-terminal regions of Serat3;1 (Met-1 to His-100) and Serat3;2 (Met-1 to Leu-100) were translationally fused to GFP, respectively. These fusion genes were introduced into Arabidopsis leaves by particle bombardment (Fig. 4). As controls, we used fusions to the N terminus of GFP with Serat1;1, the transit peptide of pea Rubisco small subunit and the transit peptide of Arabidopsis Ser hydroxymethyl transferase (SHMT) for cytosolic, plastidic, and mitochondrial localization, respectively. Results in Figure 4, E and F, indicated the cytosolic localization of the GFP fusion proteins with two proteins, indicating both Serat3;1 and Serat3;2 are localized in cytosol. These localizations were independently confirmed by particle bombardment of tobacco (Nicotiana tabacum) leaves (data not shown).
Figure 4.
Subcellular localization of GFP fused to the N-terminal sequences of Arabidopsis Serat proteins. Particle bombardment with chimeric GFP fusion genes was carried out with 3-week-old Arabidopsis leaves. The expression and localization of GFP was observed after 16-h incubation by confocal laser scanning microscopy. A, Plasmid pFF19-GFP used as a control for localization in cytosol and nucleus; B, GFP-carrying transit peptide of the pea ribulose-1,5-bisphophate carboxylase small subunit as a control for chloroplast translocation. C, SHMT-GFP carrying transit peptide of SHMT from Arabidopsis as a control for mitochondrion targeting; D, Serat1;1-GFP localized in cytosol as a control; E, Serat3;1-GFP localized in cytosol; F, Serat3;2-GFP also localized in cytosol.
Promoter Studies in Transgenic Arabidopsis
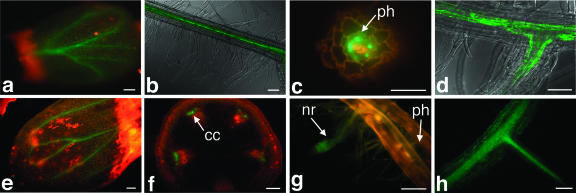
The cell type-specific expression of Serat genes was studied in transgenic Arabidopsis plants transformed with fusion gene constructs of the Serat gene promoters (2,500 bp) and GFP gene. For each construct, three to six independent transgenic lines were analyzed with fluorescent confocal microscopy. Figure 5 shows GFP expression in Arabidopsis lines transformed with the five Serat promoter-GFP constructs. GFP expression with all Serat genes was generally found in the vascular tissues of leaves and roots (Fig. 5, A, B, D, E, and H), preferentially in the phloem (Fig. 5, C and F). A relatively weak GFP expression was also observed in all photosynthetic tissues such as leaf mesophyll cells (Fig. 5, A and E). In addition, Serat1;1-GFP could be seen in the calyx of flowers and the cortex in transverse sections of mature root (data not shown). The expression of GFP in root epidermis and cotyledons, root hair, and the calyx of flowers was found with Serat2;1, Serat2;2, and Serat3;2 (data not shown). With Serat3;1, GFP fluorescence was visualized in nascent roots (Fig. 5G).
Figure 5.
Fluorescence microscopy image of Serat promoter-GFP fusion gene constructs expressed in Arabidopsis. Three-week-old plants were analyzed as described in “Materials and Methods.” A, Serat1;1, cotyledon; B, Serat1;1, longitudinal view of root; C, Serat2;1, cross-section of the mature part of root; D, Serat2;1, junction of a lateral root; E, Serat2;2, cotyledon; F, Serat2;2, cross-section of stem; G, Serat3;1, junction of a lateral root; H, Serat3;2, junction of a lateral root. ph, Phloem; cc, companion cell; nr, nascent root. Scale bars = 100 μm.
Developmental and Stress-Inducible Expression of Serat Genes
The mRNA abundance of the Serat genes was examined by northern blotting of RNA from 3-week-old leaves (data not shown). A high level of expression was observed for Serat1;1, Serat2;1, and Serat2;2. In contrast, the expression of Serat3;1 and Serat3;2 was low, suggesting distinct expression patterns among the Serat genes.
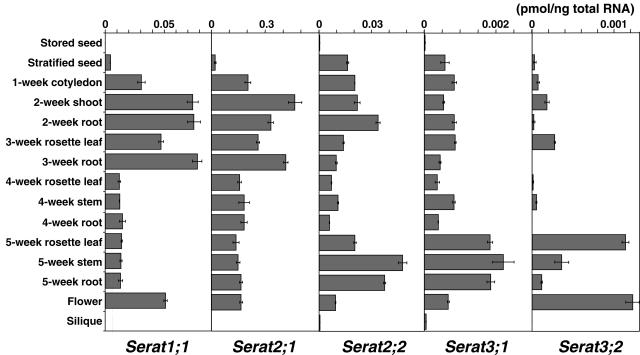
Because of the low expression of Serat3;1 and Serat3;2, further analysis was conducted with the more sensitive method of real-time quantitative PCR (Figs. 6 and 7). Serat1;1, Serat2;1, and Serat2;2 expressed higher amounts of mRNA (approximately 10-fold) compared with Serat3;1 and Serat3;2, consistent with the results of northern blotting. Among them, Serat2;1 was the dominant form in most tissues examined, followed by Serat1;1 and Serat2;2. Each gene exhibited distinct expression patterns during plant development (Fig. 6). Serat1;1 and Serat2;1 showed a similar pattern, with an increase in expression at the vegetative stage (up to 3 weeks). During the reproductive phase (after 4 weeks), the expression levels of both genes were significantly reduced. Serat2;2 showed a different pattern, in which the expression increased over the first 2 weeks of growth, then decreased after 4 weeks to a lower level than at the start of the experiment. Expression increased after 4 weeks and reached a maximum value after 5 weeks. Interestingly, Serat3;1 and Serat3;2 had similar expression patterns but were distinctly different from other genes, showing expression at low levels in young stages and increased expression at the late stage after 5 weeks.
Figure 6.
Real-time quantitative PCR analysis of Serat mRNA accumulation during development. Total RNA (5 ng) isolated from stored seeds, stratified seeds, cotyledons, leaves, stems, roots, siliques, and flower tissues was used for amplification. Data are the means of triplicate determinations ± sd (bars).
Figure 7.
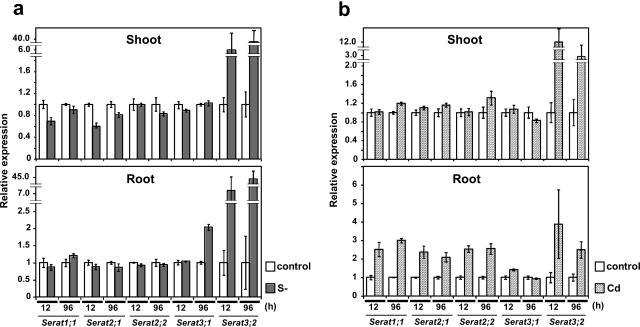
Real-time quantitative PCR analysis of Serat mRNA accumulation under stress conditions. A, Sulfur deficiency; B, Cd treatment. The experiment was performed using 5 ng of total RNA isolated from 3-week-old plants after 12 and 96 h after transferring the plants to sulfur deficient medium and in medium containing CdCl2 (50 μm). Data are the means of triplicate determinations ± sd (bars). White, Control; gray, sulfur deficiency; hatched, Cd treatment.
Expression of each Serat gene was further analyzed under the stress conditions of sulfur nutritional deficiency and cadmium (Cd) exposure (Fig. 7). Remarkably, under sulfur-deficient conditions, the mRNA levels of the three genes, Serat1;1, Serat2;1, and Serat2;2, did not change in both roots and shoots. Serat3;1 expression increased 2-fold at 96 h only in the roots. In contrast, Serat3;2 expression increased at 12 h (6–7-fold) and 96 h (40–45-fold) in both the roots and shoots, indicating that Serat3;2 may play a specific role under conditions of sulfur deficiency.
In the roots of plants treated with Cd (50 μm), expression of all the genes increased except for Serat3;1. Notably Serat3;2 expression was increased in the roots (4-fold after 12 h) and in the shoots (12-fold after 12 h) by Cd treatment. In contrast, expression of the other Serat genes did not increase in the shoots.
DISCUSSION
Completion of genome sequencing (Arabidopsis Genome Initiative, 2000) allowed the identification of all members of the Serat gene family in the Arabidopsis genome. In this study, all five SATase-like proteins encoded by Serat genes were characterized regarding different sensitivity to Cys feedback regulation, subcellular localization, and expression of mRNA during development and under stress.
The five Serat genes of Arabidopsis are each located on different chromosomes. Three of these, Serat1;1, Serat2;1, and Serat2;2, have been previously characterized regarding their differences for Cys-feedback regulation of the enzymatic activity and subcellular localization (Noji et al., 1998). The activity of cytosolic form (Serat1;1) was inhibited by l-Cys at a physiological concentration in an allosteric manner, but the plastidic/cytosolic (Serat2;1) and mitochondrial (Serat2;2) forms were not subject to this feedback regulation. The cytosolic Serat1;1 regulates the OAS concentration strictly by the feedback inhibition by Cys, since OAS is not only the key intermediate of Cys formation but also regarded as a regulatory molecule of the whole sulfur assimilation pathway (Leustek et al., 2000; Saito, 2000, 2004). In chloroplasts, however, high OAS levels have to be maintained even at high Cys concentrations for the biosynthesis of Met and glutathione by feedback-insensitive SATase isoforms in this compartment (Noji et al., 1998). Nevertheless, chloroplastic isoforms from spinach (Noji et al., 2001) and pea (Droux, 2003) are sensitive to Cys, suggesting various species-dependent relations of feedback inhibition and subcellular compartmentation. Cytosolic localization of Serat3;1 and Serat3;2 was indicated by the analyses of the GFP fusion proteins directed by the transit peptides of these proteins. Surprisingly, Serat3;2 and Serat3;1 showed contrasting properties with respect to sensitivity of Cys-feedback inhibition. However, their low substrate affinities suggest low enzyme activities in vivo. From these findings, a minor role of these two proteins in flux regulation toward Cys synthesis may be inferred under nonstressed conditions, but the strong induction, especially of the Cys-sensitive Serat3;2 in response to sulfate deficiency and Cd application, indicates a possible contribution to the regulation of Cys synthesis, while Serat3;1, the Cys-insensitive cytosolic isoform, probably does not significantly regulate OAS availability.
The 18 SATase-like proteins deduced from the DNA sequences in the database can be grouped into at least three groups as shown in Figure 1. As far as reported in the literature, the isoforms in group 1 are sensitive to feedback regulation by Cys. However, subcellular localization is not perfectly correlated to grouping, though most of group 1 isoforms are cytosolic protein and group 2 members are localized in organelles. In contrast to high-affinity property of the isoforms in groups 1 and 2 to the substrates, two proteins in group 3, Serat3;1 and Serat3;2, exhibited low affinity. Nevertheless, their enzymatic activities are sufficient to support the growth of the E. coli mutant. These results leave open the possibility that the OAS-producing activities of Serat3;1 and Serat3;2 might be side activities of those proteins that may have another unknown function(s) similar to Cys synthase and β-cyanoalanine synthase (Hatzfeld et al., 2000; Warrilow and Hawkesford, 2000).
The study of Serat promoter-GFP fusion constructs indicated for all Serat genes a more preferential expression in vascular tissues, in particular in phloem, than in photosynthetic tissues of cotyledons, in hypocotyls or in roots. There is still the possibility that cis-elements directing the chromosomal expression of Serat genes might be missing in our constructs or cis-elements of adjacent genes might be included. For more detailed investigations of the tissue-specific localization of Serat proteins, immunolocalization studies using isoform-specific antibodies or in situ hybridization experiments with specific probes will be necessary in the future. However, the expression in vascular tissues agrees with the strong synthetic activity of Cys in maize (Zea mays) bundle sheath cells (Burgener et al., 1998). Also in the dicot plants, the vascular tissues may need to produce Cys intensively. The promoter of Cys synthase-B gene (oasB) from Arabidopsis (Jost et al., 2000) fused to the β-glucuronidase gene expressed strongly in vascular tissues, though the Cys synthase-A gene (oasA) expressed ubiquitously (R. Jost and R. Hell, unpublished data). In rice (Oryza sativa), high amounts of thioredoxin-h mRNA was accumulated in the phloem sap (Sasaki et al., 1998). Thioredoxin is a disulfide-containing small protein that has been isolated from almost all organisms (Verdoucq et al., 1999) and plays a key role in a variety of redox reactions. High level of Serat mRNAs may be required to fulfill the high demand of Cys supply in phloem for the production of large amounts of thioredoxins. Cys may also be required in elevated amounts because the phloem is known to carry glutathione, one of the major downstream products of Cys, to sink tissues (Herschbach, 2003).
Expression analysis of the five Serat genes revealed the gene-specific unique regulation during the course of plant development, and by nutritional and Cd stresses. Serat genes exhibited two types of expression patterns during plant development. The three major genes, Serat1;1, Serat2;1, and Serat2;2, had a similar trend of expression increasing until 3 weeks, whereas Serat3;1 and Serat3;2 showed an opposite pattern of expression increasing even in the reproductive stage (5–6 weeks). Expression of Serat genes is therefore switched from group 1 (Serat1;1, Serat2;1, and Serat2;2) to group 2 (Serat3;1 and Serat3;2) by the developmental transition from the vegetative stage to the reproductive stage, for provision of OAS and subsequently Cys in parallel to the export of Cys and glutathione from source tissues to the growing shoots and seeds.
Under sulfur-starved conditions, the expression of Serat3;2 was induced in both roots and shoots, indicating a special role of Serat3;2 under sulfur starvation. The induction of Serat3;2 expression was also indicated by a DNA microarray study (Hirai et al., unpublished data). In addition, Serat3;1 expression was induced after long-term sulfur starvation. No induction was observed with the other genes. It is known that OAS levels increase by sulfur starvation (Kim et al., 1999). The induced expression of Serat3;1 and Serat3;2 can partially contribute to the increasing level of OAS by sulfur depletion. Similar trends of induction of gene expression were seen by Cd stress. Serat3;2 responded to Cd treatment both in roots and shoots, indicating again the particular function of Serat3;2 during the stress response. It may be speculated that the strong induction of the Serat3;2 gene compensates for the low catalytic efficiency of the encoded Serat3;2 protein, thus contributing significantly to the flux of Cys synthesis toward glutathione and ultimately to phytochelatins for Cd complexation. The other Serat genes were slightly induced in the roots. Increased heavy metal tolerance was confirmed in the transgenic plants overexpressing the Cys synthase gene (Dominguez-Solis et al., 2001; Kawashima et al., 2004). These studies suggest that the increased Cys and glutathione contents in these transgenic plants are likely to be responsible for tolerance to heavy metals. Since Serat3;2 is inducible by Cd, a more detailed investigation on the induction and mechanism of tolerance would be applicable for further study and engineering of Cd resistance in plants. It is concluded that Arabidopsis employs a family of compartment-specific Serat proteins with different kinetic properties and expression patterns that support the developmental and stress-related control of Cys biosynthesis throughout the plant.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana; Columbia ecotype) plants were grown on germination medium agar medium (Valvekens et al., 1988) at 22°C under 16-h-light (approximately 2,500 lux) and 8-h-dark cycles. After 3 to 5 weeks, the plants were used either for isolation of total RNA or for particle bombardment.
Isolation of Serat3;2 cDNA
For the isolation of Serat3;2 cDNA, the specific primers were designed based on the sequences reported in the database (GenBank accession nos. AF331847 and CAB80280) and used for 3′- and 5′-RACE PCR using a Smart RACE PCR cDNA amplification kit (Clontech, Tokyo, Japan). The first and second primer sets used for the nested PCR were respectively: 5′-CTCCTCTTCTGCTTCAGGCTTAGCTTCT-3′ and 5′-CTACCTTGTTGGCAACTCAGGTTATGGA-3′; 5′-CAGTCATGCGACAAGATACTAGATACA-3′ and 5′-TGCAACGTTATGGTACATGACAGAGTA-3′. The RACE products were cloned into pGEM-T easy vector system (Promega, Tokyo, Japan). Sequencing of full length DNA was carried out on both strands.
Functional Complementation of Escherichia coli Mutant
Functional complementation by Serat3;1 and Serat3;2 was carried out with a Cys-auxotrophic E. coli JM39/5 (F+, cysE51, recA56) lacking cysE locus encoding endogenous SATase. The primers for cDNA amplification were: AGAGCCATGGATGGCGATGAGCTTCC-3′ and 5′-CATGCCATGGTCATGATGTGCTGTTAGTG-3′ for Serat3;1; and 5′-CAGTCAGTCGACGTCATGGCTTGTATAAACGGCG-3′ and 5′-CAGTCAGGATCCCTTTTTAATGTCTCCTTTCC-3′ for Serat3;2. The amplified fragments were cloned into pTV118N for functional expression in E. coli. For genetic complementation of the Cys requirement, the transformed E. coli was cultured on M9 minimal medium (Sambrook et al., 1989) containing 100 μg/mL amplicilin and 500 μm IPTG at 37°C for 1 week. As a positive control pSAT2 carrying SATase gene of watermelon (Citrullus vulgaris) and pTV118N vector as a negative control, respectively, were used.
Isolation of Recombinant Serat3;1 and Serat3;2 Proteins
The NcoI sites were created on both ends of the coding region of Serat3;1 and Serat3;2 by PCR engineering using primers: Serat3;1, 5′-AGAGCCATGGATGGCGATGAGCTTCC-3′ and 5′-CATGCCATGGTCATGATGTGCTGTTAGTG-3′; and Serat3;2, 5′-CAGTCAGGATCCATGGCTTGTATAAACGGC-3′ and 5′-CAGTCAGTCGACTTAATGTCTCCTTTCCCT-3′. The engineered cDNA fragments were inserted into the NcoI site of pMAL-2 (New England Biolabs, Beverly, MA). The cloned Serat genes were overexpressed in E. coli NovaBlue (endA1 hsdR17 supE44 thi-1 recA1 gyrA96 relA1 lac; Novagen, Darmstadt, Germany). The gene under the tac promoter was induced by isopropyl thio-β-d-galactosidase (IPTG). Overnight cultures of E. coli NovaBlue carrying the expression vector were diluted 100-fold in a rich medium (10 g tryptone, 5 g yeast extract, 5 g NaCl, 5 g Glc) containing ampicillin (100 μg/mL), and incubated at 37°C, until the culture reached A600 approximately 0.5 (approximately 3 h). Subsequently, protein expression was induced by addition of 1 mm IPTG, followed by further 3 h incubation time at 37°C. The cells were harvested by centrifugation, resuspended in 25 mL column buffer containing 1 m Tris-HCl (pH 7.4), 11.7 g NaCl, 0.5 m EDTA, and 0.7 mL β-mercaptoethanol, and then disrupted by sonication (30 times at 60 W for 15 s each, on ice). After a second centrifugation, the supernatant was loaded on an amylose resin column for affinity purification. The fusion protein was cleaved on the column with a Factor Xa, and the completely purified protein was eluted with a buffer containing 10 mm maltose.
Assay of SATase Enzymatic Activity
The SATase reaction mixture contained 50 mm Tris-HCl (pH 8.0), 1 mm acetyl-CoA, 10 mm l-Ser, and the purified protein in a final volume of 100 μL. The reaction was initiated by the addition of l-Ser and proceeded for 1 h incubation at 30°C. The reaction was stopped by addition of 10 μL of 7.5% TCA. SATase activity was determined by the production of OAS, that was derivatized with o-phthaldialdehyde and then determined in the reverse-phase HPLC system as described previously (Droux et al., 1998).
Subcellular Localization by Fusion Protein with GFP
The first 300 bp of the ORFs were PCR amplified and cloned into the SacI/XbaI sites of pFF19-GFP (kindly provided by A. Wachter, Heidelberg Institute for Plant Sciences, Germany). In this vector the GUS gene of pFF19-GUS (Timmermans et al., 1990) was replaced by EGFP.
For the positive controls, the plasmid pFF19-GFP, without any fusion protein was used as a control for localization of GFP in cytosol and partly in nuclei. For mitochondrial localization the transit peptide (first 52 aa) of the Arabidopsis SHMT (At5g26789) was fused to GFP, and for plastidic localization the transit peptide sequence (first 36 aa) from the ribulose-1,5-bisphosphate carboxylase small subunit polypeptide of pea (Pisum sativum) was used (constructs provided by A. Wachter, Heidelberg Institute for Plant Sciences, Germany).
These plasmids were used for subsequent particle bombardment of Arabidopsis seedlings grown on MS plates (Schenk et al. 1998). Transient transformation was carried out with 5 μg of plasmid DNA on 25 mg gold particles using a PDS-1000/HE system according to the manufacturer (Bio-Rad). After bombardment seedlings were incubated for 16 h under standard light/dark conditions and GFP localization then visualized by confocal laser scanning microscopy (Zeiss LSM510 META system, excitation at 488 nm, emission GFP: 510–525 nm, Chlorophyll: 645–700 nm).
Expression Analysis by Real-Time Quantitative PCR
Arabidopsis seeds were sterilized and sown on germination medium with 1% Suc and 0.7% washed agar. For sulfur starvation experiments, sulfate ions in the medium were replaced with chloride ions. For Cd treatment experiments, CdCl2 was added to a final concentration of 50 μm. Tissue samples were harvested every week for 5 weeks. Total RNA was isolated using 100 mg of plant material from stored and stratified seeds, cotyledons, leaves, stems, roots, siliques, and flower tissues. Stratified seeds were firstly sown on agar plates and incubated for 3 d at 4°C, and then released to 22°C under continuous light (approximately 2,500 lux). Five nanograms of total RNA were used for real-time quantitative PCR analysis. The N-terminal region unique to each Serat gene was amplified by PCR using two synthetic primers together with TaqMan probe specific to each amplified fragment. TaqMan One-Step RT-PCR Master Mix kit (Applied Biosystems, Foster City, CA) was used for amplification according to the protocols provided by the supplier. The real-time quantitative PCR analysis was standardized based on equal quantities of RNA sample, and respective linearized plasmid DNA of each gene was used to draw a standard curve. The sequences are: for Serat1;1, PCR primers: 5′-TGGACACAGATCAAGGCGG-3′, 5′-ATGAGAAAGAATCGTCGAATATAGATAGC-3′, TaqMan probe: 5′-ATGCTGAGGCGGAGCCAGCTTTAGC-3′; for Serat2;1, PCR primers: 5′-CACATGCCGAACCGGTAATAC-3′, 5′-GGTGAATCTTCCGGTTTACAGAGA-3′, TaqMan probe: 5′-TGATTCCCGGTTCTGTTGCATCAAGA-3′; for Serat2;2, PCR primers: 5′-AATGGAACCCAGACCAAAACC-3′, 5′-GCCCAAACATCATCGACTTCA-3′, TaqMan probe: 5′-TCCATACTCGTCCTTTGCTTGAAGATCTCG-3′; for Serat3;1, PCR primers: 5′-ACGCTAAGGGAACTCATAAGTCAGA-3′, 5′-TCTTCTCTTATAGCATCCCAAATAGGA-3′, TaqMan probe: 5′-TTGACTCGAATTTGCTTGATCCTCGTTCTG-3′; for Serat3;2, PCR primers: 5′-CTCTTCCAATGATTGTCTCCCG-3′, 5′-CCTCTCGAAAGGAAACTCGTCA-3′, TaqMan probe: 5′-ACTTTTCTGCCAGAGACGATGGAGAGACC-3′.
Transgenic Plants of Serat Promoter-GFP
Each promoter region of 2.5-kb length in Arabidopsis genome was amplified and cloned into pUC19 to verify the sequence. Subsequently, the promoter fragment was introduced into the modified pB101 carrying the GFP-reporter gene from sGFP (65T; Chiu et al., 1996) instead of uidA. Arabidopsis plants were transformed using flower dip method (Clough and Bent, 1998) with Agrobacterium tumefaciens C58C1 (GV3101) carrying the promoter-GFP constructs. The oligonucleotides used are: Serat1;1, 5′-CAGTCAGTCGACACTCAAACTCACCCGGTCTA-3′ and 5′-CAGTCAGGATCCATTAACGCGGCGATAAGATT-3′; Serat2;1, 5′-CAGTCAGTCGACAACCTTAGACCCATTGCCTG-3′ and 5′-CAGTCAGGATCCGGTTTATGATTTGGAAGAAG-3′; Serat2;2, 5′-CAGTCATCTAGAAACTGAAAGGAAACAAAATA-3′ and 5′-CAGTCACCATGGTTTTTTTTTTTCTCGGCCAA-3′; Serat3;1, 5′-CAGTCAGTCGACGATGATCTGGACGTGCTACA-3′ and 5′-CAGTCAGTCGACATCTCTCCAAGGAGACGATA-3′; and Serat3;2, 5′-CAGTCAGTCGACAAAATCTTTGGTCTTTGCGC-3′ and 5′-CAGTCAGGATCCGACCAAAAAGAGTGAAGCTA-3′. GFP fluorescence was observed as reported previously (Ho et al., 1999).
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession numbers U30298, AJ414053, D49535, L34076, D88529, AF452452, AB040502, AB040503, AF212156, AF453837, AF453836, AP003292, L42212, Z34888, X82888, U22964, AJ414051, AJ414052, AF112303, AF331847, AC099399, and AF453838.
Acknowledgments
We thank Core Research for Evolutional Science and Technology-Akita Plant Molecular Science Satellite Laboratory in Life Research Support Center of Akita Prefectural University for analysis of DNA sequencing, Dr. Andreas Wachter (Heidelberg Institute for Plant Sciences, Germany) for kind supply of GFP constructs, and Dr. Mark Collier (Institute of Food Research, Norwich, UK) for critical comments on the manuscript.
This work was supported in part by Grants-in-Aid for Scientific Research from Ministry of Education, Culture, Sports, Science and Technology, Japan; by Core Research for Evolutional Science and Technology of Japan Science and Technology; and the German Research Foundation (grant no. SFB 363).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.045377.
References
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Ascano A, Nicholas DJD (1977) Purification and properties of O-acetyl-L-serine sulphydrylase from wheat leaves. Phytochemistry 16: 889–893 [Google Scholar]
- Bogdanova N, Bork C, Hell R (1995) Cysteine biosynthesis in plants: isolation and functional identification of a cDNA encoding a serine acetyltransferase from Arabidopsis thaliana. FEBS Lett 358: 43–47 [DOI] [PubMed] [Google Scholar]
- Brunold C, Suter M (1982) Intracellular localization of serine acetyltransferase in spinach leaves. Planta 155: 321–327 [DOI] [PubMed] [Google Scholar]
- Burgener M, Suter M, Jones S, Brunold C (1998) Cyst(e)ine is the transport metabolite of assimilated sulfur from bundle-sheath to mesophyll cells in maize leaves. Plant Physiol 116: 1315–1322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu WL, Niwa Y, Zeng W, Hirano T, Kobayashi H, Sheen J (1996) Engineered GFP as a vital reporter in plants. Curr Biol 6: 325–330 [DOI] [PubMed] [Google Scholar]
- Chronis D, Krishnan HB (2004) Sulfur assimilation in soybean (Glycine max [L.] Merr.): molecular cloning and characterization of a cytosolic isoform of serine acetyltransferase. Planta 218: 417–426 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Dominguez-Solis JR, Gutierrez-Alcala G, Romero LC, Gotor C (2001) The cytosolic O-acetylserine (thiol) lyase gene is regulated by heavy metals and can function in cadmium tolerance. J Biol Chem 276: 9297–9302 [DOI] [PubMed] [Google Scholar]
- Droux M (2003) Plant serine acetyltransferase: new insights for regulation of sulphur metabolism in plant cells. Plant Physiol Biochem 41: 619–627 [Google Scholar]
- Droux M, Martin J, Sajus P, Douce R (1992) Purification and characterization of O-acetylserine (thiol) lyase from spinach chloroplasts. Arch Biochem Biophys 295: 379–390 [DOI] [PubMed] [Google Scholar]
- Droux M, Ruffet ML, Douce R, Job D (1998) Interactions between serine acetyltransferase and O-acetylserine (thiol) lyase in higher plants—structural and kinetic properties of the free and bound enzymes. Eur J Biochem 1: 235–245 [DOI] [PubMed] [Google Scholar]
- Hatzfeld Y, Maruyama A, Schmidt A, Noji M, Saito K (2000) β-Cyanoalanine synthase is a mitochondrial cysteine synthase-like protein in spinach and Arabidopsis thaliana. Plant Physiol 123: 1163–1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hell R, Bogdanova N (1995) Characterization of a full-length cDNA encoding a serine acetyltransferase from Arabidopsis thaliana. Plant Physiol 109: 1498 [Google Scholar]
- Hell R, Hillebrand H (2001) Plant concepts for mineral acquisition and allocation. Curr Opin Biotechnol 12: 161–168 [DOI] [PubMed] [Google Scholar]
- Hell R, Jost R, Berkowitz O, Wirtz M (2002) Molecular and biochemical analysis of the enzymes of cysteine biosynthesis in the plant Arabidopsis thaliana. Amino Acids 22: 245–257 [DOI] [PubMed] [Google Scholar]
- Herschbach C (2003) Whole plant regulation of sulfur nutrition to deciduous trees: influences of the environment. Plant Biol 5: 233–244 [Google Scholar]
- Ho C-L, Noji M, Saito M, Saito K (1999) Regulation of serine biosynthesis in Arabidopsis: crucial role of plastidic 3-phosphoglycerate dehydrogenase in non-photosynthetic tissues. J Biol Chem 274: 397–402 [DOI] [PubMed] [Google Scholar]
- Howarth JR, Dominguez-Solis JR, Gutierrez-Alcala G, Wray JL, Romero LC, Gotor C (2003) The serine acetyltransferase gene family in Arabidopsis thaliana and the regulation of its expression by cadmium. Plant Mol Biol 51: 589–598 [DOI] [PubMed] [Google Scholar]
- Howarth JR, Roberts MA, Wray JL (1997) Cysteine biosynthesis in higher plants: a new member of the Arabidopsis thaliana serine acetyltransferase small gene family obtained by functional complementation of an Escherichia coli cysteine auxotroph. Biochim Biophys Acta 1350: 123–127 [DOI] [PubMed] [Google Scholar]
- Jost R, Berkowitz O, Wirtz M, Hopkins L, Hawkesford MJ, Hell R (2000) Genomic and functional characterization of the oas gene family encoding O-acetylserine (thiol) lyases, enzymes catalyzing the final step in cysteine biosynthesis in Arabidopsis thaliana. Gene 253: 237–247 [DOI] [PubMed] [Google Scholar]
- Kawashima CG, Noji M, Nakamura M, Ogra Y, Suzuki KT, Saito K (2004) Heavy metal tolerance of transgenic tobacco plants over-expressing cysteine synthase. Biotechnol Lett 26: 153–157 [DOI] [PubMed] [Google Scholar]
- Kim H, Hirai NY, Hayashi H, Chino M, Naito S, Fujiwara T (1999) Role of O-acetyl-L-serine in the coordinated regulation of the expression of a soybean seed storage-protein gene by sulfur and nitrogen nutrition. Planta 209: 282–289 [DOI] [PubMed] [Google Scholar]
- Kuske CR, Hill KK, Guzman E, Jackson PJ (1996) Subcellular location of O-acetylserine sulfhydrylase isoenzymes in cell cultures and plant tissues of Datura innoxia mill. Plant Physiol 112: 659–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leustek T, Martin MN, Bick J-A, Davies JP (2000) Pathways and regulation of sulfur metabolism revealed through molecular and genetic studies. Annu Rev Plant Physiol Plant Mol Biol 51: 141–165 [DOI] [PubMed] [Google Scholar]
- Leustek T, Saito K (1999) Sulfate transport and assimilation in plants. Plant Physiol 120: 637–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunn JE, Droux M, Martin J, Douce R (1990) Localization of ATP-sulfurylase and O-acetylserine (thiol) lyase in spinach leaves. Plant Physiol 94: 1345–1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lütcke HA, Chow KC, Mickel FS, Moss KA, Kern HF, Scheele GA (1987) Selection of AUG initiation codons differs in plants and animals. EMBO J 6: 43–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murillo M, Foglia R, Diller A, Lee S, Leustek T (1995) Serine acetyltransferase from Arabidopsis thaliana can functionally complement the cysteine requirement of a cysE mutant strain of Escherichia coli. Cell Mol Biol Res 41: 425–433 [PubMed] [Google Scholar]
- Noji M, Inoue K, Kimura N, Gouda A, Saito K (1998) Isoform-dependent differences in feedback regulation and subcellular localization of serine-acetyltransferase involved in cysteine biosynthesis from Arabidopsis thaliana. J Biol Chem 273: 32739–32745 [DOI] [PubMed] [Google Scholar]
- Noji M, Takagi Y, Kimura N, Inoue K, Saito M, Horikoshi M, Saito F, Takahashi H, Saito K (2001) Serine acetyltransferase involved in cysteine biosynthesis from spinach: molecular cloning, characterization and expression analysis of cDNA encoding a plastidic isoform. Plant Cell Physiol 42: 627–634 [DOI] [PubMed] [Google Scholar]
- Roberts MA, Wray JL (1996) Cloning and characterization of an Arabidopsis cDNA clone encoding an organellar isoform of serine acetyltransferase. Plant Mol Biol 30: 1041–1049 [DOI] [PubMed] [Google Scholar]
- Rolland N, Droux M, Douce R (1992) Subcellular distribution of O-acetylserine (thiol) lyase in cauliflower (Brassica oleracea L.) inflorescence. Plant Physiol 98: 927–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruffet M-L, Lebrun M, Droux M, Douce R (1995) Subcellular distribution of serine acetyltransferase from Pisum sativum and characterization of an Arabidopsis thaliana putative cytosolic isoform. Eur J Biochem 227: 500–509 [DOI] [PubMed] [Google Scholar]
- Saito K (2000) Regulation of sulfate transport and synthesis of sulfur-containing amino acids. Curr Opin Plant Biol 3: 188–195 [PubMed] [Google Scholar]
- Saito K (2004) Sulfur assimilatory metabolism: the long and smelling road. Plant Physiol 136: 2443–2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Yokohama H, Noji M, Murakoshi I (1995) Molecular cloning and characterization of a plant serine acetyltransferase playing a regulatory role in cysteine biosynthesis from watermelon. J Biol Chem 270: 16321–16326 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York
- Sasaki T, Chino M, Hayashi H, Fujiwara T (1998) Detection of several mRNA species in rice phloem sap. Plant Cell Physiol 39: 895–897 [DOI] [PubMed] [Google Scholar]
- Sasaki T, Matsumoto T, Yamamoto K, Sakata K, Baba T, Katayose Y, Wu J, Niimura Y, Cheng Z, Nagamura Y, et al (2002) The genome sequence and structure of rice chromosome 1. Nature 420: 312–316 [DOI] [PubMed] [Google Scholar]
- Schenk PM, Elliott AR, Manners JM (1998) Assessment of transient gene expression in plant tissue using the green fluorescent protein as a reference. Plant Mol Biol Rep 16: 313–322 [Google Scholar]
- Smith IK (1972) Studies of L-cysteine biosynthetic enzymes in Phaseolus vulgaris L. Plant Physiol 50: 477–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, Yamazaki M, Sasakura N, Watanabe A, Leustek T, de Almeida Engler J, Engler G, Van Montagu M, Saito K (1997) Regulation of sulfur assimilation in higher plants: a sulfate transporter induced in sulfate-starved roots plays a central role in Arabidopsis thaliana. Proc Natl Acad Sci USA 94: 11102–11107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmermans MCP, Maliga P, Vieira J, Messing J (1990) The pFF plasmids: cassettes utilizing CaMV sequences for expression of foreign genes in plants. J Biotechnol 14: 333–344 [DOI] [PubMed] [Google Scholar]
- Urano Y, Manabe T, Noji M, Saito K (2000) Molecular cloning and functional characterization of cDNAs encoding cysteine synthase and serine acetyltransferase that may be responsive for high cellular cysteine content in Allium tuberosum. Gene 257: 269–277 [DOI] [PubMed] [Google Scholar]
- Valvekens RD, Van Montagu M, Van Lijsegettens M (1988) Agrobacterium-mediated transformation of Arabidopsis thaliana root explants by using kanamycin selection. Proc Natl Acad Sci USA 85: 5536–5540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdoucq L, Florence V, Jacquot J-P, Chartier Y, Meyer Y (1999) In vivo characterization of a thioredoxin h target protein defines a new peroxiredoxin family. J Biol Chem 274: 19714–19722 [DOI] [PubMed] [Google Scholar]
- Warrilow A, Hawkesford M (2000) Cysteine synthase (O-acetyl-serine (thiol) lyase) substrate specificities classify the mitochondrial isoform as a cyanoalanine synthase. J Exp Bot 51: 985–993 [DOI] [PubMed] [Google Scholar]
- Wirtz M, Hell R (2003) Production of cysteine for bacterial and plant biotechnology: application of cysteine feedback-insensitive isoforms of serine acetyltransferase. Amino Acids 24: 195–203 [DOI] [PubMed] [Google Scholar]
- Yamaguchi T, Masada M (1995) Comparative studies on cysteine synthase isozymes from spinach leaves. Biochim Biophys Acta 1251: 91–98 [DOI] [PubMed] [Google Scholar]