Abstract
The present study has reported a novel method for producing induced pluripotent stem (iPS) cells. Primary human amniotic epithelial cells (HuAECs) were isolated from the amniotic membranes of pregnant women who received Cesarean sections. These cells were infected with retroviruses carrying octamer-binding transcription factor 4 (Oct4), (sex determining region Y)-box 2 (Sox2) and Yes-associated protein (Yap) (OSY). Following in vitro culture for ~14 days, epithelial-like HuAECs exhibited several iPS clone-like cell colonies (OSY-iPS). These cell clones presented positive alkaline phosphatase features and expressed high levels of embryonic stem cell-like markers (Nanog homeobox, Sox2, Oct4, reduced expression protein 1, and SSES3/4). Additionally, epigenetic analysis results indicated that the methylation of CpG islands on endogenous Oct4 and Sox2 promoters was reduced in OSY-iPS cells. Furthermore, the majority of the histone H3 at lysine 9 sites that interacted with the Oct4 and Sox2 promoters were acetylated, suggesting that the transcription activities of the above two transcription factors significantly increased. In vivo and in vitro induced differentiation experiments demonstrated that OSY-iPS could develop into embryoid bodies in vitro, and express numerous cellular markers in the three germ layers. Furthermore, OSY-iPS could form teratomas in immunodeficient mice. The pathological detection results suggest that these teratomas contain numerous types of cells from the three germ layers. However, the results from the quantitative polymerase chain reaction and western blot analyses suggest that the Hippo-Yap signaling pathway was significantly activated in OSY-iPS cells. In conclusion, a novel method for iPS induction was established in the present study. HuAECs were successfully induced to reprogram iPS cells through the introduction of OSY to activate the Hippo-Yap signaling pathway.
Keywords: human amniotic epithelial cells, induced pluripotent stem cells, Yes-associated protein, reprogramming, Hippo-Yap pathway
Introduction
Adult cells generally cannot undergo chromatin remodeling (i.e., cell reprogramming reaction) under normal conditions. However, previous research groups separately used four exogenous transcription factors (octamer-binding transcription factor 3/4, (Oct3/4), (sex determining region Y)-box 2 (Sox2), c-Myc, and Kruppel-like factor 4 (Klf4) by Yamanaka, and Oct3/4, Sox2, Nanog, and Lin28 by Thomson) to induce complex reprogramming reactions in adult cells to obtain induced pluripotent stem (iPS) cells comparable with embryonic stem cells (ESCs) (1–3). These iPS cells were very similar to ESCs in many respects, including pluripotent differentiation, self-proliferation capacity, and the ability to form teratomas in vivo (4,5). Because these seed cells of the iPS cells were adult cells sourced from humans or animals, the preparation of the iPS cells did not involve ethical constraints. Additionally, iPS cells advantageously have low immunogenicity and diverse sources. Thus, they could be ideal materials for cell and gene therapies (4,5).
The Yes-associated protein (Yap) is a downstream transcriptional coactivator of the Hippo-Yap pathway (6,7). During normal growth and development, activated Yap can induce the transcription of downstream genes and maintain organ development and cell growth. Yap loses activity after phosphorylation by large tumor suppressor 1/2 (Lats1/2) kinases, resulting in the inhibition of the transcription of downstream genes and the subsequent termination of cell proliferation and organ hyperplasia. Therefore, Yap activation directly influences the growth and development of tissues and organs (7–9). The gene for human Yap is localized on chromosome 11q12. Except for peripheral white blood cells that do not express Yap protein, other tissues and organs extensively express Yap (6,10,11). Additionally, many studies have indicated that Yap plays important roles in maintaining stem cell pluripotency, promoting stem cell proliferation, and regulating stem cell differentiation (9–11).
In this study, we combined two Yamanaka factors, Oct4 and Sox2, and a key factor in the Hippo-Yap pathway, Yap, to investigate whether human amniotic epithelial cells (HuAECs) could be induced for reprogramming into iPS cells.
Materials and methods
Isolation and culture of HuAECs
According to previously reported methods (4,12), amniotic membranes were washed with 4°C phosphate-buffered saline (PBS; Gibco, Gaithersburg, MD, USA) three times and cut into small pieces. The pieces were then digested in 20 ml 0.125% Trypsin-ethylenediaminetetraacetic acid (EDTA; Gibco) at 37°C for 30 min and mixed thoroughly in 20 ml Dulbecco's modified Eagle's medium (DMEM):F12 (1:1) cell culture medium (containing 15% fetal bovine serum, FBS). The cell suspension was filtered through a 200-mesh filter (Millipore, Bedford, MA, USA). The cell filtrate solution was collected and centrifuged at 1,500 rpm for 10 min. The supernatant was discarded, and the cell pellet was resuspended in DMEM:F12 (1:1) cell culture medium (containing 15% FBS; Gibco). The cell density was adjusted to 1×105/ml and directly inoculated onto 6-cm cell culture dishes. Cells were cultured in a cell incubator set at 37°C and 5% CO2. The cell culture medium was replaced after 48 hr.
Preparation of iPS cells
According to previously reported methods (1,3,13), HuAECs in the logarithmic growth phase were used at a cell density of 1×106/ml. The original culture medium was discarded, and 2 ml Opti-MEM (Gibco) culture medium was added. pLVX-Oct3/4, pLVX-Sox2, and pLVX-Yap1 lentiviruses were added (virus concentrations were 1×108 infectious units [IFU]/plaque-forming units [PFU]; Novobio, Shanghai, China), gently but thoroughly mixed, and reacted in a 37°C water bath for 120 min. After the reactions were finished, 4 ml mTeSR™1 medium (STEMCELL Technologies, Inc., MA, USA) was added. The cells were cultured in a 37°C and 5% CO2 cell incubator. The cell culture medium was replaced after 24 h.
Preparation of embryoid bodies
In accordance with previously reported methods (1,2), the concentration of iPS cells was adjusted to 1×105/ml using cell differentiation culture medium (DMEM, 15% FBS, 0.1 mmol/l non-essential amino acids, 2 mmol/l glutamate, and 0.1 mmol/l β-mercaptoethanol; all from Gibco). Cell suspensions at 2 µl were dropped onto the covers of cell culture dishes. After the covers were fully covered with cell suspension, they were placed onto the bottom of the dishes. Cells were continuously cultured for 48 h.
RNA extraction and qPCR
According to previously reported methods (4,12), the total RNA from cells in all groups were extracted based on the manufacturer instructions for the Trizol reagent (Invitrogen Life Technologies, Carlsbad, CA, USA). The total RNA was treated with DNase I (Sigma-Aldrich, St. Louis, USA), quantified, and reverse transcribed into cDNA using a ReverTra Ace-α First Strand cDNA Synthesis kit (Toyobo, Shanghai, China; Biotech Co., Ltd., Shanghai, China). Quantitative polymerase chain reaction (qPCR) was performed using a RealPlex4 real-time PCR detection system (Eppendorf Co., Ltd., Hamburg, Germany). A SYBR-Green Real-Time PCR Master Mix (Toyobo) was used as the fluorescence dye for nucleic acid amplification. qRT-PCR was performed for 40 amplification cycles of the following steps: 95°C denaturation for 15 sec, 58°C annealing for 30 sec, and 72°C extension for 42 sec. The relative gene expression levels were calculated and determined using the 2−ΔΔCt method as follows: ΔCt = Ct_genes - Ct_18sRNA and ΔΔCt = ΔCt_all_groups - ΔCt_blank control_group. The mRNA expression levels were calibrated based on the expression level of 18 s rRNA. The primers used are shown in Table I.
Table I.
qRT-PCR primers.
| Gene product | Forward (F) and reverse (R) primers (5′→3′) | Size (bp) |
|---|---|---|
| Oct4 | F: GTGGAGGAAGCTGACAACAA | 118 |
| R: TCTCCAGGTTGCCTCTCACT | ||
| Sox2 | F: AGAAAAACGAGGGAAATGGG | 120 |
| R: GTCATTTGCTGTGGGTGATG | ||
| Rex1 | F: GGTGGCATTGGAAATAGCAG | 148 |
| R: TGCCTAGTGTGCTGGTGGT | ||
| Nanog | F: GATTTGTGGGCCTGAAGAAA | 119 |
| R: CAGGGCTGTCCTGAATAAGC | ||
| Ssea3/4 | F: CTTTGAGGCTCTGCAGCTTA | 150 |
| R: CTGGTTCGCTTTCTCTTTCG | ||
| Mst | F: AGAAGGATGGGGTGGCTC | 117 |
| R: CAGGTGCTGTAGCTCTGTGC | ||
| Lats1 | F: TTTCTTGGCACAAACACCAT | 130 |
| R: GGGTCCTCGGCAAAGTTTA | ||
| Mob1 | F: TGACTTGGGTTCAAGATCAGC | 128 |
| R: ATGGGCATAAACCCTGAACA | ||
| Yap | F: TTGGGAGATGGCAAAGACAT | 113 |
| R: CTGTGACGTTCATCTGGGAC | ||
| 18S rRNA | F: CAGCCACCCGAGATTGAGCA | 223 |
| R: TAGTAGCGACGGGCGGTGTG |
Semi-quantitative RT-PCR
According to a previously reported method (1), the total RNA from cells in all groups were extracted based on the manufacturer instructions for the TRIzol reagent (Invitrogen). The total RNA was treated with DNase I (Sigma-Aldrich, St. Louis, USA), quantified, and reverse transcribed into cDNA using a ReverTra Ace-α First Strand cDNA Synthesis kit (Toyobo). Semi-quantitative RT-PCR was performed in a PTC-200 PCR machine (MJ Research Inc., Waltham, MA, USA). Each sample used 100 ng cDNA template. Additionally, 5 pmoles PCR forward and reverse primers, 200 µM dNTP, 1 unit RED-Taq Polymerase, and 1X RED-Taq polymerase buffer were added (all reagents were purchased from Sigma-Aldrich). The reaction volume was adjusted to 20 µl using nuclease-free deionized water. The qRT-PCR was performed for 32 amplification cycles of the following steps: 95°C denaturation for 15 sec, 58°C annealing for 30 sec, and 72°C extension for 42 sec. The PCR product was subjected to 1.5% agarose gel electrophoresis (Bio-Rad Laboratories, Inc., Hercules, CA, USA). The mRNA expression levels were calibrated based on the expression level of 18s rRNA. The primers used are shown in Table I.
Detection of alkaline phosphatase
According to the manufacturer instructions of the BCIP/NBT Alkaline Phosphatase Color Development kit (Beyotime Biotechnology Co., Ltd., Zhejiang, China), cell samples were fixed in 1 ml 4% paraformaldehyde (Sigma-Aldrich) at room temperature for 30 min. The fixative solution was discarded, and the 5-bromo-4-chloro-3-indolyl phosphate/nitroblue tetrazolium (BCIP/NBT) staining working solution was added and incubated at room temperature for 30 min. The BCIP/NBT staining working solution was subsequently discarded, and the cells were washed with distilled water twice to terminate the coloring reaction.
Western blot analysis
According to previously reported methods (4,12), the total protein samples of all groups were subjected to 12% SDS-PAGE denaturing gel electrophoresis (Bio-Rad) and transferred onto polyvinylidene difluoride PVDF membranes (Millipore, Bedford, MA, USA). After the membranes were blocked and washed, primary antibodies were added and incubated at 37°C for 45 min (Table II). Then, after the membranes were fully washed, secondary antibodies were added and incubated at 37°C for 45 min (Table II). The membranes were washed with Tris-buffered saline containing Tween-20 (TBST, Bio-Rad) at room temperature four times for 14 min per wash. The results were developed using the enhanced chemiluminescence (ECL) method (Bio-Rad). The membranes were exposed using Kodak XAR-5 films (Sigma-Aldrich).
Table II.
Primary antibodies, their source and dilutions.
| Antibodies | Companies | Applications |
|---|---|---|
| Rabbit anti-human Oct4 (no. 2890) | Cell Signaling Technology, Danvers, MA, USA | IF (1:100) |
| WB (1:1,000) | ||
| Rabbit anti-human Sox2 (no. 3579) | Cell Signaling Technology, Danvers, MA, USA | WB (1:1,000) |
| Rabbit anti-mouse SSEA3/4 (no. 4755) | Cell Signaling Technology, Danvers, MA, USA | IF (1:100) |
| Rabbit anti-human MST (no. 14946) | Cell Signaling Technology, Danvers, MA, USA | WB (1:1,000) |
| Rabbit anti-human p-MST (no. 3681) | Cell Signaling Technology, Danvers, MA, USA | WB (1:1,000) |
| Rabbit anti-human LATS1 (no. 3477) | Cell Signaling Technology, Danvers, MA, USA | WB (1:1,000) |
| Rabbit anti-human MOB1 (no. 13730) | Cell Signaling Technology, Danvers, MA, USA | WB (1:1,000) |
| Rabbit anti-human Yap (no. 14074) | Cell Signaling Technology, Danvers, MA, USA | WB (1:1,000) |
| Rabbit anti-human p-Yap (no. 13008) | Cell Signaling Technology, Danvers, MA, USA | WB (1:1,000) |
| Rabbit anti-human GAPDH (no. 5174) | Cell Signaling Technology, Danvers, MA, USA | WB (1:1,000) |
Chromatin immunoprecipitation (ChIP)-PCR
In accordance with previously reported methods (4,12), the manufacturer instructions of the EZ-ChIP kit (Millipore, Bedford, MA, USA) were followed. Briefly, cells were fixed in 1% paraformaldehyde at 37°C for 30 min and incubated in 125 mM glycine at room temperature for 10 min to terminate cross-linking. Cells were sonicated on ice until the DNA was broken into chromatin fragments of 200–1000 bp. The primary antibody was added, and the samples were incubated at 4°C overnight. Protein A/G and agarose were added for adsorption, and a final immune precipitate was obtained. PCR amplification was then performed for 33 amplification cycles of the following steps: 95°C denaturation for 30 sec, 55°C annealing for 30 sec, and 72°C extension for 30 sec. The 2-ΔCt calculation method was performed to determine the relative expression levels of the PCR products as follows: ΔCt = Ct_all_groups - Ct_Input_group. The primers used are listed in reference (2).
Analysis of chromosome karyotype
According to a previously reported method (4), iPS cells with excellent growth status were incubated with 0.1 µg/ml colchicine (Sigma-Aldrich) for 30 min. The supernatant was discarded, and the cells were collected and mixed thoroughly in 9 ml 0.075% KCl (Sigma-Aldrich) by pipetting. Hypotonic treatment was performed at 37°C for 30 min. The recovered cell pellet was fixed in a fixative solution (Beyotime Biotechnology) at room temperature three times for 15 min per fix. The cell suspension was dropped onto slides and baked at 80°C for 2 h. Cells were digested with 0.25% Trypsin-EDTA (Gibco) for 3 min, washed with deionized water three times, and stained with Giemsa staining solution (Beyotime Biotechnology) at room temperature for 10 min. After washing again with deionized water three times, the slides were mounted in neutral balsam (Beyotime Biotechnology).
Preparation of the teratoma
In accordance with previously reported methods (4,12), iPS cells with excellent growth status were inoculated into back subcutaneous tissues of nude mice in a sterile environment. Each nude mouse was inoculated at one point with approximately 1×108/ml iPS cells. Nude mice were fed under normal conditions until tumor formation.
H&E staining
According to a previously reported method (4), tissues were fixed in 4% paraformaldehyde at room temperature for 12 h. Frozen tissue sections were prepared at thicknesses of approximately 5 µm. Sections were fixed in 95% anhydrous ethanol for 2 min, stained in hematoxylin for 5 min, and differentiated in differentiation solution for 2 min. Sections were immersed in weak ammonia solution for 3 min, washed with deionized water for 5 min, stained with eosin for 5 min, and washed with deionized water for 5 min. Tissue sections were immersed in 70, 80, and 90% alcohol solution once for 1 min, washed with anhydrous ethanol twice for 1 min each wash, cleared in xylene twice for 1 min each wash, and mounted using neutral balsam. These reagents and materials were all purchased from Beyotime Biotechnology Co., Ltd., Zhejiang, China.
Immunofluorescence staining
According to previously reported methods (4,12), cell samples were fixed in 1 ml 4% paraformaldehyde (Sigma-Aldrich) at room temperature for 30 min and blocked in blocking solution (Beyotime Biotechnology) at 37°C for 60 min. The blocking solution was discarded, and the cells were washed with an immunohistochemistry washing solution (Beyotime Biotechnology) at room temperature three times for 5 min each wash. Primary antibodies (Table II) were added and incubated at 37°C for 45 min. The antibodies were discarded, and the cells were washed with the immunohistochemistry washing solution (Beyotime Biotechnology) at room temperature three times for 5 min each wash. Secondary antibodies (Table II) were added and incubated at 37°C for 45 min. The antibodies were discarded, and the cells were washed with the immunohistochemistry washing solution (Beyotime Biotechnology) at room temperature three times for 5 min each. Finally, the cells were mounted in immunofluorescence mounting fluid (Sigma-Aldrich).
Results
Overexpression of Yap, Oct4, and Sox2 induced HuAECs to express high levels of ESC markers
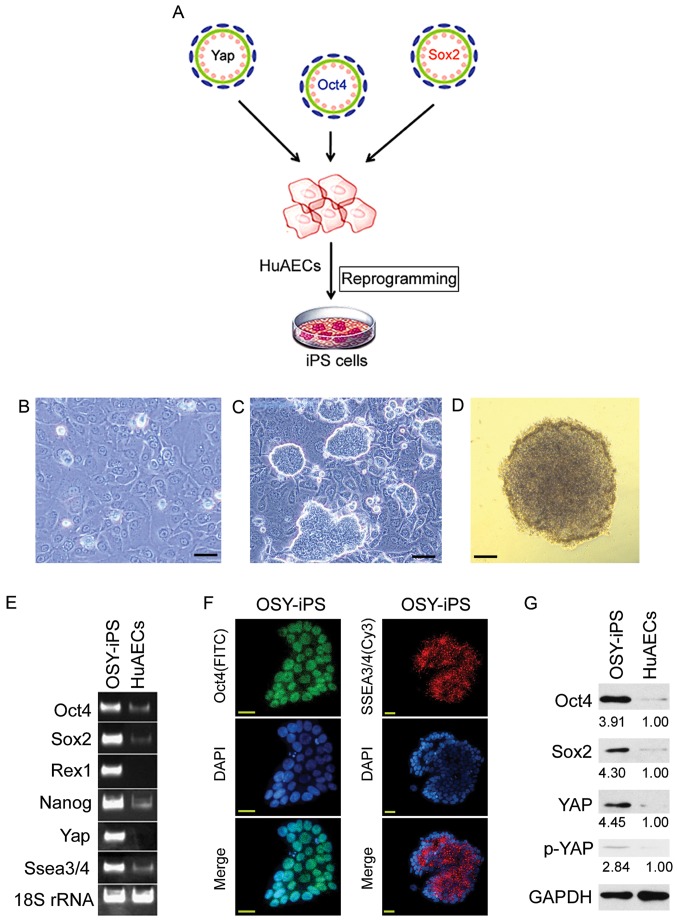
HuAECs were isolated from fetal amniotic membrane and infected with lentiviruses carrying Oct4, Sox4, and Yap (OSY) coding sequences. These cells were cultured using iPS cell culture methods to investigate whether the OSY factors could induce HuAEC reprogramming into iPS cells (Fig. 1A). Microscopy showed that HuAECs showed typical epithelial cell characteristics and had cobblestone morphologies (Fig. 1B). After transducing with OSY factors, the cells were cultured for 14 days. Microscopy showed the gradual development of clone-like cell masses (Fig. 1C). These clones were identified as OSY-iPS. The alkaline phosphatase assays suggested these clone-like cells exhibited dark-purple, positive reactions (Fig. 1D). The semi-quantitative PCR results indicated the mRNA levels of ESC markers (Oct4, Sox2, Nanog, Rex1, and Ssea3/4) and Yap in OSY-iPS cells were higher than those in the HuAECs (Fig. 1E). Additionally, immunofluorescence staining results suggested that the expression of Oct4 and SSEA3/5 proteins in OSY-iPS cells was positive (Fig. 1F). Finally, Western blot results showed that the expression levels of Oct4, Sox2, and YAP proteins in the OSY-iPS cells were significantly higher than those in the HuAECs (Fig. 1G). Therefore, the three OSY factors induced HuAECs to express high levels of ESC markers.
Figure 1.
Overexpression the three factors, OSY, induced HuAECs to express high levels of ESC markers. (A) The process of induction of HuAEC reprogramming into iPS cells by OSY. (B) Cell morphology of HuAECs; scale, 30 µm. (C) OSY-iPS cells had clone-like morphology; scale, 30 µm. (D) Alkaline phosphatase staining identification of OSY-iPS cells was positive; scale, 30 µm. (E) Semi-quantitative PCR results indicated that OSY-iPS cells expressed high levels of ESC markers (Oct4, Sox2, Nanog, Rex1, and Ssea3/4) and Yap. (F) Immunofluorescence staining results suggested that OSY-iPS cells expressed high levels of Oct4 and SSEA3/4 proteins; scale, 30 µm. (G) Western blot results showed that OSY-iPS cells expressed high levels of Oct4, Sox2, and Yap proteins.
Overexpression of Yap, Oct4, and Sox2 induced HuAECs to undergo chromatin reprogramming
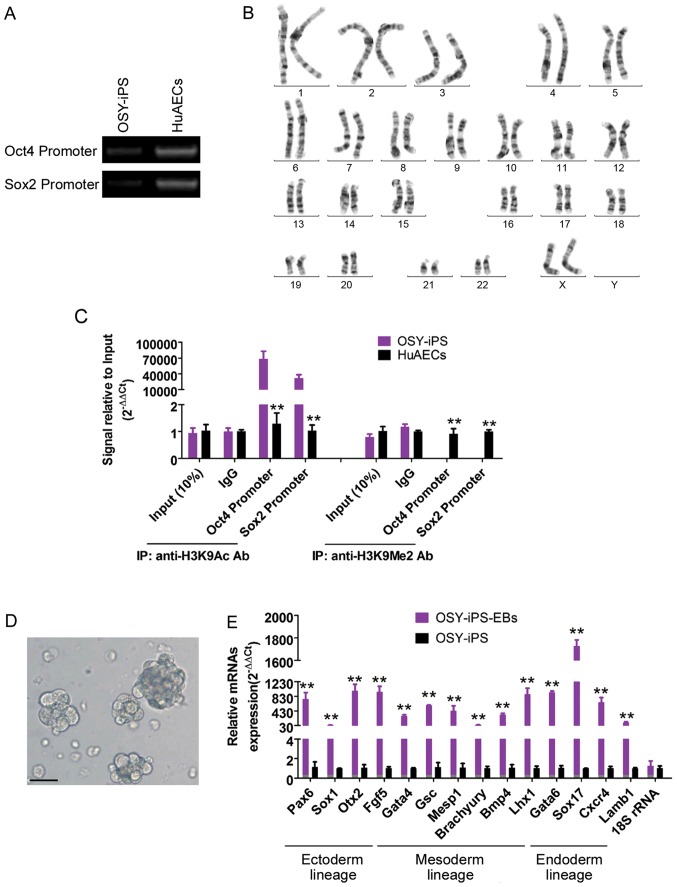
The methylation PCR results showed that the methylation levels of endogenous Oct4 and Sox2 gene promoter regions were significantly higher than those in HuAECs (Fig. 2A). Additionally, the ChIP results showed that the Oct4 and Sox2 gene promoter regions in the OSY-iPS cells primarily interacted with H3K9 acetylation sites, whereas the Oct4 and Sox2 gene promoter regions in the HuAECs primarily interacted with H3K9 dimethylation sites (Fig. 2B). H3K9 acetylation can activate gene transcription, and H3K9 methylation can inhibit the transcription activities of genes. Additionally, chromosome karyotype analyses showed that these iPS cells had a normal female chromosome core (46XX), indicating that chromatin reprogramming did not cause chromosome abnormalities in the cells (Fig. 2C). Furthermore, the natural differentiation of the OSY-iPS cells was induced in vitro using the embryoid body culture method. The expression of the makers of the three germ layers was identified using qPCR. These results indicated that the OSY-iPS cells expressed high levels of markers associated with the three germ layers after six days of natural, induced differentiation. These results indicated that the OSY factors induced chromatin reprogramming in HuAECs.
Figure 2.
Overexpression of the three factors, OSY, induced chromatin reprogramming in HuAECs. (A) Methylation PCR results showed that the methylation levels of the promoter regions of endogenous Oct4 and Sox2 genes in OSY-iPS cells were significantly higher than those in HuAECs. (B) Chromosome karyotype analysis results indicated that the iPS cells had the normal female chromosome core (46XX). (C) ChIP results showed that Oct4 and Sox2 gene promoter regions mainly interacted with H3K9 acetylation sites in OSY-iPS cells, while Oct4 and Sox2 gene promoter regions in HuAECs mainly interacted with H3K9 dimethylation sites. **P<0.01, OSY-iPS vs. HuAECs; n=3. (D) Embryoid bodies from OSY-iPS after natural differentiation for six days; scale, 30 µm. **P<0.01, OSY-iPS-EB vs. OSY-iPS; n=3. (E) qPCR results indicated that embryoid bodies derived from OSY-iPS cells expressed high levels of markers associated with three germ layers.
OSY-iPS cells have pluripotency
To confirm that the OSY-iPS cells had ESC-like pluripotent differentiation capacity, the OSY-iPS cells were injected into nude mice. After a certain time, the left back sides of the nude mice developed tumor bodies (Fig. 3A). The surfaces of these tumor bodies were smooth and had a soft texture. Obvious blood vessel distribution could be observed on the surfaces (Fig. 3B). Pathological identification showed that these tumor bodies contained many types of tissues and cells, including glands and intestinal epithelia of the endoderm, striated muscles of the mesoderm, and neural tubes and naïve neurons of the ectoderm (Fig. 3C). Therefore, these tumor bodies exemplified typical teratoma. The results of in vivo experiments showed that the OSY-iPS cells had pluripotent differentiation capacity and could form teratomas containing cells from three germ layers in nude mice.
Figure 3.
OSY-iPS cells showed pluripotency. (A) OSY-iPS cells could form a teratoma in the body of nude mice. The black arrow indicated tumor tissues. (B) Teratoma tissues isolated from the body of nude mice. (C) Pathological identification results in hematoxylin and eosin (H&E) staining showed that this tumor body contained many types of tissues and cells such as glands and intestinal epithelia (blue arrows), striated muscles (black arrows), neural tubes (green arrows), and naïve neurons (yellow arrows). Scale, 30 µm.
Overexpression of Yap, Oct4, and Sox2 activated the Hippo-Yap pathway in OSY-iPS cells
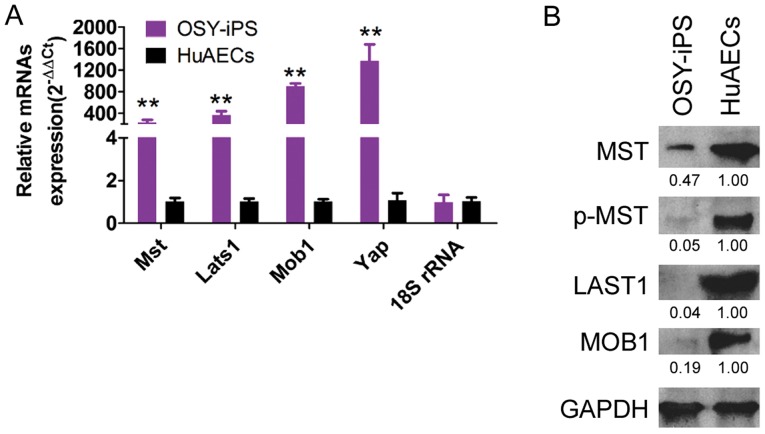
qPCR results indicated that the mRNA expression levels of the Mst, Lats1, Mob1, and Yap genes in OSY-iPS cells were significantly higher than those in the HuAECs (Fig. 4A). Additionally, the western blot results indicated that the expression levels of the important proteins in the Hippo-Yap pathway, Mst, Lats1, and Mob1, in the OSY-iPS cells were significantly higher than those in the HuAECs. Furthermore, the level of phosphorylation of Mst protein (p-Mst) in the OSY-iPS cells was also significantly higher than that in the HuAECs (Fig. 4B). These experimental results indicated that the OSY factors could activate the Hippo-Yap pathway.
Figure 4.
Overexpression of the three factors, OSY, activated the Hippo-Yap pathway in OSY-iPS cells. (A) Results of qPCR experiments indicated that the mRNA expression levels of Mst, Lats1, Mob1, and Yap genes in OSY-iPS cells were significantly higher than those in HuAECs. **P<0.01, OSY-iPS vs. HuAECs; n=3. (B) Results of western blot experiments indicated that the expression levels of Mst, Lats1, and Mob1 in OSY-iPS cells were significantly higher than those in HuAECs. In addition, the phosphorylation modification of Mst protein (p-Mst) in OSY-iPS cells was also significantly higher than that in HuAECs.
Discussion
Since Yamanaka, Takahashi, and others prepared the first strain of iPS cells in 2006, this methodology rapidly developed (1–3). More methods have been reported for establishing iPS cells from different sources (4,5). We used HuAECs as seed cells and transduced two Yamanaka factors, Oct4 and Sox2, and a key protein in the Hippo-Yap pathway, Yap, into these cells to investigate whether iPS cells could be prepared. Surprisingly, the production of typical human ESC-like clones was observed under a microscope at approximately 2 weeks after transduction. Further studies showed that OSY-iPS cells expressed high levels of pluripotent markers of ESCs and could be differentiated into cells of the three germ layers in vivo and in vitro. iPS cells have been applied in transplantation therapy studies in many clinical disease models (5,14,15). If iPS cells could be readily applied in clinical therapy, this technology would guarantee high efficiency and safety (4,12). In this study, the preparation of iPS cells using Oct4, Sox2, and Yap had the advantage of safety than other methods. In the past, the four Yamanaka factors (Oct4, Sox2, Klf4, and c-Myc) have been typically used to induce the reprogramming of adult cells into iPS cells (1,2). However, c-Myc and Klf4 are both proto-oncogenes. iPS cells carrying c-Myc gene have been reported to develop malignant tumors in vivo, whereas Klf4 transmits tumors to offspring (13,16). However, as major regulators, the transcription factors Oct4 and Sox2 maintain the pluripotency and self-renewal of ESCs (4,12). The use of the Yap factor could control the balance of cells between self-proliferation and differentiation. This combination is relatively reasonable and efficient. Experimental results have confirmed that the combination of the three factors, Oct4, Sox2, and Yap, could also induce the reprogramming of general epithelial cells into iPS cells without the involvement of c-Myc or Klf4. However, we considered that the activation of the Hippo-Yap pathway also promoted iPS reprogramming. Lian et al showed that during the preparation of iPS cells using the four Yamanaka factors, the overexpression of Yap increased the iPS cell production efficiency by two-fold compared with the control group (10). Qin et al found that the use of Lats2 knockout cells to prepare iPS cells shortened the reprogramming time by approximately 5 days (11). We referenced their study results and only used two Yamanaka factors (Oct4 and Sox2) combined with overexpression of the key factor in the Hippo-Yap pathway, Yap, to achieve HuAEC reprogramming. Additionally, the preparation of iPS reprogramming using the four Yamanaka factors usually requires approximately 1 month (1,2). However, our iPS reprogramming only required 2 weeks. Experimental results indicated that the activation of the endogenous Hippo-Yap pathway in cells by overexpressing Yap could greatly shorten the iPS reprogramming time. Overall, the combination of the three factors, Oct4, Sox2, and Yap, could efficiently induce the reprogramming of HuAECs into iPS cells.
In conclusion, we established a new method for iPS induction. Through the introduction of Oct4, Sox2 and Yap to activate the Hippo-Yap pathway, HuAECs were successfully induced to reprogram iPS cells. And, using this method, it is possible to shorten the time required for iPS cells reprogramming.
Acknowledgements
This study was supported by grant from the Shanghai Natural Science Foundation (no. 16ZR1434000), and project funded by the China Postdoctoral Science Foundation (nos. 2014M550250 and 2015T80455).
References
- 1.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 2.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 3.Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313–317. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- 4.Liu T, Zou G, Gao Y, Zhao X, Wang H, Huang Q, Jiang L, Guo L, Cheng W. High efficiency of reprogramming CD34+ cells derived from human amniotic fluid into induced pluripotent stem cells with Oct4. Stem Cells Dev. 2012;21:2322–2332. doi: 10.1089/scd.2011.0715. [DOI] [PubMed] [Google Scholar]
- 5.Wu J, Ocampo A, Belmonte JC Izpisua. Cellular metabolism and induced pluripotency. Cell. 2016;166:1371–1385. doi: 10.1016/j.cell.2016.08.008. [DOI] [PubMed] [Google Scholar]
- 6.Zhang H, Pasolli HA, Fuchs E. Yes-associated protein (YAP) transcriptional coactivator functions in balancing growth and differentiation in skin; Proc Natl Acad Sci USA; 2011; pp. 2270–2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu FX, Zhao B, Panupinthu N, Jewell JL, Lian I, Wang LH, Zhao J, Yuan H, Tumaneng K, Li H, et al. Regulation of the Hippo-YAP pathway by G-protein-coupled receptor signaling. Cell. 2012;150:780–791. doi: 10.1016/j.cell.2012.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoa L, Kulaberoglu Y, Gundogdu R, Cook D, Mavis M, Gomez M, Gomez V, Hergovich A. The characterisation of LATS2 kinase regulation in Hippo-YAP signalling. Cell Signal. 2016;28:488–497. doi: 10.1016/j.cellsig.2016.02.012. [DOI] [PubMed] [Google Scholar]
- 9.Qi YF, Yu J, Han W, Fan X, Qian H, Wei H, Tsai YH, Zhao J, Zhang W, Liu Q, et al. A splicing isoform of TEAD4 attenuates the Hippo-YAP signalling to inhibit tumour proliferation. Nat Commun. 2016;7:ncomms11840. doi: 10.1038/ncomms11840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lian I, Kim J, Okazawa H, Zhao J, Zhao B, Yu J, Chinnaiyan A, Israel MA, Goldstein LS, Abujarour R, et al. The role of YAP transcription coactivator in regulating stem cell self-renewal and differentiation. Gene Dev. 2010;24:1106–1118. doi: 10.1101/gad.1903310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qin H, Blaschke K, Wei G, Ohi Y, Blouin L, Qi Z, Yu J, Yeh RF, Hebrok M, Ramalho-Santos M. Transcriptional analysis of pluripotency reveals the Hippo pathway as a barrier to reprogramming. Hum Mol Genet. 2012;21:2054–2067. doi: 10.1093/hmg/dds023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu T, Cheng W, Huang Y, Huang Q, Jiang L, Guo L. Human amniotic epithelial cell feeder layers maintain human iPS cell pluripotency via inhibited endogenous microRNA-145 and increased Sox2 expression. Exp Cell Res. 2012;318:424–434. doi: 10.1016/j.yexcr.2011.12.004. [DOI] [PubMed] [Google Scholar]
- 13.Hamanaka S, Yamaguchi T, Kobayashi T, Kato-Itoh M, Yamazaki S, Sato H, Umino A, Wakiyama Y, Arai M, Sanbo M, et al. Generation of germline-competent rat induced pluripotent stem cells. Plos One. 2011;6:e22008. doi: 10.1371/journal.pone.0022008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lengner CJ. iPS cell technology in regenerative medicine. Ann NY Acad Sci. 2010;1192:38–44. doi: 10.1111/j.1749-6632.2009.05213.x. [DOI] [PubMed] [Google Scholar]
- 15.Rowntree RK, McNeish JD. Induced pluripotent stem cells: Opportunities as research and development tools in 21st century drug discovery. Regen Med. 2010;5:557–568. doi: 10.2217/rme.10.36. [DOI] [PubMed] [Google Scholar]
- 16.Wang X, Zhao Y, Xiao Z, Chen B, Wei Z, Wang B, Zhang J, Han J, Gao Y, Li L, et al. Alternative translation of OCT4 by an internal ribosome entry site and its novel function in stress response. Stem cells. 2009;27:1265–1275. doi: 10.1002/stem.58. [DOI] [PubMed] [Google Scholar]