Abstract
Phytoprostanes are prostaglandin/jasmonate-like products of nonenzymatic lipid peroxidation that not only occur ubiquitously in healthy plants but also increase in response to oxidative stress. In this work, we show that the two naturally occurring B1-phytoprostanes (PPB1) regioisomers I and II (each comprising two enantiomers) are short-lived stress metabolites that display a broad spectrum of biological activities. Gene expression analysis of Arabidopsis (Arabidopsis thaliana) cell cultures treated with PPB1-I or -II revealed that both regioisomers triggered a massive detoxification and defense response. Interestingly, expression of several glutathione S-transferases, glycosyl transferases, and putative ATP-binding cassette transporters was found to be increased by one or both PPB1 regioisomers, and hence, may enhance the plant's capacity to inactivate and sequester reactive products of lipid peroxidation. Moreover, pretreatment of tobacco (Nicotiana tabacum) suspension cells with PPB1 considerably prevented cell death caused by severe CuSO4 poisoning. Several Arabidopsis genes induced by PPB1, such as those coding for adenylylsulfate reductase, tryptophan synthase β-chain, and PAD3 pointed to an activation of the camalexin biosynthesis pathway that indeed led to the accumulation of camalexin in PPB1 treated leaves of Arabidopsis. Stimulation of secondary metabolism appears to be a common plant reaction in response to PPB1. In three different plant species, PPB1-II induced a concentration dependent accumulation of phytoalexins that was comparable to that induced by methyl jasmonate. PPB1-I was much weaker active or almost inactive. No differences were found between the enantiomers of each regioisomer. Thus, results suggest that PPB1 represent stress signals that improve plants capacity to cope better with a variety of stresses.
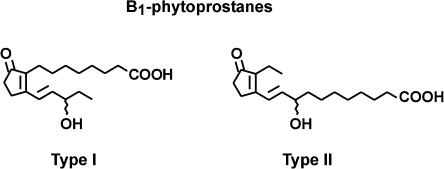
Phytoprostanes belong to a novel family of plant effectors that are formed nonenzymatically by a free radical catalyzed biochemical mechanism from α-linolenic acid. Via an identical nonenzymatic mechanism, isoprostanes (isomers of prostaglandins) are formed in animals from arachidonic acid. Nomenclature used to name different phytoprostane classes conforms with the general isoprostane/prostaglandin terminology (Thoma et al., 2004). Due to the nonenzymatic formation of phytoprostanes, two racemic regioisomers (type I and II) of each class can be generated. One pathway leads to the formation of B1-phytoprostanes (PPB1; Fig. 1) that are chemically stable end products of lipid peroxidation. In animals, isoprostanes are not only extremely reliable markers of oxidative stress but also display potent (prostaglandin-) receptor-mediated biological activities in the nanomolar concentration range (Cracowski et al., 2002). Therefore, it is postulated that isoprostanes represent mediators of oxidant injury in animals.
Figure 1.
PPB1. Free radical catalyzed oxidation of α-linolenate yields two racemic PPB1 regioisomers (type I and type II).
In plants, several classes of phytoprostanes are constitutively present, and, notably, their levels increase in a variety of conditions associated with enhanced free radical generation (Thoma et al., 2003). Previously, it has been shown that PPB1 formation, together with the synthesis of a series of other phytoprostane classes, can be triggered in vivo by fungal pathogens (Botrytis cinerea) or peroxides (Thoma et al., 2003). Interestingly, endogenous accumulation of PPB1 or F1-phytoprostanes after peroxide, heavy metal challenge, or wounding is a transient process, suggesting that phytoprostanes, similar to isoprostanes in animals, can be rapidly generated and metabolized in vivo (Imbusch and Mueller, 2000a; Thoma et al., 2003). In this study, we show that not only endogenous but also exogenously administered PPB1 are rapidly metabolized by tobacco (Nicotiana tabacum) cell cultures, suggesting that PPB1 are continuously synthesized de novo as a consequence of free radical formation during aerobic metabolism. So far, little is known about the spectrum of biological activities displayed by phytoprostanes. Previously, it has been shown that cyclopentenone phytoprostanes with the deoxy-J1-prostaglandin, A1-prostaglandin (PPA1), and PPB1 ring system activate mitogen-activated protein kinase activity in tomato (Lycopersicon esculentum) cell suspension cultures. In addition, in tomato cell cultures, a gene involved in primary metabolism, extracellular invertase, was induced by PPB1 but not by PPA1 (Thoma et al., 2003). There is also evidence that several classes of phytoprostanes, including PPB1, trigger secondary metabolism in taxonomically distant plant species (Thoma et al., 2003, 2004; Iqbal et al., 2005). PPB1 are structurally related to jasmonates and in fact, biological effects on invertase and secondary metabolism appear to be qualitatively similar to jasmonic acid (JA) or methyl jasmonate (MeJA). However, it has also been shown that biological activities of JA/MeJA and PPB1 differ in various bioassay systems (Thoma et al., 2003).
It is to be expected that even different isomers of one class of phytoprostanes exhibit a different qualitative and quantitative profile of biological activities similar to isoprostane isomers in mammals (Cracowski et al., 2002; Cracowski, 2003, 2004; Janssen, 2004). For studying the biological properties of isoprostanes, chemically pure isoprostane isomers are required that usually can be provided only by total synthesis. Recently, synthetic strategies have been developed for deoxy-J1-, E1-, and F1-phytoprostanes. Due to the large number of possible isomers (32 of each class), only a few isomers are currently available (Rodriguez and Spur, 2003; El Fangour et al., 2004; Iqbal et al., 2005) and under investigation. However, selected isomers may not be representative for any one of the classes.
Although PPB1 are not the most abundant phytoprostanes in vivo, the possibility to isolate large quantities of all theoretically possible PPB1 isomers from linolenate autoxidation mixtures (as demonstrated in this work) allows us to study the biological properties of one class of phytoprostanes. We have prepared methyl esters of all four isomers of PPB1 and compared their effect on secondary metabolism with MeJA in three different plant species (Eschscholzia californica Cham., Crotalaria cobalticola Duvign. & Plancke, tobacco cv Xanthi L.). Moreover, we also probed the spectrum of biological activity of PPB1 using an Arabidopsis (Arabidopsis thaliana) L. Heyn., ecotype Columbia (Col-0) DNA array approach that indicates that PPB1 might induce enzymes that protect cells from the consequences of oxidative stress. To this end, cell death of PPB1 primed tobacco cells in response to severe heavy metal stress was investigated.
RESULTS
cDNA Array Analysis of PPB1 Induced Gene Expression in Arabidopsis Cell Cultures
Little is known about phytoprostane-responsive genes in plants. We first studied gene expression in phytoprostane-treated Arabidopsis cell cultures by using a custom-designed cDNA array that included 626 defense-related genes encoding pathogenesis-related proteins or proteins induced by oxidative stress, cold, UV, ozone, or heavy metals and 50 genes associated with primary metabolism. We defined induction or repression of a gene as a minimum 2.0 change in its transcript level. Array hybridizations were based on four replicates and a dye-swap. As described previously (Huang et al., 2002), we applied the following selection procedure to our expression data: (1) only signals more than 2-fold above local background level were considered; (2) only expression ratios higher than 2.0 were regarded as significant; and (3) values with cv values over 10 were emitted. These very rigorous criteria implicate that our procedure ignores such genes with relatively low basal expression ratios (Table I). Acuity 3.1 (Axon Instruments, Union City, CA) was used for statistical analysis. Induction of several enzymes involved in plant stress responses could be observed after treatment with phytoprostanes. Analysis of these data revealed that 60 genes showed significant differential expression in response to 1 or more of the treatments. The response to PPB1-I was stronger than that to PPB1-II. Treatment with both phytoprostanes led to an induction of genes from different pathways including detoxification and plant defense. Table I gives a short survey over some interesting genes induced by PPB1. Complete sets of array data can be found in the supplemental material. Notably, detoxification genes that were induced by PPB1 include 17 putative glutathione S-transferases, 8 glycosyltransferases, 2 glutathione S-conjugate ABC transporters, and 3 ABC transporters. Induction of these genes most likely is important for conjugation and storage of toxic lipid peroxidation products and phenols generated endogenously during oxidative stress, and thus, might protect cellular proteins from irreversible damage. In addition, induction of genes coding for adenylylsulfate reductase, Trp synthase β-chain, and cytochrome P450 enzymes including PAD3 indicate that secondary metabolism, i.e. camalexin synthesis, can be stimulated by PPB1. Biosynthesis of camalexin can be both triggered by abiotic (i.e. copper ions) and biotic (i.e. pathogens) stresses associated with enhanced formation of reactive oxygen species (ROS); however, endogenous signals that trigger camalexin biosynthesis are not known.
Table I.
Short survey of DNA array analysis of transcripts in Arabidopsis suspension cells in response to phytoprostane treatment
Genes on the array were gene-specific 3′ UTRs (Glombitza et al., 2004) or expressed sequence tags available from ABCR, The Ohio State University, Rightmire Hall, 1060 Carmack Road, Columbus, OH 43210, or designed at the GSF-National Research Center for Environment and Health, 85764 Oberschleissheim, Germany. Only genes with detectable activity are shown. More than 3-fold activation of gene expression is in bold type. sd, Standard deviation; CV, coefficient of variance; detox, detoxification; P450, cytochrome p450 family enzyme; OS, oxidative stress; PI, pathogen induced; SM, secondary metabolism; SR, stress response; *, value not statistically significant due to weak signal or high cv value; **, one or more weak signal replicates.
| ID
|
Source
|
Locus
|
Name
|
Pathway
|
PPB1-I
|
PPB1-II
|
||||
|---|---|---|---|---|---|---|---|---|---|---|
| Median | sd | CV | Median | sd | CV | |||||
| Gene-specific probe | GSF | At4g10380 | Major intrinsic protein (MIP)-like | Aquaporin | 4.35 | 0.18 | 4.03 | 0.74* | ** | ** |
| Gene-specific probe | GSF | At1g02920 | Glutathione transferase, putative (GST 11) | Detox | 3.32 | 0.1 | 3.1 | 1.94 | 0.02 | 0.98 |
| Gene-specific probe | GSF | At1g02930 | Glutathione transferase, putative (GST 1) | Detox | 3.52 | 0.09 | 2.61 | 2.03 | 0.04 | 2.1 |
| Gene-specific probe | GSF | At1g10370 | Glutathione transferase, putative (GST 30) | Detox | 3.25 | 0.16 | 4.98 | 1.91* | ** | ** |
| Gene-specific probe | GSF | At1g22340 | UDP-Glc glucosyltransferase, putative | Detox | 2.31 | 0.03 | 1.07 | 2.18 | 0.12 | 5.54 |
| Gene-specific probe | GSF | AT1g22350 | GT-84 | Detox | 2.22 | 0.01 | 0.43 | 1.94 | 0.16 | 8.36 |
| Gene-specific probe | GSF | At1g22400 | UDP-Glc glucosyltransferase, putative | Detox | 2.02 | 0.02 | 0.78 | 1.65 | 0.08 | 4.56 |
| Gene-specific probe | GSF | At1g27130 | Glutathione transferase, putative (GST 12) | Detox | 1.66* | 0.18 | 10.59 | 2.33 | 0.07 | 2.86 |
| Gene-specific probe | GSF | At1g27140 | Glutathione transferase, putative (GST 13) | Detox | 3.86 | 0.14 | 3.49 | 2.15* | ** | ** |
| Gene-specific probe | GSF | At1g30400 | Glutathione S-conjugate ABC transporter (AtMRP1) | Detox | 3.1 | 0.01 | 0.2 | 3.02 | 0.05 | 1.79 |
| Gene-specific probe | GSF | At1g30530 | Glycosyltransferase family | Detox | 2.01 | 0 | 0.15 | 1.75 | 0.11 | 6.23 |
| Gene-specific probe | GSF | At1g59670 | GST-40 | Detox | 2.69 | 0.08 | 2.88 | 1.37 | 0.05 | 3.38 |
| Gene-specific probe | GSF | At1g59870 | ABC transporter family protein | Detox | 3.07 | 0.07 | 2.34 | 1.38 | 0.01 | 0.63 |
| Gene-specific probe | GSF | At1g65820 | Microsomal glutathione S-transferase, putative | Detox | 3.81 | 0 | 0.04 | 3.02 | 0.08 | 2.74 |
| Gene-specific probe | GSF | At1g78270 | UDP-Glc glucosyltransferase, putative | Detox | 2.27 | 0.01 | 0.33 | 1.79 | 0.15 | 8.59 |
| Gene-specific probe | GSF | At2g02930 | Glutathione transferase, putative (GST 16) | Detox | 3.52 | 0.01 | 0.36 | 2.45 | 0.04 | 1.64 |
| Gene-specific probe | GSF | At2g15490 | Putative glucosyltransferase | Detox | 6.47 | 0.2 | 3.02 | 2.92 | 0.05 | 1.7 |
| Gene-specific probe | GSF | At2g29420 | Glutathione transferase, putative (GST 25) | Detox | 5.47 | 0.1 | 1.73 | 1.82* | ** | ** |
| Gene-specific probe | GSF | At2g29460 | Glutathione transferase, putative (GST 22) | Detox | 3.5 | 0.04 | 1.26 | 1.3 | 0.13 | 9.71 |
| Gene-specific probe | GSF | At2g29470 | Glutathione transferase, putative (GST 21) | Detox | 7.1 | 0.29 | 4.07 | ** | ** | ** |
| Gene-specific probe | GSF | At2g29490 | Glutathione transferase, putative (GST 19) | Detox | 2.59 | 0.03 | 1.01 | 1.62* | ** | ** |
| Gene-specific probe | GSF | At2g29710 | Putative flavonol 3-O-glucosyltransferase | Detox | 3.8 | 0.01 | 0.17 | 1.22* | 0.33 | 26.58 |
| Gene-specific probe | GSF | At2g30870 | Glutathione transferase, putative (ERD 13) | Detox | 1.34 | 0.09 | 7 | 2.05 | 0.08 | 3.84 |
| Gene-specific probe | GSF | At2g34660 | Glutathione S-conjugate ABC transporter (AtMRP2) | Detox | 5.07 | 0.06 | 1.2 | 2.9 | 0.09 | 3.03 |
| Gene-specific probe | GSF | At2g47000 | ABC transporter family protein | Detox | 2.35 | 0.03 | 1.09 | 1.42 | 0.04 | 2.9 |
| Gene-specific probe | GSF | At2g47730 | Glutathione transferase, putative (GST6) | Detox | 5.18 | 0.12 | 2.38 | 1.34* | ** | ** |
| Gene-specific probe | GSF | At3g21750 | UDP-glycosyltransferase family | Detox | 2.52 | 0 | 0.1 | 1.19* | ** | ** |
| Gene-specific probe | GSF | At3g55710 | Glucuronosyl transferase-like protein (GT) | Detox | 2.37 | 0.02 | 0.77 | 2.01 | 0.06 | 2.87 |
| Gene-specific probe | GSF | At4g02520 | Glutathione transferase, putative (GST 2) | Detox | 2.05 | 0 | 0 | 1.31* | 0.18 | 13.14 |
| Gene-specific probe | GSF | AT5g03490 | GT-88 | Detox | 2.11 | 0.01 | 0.54 | 1.54 | 0.03 | 2.02 |
| Gene-specific probe | GSF | At5g05880 | Glucuronosyl transferase-like protein (GT) | Detox | ** | ** | ** | 2.13 | 0.07 | 3.37 |
| Gene-specific probe | GSF | At5g41240 | Glutathione transferase, putative (GST 10B) | Detox | 4.67 | 0.06 | 1.28 | 2.01* | ** | ** |
| Gene-specific probe | GSF | AT5g58270 | ABC transporter family protein | Detox | 2.15 | 0.03 | 1.35 | 2.4 | 0.02 | 0.68 |
| AA394533 | ABCR | At1g67980 | Caffeoyl-CoA 3-O-methyltransferase, putative | Liginfication | 2.69 | 0.04 | 1.45 | 1.34 | 0.05 | 3.94 |
| AA395336 | ABCR | At1g75280 | Isoflavone reductase homolog P3 | Metabolism | 3.14 | 0.01 | 0.39 | 1.00* | ** | ** |
| R64825 | ABCR | At2g14890 | Arabinogalactan-protein (AGP9) | Metabolism | 0.36 | 0.04 | 9.88 | 0.98 | 0.08 | 8.45 |
| Z35366 | ABCR | At4g04610 | 5-Adenylylsulfate reductase | Metabolism | 7.88 | 0.28 | 3.6 | 1.21* | 0.21 | 17.13 |
| R30627 | ABCR | At2g36460 | Fru bisphosphate aldolase | Metabolism | 2.6 | 0.04 | 1.62 | 1.13 | 0.02 | 1.45 |
| T88134 | ABCR | At2g04780 | Fasciclin-like arabinogalactan-protein 7 | Metabolism | 4.12 | 0.17 | 4.2 | 1.74 | 0.05 | 2.7 |
| AA713297 | ABCR | At4g30530 | GMP synthase-like protein | Metabolism | 3.27 | 0.01 | 0.3 | 2.56 | 0.09 | 3.57 |
| N37983 | ABCR | At3g22370 | Alternative oxidase 1a precursor | OS | 2.88 | 0.11 | 3.89 | 0.94 | 0.03 | 3.18 |
| Gene-specific probe | GSF | At2g24180 | Cytochrome p450 family | P450 | 2.59 | 0 | 0.14 | 2.34 | 0.07 | 2.98 |
| Gene-specific probe | GSF | At3g26830 | Cytochrome p450 family (PAD 3) | P450 | 10 | 0.14 | 1.41 | 2.99 | 0.08 | 2.74 |
| Gene-specific probe | GSF | At4g31500 | CYP83B1 | P450 | 1.56 | 0.02 | 1.5 | 4.56 | 0.34 | 7.35 |
| Gene-specific probe | GSF | At4g37370 | Cytochrome p450 family | P450 | 11.4 | 0.05 | 0.48 | 3.3 | 0.1 | 2.92 |
| AA712693 | ABCR | At2g02100 | Plant defensin protein, putative (PDF2.2) | PD | 0.36 | 0.01 | 2.51 | 0.77 | 0.04 | 5.35 |
| AA597528 | ABCR | At3g04720 | Hevein-like protein precursor (PR-4) | PD | 2.48 | 0.05 | 2.09 | 1.66 | 0.16 | 9.53 |
| T21313 | ABCR | At3g20600 | NDR1 | PD | 1.33 | 0.11 | 8.38 | 2.02 | 0.13 | 6.55 |
| T45680 | ABCR | At5g06870 | Polygalacturonase inhibiting protein (PGIP2) | PD | 2.58 | 0.01 | 0.19 | 1.51 | 0.06 | 4.1 |
| T44910 | ABCR | At3g45140 | Ser/Thr kinase-like protein | PD | 3.16 | 0.15 | 4.8 | 1.46 | 0.18 | 11.98 |
| AL161590 nt 130695–131312 | GSF | At4g37000 | Red chlorophyll catabolite reductase | Senescence | 9.12* | 1.03 | 11.29 | 2.02 | 0.08 | 3.84 |
| T44308 | ABCR | At3g51240 | Flavanone 3-hydroxylase (F3H) | SM | 0.35 | 0.03 | 9.91 | 0.42* | ** | ** |
| T44580 | ABCR | At5g54810 | Trp synthase β-chain 1 precursor | SM | 3.19 | 0.04 | 1.18 | 1.61* | ** | ** |
| Gene-specific probe | GSF | At1g08110 | Glyoxalase I, putative | SR | 0.47 | 0.02 | 3.94 | 0.73* | ** | ** |
| T88481 | ABCR | At1g10460 | Germin-like oxalate oxidase | SR | 0.46 | 0.02 | 4.97 | 0.91* | 0.29 | 31.61 |
| T22626 | ABCR | At3g48100 | Response reactor 2 (ATRR2) | SR | 0.37 | 0.01 | 1.85 | 0.79* | 0.1 | 13.15 |
| T46224 | ABCR | At5g62920 | Response regulator 6 (ARR6) | SR | 0.48 | 0.03 | 6.2 | 0.85 | 0.06 | 7.21 |
| T21765 | ABCR | At5g61820 | MtN19 | Unknown | 2.27 | 0.09 | 3.78 | 1.83* | 0.2 | 10.83 |
Priming of Tobacco Cells with Either Copper Ions or PPB1 Augments the Resistance toward Severe Oxidative Stress (Copper Ion Poisoning)
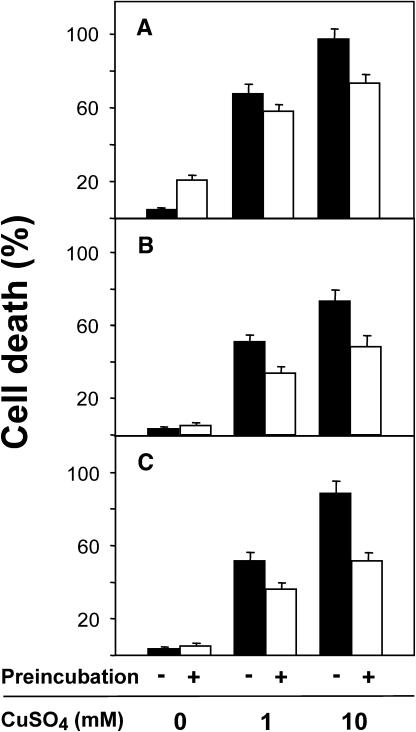
Results obtained from the DNA array analysis of PPB1 induced gene expression suggest that PPB1-I increase the capability of plant cells to detoxify lipid peroxidation products and, thus, may increase the resistance toward oxidative stress. To test this hypothesis, tobacco suspension cells were pretreated with PPB1-I (75 μm) or CuSO4 (75 μm) for 17 h to induce an adaptive response. Preincubation of cells with low concentrations of CuSO4 (75 μm) increased the number of dead cells to 20% (Fig. 2A) when counted after 41 h, while PPB1-I did not increase the mortality of the cells (Fig. 2, B and C). Severe stress, addition of 1 mm or 10 mm CuSO4 to the culture medium of cells that have not received any pretreatment, resulted in cell death of nearly 70% up to 95% of the treated cells after 24 h, respectively (Fig. 2). However, when cells that were either preincubated with 75 μm CuSO4 or PPB1-I for 17 h were subsequently treated with 1 or 10 mm CuSO4, the number of dead cells was significantly reduced. Both PPB1-I isomers were more effective than CuSO4 in adapting tobacco cells to severe oxidative stress (1 or 10 mm CuSO4). After preincubation with PPB1 (16(S)-PPB1-I or 16(R)-PPB1-I), more than 50% of the cells survived, indicating that PPB1 pretreatment indeed triggers an adaptive response in tobacco cells.
Figure 2.
Priming of tobacco cells with CuSO4 or PPB1 reduces cell death caused by severe CuSO4 intoxication. Tobacco cells were preincubated with 75 μm CuSO4 (A), 75 μm 16(S)-PPB1-I (B), and 16(R)-PPB1-I (C) for 17 h (white bars) or water (controls, black bars). Thereafter, cells were incubated with 0 mm, 1 mm, or 10 mm CuSO4 for 24 h and the percentage of dead cells was determined.
Rapid Metabolism of PPB1
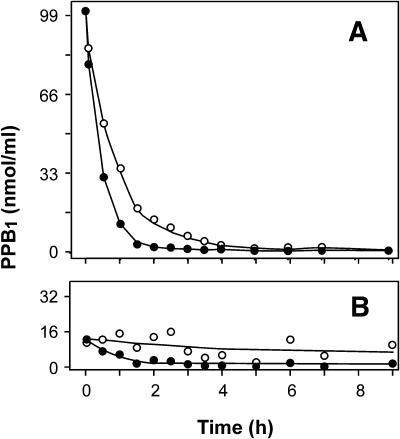
Accumulation of PPB1 in response to certain oxidative stresses appears to be a transient process, suggesting that PPB1 are rapidly metabolized in vivo (Thoma et al., 2003). To investigate the metabolic stability of PPB1, PPB1 were applied to tobacco suspension cells, and PPB1 levels were subsequently determined in the medium and in the cells after different time points. PPB1 regioisomers of type I and II were prepared by autoxidation of α-linolenic acid in vitro as described previously (Parchmann and Mueller, 1998). A tobacco cell culture was incubated with either 100 μm PPB1 type I or type II. At different time points, cells were separated from the medium, shock frozen, and stored in liquid nitrogen until analysis. Levels of PPB1 in medium and cells were determined by HPLC (see “Materials and Methods” for details). As shown in Figure 3, PPB1 type I and II rapidly disappeared from the cell culture medium in the presence of tobacco cells. In contrast, PPB1 were perfectly stable in either culture medium alone or the cell supernatant (data not shown). These experiments indicate that 50% of the PPB1 type I and type II are cleared from the medium by the cells within 36 and 22 min, respectively. Five minutes after PPB1 addition, 13% of the applied PPB1 was removed from the medium and found cell associated. However, further decrease of PPB1 in the medium did not correspond to an increase of PPB1 levels in the cellular fraction. Instead, it was observed that PPB1 levels in the cell associated fraction decreased over time. Thus, PPB1 were either rapidly taken up and instantaneously metabolized within the cells or metabolized by cell wall associated enzymes. Previously, it has been shown that levels of endogenous PPB1 in tobacco cell cultures could be transiently increased by peroxide treatment, suggesting that the half-life of endogenous PPB1 is also well below 60 min (Thoma et al., 2003). Apparently, exogenously administered as well as endogenous PPB1 are rapidly metabolized, suggesting that levels of PPB1 in untreated cells reflect steady-state levels in a high turnover situation (i.e. continuous formation keeps pace with rapid metabolism).
Figure 3.
Metabolism of PPB1 by tobacco cell cultures. Tobacco cell cultures were treated with either PPB1-I or PPB1-II (30.8 μg/mL, 100 μm final concentration). The time course of levels of PPB1-I (○) and PPB1-II (•) in the medium (A) and associated with cells (B) was determined by HPLC.
Delayed Formation of Phytoprostanes in Metabolically Inactive, Dead Cells
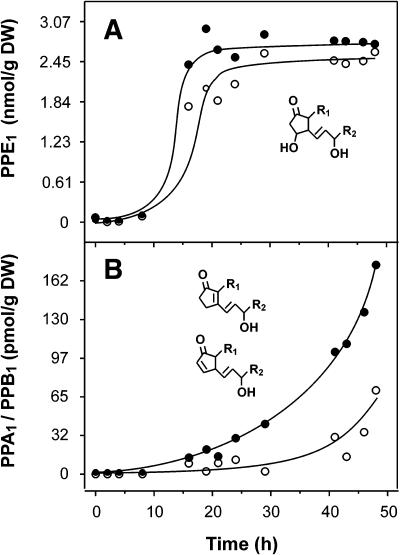
The only requirements for phytoprostane formation are the presence of linolenic acid and molecular oxygen, suggesting that phytoprostane formation not necessarily requires metabolic activity of living cells. Thus, when plain linolenic acid is autoxidized in vitro, PPE1 formation starts instantaneously and increases in an almost linear fashion until α-linolenate is completely autoxidized after 4 d (Thoma et al., 2004). However, in living cells, phytoprostane formation can be catalyzed by free radicals derived from ROS formed by metabolic processes. In vivo, endogenous ROS are produced continuously as by-products of aerobic metabolism at many sites. To estimate the rate of phytoprostane formation by autoxidation in the absence of aerobic metabolism, cultured tobacco cells were killed by shock freezing in liquid nitrogen and thawing. Thereafter, cells were stored at room temperature under sterile conditions and phytoprostane accumulation was monitored. As shown in Figure 4A, E1-phytoprostane (PPE1) levels remained constant for at least 8 h. Thereafter, a more than 40-fold increase of PPE1 levels was observed, reaching high levels after 20 h. Both regioisomers remained at this level (about 800 ng/g of dry weight) for at least 28 h. Since E-ring containing compounds such as PPE1 are relatively stable in the physiological pH range, (Stehle, 1982) nonenzymatic degradation to PPA1/PPB1 is a slow process leading to a delayed accumulation and low levels of these cyclopentenones (Fig. 4B). Similar kinetics of oxidized lipids have been described for the autoxidation of native vegetable oils. Typically, autoxidation of vegetable oils is prevented by endogenous tocopherols and proceeds exponentially only after a delay time (known as the induction period) during which tocopherols are consumed by autoxidation (Frankel, 1998). Initially, lipids in metabolically inactive (dead) cells appear also to be protected from autoxidation by endogenous antioxidants.
Figure 4.
Time course of phytoprostane formation in freeze-killed tobacco cells. Tobacco cells were harvested, shock frozen, and stored at room temperature in a water saturated atmosphere under sterile conditions. At the time points indicated, PPE1 (A) and PPA1/B1 (B) levels were determined in the dead cells. Levels of type I (○) and type II (•) compounds were monitored.
Notably, catalytic amounts of free radicals dramatically increase the lipid peroxidation rate and decrease the induction period (Frankel, 1998). For instance, treatment of tobacco cell cultures with peroxides led to a transient and parallel accumulation of PPE1, PPA1, and PPB1 within 1 h (Thoma et al., 2003). Thus, phytoprostane formation and metabolism (i.e. steady-state levels) of phytoprostanes appear to be regulated in living cells.
Resolution of PPB1 Enantiomers
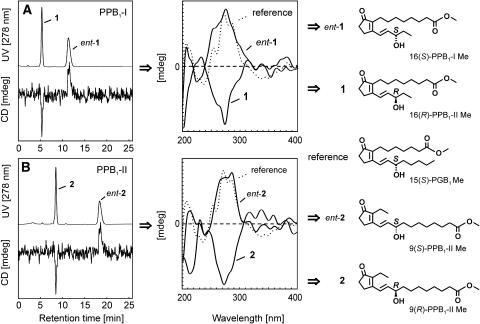
Previously, it has been shown that several classes of phytoprostanes induce secondary metabolism in different plant species (Thoma et al., 2003, 2004). To clarify structure-activity and dose-response relationships, all of the possible regio- and stereoisomeric forms of PPB1 were prepared and their activities were compared with that of MeJA. PPB1 (Fig. 1) consists of two regioisomers, PPB1 type I and II, which were separated by reverse phase (RP)-HPLC as described (Parchmann and Mueller, 1998). The pure PPB1 regioisomers were then methylated and resolved into their enantiomers on a Chiralpak AD column. As shown in Figure 5, baseline separation was achieved for both, the two PPB1-I and the two PPB1-II enantiomers. Assignment of the absolute configuration of the respective enantiomers was attained by hyphenation of the above mentioned chiral HPLC analysis with circular dichroism (CD) spectroscopy. This HPLC-CD coupling, which has recently been introduced into phytochemical analysis (Bringmann et al., 1999), permitted measurement of the full CD spectra of all four isomers in the stop-flow mode without prior isolation. The on-line CD spectra of PPB1 methyl esters were interpreted by comparison with that of authentic 15(S)-prostaglandin B1 methyl ester, which has a similar chromophore (Fig. 5), as a reference. Peaks 1 and 2 showed opposite CD spectra as compared to that of the reference, assigning these to be the (R)-configured PPB1 regioisomers. The corresponding enantiomers, i.e. peaks ent-1 and ent-2, which were eluted more slowly, showed similar CD behaviors, and were thus unambiguously assigned to be (S)-configured. The CD trace recorded at one single wavelength (here at 278 nm, where the CD spectrum shows a maximum) gives clear negative peaks for 1 and 2, and positive ones for ent-1 and ent-2; this permits a rapid and unambiguous assignment of the enantiomers, even in the on-flow mode, in future investigations.
Figure 5.
Resolution of PPB1 enantiomers and determination of their absolute configuration. Racemic methyl esters of PPB1 regioisomers of (A) type I (isomers 1 and ent-1) and (B) type II (isomers 2 and ent-2) were resolved into the enantiomers by chromatography on a Chiralpak AD column. The absolute configuration of all four PPB1 isomers was determined on-line, by circular dichroism spectroscopy hyphenated to HPLC at a chiral stationary phase using 15(S)-prostaglandin B1 as the reference. Compounds ent-1 and ent-2 were assigned to be (S)-configured due to their similar CD spectra compared to those of the reference, while 1 and 2, whose CD are opposite, represent their (R)-configured counterparts.
Induction of Secondary Metabolism by PPB1
For testing the potency of PPB1 isomers in inducing secondary metabolism, three different cell culture systems and the model plant Arabidopsis L. Heyn., ecotype Col-0 were employed.
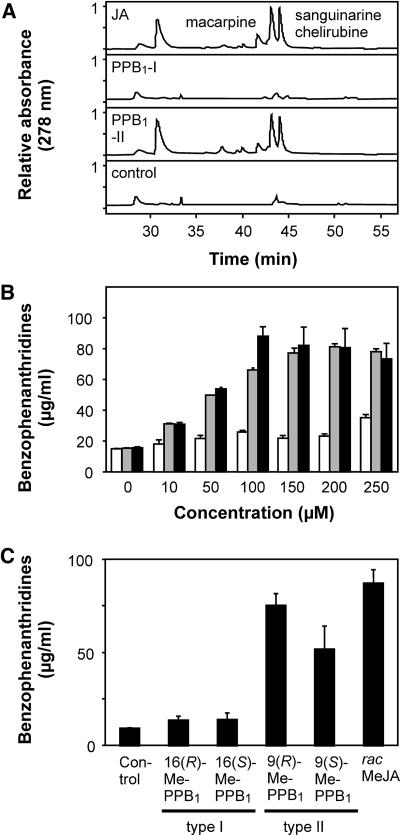
E. californica cell cultures produce only small amounts of benzophenanthridine alkaloids. Elicitation of the cell culture with fungal cell wall fragments as well as jasmonates (JA or MeJA) has been shown to induce defense genes, leading to a dramatic accumulation of these alkaloids with maximum alkaloid levels after 3 to 5 d (Mueller et al., 1993). This bioassay system has found a wide application to evaluate the relative potency of various oxylipins in stimulating secondary metabolism (Blechert et al., 1997). To investigate whether the two regioisomers of PPB1 induce a similar spectrum of secondary metabolites as JA, E. californica cultures were treated with 50 μm of JA, PPB1-I, PPB1-II, or methanol/water (0.1%, v/v, control). After 5 d, cells were harvested by centrifugation and extracted with methanol. Extracts were reduced with sodium borohydride (reduction of benzophenanthridines to their corresponding dihydro-derivatives) and analyzed by HPLC at 278 nm. As shown in Figure 6A, PPB1-II and MeJA induced a similar metabolite pattern. In contrast, PPB1-I (50 μm) did not induce accumulation of any of these alkaloids. Main metabolites were identified as macarpine, sanguinarine, and chelirubine by HPLC (Fig. 6A) and gas chromatography-mass spectrometry after reduction to their dihydro-analogs (data not shown). When comparing the effects of MeJA, PPB1-I, and PPB1-II over a concentration range from 10 to 250 μm, it could be established that PPB1-II was as potent as MeJA in this bioassay system. PPB1-I was almost inactive and elicited only a small alkaloid increase at the highest concentration (Fig. 6B). Alkaloid inducing activity of the methylated PPB1 enantiomers was evaluated at a concentration of 100 μm. The (S)-configured PPB1-II enantiomer was almost as active as the (R)-configured counterpart, while both enantiomers of the type I compound were inactive (Fig. 6C).
Figure 6.
Induction of benzophenanthridine alkaloids in E. californica (Papaveraceae) cell cultures. A, HPLC chromatograms of secondary metabolites induced by 50 μm of JA, PPB1-I, and PPB1-II or 1% (v/v) methanol/water (control). The spectrum of alkaloids induced by JA and PPB1-II is similar and comprises macarpine, sanguinarine, and chelirubine, the principal phytoalexins of E. californica. Alkaloids were determined by HPLC after reduction to their corresponding dihydro-derivatives (see “Materials and Methods”). B, Benzophenanthridine levels determined 5 d after addition of different concentrations of oxylipins (PPB1-I, white bars; PPB1-II, gray bars; JA, black bars). C, Alkaloid accumulation was also determined 5 d after application of methyl esters of PPB1 enantiomers or racemic MeJA (100 μm each).
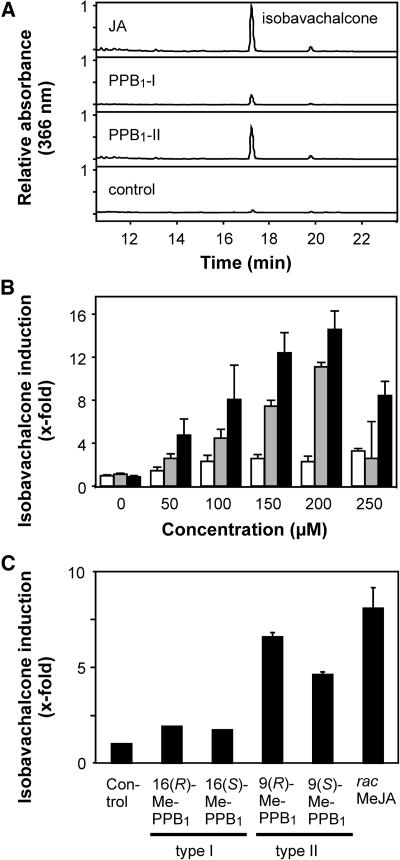
C. cobalticola cell cultures accumulate the chalcone isobavachalcone in response to MeJA (Thoma et al., 2004). As shown in Figure 7, A and B, JA and PPB1-II induce isobavachalcone accumulation in a concentration dependent manner, while PPB1-I displayed only weak activity. Notably, PPB1-II was almost as active as JA. When the four enantiomers of PPB1 were tested as methyl esters, again the (S)-enantiomer of PPB1-II was almost as active as the (R)-configured counterpart.
Figure 7.
Induction of isobavachalcone in C. cobalticola (Fabaceae) cell cultures. HPLC chromatograms of secondary metabolites induced by 100 μm of JA, PPB1-I, and PPB1-II are shown. A, The major metabolite induced by PPB1 and JA was identified as isobavachalcone. B, Isobavachalcone levels were determined 36 h after addition of different concentrations of oxylipins (PPB1-I, white bars; PPB1-II, gray bars; JA, black bars). C, Isobavachalcone accumulation was also determined 36 h after application of methyl esters of PPB1 enantiomers or racemic MeJA (100 μm each).
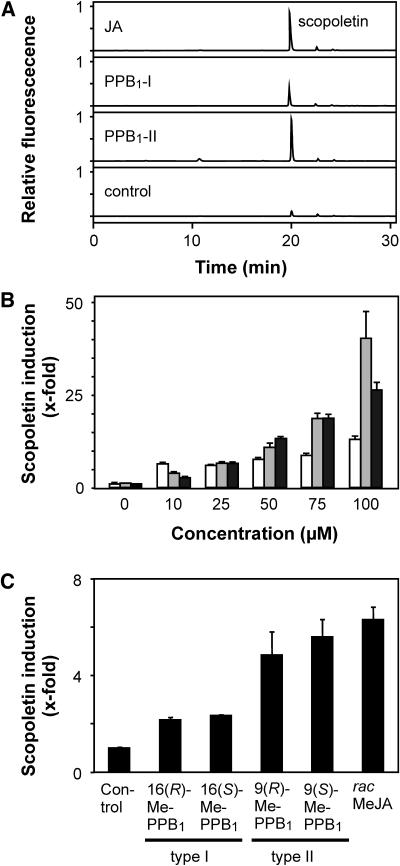
Previously, it has been shown that in tobacco cell cultures several classes of phytoprostanes including PPB1 and JA induce a transient accumulation of scopoletin in the medium (Thoma et al., 2003). Maximum scopoletin levels were observed 4 h after addition of the lipids. When PPB1-I, PPB1-II, and JA were tested at different concentrations (Fig. 8, A and B), it was found that the scopoletin inducing potency increased in the order PPB1-I < PPB1-II = JA. Again, there was no difference between the corresponding (R)- and (S)-enantiomers of the PPB1-methyl esters (Fig. 8C). It is unlikely that racemization of PPB1 takes place in vivo, since PPB1 do not epimerize in the physiological pH range even after prolonged storage. There is also no evidence indicating that epimerization of prostaglandin B1 takes place in mammals.
Figure 8.
Induction of scopoletin in tobacco (Solanaceae) cell cultures. A, Scopoletin accumulation in the medium of tobacco cell suspension cultures induced by 10 μm of JA, PPB1-I, and PPB1-II was determined after an incubation period of 4 h by HPLC. B, Scopoletin levels in the culture medium were determined 4 h after addition of different concentrations of oxylipins (PPB1-I, white bars; PPB1-II, gray bars; JA, black bars). C, Scopoletin accumulation was also determined 4 h after application of methyl esters of PPB1 enantiomers or racemic MeJA (10 μm each).
Taken together, it can be concluded that in the three bioassay systems the potency of the lipids followed generally the order PPB1-I < PPB1-II ≈ JA/MeJA and is independent of the configuration of the side chain hydroxyl group of the two PPB1 regioisomers.
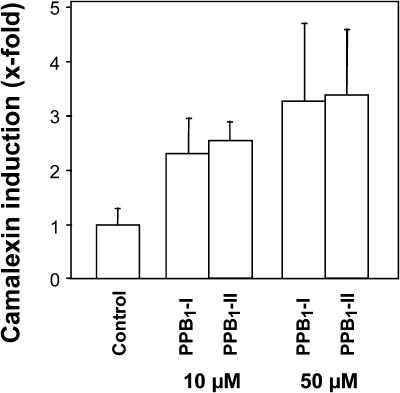
In Arabidopsis, the sulfur-containing indole derivative camalexin is the predominant phytoalexin that accumulates after infection with bacteria and fungi (Rogers et al., 1996). Camalexin biosynthesis is also induced following PPB1 treatment (Fig. 9). After infiltration of Arabidopsis L. Heyn., ecotype Col-0 leaves with 10 μm or 50 μm of PPB1-I and PPB1-II, respectively, an increase of camalexin was found showing maximum phytoalexin levels after 5 d. In contrast to the cell suspension bioassay systems described above, but in agreement with the gene expression data for PAD3 (believed to be involved in camalexin biosynthesis), both PPB1 regioisomers induced camalexin biosynthesis. Nevertheless, the response of Arabidopsis plants following treatment with 10 μm and 50 μm PPB1 was much less pronounced than that observed after infection with Pseudomonas syringae pv tomato DC3000 (55-fold camalexin induction 5 d after infiltration of the virulent bacteria into Arabidopsis leaves; data not shown), various other phytopathogenic bacteria, fungi, or oxidative stress (Thomma et al., 2001), suggesting that in addition to PPB1 other yet to be identified mediators participate in the camalexin response in vivo. In this context, it should be noted that PPB1 are components of a much larger pattern of biologically active phytoprostanes and other oxidized lipids that are generated simultaneously during oxidative stress (Mueller, 2004).
Figure 9.
Induction of camalexin in Arabidopsis plants. Camalexin accumulation in leaves of Arabidopsis induced by 10 μm or 50 μm of PPB1-I and PPB1-II was determined after an incubation time of 5 d by HPLC.
DISCUSSION
Living plant cells continuously produce ROS at many intracellular sites during normal aerobic metabolism. ROS such as hydrogen peroxide and superoxide anions can be either inactivated by antioxidative enzymes or converted in vivo into the highly detrimental hydroxyl radical that in turn may initiate free radical chain reactions leading to the accumulation of a great variety of oxidized lipids. Nonenzymatically generated oxidized lipids such as 12- and 16-hydroperoxy fatty acids (Berger et al., 2001), malondialdehyde (Weber et al., 2004), and phytoprostanes (Thoma et al., 2004) are therefore present in healthy plants throughout their life cycle. These findings indicate that in vivo, the plant antioxidative capacity is not sufficient to suppress free radical formation entirely. In metabolic inactive (dead) cells, however, endogenous antioxidants are sufficient to prevent nonenzymatic lipid peroxidation for at least several hours (Fig. 4). Hence, formation of oxidized lipids (i.e. PPB1) in vivo reflects most likely free radical generation by metabolic processes.
After an induction period, continuous autoxidation of linolenate in dead cells leads to steadily increasing levels of chemically stable lipid peroxidation end products such as PPB1 as long as precursors are present (Fig. 4). This, however, is not observed in living cells. Instead, we show (Fig. 3) that PPB1 are short-lived molecules that are rapidly metabolized in vivo, suggesting that basal levels of PPB1 in healthy plants reflect steady-state levels in a high turnover situation. It remains to be clarified how PPB1 are metabolized. In analogy to JA, metabolism may include conjugation to amino acids or Glc, reduction of the ring double bond and/or the ring carbonyl, as well as β-oxidation of the carboxyl side chain.
Notably, levels of several classes of phytoprostanes including PPB1 can be transiently increased by a variety of oxidative stresses such as exogenous peroxides or heavy metals and pathogens (Imbusch and Mueller, 2000b; Thoma et al., 2003). Actual levels of short-lived oxidized lipids therefore indicate severe oxidant injury or metabolic disturbance and can be used as markers of oxidative stress in vivo. However, isoprostanes/phytoprostanes and malondialdehyde are not only excellent biomarkers but also biologically active molecules.
To this end, we observed that both PPB1 regioisomers (occurring in plants in a 1:1 ratio) may induce a variety of genes (Table I). Analysis revealed that the two regioisomers of PPB1 display similar effects on induction and repression of a variety of genes, albeit the PPB1-I regioisomer was generally more potent than the PPB1-II regioisomer. Notably, strong induction of genes involved in detoxification and secondary metabolism was observed.
PPB1 increased the expression of at least 17 glutathione S-transferases including GST1 (which was reported previously to be triggered by oxidative stress), deoxy-J1-prostaglandin, PPA1, and PPB1 (Thoma et al., 2003; Iqbal et al., 2005). Glutathione S-transferases are thought to play a role in the detoxification of reactive electrophiles, which may enter cells from the outside (i.e. herbicides, xenobiotics) or are generated endogenously by lipid peroxidation (Wagner et al., 2002). Furthermore, several putative ABC transporters and the vacuolar glutathione conjugate ABC transporters AtMRP1 and AtMRP1, which belong to the multidrug resistance-associated protein family (Lu et al., 1998; Marin et al., 1998; Liu et al., 2001), are also induced by PPB1. These transporters are thought to transport a broad spectrum of glutathione conjugates and chlorophyll breakdown products into the vacuole. Thus, induction of an array of glutathione S-transferases and glutathione conjugate transporters by lipid peroxidation end products such as PPB1 may serve to detoxify protein damaging electrophiles. Results suggest that PPB1 may induce defense mechanisms that reduce protein damage by electrophiles and thus enable plants to cope better with the consequences of oxidative stress. This hypothesis was brought to experimental testing.
We show that pretreatment of tobacco cell cultures with PPB1 primed the cells to resist severe oxidative stress caused by copper intoxication. Pretreatment of tobacco cells with low concentrations of copper ions also rendered the cells more resistant to severe copper poisoning. Notably, PPB1 are neither reactive electrophiles (see below) nor toxic molecules (Fig. 2). Thus, weak oxidative stress as well as low levels of PPB1 triggers an adaptive response that partially prevents cell death. It remains to be shown if this effect is displayed only by PPB1 or shared by other phytoprostanes.
In plant pathogen interactions, plants seemingly produce oxidants such as ROS willingly. ROS are established plant signals that may induce defense genes via different mechanisms (Apel and Hirt, 2004; Mueller, 2004). One arm of ROS signal transduction apparently includes the nonenzymatic formation of oxidized lipids that in turn may act as intracellular second messengers that limits oxidative cell injury (Fig. 2) and induces the biosynthesis of antimicrobial secondary metabolites (Figs. 6–9).
Interestingly, induction of secondary metabolism is a well-known plant defense reaction in response to conditions associated with enzymatic ROS or jasmonate formation. This study indicates that ROS-inducible PPB1 are as active as MeJA or JA in inducing secondary metabolism in taxonomically distant plant species. PPB1 comprise two regioisomers, each of which is composed of two enantiomers. Notably, the type II regioisomers were considerably more active than the type I compounds, suggesting that the secondary metabolite inducing effect is not due to the almost similar physicochemical properties of PPB1 and apparently involves specific recognition mechanisms yet to be identified.
Recently, it has been suggested by Farmers group that molecules containing an α,β-unsaturated carbonyl group such as cyclopentenones activate a set of electrophile responsive genes (not activated by JA) due to their inherent chemical reactivity (Almeras et al., 2003). Although PPB1 are cyclopentenones, they are only weak electrophiles that do not fall into that category. In contrast to 12-oxo-phytodienoic acid, malondialdehyde, A-, J-, and deoxy-J-ring prostanoids, as well as cyclopentenone, PPB1 are unreactive, do not conjugate to free thiol groups (i.e. glutathione; data not shown), and, hence, are unable to chemically modify proteins. In animals, chemical modification of transcription factors and regulatory proteins by reactive cyclopentenones/electrophiles has been proposed to represent a common signaling mechanism triggering apoptosis as well as antiinflammatory and antiviral cell responses (Straus and Glass, 2001). However, B-ring compounds do not display any of these activities.
The finding that the PPB1-I isomers were more active than the PPB1-II isomers in inducing certain genes while the reverse was observed for the secondary metabolite inducing activity suggests that more than one recognition mechanism exists for PPB1 (or its metabolites). However, PPB1 isomers differ in their relative potency but are not qualitatively different. The same is apparently true for the perception of structurally related jasmonates. For instance, Arabidopsis jar1 mutants lack an enzyme conjugating amino acids to the free carboxyl group of JA. It has been shown that this enzyme displays high specificity for JA and is essential for some but not all JA-mediated responses such as root growth inhibition and resistance toward certain microorganisms and ozone (Staswick and Tiryaki, 2004). In contrast, secondary metabolites can be induced by a surprisingly great variety of natural and synthetic JA analogs, even those lacking a free carboxyl group such as jasmone (Birkett et al., 2000) or coronatine (Haider et al., 2000; Schüler et al., 2004). In addition, a broad spectrum of jasmonate analogs has been compared in several JA bioassay systems, revealing that small structural changes do not only change their relative potency but also that a structural related compound may display increased activity in one bioassay and decreased activity in another bioassay (Blechert et al., 1997). Apparently, PPB1, 12-oxo-phytodienoic acid, and JA all display their own spectrum of activities, which, however, partially overlap. The different mechanisms involved in recognition of each compound remain to be clarified.
Notably, phytoprostanes have prevailed throughout the evolution of the enzymatic jasmonate pathway and might have an evolutionary ancient function as mediators of oxidative stress in host defense, while jasmonates have more advanced and specific functions in development, defense, and reproduction (Mueller, 2004).
MATERIALS AND METHODS
DNA Array
The Arabidopsis (Arabidopsis thaliana) DNA array currently being used for examining stress and/or redox-regulated gene expression involves longer fragments of synthetic or complementary DNA. Sequences are derived from databanks, as PCR-amplified partial open reading frames or specific 3′ untranslated region (UTR)-sequences. The array consists of about 626 gene fragment cDNAs involved in or associated with plant defense and includes 50 cDNAs associated with primary metabolism and/or housekeeping. Specific 3′ UTR sequences are for members of the family of ABC transporters, P450 monooxygenases, glucosyltransferases, glutathione-S-transferases, and aquaporins (Glombitza et al., 2004). Members of other gene families are represented by parts (at least 450 bp) or complete coding sequence. These expressed sequence tag clones are available from Arabidopsis Biological Resource Center (ABCR); The Ohio State University, Rightmire Hall, 1060 Carmack Road, Columbus, OH 43210; or designed at the GSF-National Research Center for Environment and Health, 85764 Oberschleissheim, Germany. Analysis was performed with GenePix 4.1 and Acuity 3.1 from Axon. Normalization was over all normalization features. Normalization features were all features printed on the array for which the following quality criteria applied: at least 55% of the pixels in both signals (635 nm and 532 nm) of a given spot are stronger than the background, plus sd. Feature of background uniformity: [Rgn R2 (635/532)] was higher than 0.5; only spots with less than 3% saturated pixels were considered, and undetected spots or weak signals (sum of medians >500) were excluded.
PPB1 Treatment of Arabidopsis Suspension Cells and DNA Array Preparation
Arabidopsis callus culture was derived from Col-0 seeds (L. Heynh.; Lehle Seeds, Round Rock, TX) and grown on media containing 0.2% agar-substitute Phytagel (Sigma, Steinheim, Germany), 1× Murashige and Skoog + MES salts (Duchefa, Haarlem, The Netherlands), 0.56 mm myoinositol, 0.1 mm FeSO4, 0.13 mm EDTA, 2.26 μm 2,4-dichlorophenoxyacetic acid, 4.06 μm nicotinic acid, 2.5 μm pyridoxal hydrochloride, 0.3 μm thiamine hydrochloride, and 2% d-Suc, pH 5.7. Arabidopsis suspension-cultured cells derived from callus were grown at 26°C on a rotary shaker in callus medium without agar (Deeken et al., 2003). Cells were subcultured weekly by transferring 20 mL of cells into 50 mL of fresh medium. Suspension cells (10 mL) were treated with 75 μm PPB1-I, 75 μm PPB1-II, or methanol/water (controls). Experiments were conducted in four replicates. After an incubation of 3 h, cells were harvested by filtration and quickly frozen in liquid nitrogen. RNA of the 4 replicates was extracted, pooled, and split into 4 vials each containing 40 μg of RNA.
Amino-modified PCR products (200 μL) were cleaned using 96-well multiscreen filter plates (Millipore, Bedford, MA; catalog no. MANU03050) and suspended in 20 μL spotting solution (3× SSC supplemented with 1.5 m betaine) and arrayed from 384-well DNA array plates onto silylated microscope slides (CSS-100 silylated slides; CEL Associates, Houston) using a DNA array robot (model GMS 417 from Genetic Microsystems). Printed arrays were incubated at room temperature overnight and rinsed twice in 0.1% (w/v) SDS with vigorous agitation for 2 min, twice in double distilled water for 2 min, and once for 5 min in sodium borhydride solution (0.75 g NaBH4 dissolved in 200 mL of PBS and 75 mL of 100% ethanol). The arrays were submerged in water for 2 min at 95°C, transferred quickly into 0.1% SDS for 1 min, rinsed twice in double distilled water, air dried, and stored in the dark at room temperature.
Fluorescent Probes
Target RNA from PPB1-treated Arabidopsis cells was extracted using the TRIzol reagent according to the supplier's instructions (GIBCO/BRL). Probes were made using an indirect aminoallyl labeling method as described (http://www.tigr.org/tdb/microarray/protocols.shtml). Each mRNA sample (one control and one treated sample) was reverse-transcribed in the presence of Cy3-dCTP or Cy5-dCTP (Amersham Pharmacia Biotech, Freiburg, Germany) and purified according to standard protocols.
Hybridization and Scanning
Following reverse transcription, labeling, and purification steps, each of Cy3- and Cy5-labeled probes were combined, added to 1 μL salmon sperm DNA (20 μg/μL) and 1 μL poly(A+)-DNA (20 μg/μL), dried in speedvac, and dissolved in 50 μL hybridization buffer (50% formamide, 6× SSC, 0.5% SDS, 5× Denhardt's). The probes were heated at 95°C for 3 min to denature and cooled on ice. The slides were immersed into prehybridization buffer (6× SSC, 0.5% SDS, 1% bovine serum albumin, and 1 μg/mL ssDNA) at 42°C for 45 min, thoroughly washed with double distilled water, and then air dried. Probes were hybridized to 1.0 cm2 microarrays in 14 × 14 Abgene gene frames in hybridization chambers (Genemachines) overnight. Subsequently, the arrays were washed for 10 min at low stringency (2× SSC/0.1% SDS), then for 5 min in 1× SSC/0.1% SDS, and finally for 5 min in high stringency wash buffer (0.1× SSC/0.1% SDS). After a brief wash in double distilled water, the arrays were air dried and scanned using an AXON GenePix 4000 scanner. Separate images were acquired for each fluorophore at a resolution of 10 μm/pixel. To identify differentially expressed genes, we used the GenePix Pro 4.1 software. Background fluorescence was calculated as the median fluorescence signal of nontarget pixels around each gene spot. Less than 50% difference between background and signal resulted in elimination of the corresponding spot.
Preparation and Analysis of PPB1
PPB1 were prepared by autoxidation of linolenic acid. The two PPB1 regioisomers were separated and isolated by RP-HPLC as described (Parchmann and Mueller, 1998).
Copper Sulfate Treatments and Cell Death Assays in Tobacco Suspension Cells
Tobacco (Nicotiana tabacum) cv Xanthi cell suspensions were used for the experiments 3 d after subculturing. Aliquots of 10 mL (about 0.5 g cell fresh weight) were transferred to 30 mL of fresh Linsmaier and Skoog (LS) medium. The cell suspensions were incubated under normal growth conditions for 12 h before addition of either 75 μm CuSO4, 75 μm 16(S)-PPB1-I or 75 μm 16(R)-PPB1-I, or methanol (control). Lipids were applied in methanol (final methanol concentration did not exceed 0.1%, v/v). After 17 h preincubation, cells were treated with CuSO4 (1 mm and 10 mm) or water and incubated for another 24 h. Subsequently, dead cells were stained with trypan blue (0.15%, v/v) and counted under the microscope.
PPB1 Metabolism in Tobacco Cell Cultures
Tobacco cv Xanthi L. cells (8 g) were grown in 100 mL of LS medium for 3 d before PPB1 regioisomers (dissolved in 100 μL of methanol) were added to yield a final concentration of 100 μm (30.8 μg/mL). At the times indicated, 1-mL aliquots of the cell suspension were centrifuged (1 min, 2,000g) to separate cells from the supernatant. Cells were washed twice with free medium prior extraction. One microgram of prostaglandin B1 was added to the cells and the supernatant as internal standard. The supernatant was acidified with citric acid (100 mm) to pH 3.5 and extracted twice with 1 mL of diethyl ether. The organic phases were combined, evaporated, and the residue analyzed by HPLC as described above.
Washed cells were extracted with 1 mL of ethanol:water:37% HClaq (80:19.8:0.2, v/v) for 2 h at 60°C. After centrifugation (5 min, 10,000g), the supernatant was dried under a stream of nitrogen and analyzed by HPLC.
Phytoprostane Formation in Freeze-Killed Tobacco Cells
Tobacco cells (80 g) were grown in 1 L of LS medium for 3 d. Subsequently, cells were collected by suction filtration. Cells were shock frozen in liquid nitrogen, spread out in a sterile container, and allowed to thaw. Cells were stored in the dark at room temperature and aliquots were analyzed for phytoprostanes at the times indicated by gas chromatography-mass spectrometry as described (Thoma et al., 2003). Since PPA1 isomerize almost quantitatively to PPB1 in the injector of the GC, PPA1/PPB1 were determined together.
Resolution and Determination of the Absolute Configuration of PPB1 Enantiomers
For resolution of the enantiomers of PPB1-I and II, each regioisomer was methylated with an etheral solution of diazomethane and applied to a Chiralpak AD 250 × 4.6-mm HPLC column (Daicel Chemical Industries, Japan). The column was eluted isocratically with a mixture of hexane:methanol:ethanol (8:1:1, v/v) at a flow rate of 1 mL min−1 and monitored at 278 nm. Circular dichroism spectra for each enantiomer were recorded on-line in the stop-flow mode on a Jasco J-715 spectropolarimeter, using a 5-mm flow cell. The CD spectrum of authentic 15(S)-prostaglandin B1 methyl ester, whose (S)-configuration causes a positive cotton effect at 278 nm, was used as the reference. The CD spectra of peaks ent-1 and ent-2 were similar to that of the reference, leading to the assignment of the absolute configuration to be 16(S)-PPB1-I and 9(S)-PPB1-II. In contrast, the (R)-configured PPB1 enantiomers, i.e. peaks 1 and 2, showed opposite CD spectra with negative cotton effects at 278 nm.
Induction of Secondary Metabolites by Phytoprostanes
Cell suspension cultures were obtained from the departmental culture collection and grown as described (Gundlach et al., 1992). Cells were harvested under sterile conditions by suction filtration, resuspended in 1-L flasks containing 250 mL of medium or 300-mL flasks containing 80 mL of medium, and used after a growth period of 3 d directly for elicitation experiments. Lipids (dissolved in 100 μL of methanol) were added to yield the final concentration indicated. Methanol (100 μL) was added to control cells.
Benzophenanthridine alkaloids levels in Eschscholzia californica Cham. (Papaveraceae) cell cultures were determined 5 d after application of lipids. Cells were harvested and extracted as described (Gundlach et al., 1992) and benzophenanthridine alkaloid levels were quantitated photometrically (Blechert et al., 1995). Extracts were also analyzed by HPLC. Therefore, extracts were dissolved in water adjusted with ammonia to pH 12 and extracted with diethyl ether. The organic phase containing dihydro-benzophenanthridine alkaloids was discarded. The water phase was treated with sodium borhydride to reduce quaternary alkaloids to their corresponding dihydro-derivatives and extracted with diethyl ether. The ether phase was dried under a stream of nitrogen and the residue was analyzed on a LiChrospher RP18ec column (5 μm particle size, 250 × 4 mm; Merck, Darmstadt, Germany). The column was eluted with a mixture of water:acetonitrile: 85% phosphoric acid (95:5:0.1, v/v, solvent A) and water:acetonitrile:85% phosphoric acid (10:90:0.1, v/v, solvent B) at a flow rate of 1.5 mL min−1. Solvent B was linearly increased from 0% (0 min) to 25% (10 min), 25% (20 min), 50% (35 min), 70% (40 min), 70% (46 min), and 75% (56 min). Dihydro-benzophenantridine alkaloids were detected at 278 nm.
Crotalaria cobalticola cell culturing and isobavachalcone quantitations were performed as described (Gundlach et al., 1992). Isobavachalcone levels increase transiently in C. cobalticola cells and were determined 36 h after addition of oxylipins (Thoma et al., 2004).
Tobacco cell culture treatments and scopoletin analysis were performed as described (Thoma et al., 2003). Scopoletin levels in the medium increased transiently and were determined after an incubation time of 4 h (Thoma et al., 2003).
PPB1 Treatment of Arabidopsis Leaves and Camalexin Determinations
Leaves of 6-week-old soil-grown Arabidopsis plants were infiltrated with 10 μm or 50 μm PPB1-I (dissolved in methanol/water, 0.1%, v/v). Control leaves were treated with methanol/water (0.1%, v/v). After an incubation of 5 d, leaves were harvested and immediately frozen in liquid nitrogen. For camalexin analyses, 50 μg of 6-fluoroindole-3-carboxaldehyde (internal standard, purchased from Sigma) was added to leaves (250 mg) prior extraction with 500 μL of methanol/water (80%, v/v) in an ultrasonic water bath for 10 min. Extraction was repeated and the combined extracts were partitioned against 3 × 1 mL of petrol ether. The upper petrol ether phases were discarded and the remaining methanol/water phase was subjected to HPLC analysis on a Purospher STAR RP-18 ec column (250 × 4.6 mm; 5 μm; Merck, Darmstadt, Germany). The solvents were water (A) and acetonitrile (B). Solvent B was linearly increased from 0% (0 min) to 10% (1 min), 20% (6 min), 20% (16 min), 55% (33.5 min), 55% (34 min), and 100% (45 min) at a flow rate of 1 mL min−1. Camalexin was monitored using a fluorescent detector (λex = 305 nm, λem = 364 nm).
Supplementary Material
Acknowledgments
We thank Barbara Dierich and Beate Hilbert for excellent technical assistance.
This work was supported by the Deutsche Forschungsgemeinschaft, Bonn, Germany (grant no. SFB 567 to S.B., G.B., and M.J.M.) and by the Fonds der Chemischen Industrie.
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.051714.
References
- Almeras E, Stolz S, Vollenweider S, Reymond P, Mene-Saffrane L, Farmer EE (2003) Reactive electrophile species activate defense gene expression in Arabidopsis. Plant J 34: 205–216 [DOI] [PubMed] [Google Scholar]
- Apel K, Hirt H (2004) Reactive oxygen species: metabolism, oxidative stress and signal transduction. Annu Rev Plant Biol 55: 373–399 [DOI] [PubMed] [Google Scholar]
- Berger S, Weichert H, Porzel A, Wasternack C, Kühn H, Feussner I (2001) Enzymatic and nonenzymatic lipid peroxidation in leaf development. Biochim Biophys Acta 1533: 266–276 [DOI] [PubMed] [Google Scholar]
- Birkett MA, Campbell CAM, Chamberlain K, Guerrieri E, Hick AJ, Martin JL, Matthes M, Napier JA, Petterson J, Pickett JA, et al (2000) New roles for cis-jasmone as an insect semiochemical and in plant defense. Proc Natl Acad Sci USA 97: 9329–9334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blechert S, Bockelmann C, Brümmer O, Füßlein M, Gundlach H, Haider G, Hölder S, Kutchan TM, Weiler EW, Zenk MH (1997) Structural separation of biological activities of jasmonates and related compounds. J Chem Soc, Perkin Trans 1 1: 3549–3559 [Google Scholar]
- Blechert S, Brodschelm W, Hölder S, Kammerer L, Kutchan TM, Xia Z-Q, Zenk MH (1995) The octadecanoic pathway: signal molecules for the regulation of secondary pathways. Proc Natl Acad Sci USA 92: 4099–4105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bringmann G, Messer K, Wohlfarth M, Kraus J, Dumbuya K, Rückert M (1999) HPLC-CD on-line coupling in combination with HPLC-NMR and HPLC-MS/MS for the determination of the full absolute stereostructure of new metabolites in plant extracts. Anal Chem 71: 2678–2686 [Google Scholar]
- Cracowski JL (2003) The putative role of isoprostanes in human cardiovascular physiology and disease: following the fingerprints. Heart 89: 821–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cracowski JL (2004) Isoprostanes: an emerging role in vascular physiology and disease? Chem Phys Lipids 128: 75–83 [DOI] [PubMed] [Google Scholar]
- Cracowski JL, Durand T, Bessard G (2002) Isoprostanes as a biomarker of lipid peroxidation in humans: physiology, pharmacology and clinical implications. Trends Pharmacol Sci 23: 360–366 [DOI] [PubMed] [Google Scholar]
- Deeken R, Ivashikina N, Czirjak T, Philippar K, Becker D, Ache P, Hedrich R (2003) Tumour development in Arabidopsis thaliana involves the Shaker-like K+ channels AKT1 and AKT2/3. Plant J 34: 778–784 [DOI] [PubMed] [Google Scholar]
- El Fangour S, Guy A, Despres V, Vidal JP, Rossi JC, Durand T (2004) Total synthesis of the eight diastereomers of the syn-anti-syn phytoprostanes F1 types I and II. J Org Chem 69: 2498–2503 [DOI] [PubMed] [Google Scholar]
- Frankel EN (1998) Lipid Oxidation. Oily Press, Dundee, UK
- Glombitza S, Dubuis P-H, Thulke O, Welzl G, Bovet L, Götz M, Affenzeller M, Geist B, Hehn A, Asnaghi C, et al (2004) Crosstalk and differential response to abiotic and biotic stressors reflected at the transcriptional level of effector genes from secondary metabolism. Plant Mol Biol 54: 817–835 [DOI] [PubMed] [Google Scholar]
- Gundlach H, Müller MJ, Kutchan TM, Zenk MH (1992) Jasmonic acid is a signal transducer in elicitor-induced plant cell cultures. Proc Natl Acad Sci USA 89: 2389–2393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haider G, von Schrader T, Füßlein M, Blechert S, Kutchan TM (2000) Structure-activity relationships of synthetic analogs of jasmonic acid and coronatine on induction of benzo[c]phenanthridine alkaloid accumulation in Eschscholzia californica cell cultures. Biol Chem 381: 741–748 [DOI] [PubMed] [Google Scholar]
- Huang X, von Rad U, Durner J (2002) Nitric oxide induces transcriptional activation of the nitric oxide-tolerant alternative oxidase in Arabidopsis suspension cells. Planta 215: 914–923 [DOI] [PubMed] [Google Scholar]
- Imbusch R, Mueller MJ (2000. a) Analysis of oxidative stress and wound-inducible dinor isoprostanes F1 (phytoprostanes F1) in plants. Plant Physiol 124: 1293–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imbusch R, Mueller MJ (2000. b) Formation of isoprostane F2-like compounds (phytoprostanes F1) from α-linolenic acid in plants. Free Radic Biol Med 28: 720–726 [DOI] [PubMed] [Google Scholar]
- Iqbal M, Evans P, Lledó A, Verdaguer X, Pericàs MA, Riera A, Loeffler C, Sinha AK, Mueller MJ (2005) Total synthesis and biological activity of 13,14-dehydro-12-oxo-phytodienoic acids (deoxy-J1-phytoprostanes). ChemBioChem (in press) [DOI] [PubMed]
- Janssen LJ (2004) The pulmonary biology of isoprostanes. Chem Phys Lipids 128: 101–116 [DOI] [PubMed] [Google Scholar]
- Liu G, Sanchez-Fernandez R, Li ZS, Rea PA (2001) Enhanced multispecificity of arabidopsis vacuolar multidrug resistance-associated protein-type ATP-binding cassette transporter, AtMRP2. J Biol Chem 276: 8648–8656 [DOI] [PubMed] [Google Scholar]
- Lu YP, Li ZS, Drozdowicz YM, Hortensteiner S, Martinoia E, Rea PA (1998) AtMRP2, an Arabidopsis ATP binding cassette transporter able to transport glutathione S-conjugates and chlorophyll catabolites: functional comparisons with Atmrp1. Plant Cell 10: 267–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin E, Leonhardt N, Vavasseur A, Forestier C (1998) Cloning of AtMRP1, an Arabidopsis thaliana cDNA encoding a homologue of the mammalian multidrug resistance-associated protein. Biochim Biophys Acta 1369: 7–13 [DOI] [PubMed] [Google Scholar]
- Mueller MJ (2004) Archetype signals in plants: the phytoprostanes. Curr Opin Plant Biol 7: 441–448 [DOI] [PubMed] [Google Scholar]
- Mueller MJ, Brodschelm W, Spannagl E, Zenk MH (1993) Signaling in the elicitation process is mediated through the octadecanoid pathway leading to jasmonic acid. Proc Natl Acad Sci USA 90: 7490–7494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parchmann S, Mueller MJ (1998) Evidence for the formation of dinor isoprostanes E1 from α-linolenic acid in plants. J Biol Chem 273: 32650–32655 [DOI] [PubMed] [Google Scholar]
- Rodriguez AR, Spur BW (2003) First total synthesis of the E type I phytoprostanes. Tetrahedron Lett 44: 7411–7415 [Google Scholar]
- Rogers EE, Glazebrook J, Ausubel FM (1996) Mode of action of the Arabidopsis thaliana phytoalexin camalexin and its role in Arabidopsis-pathogen interactions. Mol Plant Microbe Interact 9: 748–757 [DOI] [PubMed] [Google Scholar]
- Schüler G, Mithöfer A, Baldwin IT, Berger S, Ebel J, Santos JG, Herrmann G, Hölscher D, Kramell R, Kutchan TM, et al (2004) Coronalon: a powerful tool in plant stress physiology. FEBS Lett 563: 17–22 [DOI] [PubMed] [Google Scholar]
- Staswick PE, Tiryaki I (2004) The oxylipin signal jasmonic acid is activated by an enzyme that conjugates it to isoleucine in Arabidopsis. Plant Cell 16: 2117–2127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stehle RG (1982) Physical chemistry, stability, and handling of prostaglandins E2, F2α, D2, and I2: a critical summary. Methods Enzymol 86: 436–458 [Google Scholar]
- Straus DS, Glass CK (2001) Cyclopentenone prostaglandins: new insights on biological activities and cellular targets. Med Res Rev 21: 185–210 [DOI] [PubMed] [Google Scholar]
- Thoma I, Krischke M, Loeffler C, Mueller MJ (2004) The isoprostanoid pathway in plants. Chem Phys Lipids 128: 135–148 [DOI] [PubMed] [Google Scholar]
- Thoma I, Loeffler C, Sinha AK, Gupta M, Steffan B, Krischke M, Roitsch T, Mueller MJ (2003) Cyclopentenone isoprostanes induced by reactive oxygen species trigger defense gene activation and phytoalexin accumulation in plants. Plant J 34: 363–375 [DOI] [PubMed] [Google Scholar]
- Thomma BPHJ, Tierens KFM, Penninckx IAMA, Mauch-Mani B, Broekaert WF, Cammue BPA (2001) Different micro-organisms differentially induce Arabidopsis disease response pathways. Plant Physiol Biochem 39: 673–680 [Google Scholar]
- Wagner U, Edwards R, Dixon DP, Mauch F (2002) Probing the diversity of the Arabidopsis glutathione S-transferase gene family. Plant Mol Biol 49: 515–532 [DOI] [PubMed] [Google Scholar]
- Weber H, Chetelat A, Reymond P, Farmer EE (2004) Selective and powerful stress gene expression in Arabidopsis in response to malondialdehyde. Plant J 37: 877–888 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.