Abstract
During seed maturation and germination, major changes in physiological status, gene expression, and metabolic events take place. Using chlorophyll sorting, osmopriming, and different drying regimes, Brassica oleracea seed lots of different maturity, stress tolerance, and germination behavior were created. Through careful physiological analysis of these seed lots combined with gene expression analysis using a dedicated cDNA microarray, gene expression could be correlated to physiological processes that occurred within the seeds. In addition, gene expression was studied during early stages of seed germination, prior to radicle emergence, since very little detailed information of gene expression during this process is available. During seed maturation expression of many known seed maturation genes, such as late-embryogenesis abundant or storage-compound genes, was high. Notably, a small but distinct subgroup of the maturation genes was found to correlate to seed stress tolerance in osmoprimed and dried seeds. Expression of these genes rapidly declined during priming and/or germination in water. The majority of the genes on the microarray were up-regulated during osmopriming and during germination on water, confirming the hypothesis that during osmopriming, germination-related processes are initiated. Finally, a large group of genes was up-regulated during germination on water, but not during osmopriming. These represent genes that are specific to germination in water. Germination-related gene expression was found to be partially reversible by physiological treatments such as slow drying of osmoprimed seeds. This correlated to the ability of seeds to withstand stress.
Reproduction through seeds is a prominent feature of higher plants. Seeds are adapted to survive for periods of time under adverse conditions until conditions favorable for seedling establishment are encountered. Usually, mature seeds have low moisture contents, reduced metabolic activity, and have accumulated protective compounds to help them survive under rather severe conditions. In the course of seed maturation, various events happen including the accumulation of storage products, the suppression of precocious germination, the acquisition of desiccation tolerance, and often the induction of dormancy (for review, see Bewley and Black, 1994). Seeds become quiescent at desiccation and can often be stored for a long time. When nondormant dry seeds imbibe water, they can germinate to start a new lifecycle. During germination, a series of events occurs, such as the activation of respiration (Bewley and Black, 1994), the repair of macromolecules (Osborne, 1993), reserve mobilization (Gallardo et al., 2001), reinitiation of the cell cycle (De Castro et al., 1995; Vásquez-Ramos and Sánchez, 2004), and weakening of covering structures to allow radicle protrusion (Groot and Karssen, 1987). At the same time, seeds lose longevity during germination and desiccation tolerance upon radicle protrusion (Hong and Ellis, 1992). Our study focuses on these early events during germination until just after radicle protrusion (germination sensu strictu) and does not address later events during germination that are related to seedling growth and establishment.
From an economic point of view, the quality of dry seeds is important in agriculture, since seeds are often starting material for crop production and crucial for achieving a good harvest. Several aspects of seed quality influence agricultural performance, such as total emergence, the rate and uniformity of emergence, emergence under suboptimal conditions, and seed longevity. To improve field emergence and uniformity, priming treatments may be applied. During priming treatments seeds are allowed to take up water and start (part of) their germination-related processes, but emergence of the radicle is prevented to avoid the loss of desiccation tolerance that is needed for subsequent drying, storage, and marketing of the treated seeds. Priming treatments are used to synchronize the germination of individual seeds (Heydekker et al., 1973). Since certain germination-related processes are initiated, priming generally causes faster germination and field emergence, especially under adverse field conditions (McDonald, 2000). To prevent radicle protrusion, water uptake may either be limited by imbibition in an osmotic solution (osmopriming) instead of water or by restricting the period of germination on water and drying the seeds prior to radicle protrusion. During osmopriming only a subset of events occurs, compared to germination on water, as previously demonstrated at the protein level (Gallardo et al., 2001).
A negative side effect of priming is that the longevity of primed seeds can be considerably less compared to that of the nonprimed seed lot. For several species a partial restoration of longevity could be obtained by keeping the seeds, after the priming treatment, under a mild water and/or temperature stress for a period of several hours to days (Bruggink et al., 1999). The seed industry is in need of tests that can provide information on the physiological quality of seed lots. Especially, the identification of markers to predict longevity should prove useful for the seed industry because, using current standard procedures, it takes a long time to measure seed longevity accurately.
The expression of certain genes during maturation and seed processing, like priming, results in an altered physiological state and affects seed quality. Therefore, the genes whose expression levels are different among seed lots of different quality could be used as seed quality markers. To more fully understand the complex interplay of genes during maturation and germination high-throughput methods to analyze the expression of many genes are needed. DNA microarray technology and proteomic analysis are now available as quite powerful techniques for such purposes. As mentioned above, Gallardo et al. (2001) studied the change of protein patterns during Arabidopsis (Arabidopsis thaliana) seed priming and germination using proteomic analysis. They showed that the transition of the quiescent seed to germination is accompanied by a diminution or increase of certain proteins. Among the about 1,300 proteins resolved, only about 74 showed a clear change in their abundance during germination and radicle protrusion. During osmopriming the level of only a subset of these proteins changed.
Seed development can be divided into distinct stages such as pattern formation, growth, and seed filling, followed by desiccation drying prior to dispersal by the plant. Gene expression during early Arabidopsis seed development (up to 12 days after pollination [DAP]) was studied using cDNA microarrays (Girke et al., 2000; Ruuska et al., 2002). Seed expressed sequence tags (ESTs) were used to create a cDNA microarray of 2,600 elements, 25% of which had preferential expression in seeds (Girke et al., 2000). Later work using an array with approximately 3,500 unique genes illustrated the vast gene expression changes that accompany seed filling. Distinct expression patterns related to photosynthetic processes, carbohydrate metabolism, lipid biosynthesis genes, and seed storage proteins were found (Ruuska et al., 2002). That study focused on gene expression during early stages of seed development and filling, up to 12 DAP, when seeds have attained their final size but have not yet started desiccation drying. Although much is known about factors controlling seed maturation and the transition to germination (for review, see Holdsworth et al., 1999; Wobus and Weber, 1999; Koornneef et al., 2002; Peng and Harberd, 2002), little is known about global gene expression programs during seed desiccation and subsequent germination.
To achieve a better molecular understanding of maturation, germination, and seed quality, we analyzed gene expression during germination and priming using cDNA microarrays containing genes that were isolated from almost mature (AM) or germinating Brassica napus seeds. For this purpose, we used Brassica oleracea seeds that differed in maturity, quality, and germinability and subjected these to germination, osmopriming, and drying treatments. We used B. oleracea since this crop combines the benefits of being closely related to the molecular plant model species Arabidopsis, which has a fully sequenced and annotated genome, while at the same time the results obtained can be directly applied in the analysis of seed quality problems encountered in agronomic practice with this or related crops. An additional advantage is the relatively large size of the seed compared to Arabidopsis, which is more suitable for the type of studies performed and allows sorting of the seeds into different maturity classes out of a single large seed lot.
In this report, we describe new molecular markers for seed quality and demonstrate the coordinated expression of gene sets during acquisition, loss, and regaining of stress and desiccation tolerance of seeds.
RESULTS
Physiological Characterization of the Seed Samples
Small Differences in Maturity Have Large Effects on Seed Quality
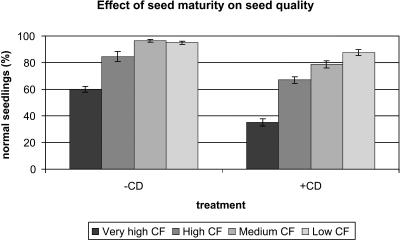
Brassica plants flower over an extended period of time, and as a consequence, seeds of different ages are present on the plant. During maturation, chlorophyll levels in Brassica seeds decline. When seeds are harvested and processed, small differences in maturity level remain, which can be detected only by using fluorescence-based detection methods. Dark brown seeds that are visually indistinguishable can therefore be sorted based on chlorophyll content (Jalink et al., 1998). A nonuniform seed batch of Bartolo (purposely chosen for this study to be of noncommercial quality) was sorted into fractions based on their chlorophyll content. The highest and lowest chlorophyll-containing fractions, representing the least and the most mature seeds, were selected for further analysis. For clarity, the high chlorophyll fraction will be referred to as AM seeds, whereas the low chlorophyll fraction will be referred to as fully mature (FM) seeds in the remainder of the article. Germination and a controlled deterioration (CD) stress treatment revealed the quality differences between mature and near mature seeds in a single seed lot, as the FM fraction displayed higher germination rates and speed than the AM fraction. Additionally the FM seeds performed better after a CD test (Fig. 1), illustrating a higher stress tolerance.
Figure 1.
Effect of seed maturity on stress tolerance and germination performance. Seeds were sorted based on chlorophyll (CF) content and subjected to a CD test to assess stress tolerance. More mature seeds have lower chlorophyll levels. The left section shows the number of normal seedlings obtained from the various seed fractions in a standard germination test, and the right section shows the number of normal seedlings obtained when seeds are first subjected to CD for 5 d and then germinated. The very high chlorophyll fraction (very high CF; AM) and the low chlorophyll fraction (FM) were used for microarray experiments.
The Decline in Longevity That Occurs during Priming Can Be Partially Restored by Slow Drying
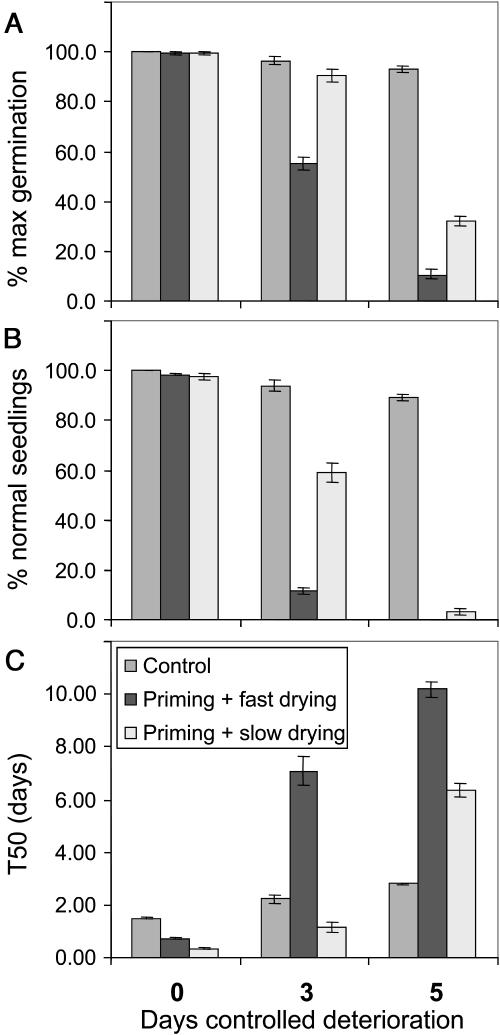
Priming treatments are commonly used to improve the germination speed and uniformity of commercial seed lots. During osmopriming, seeds are limited in their uptake of water, allowing some germination-related processes to occur without resulting in radicle protrusion and loss of desiccation tolerance. Osmopriming was done using a polyethylene glycol (PEG) solution with an osmotic potential of −1.0 MPa for 7 d. After the osmopriming treatment seed moisture content was about 40%. To store primed seeds, they need to be dried back to a lower moisture content, which was done by one of two alternative methods, fast or slow drying. Fast drying at 20°C reduced the moisture content of the primed seeds to 7% within 24 h, while the slow-drying method at 30°C reduced the moisture content to 25% in the first 72 h of drying, followed by fast drying at 20°C to 7% as in the fast-drying method. Figure 2 illustrates the beneficial effects of osmopriming on germination characteristics. The left sections of Figure 2, A to C (no CD), show almost 100% germination and normal seedlings in primed seeds, accompanied by a considerably faster germination of primed seeds relative to nonprimed seeds. Although osmopriming improved germination behavior of seeds subjected to both drying treatments, slow-dried primed seeds germinated even faster than fast-dried primed seeds, as seen by the change in T50 from 1.49 d (control seeds) to 0.73 d (fast-dried osmoprimed seeds) or 0.34 d (slow-dried osmoprimed seeds).
Figure 2.
Effect of priming and drying treatments on germination performance. Germination performance was scored based on the percentage germination (A), the percentage normal seedlings (B), and the germination speed (T50; C). Prior to the germination test seeds were subjected to 0 (left), 3 (middle), or 5 (right) d of CD.
To assess the effect of osmopriming on seed longevity and stress tolerance, control, primed, and dried seeds were subjected to a CD test. Unprimed control seeds were best able to withstand the CD test. Osmopriming followed by fast drying severely decreased longevity of B. oleracea seeds, whereas osmopriming followed by slow drying had less severe effects on seed longevity. Both total germination and the proportion of normal seedlings were significantly higher in slow-dried primed seeds than in fast-dried primed seeds (Fig. 2, A and B). Another effect of the CD treatment was the reduction in germination speed. Again, control seeds suffered the least from the CD test, while slow-dried primed seeds performed better than fast-dried primed seeds (Fig. 2C). Other tests confirmed the improved stress tolerance and storability of slow-dried primed seeds compared to fast-dried primed seeds, as slow-dried primed seeds were better able to withstand an ultra-drying treatment or 8-month milder storage than fast-dried primed seeds (Groot et al., 2003).
Gene Expression Analysis
EST Analysis and Microarray Preparation
Random cDNAs from three different B. napus seed libraries were used to prepare a cDNA microarray. Hybridizations were done with RNA from B. oleracea seeds. B. napus is an amphidiploid species with 38 chromosomes (20 from Brassica rapa and 18 from B. oleracea genomes). The first B. napus cDNA library was prepared from mRNA isolated from bent-cotelydon stage immature seeds (harvested 28 DAP). For the other two libraries germinating seeds from a very mature fraction (harvested 70 DAP, sorted for low chlorophyll content) were used. The second library was prepared from root tips, isolated from seeds that had been germinated on water for 7 h, while the third library was prepared using mRNA from whole seeds that had been germinated on water for 15 h. A total of 238 cDNA clones (149 sequenced) from the bent-cotyledon stage seed library, 599 cDNA clones (373 sequenced) from the 7-h germination seed root tip library, and 600 cDNA clones (457 sequenced) from the 15-h germination seed library were spotted. In addition, 33 Arabidopsis ESTs (or PCR products) and 12 tomato (Lycopersicon esculentum) ESTs related to stress, germination, cell cycle, or seed development, yeast (Saccharomyces cerevisiae) genes that do not hybridize with plant cDNA (negative controls), and luciferase cDNA (for normalization) were spotted on the microarray.
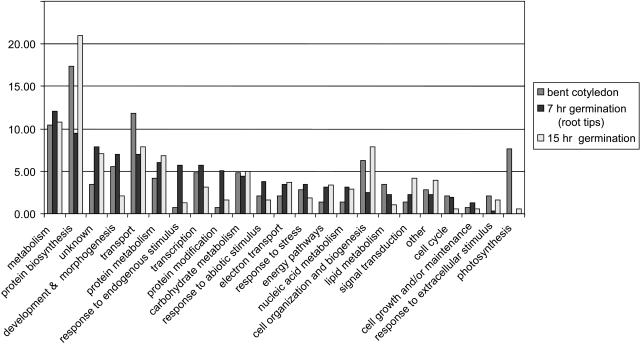
Due to the high sequence homology between Brassica and Arabidopsis (average sequence identity in the coding region between Arabidopsis and Brassica species is 87%; http://ukcrop.net/brassica.html), we were able to classify the unique genes from each library using the annotated Arabidopsis genome (Fig. 3). The homology between B. napus and B. oleracea cDNAs is expected to be higher, since they share a common genome. This was confirmed by the fact that signal intensities were much higher when B. oleracea RNA was used for target preparation than when Arabidopsis RNA was used for hybridizations (data not shown). Although the number of ESTs is relatively low, the relative abundance of the different gene classes can be seen as an indicator of their expression in the tissues used to prepare the libraries.
Figure 3.
EST classification of the cDNA libraries. Distribution of the cDNAs identified in the three libraries over the various gene ontology classes.
In the cDNA library from developing seeds photosynthesis-related genes were relatively more abundant than in the other two libraries, indicating that these seeds were photosynthetically active at this stage (Fig. 3). After germinating mature seeds in water for 7 h, ESTs normally present in dry seeds (including storage component biosynthesis genes and late-embryogenesis abundant [LEA] genes), as well as ESTs related to active metabolism (translation, carbon metabolism) were found in the root tip library, representing stored mRNA and newly synthesized mRNA. The 7-h germination seed root tip library shows a greater diversity in gene classes. This is reflected by the fact that fewer genes (9.5%) fall in the protein biosynthesis class (Fig. 3) and that the distribution over different classes is more even. This could be related to the fact that this library contains both dry-seed-abundant mRNAs (not yet degraded) and germination-related mRNAs (already expressed). Another striking difference between this library and the others is the relative abundance of genes involved in response to internal or external stimulus (such as stress and abiotic factors). This illustrates either a remaining presence of stored mRNA related to the acquisition of stress tolerance during seed maturation or de novo gene expression induced by fast water uptake that occurs during the transition from a dry quiescent state to an actively growing seedling. This is especially clear in the 7-h germination seed library, since for this library RNA was isolated from root tips, the part of the seed where the cell cycle starts first (Bino et al., 1992).
After 15 h germination in water, gene classes related to reserve mobilization and growth become more dominant (e.g. protein biosynthesis and metabolism, cell organization and biogenesis, and signal transduction), since during this phase cell divisions have started and radicle protrusion occurs within hours (Fig. 3).
Microarray Data Analysis
Each clone was spotted in duplicate onto the array, and hybridizations of most samples were done twice (using swapped dyes), yielding four measurements for each clone. Independent clones from the same gene that were spotted on the microarray were used to verify reliability of the hybridization data. To ensure that only reproducible differences in gene expression were included in the analysis the following criteria were used. Only data points from cDNAs with a hybridization intensity more than 1.5 times background in all 4 spots of a given sample (representing swapped-dye analysis in duplo) were used. From the 1,500 cDNAs on the array, an average of 247 cDNAs were thus excluded per sample. This included most of the tomato ESTs and about one-half the Arabidopsis ESTs that had been spotted. Furthermore, in rare cases single outliers (individual spots that differed from the other three spots for the same cDNA/sample combination) were removed using the two-sided Dixon's test at α = 0.05 with exclusion criterion 0.829 (Massart et al., 1997). This resulted in the exclusion of an average of 50 outliers per sample. Finally, when variation among the 4 data points exceeded a preset limit in the absence of a clear outlier (CV > 1.1), all 4 data points were left out of the analysis, removing 3 cDNAs per sample. For cluster analysis, only clones that gave a reliable hybridization result (3 or 4 data points with acceptable variance) in all samples (1,006 clones), all samples but 1 (95 clones), or all samples but 2 (15 clones) were included in the analysis. Thus, out of 1,500 cDNAs spotted onto the chip, 1,100 cDNAs fit these criteria and were used in the analysis. When redundancy on the chip is taken into account, these 1,100 cDNAs represent approximately 850 individual genes.
Overall Expression Patterns
The different B. oleracea seed samples analyzed in this study represent three different maturity levels (AM, nonsorted dry, and FM), two types of priming/drying treatment, differing in germination speed and stress tolerance, and three germination stages. Germination of B. oleracea seeds on water-soaked filter paper results in relatively fast and uniform radicle protrusion (90% germination within 48 h; t50 = 38 h, U90−10 = 17 h), so the three germination time points represent gene expression before (15 and 30 h) and after (45 h) radicle protrusion. Osmoprimed seeds prior to drying were used as a common reference in all hybridizations since these represent an intermediate state between dry and germinating seeds, facilitating comparisons to both dry and germinating seeds. Therefore, in all figures and tables mRNA levels in the various seed samples are expressed as a 2-log ratio compared to RNA levels in osmoprimed seeds prior to drying.
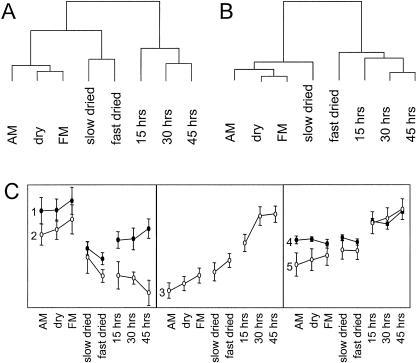
When the expression pattern of all 1,100 genes included in the analysis is used to cluster the seed samples, the physiological similarities in the different seed samples can be seen. The cluster analysis shows these three main groups, reflecting dry seeds, primed seeds, and seeds germinating on water, respectively (Fig. 4A). Repeating the clustering of samples after excluding redundant spots (taking the average expression of all spots representing a single gene) and selecting those genes that show expression patterns that are indicative for seed maturation, stress tolerance, or seed germination (the genes found in Tables I–IV), a different cluster pattern emerges. Now only two main groups are visible, one representing stress-tolerant samples (including dry and slow-dried seeds) and one representing stress-sensitive samples (including germinating and fast-dried seeds; Fig. 4B). This illustrates that the choice of genes included in the analysis influences the outcome of the hierarchical clustering of the samples, since they are clustered on the basis of the expression profiles of the selected genes. Based on their overall expression patterns, we divide the genes into five main groups. These are shown in Figure 4C. The significance of the individual groups is discussed below, and the genes they contain can be found in Tables I to IV.
Figure 4.
Clustering the samples based on their overall gene expression programs. A, Clustering based on all genes used in the analysis (1,100 random genes). B, Clustering based on the differentially expressed genes after averaging spots representing the same gene as listed in Tables I to IV. C, Average expression profiles of gene groups obtained by hierarchical clustering. Groups 1 to 5 are described in the text and Tables I to IV and contained 10, 13, 38, 33, and 10 genes, respectively.
Table I.
Genes preferentially expressed during seed maturation, in which RNA levels decrease during priming and/or germination
Predicted gene function, number of independent cDNAs on the array that represent that gene (between parentheses), the closest Arabidopsis gene homolog, and the average 2-log ratios of swapped dye duplo hybridizations relative to osmoprimed seed are shown. Genes discussed in the text are indicated in bold font. RNA levels from group 1 genes are relatively high in dry seeds and during later germination, whereas group 2 gene RNA levels decline during both priming and germination (Fig. 4C).
| Predicted Gene Function | Arabidopsis Homolog | AM | Dry | FM | Slow | Fast | 15 h | 30 h | 45 h |
|---|---|---|---|---|---|---|---|---|---|
| Group 1 | |||||||||
| Napin (2S storage protein) (15) | AT4G27140, 150, 160 | 4.11 | 2.79 | 0.82 | −0.34 | −0.16 | 1.52 | 1.54 | 1.10 |
| Oleosin (4) | AT3G27660 | 3.08 | 2.23 | 2.84 | 0.52 | 0.24 | 1.48 | 1.39 | 1.83 |
| Jacalin lectin family protein (2) | AT3G21380 | 2.66 | 3.82 | 4.70 | 0.62 | −1.16 | −0.01 | −0.72 | 0.12 |
| Hypothetical protein | AT1G08220 | 1.24 | 1.50 | 2.12 | 1.31 | −0.59 | 0.15 | 0.61 | 0.90 |
| HSP17.6A, cytoplasmic | AT5G12020 | 2.78 | 1.96 | 3.57 | −0.42 | −0.06 | 0.68 | 0.92 | 1.22 |
| HSP23.6, mitochondrial | AT4G25200 | 2.56 | 2.66 | 3.29 | −0.29 | −1.11 | 0.58 | −0.08 | 1.07 |
| B. napusβ-glucosidase | AT3G21370 | 2.71 | 2.82 | 3.36 | 0.31 | 0.06 | 0.79 | 0.66 | 1.28 |
| Hypothetical protein | AT1G05510 | 2.02 | 2.23 | 2.84 | −0.36 | −0.25 | −0.17 | 0.36 | 1.40 |
| E3 ubiquitin ligase APC1, putative | AT5G05560 | 2.56 | 2.16 | 2.46 | −0.09 | −0.01 | 1.95 | 2.01 | 2.88 |
| Expressed protein | AT1G63000 | −0.39 | 0.46 | 1.75 | −0.17 | −0.77 | 0.18 | 1.86 | 2.48 |
| Group 2 | |||||||||
| Cruciferin (12S storage protein) (4) | AT4G28520 | 1.02 | 0.24 | −0.46 | −0.37 | −1.89 | −1.61 | −2.01 | −2.36 |
| RAB18 | AT5G66400 | 2.39 | 2.51 | 3.34 | 2.02 | −0.96 | −1.29 | −2.01 | −3.56 |
| Em6 (10) | AT2G40170 | 0.67 | 2.12 | 3.28 | 0.80 | −1.24 | 0.69 | −0.78 | −1.73 |
| Seed maturation protein (group 5 LEA) | AT3G22490 | 0.01 | 1.05 | 2.04 | −2.30 | −1.46 | −1.84 | −2.38 | −1.92 |
| M10 LEA | AT2G41280 | 1.95 | 2.08 | 2.63 | 0.18 | −1.16 | −0.18 | −1.30 | −1.68 |
| M17 LEA (3) | AT2G41260 | 1.07 | 0.68 | 1.33 | −2.14 | −1.39 | −1.16 | −1.97 | −4.33 |
| LEA76 gene (3) | AT3G15670 | 0.16 | 0.01 | 0.53 | −1.27 | −1.45 | −3.47 | −0.77 | −3.12 |
| Group 1 LEA like (2) | AT5G06760 | −0.70 | 0.36 | 0.94 | −0.84 | −1.76 | −2.58 | −1.38 | −2.09 |
| Group 3 LEA-like protein | AT3G17520 | −0.07 | 0.92 | 1.42 | −1.28 | −1.24 | −2.26 | −1.02 | −1.37 |
| Dormancy related (short-chain dehydrogenase) | AT1G54870 | 1.95 | 2.01 | 2.76 | −0.91 | −2.36 | −0.09 | −1.44 | −2.61 |
| Histone H1-1 | AT1G06760 | 1.82 | 1.54 | 2.74 | 0.84 | −0.09 | −1.95 | −1.71 | 0.16 |
| Squalene epoxidase | AT4G37760 | 0.98 | 0.68 | 1.42 | −2.47 | −1.70 | −1.24 | −1.96 | −3.88 |
| Average peroxiredoxin (2) | AT1G48130 | 0.49 | 1.12 | 1.71 | −0.52 | −2.07 | −2.58 | −1.73 | −2.78 |
Table II.
Genes in which RNA levels increase during priming and germination
Predicted gene function, number of independent cDNAs on the array that represent that gene (between parentheses), the closest Arabidopsis gene homolog, and the average 2-log ratios of swapped dye duplo hybridizations relative to osmoprimed seed are shown. Genes discussed in the text are indicated in bold font. Average expression of group 3 genes is shown in Figure 4C.
| Predicted Gene Function | Arabidopsis Homolog | AM | Dry | FM | Slow | Fast | 15 h | 30 h | 45 h |
|---|---|---|---|---|---|---|---|---|---|
| S-Adenosyl-Met synthetase (2) | AT3G17390 | −1.60 | −2.26 | −1.64 | −0.93 | 0.33 | 0.52 | 2.65 | 2.59 |
| Gap C (2) | AT3G04120 | −2.52 | −1.88 | −1.61 | −1.39 | −1.21 | 0.84 | 2.82 | 2.64 |
| Sugar epimerase (2) | AT5G28840 | −2.09 | −1.94 | −1.36 | −1.16 | −0.43 | 0.32 | 2.37 | 2.55 |
| Dioxygenase (4) | AT2G25450 | −3.62 | −2.64 | −2.51 | −2.60 | −0.91 | 0.37 | 2.94 | 2.49 |
| Sus malate dehydrogenase | AT5G43330 | −2.06 | −2.31 | −2.06 | −1.59 | −1.20 | −0.07 | 1.42 | 1.72 |
| Cytochrome P450 83B1 | AT4G31500 | −1.69 | −1.80 | −1.35 | −0.45 | 0.46 | −0.10 | 1.51 | 2.16 |
| Probable fibrillarin | AT4G25630 | −2.11 | −1.88 | −1.75 | −1.18 | −0.80 | −0.02 | 1.70 | 1.86 |
| Vacuolar membrane ATPase subunit G | AT3G01390 | −2.12 | −1.69 | −1.09 | −0.54 | −0.16 | −0.36 | 0.60 | 1.04 |
| Xyloglucan endo-1,4-β-d-glucanase | AT4G30280 | −1.79 | −2.31 | −0.98 | −0.38 | −0.24 | −0.93 | 0.10 | 1.87 |
| Ser carboxypeptidase II-3 | AT5G08260 | −3.92 | −2.90 | −2.43 | −1.12 | −1.23 | 0.67 | 0.16 | |
| Putative imbibition protein | AT3G57520 | −2.12 | −2.12 | −1.77 | −2.19 | −0.93 | 0.78 | 1.28 | 0.67 |
| AtGRP2 (3) | AT4G13850 | −1.76 | −1.31 | −0.67 | −0.57 | −0.30 | 1.48 | 2.92 | 2.81 |
| Glutathione transferase | AT2G29450 | −1.60 | −1.35 | −1.24 | −0.85 | −0.49 | 1.09 | 2.63 | 3.02 |
| Stress related protein | AT3G05500 | −2.32 | −1.16 | −0.95 | −0.30 | −0.22 | 2.70 | 3.54 | 2.04 |
| l-Ascorbate peroxidase | AT4G35000 | −1.91 | −1.29 | −1.11 | −0.30 | 0.03 | 1.01 | 2.43 | 3.02 |
| Putative allergen (3) | AT1G78040 | −1.99 | −1.24 | −0.85 | −0.79 | −0.08 | 1.18 | 3.27 | 3.22 |
| UbiEP (2) | AT1G23410 | −1.30 | −1.11 | −0.57 | −0.36 | −0.10 | 1.54 | 2.85 | 2.96 |
| Cytochrome c | AT1G22840 | −1.31 | −1.41 | −0.89 | −0.33 | −0.02 | 0.83 | 2.21 | 2.89 |
| Expressed protein | AT3G08030 | −1.24 | −1.09 | −0.57 | 0.48 | −0.24 | 2.08 | 3.21 | 1.30 |
| Expressed protein | AT1G74730 | 1.10 | 0.83 | 0.39 | 0.92 | 0.43 | 2.22 | 1.55 | 2.44 |
| Putative protein | AT4G30010 | −2.66 | −2.08 | −1.66 | −1.33 | −0.62 | −0.10 | 1.01 | 1.52 |
| Expressed protein | AT5G25460 | −3.10 | −2.54 | −2.29 | −1.52 | −1.32 | 1.03 | 1.85 | 1.13 |
| Expressed protein | AT1G56580 | −2.06 | −1.39 | −1.13 | −0.47 | 0.59 | 0.64 | 2.47 | 2.95 |
| Vacuolar H+-ATPase subunit | AT1G19910 | −2.23 | −1.06 | −0.35 | −0.95 | −0.87 | 0.63 | 1.54 | 1.94 |
| Gly-rich protein | AT4G02450 | −2.01 | −0.93 | −0.17 | 0.13 | −0.31 | 1.03 | 2.44 | 1.64 |
| Nucleotide translocator (3) | AT5G13490 | −2.70 | −2.37 | −2.04 | −1.89 | −1.49 | 0.17 | 1.28 | 1.72 |
| Initiation factor 5A-4 | AT1G26630 | −1.69 | −1.68 | −0.57 | −1.16 | −0.47 | 0.94 | 2.50 | 2.81 |
| Elongation factor EF-1 α (5) | AT1G07940 AT5G60390 | −2.76 | −1.77 | −1.75 | −1.79 | −1.43 | 0.52 | 2.14 | 2.00 |
| Elongation factor 1B α-subunit | AT5G19510 | −2.84 | −2.05 | −1.57 | −1.88 | −1.06 | −0.93 | 0.77 | 1.15 |
| 60S rib L18A (5) | AT2G34480 | −2.99 | −2.13 | −1.56 | −0.83 | −0.62 | 0.06 | 1.61 | 1.79 |
| 60S ribosomal protein L8 (RPL8C) | AT4G36130 | −3.46 | −2.64 | −2.24 | −2.50 | −1.73 | 0.30 | 1.59 | 2.29 |
| 60s rib. prot. L36 | AT2G37600 | −1.84 | −2.13 | −1.51 | −1.85 | −0.73 | 0.65 | 2.07 | 2.25 |
| Ribosomal protein GL41-like | AT3G56020 | −2.39 | −1.78 | −1.33 | −1.00 | −0.26 | 0.12 | 2.04 | 2.47 |
| 60S rib L27A (3) | AT1G70600 | −2.81 | −1.81 | −1.35 | −1.79 | −1.01 | 0.44 | 1.99 | 2.28 |
| 60s ribosomal protein L3 (3) | AT1G43170 | −3.17 | −2.44 | −1.84 | −2.48 | −1.70 | −0.16 | 1.75 | 1.81 |
| 40S rib L17 (3) | AT5G15200 | −2.11 | −1.39 | −1.07 | −1.02 | −0.53 | 0.55 | 1.97 | 2.18 |
| 60S acidic ribosomal protein | AT5G47700 | −1.85 | −1.78 | −0.86 | −0.96 | −0.82 | 0.24 | 1.92 | 2.01 |
| Ribosomal protein L18a (5) | AT2G34480 | −2.99 | −2.13 | −1.56 | −0.83 | −0.62 | 0.06 | 1.61 | 1.79 |
Table III.
Genes in which RNA levels increase primarily during germination
Predicted gene function, number of independent cDNAs on the array that represent that gene (between parentheses), the closest Arabidopsis gene homolog, and the average 2-log ratios of swapped dye duplo hybridizations relative to osmoprimed seed are shown. Genes discussed in the text are indicated in bold font. Average expression of group 4 and 5 genes is shown in Figure 4C.
| Predicted Gene Function | Arabidopsis Homolog | AM | Dry | FM | Slow | Fast | 15 h | 30 h | 45 h |
|---|---|---|---|---|---|---|---|---|---|
| Group 4 | 1.62 | 2.67 | |||||||
| Superoxide dismutase | AT4G25100 | 0.57 | 0.69 | 0.36 | 0.58 | 0.57 | 1.60 | 1.37 | 2.05 |
| Glutathione S-transferase | AT2G47730 | 1.07 | 0.85 | 0.79 | 0.85 | 0.74 | 1.70 | 1.48 | 2.03 |
| Basic endochitinase (2) | AT1G05850 | 0.53 | 0.50 | 0.30 | 0.75 | 0.31 | 1.66 | 1.52 | 2.22 |
| β-1,3-Glucanase-like | At4g29360 | 0.62 | 0.61 | 0.88 | 0.80 | 0.83 | 1.47 | 1.52 | 2.11 |
| Tetrahydrofolate dehydrogenase | AT3G12290 | 0.68 | 0.56 | 0.44 | 0.79 | 0.63 | 1.69 | 1.53 | 2.35 |
| Dormancy related (DRM1) | AT1G28330 | 0.42 | 0.63 | 0.22 | 0.75 | 0.60 | 1.75 | 1.43 | 2.24 |
| Porin-like protein | AT5G57490 | 0.82 | 0.90 | 0.53 | 0.73 | 0.60 | 1.88 | 1.81 | 2.33 |
| Mago Nashi-like protein | AT1G02140 | 0.67 | 0.86 | 0.71 | 1.04 | 0.74 | 1.92 | 1.88 | 2.39 |
| Putative RNA-binding protein | AT1G51510 | 0.68 | 0.87 | 0.50 | 0.93 | 0.55 | 1.97 | 1.78 | 2.36 |
| Expressed protein | AT5G22320 | 0.87 | 0.83 | 0.88 | 0.96 | 0.81 | 1.56 | 1.65 | 2.30 |
| Hypothetical protein | AT3G26932 | 0.42 | 0.40 | 0.09 | 0.46 | 0.22 | 1.64 | 1.40 | 2.30 |
| Putative protein | AT4G32330 | 0.62 | 0.61 | 0.51 | 0.95 | 0.67 | 1.69 | 1.72 | 2.30 |
| Expressed protein | AT3G50900 | 0.70 | 0.71 | 0.49 | 0.70 | 0.67 | 1.72 | 1.50 | 2.12 |
| Expressed protein | AT2G40280 | 0.74 | 0.83 | 0.59 | 1.04 | 0.71 | 2.04 | 1.73 | 2.42 |
| Expressed protein (2) | AT1G21090 | 0.66 | 0.78 | 0.70 | 0.99 | 0.53 | 1.49 | 1.49 | 2.32 |
| Group 5 | |||||||||
| Cytochrome b (2) | AT2G07727 | −2.90 | −2.40 | −1.84 | −1.35 | −2.07 | 0.26 | 0.59 | −0.12 |
| Glu-ammonia ligase | AT5G37600 | −1.14 | −0.06 | 0.56 | 1.05 | 0.42 | 4.03 | 5.03 | 4.61 |
| Elicitor-like protein | AT4G14420 | −0.75 | −1.45 | −0.91 | 0.22 | 0.37 | 1.75 | 2.20 | 2.68 |
| HSP81.2 | AT5G56030 | −1.11 | −0.83 | −0.44 | 0.23 | 0.13 | 1.75 | 1.56 | 2.89 |
| dnaK-type molecular chaperone hsc70.1 | AT5G02490 | −2.13 | −1.54 | −0.84 | −0.24 | −0.65 | 1.91 | 1.79 | 2.37 |
| Ubiquitin precursor UBQ4 | AT5G20620 | −0.76 | −0.59 | 0.27 | −0.66 | −0.13 | 3.02 | 2.39 | 2.55 |
| ubq14 (5) | AT4G02890 | −0.76 | −0.17 | 0.66 | −0.41 | 0.19 | 3.15 | 2.26 | 2.74 |
| Rab-type small GTP-binding protein | AT3G18820 | −0.16 | 0.12 | −0.03 | 0.47 | 0.40 | 1.54 | 1.68 | 2.59 |
| GAPDH | AT1G13440 | −0.86 | −0.32 | −0.49 | 0.09 | −0.08 | 1.30 | 2.30 | 2.61 |
| Putative histone deacetylase | AT5G22650 | 0.07 | 0.26 | 0.15 | 0.70 | 0.33 | 1.36 | 1.58 | 1.87 |
| Inorganic pyrophosphatase (2) | AT1G15690 | −0.49 | −0.07 | −0.49 | 0.04 | 0.08 | 1.28 | 1.26 | 2.40 |
| Unknown protein | AT1G28135 | 0.21 | 0.59 | 0.36 | 1.00 | 0.70 | 1.40 | 1.49 | 2.30 |
| Unknown protein | AT3G21865 | −0.30 | 0.03 | 0.14 | 0.17 | 0.16 | 1.41 | 1.47 | 2.64 |
| Putative protein | AT5G06550 | −0.58 | −0.62 | −0.61 | 0.18 | 0.51 | 1.42 | 2.17 | 3.06 |
| Putative protein (2) | AT4G32460 | −1.10 | −0.77 | −0.73 | −0.02 | 0.09 | 1.28 | 2.26 | 2.92 |
| Putative protein | AT5G63420 | 0.71 | 0.62 | 0.72 | 0.95 | 0.85 | 1.50 | 1.69 | 2.27 |
Table IV.
Genes in which RNA levels differ between fast- and slow-dried osmoprimed seeds
Predicted gene function, number of independent cDNAs on the array that represent that gene (between parentheses), the closest Arabidopsis gene homolog, and the average 2-log ratios of swapped dye duplo hybridizations relative to osmoprimed seed are shown. Genes discussed in the text are indicated in bold font. These genes belong to different groups (Fig. 4C) and are also included in Tables I to III.
| Predicted Gene Function | Arabidopsis Homolog | AM | Dry | FM | Slow | Fast | 15 h | 30 h | 45 h | Group |
|---|---|---|---|---|---|---|---|---|---|---|
| Jacalin lectin family protein (2) | AT3G21380 | 2.66 | 3.82 | 4.70 | 0.62 | −1.16 | −0.01 | −0.72 | 0.12 | 1 |
| Cruciferin (4) | AT4G28520 | 1.02 | 0.24 | −0.46 | −0.37 | −1.89 | −1.61 | −2.01 | −2.36 | 2 |
| Peroxiredoxin (2) | AT1G48130 | 0.49 | 1.12 | 1.71 | −0.52 | −2.07 | −2.58 | −1.73 | −2.78 | 2 |
| Em6 (10) | AT2G40170 | 0.67 | 2.12 | 3.28 | 0.80 | −1.24 | 0.69 | −0.78 | −1.73 | 2 |
| RAB18 | AT5G66400 | 2.39 | 2.51 | 3.34 | 2.02 | −0.96 | −1.29 | −2.01 | −3.56 | 2 |
| Dioxygenase (4) | AT2G25450 | −3.62 | −2.64 | −2.51 | −2.60 | −0.91 | 0.37 | 2.94 | 2.49 | 5 |
| Putative RNA polB transcription factor 3 | AT1G17880 | −2.89 | −2.36 | −1.44 | −1.68 | −0.72 | 1.28 | 2.42 | 2.67 | 5 |
| Fatty acid hydroxylase-like protein | AT4G20870 | −2.08 | −1.49 | −0.90 | 0.71 | −0.04 | 1.46 | 2.23 | 5 | |
| S-Adenosyl-Met synthetase (2) | AT3G17390 | −1.60 | −2.26 | −1.64 | −0.93 | 0.33 | 0.52 | 2.65 | 2.59 | 5 |
RNA Species That Are Abundant in Dry Seeds Rapidly Decline during Osmopriming and Germination
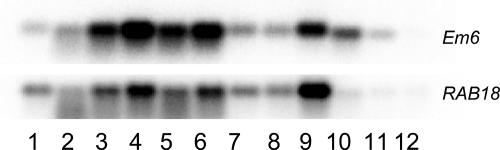
Three different dry-seed samples from the same B. oleracea variety were analyzed for gene expression. These were a commercial seed lot and two fractions from a noncommercial seed lot of the same genotype that were sorted for chlorophyll level. Two main groups of genes that were preferentially expressed in dry seeds compared to osmoprimed or germinating seeds could be distinguished (Fig. 4C, groups 1 and 2; Table I). Genes in group 1 were high in all three dry-seed fractions, rapidly decreased during priming, and moderately increased during germination. Genes in group 2 decreased both during priming and germination. RNA levels from some genes that increased during late seed maturation were also elevated in slow- compared to fast-dried primed seeds (Tables I and IV). This group includes the B. oleracea Em6, RAB18 genes, and a seed maturation gene homolog. These genes are members of the LEA gene family, which together with other stress-related genes that are expressed here could contribute to the increased stress tolerance of the FM seeds. During germination on water, their RNA levels rapidly declined. Interestingly, in slow-dried seeds these mRNAs were present at 2- to 4-fold higher levels than in fast-dried seeds, also implicating these genes in the stress-tolerance potential of slow-dried seeds. RNA blots from Em6 and RAB18 of these genes are presented in Figure 5.
Figure 5.
Expression of maturation-related gene RNA blots of two maturation/stress tolerance-related genes, Em6 (AT2G40170) and RAB18 (AT5G66400). Lane 1, Bent cotyledon stage seeds; lane 2, AM seeds; lane 3, dry seeds; lane 4, FM seeds; lane 5, CD-treated seeds; lane 6, seeds after 9 months storage; lane 7, osmoprimed seeds before drying; lane 8, osmoprimed seeds after fast drying; lane 9, osmoprimed seeds after slow drying; lane 10, germinating seeds 15 h after start of imbibition on water; lane 11, germinating seeds 30 h after start of imbibition on water; and lane 12, germinating seeds 45 h after start of imbibition on water.
The stress tolerance developed in the last phase of seed maturation is dramatically reduced during germination and upon radicle protrusion, as indicated by the poor storability of primed seeds and the inability of tomato seeds to survive drying after radicle protrusion (Bruggink et al., 1999; Buitink et al., 2000). The decline in RNA levels of group 2 genes during 7-d osmopriming is less than that during germination on water, reflecting the fact that germination on water more fully removes stress tolerance than osmopriming.
For many genes RNA levels declined during germination on water or osmopriming, reflecting degradation of RNA during these processes. All these genes have in common that during 7-d osmopriming their RNA is degraded, but they differ in the extent to which their RNA is degraded during germination on water. For example, RNAs encoding RAB18, seed maturation protein, and dormancy-related protein (group 2) were degraded even further during germination on water, whereas members of group 1 (such as small heat shock proteins [sHSPs]) had higher RNA levels in germinating seeds than in osmoprimed seeds. Histone H1-1 RNA was degraded during osmopriming and early germination on water, but its RNA levels increased after radicle protrusion.
Genes representing three different groups of storage compounds were included on the array. During the last stage of maturation, RNA levels of napin and cruciferin declined (Table I), whereas oleosin RNA levels were high in all three dry-seed samples. Interestingly, the degradation kinetics during 7-d osmopriming and 15- to 45-h germination on water also differed among the storage compound-related genes. Both napin and oleosin RNAs seemed to be degraded preferentially during 7-d osmopriming, while in seeds that had germinated in water for 45-h levels were higher than in the reference sample (primed seed). Cruciferin mRNA levels appeared to be more stable during osmopriming, as dry and primed seeds contain similar levels of cruciferin RNA, while seeds germinating on water have much lower levels.
Gene Expression Profiles during Osmopriming and Germination
Osmopriming treatments intentionally provide only limited water to seeds, resulting in the initiation of a number of germination-related processes, such as the cell cycle in tomato (De Castro et al., 2000) and breakdown of storage components in Arabidopsis (Gallardo et al., 2001) but preventing radicle protrusion. Osmopriming for 7 d in PEG solution followed by redrying resulted in faster and more uniform germination of primed seeds compared to nonprimed seeds but reduced longevity of the seeds. From a physiological point of view, osmoprimed seeds therefore resemble a transition state between dry seeds and seeds germinating on water. The microarray hybridizations allowed the comparison of transcriptional events during osmopriming and germination on water.
The majority of the genes on the chip (approximately 75%) were found to be up-regulated in germinating and/or in primed seeds. This was to be expected since most of the genes on the chip were obtained from cDNA libraries prepared using mRNA from germinating B. napus seeds (that were soaked in water). RNA levels of these genes either increased during osmopriming and further increased during germination on water (group 3, Fig. 4C; Table II), or they mainly increased during germination on water and not so much during osmopriming (groups 4 and 5, Fig. 4C; Table III).
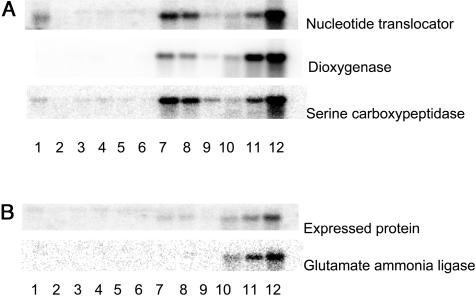
A typical expression pattern for genes in group 3 was that their RNA levels in primed seeds (the common reference) was higher than in dry seeds (regardless of maturity level), resulting in a −2-log ratio for dry seeds. Fifteen hours after adding water to the seeds, their RNA levels were similar to those found in primed seeds, while during longer periods of germination on water, their expression level rose progressively further, reflecting the advancement of germination processes in both samples. This is illustrated by the RNA blot of the nucleotide translocator and dioxygenase genes (Fig. 6A), where the amount of RNA after 45-h germination on water (lane 12) is much higher than after osmopriming (lanes 7–9). The genes with the most dramatic increase in expression are shown in Table II. Many of these are involved in protein biosynthesis, while another group contains metabolic genes such as S-adenosyl-Met synthetase, sugar epimerase, and Gap C. Only a few genes had similar RNA levels after 7-d osmopriming and after 45-h germination on water, indicating that during osmopriming their expression reached levels resembling those found in germinated seeds. These genes encode a Ser carboxypeptidase, a putative imbibition protein, and a cytochrome B gene. The RNA blot for the Ser carboxypeptidase gene (Fig. 6A) confirms this expression pattern, since RNA levels in primed seeds (prior to drying, lane 7) are similar to RNA levels after 45-h germination on water (lane 12).
Figure 6.
Gene expression during priming and germination. A, RNA blots of three genes that are expressed during osmopriming and water germination: dioxygenase (AT2G25450), nucleotide translocator (AT5G13490), and Ser carboxypeptidase (AT5G08260). B, RNA blots of two genes that are preferentially expressed during water germination: expressed protein (AT3G08030) and Glu ammonia ligase (AT5G37600). Lane 1, Bent cotyledon stage seeds; lane 2, AM seeds; lane 3, dry seeds; lane 4, FM seeds; lane 5, CD-treated seeds; lane 6, seeds after 9 months storage; lane 7, osmoprimed seeds before drying; lane 8, osmoprimed seeds after fast drying; lane 9, osmoprimed seeds after slow drying; lane 10, germinating seeds 15 h after start of imbibition on water; lane 11, germinating seeds 30 h after start of imbibition on water; and lane 12, germinating seeds 45 h after start of imbibition on water.
Genes that differ in their expression between osmopriming and germination on water are especially interesting since they may shed light on the processes that take place during these treatments. During osmopriming in −1-Mpa PEG, water uptake is limited, preventing radicle protrusion. During germination on water, water uptake is much more rapid and radicle protrusion occurs within 48 h. Most of the genes in Tables II and III have RNA levels that were more than 4-fold higher in one or more samples from seeds germinating on water compared to primed seeds, reflecting their involvement in the germination process. Interestingly, a number of the genes up-regulated by germination on water are also known to be induced by various abiotic or biotic stresses, such as cyclophyllin, superoxide dismutase, GRP2, glutathione-S-transferase, and stress-related protein. Some of these genes encode enzymes that may be involved in the degradation of the cell walls, such as a putative β-1-3-glucanase and an endochitinase. Several hypothetical or putative proteins that have been annotated in the Arabidopsis genome but that await further characterization are also expressed during germination on water. Analyzing the predicted proteins from these genes for conserved motifs reveals the presence of a Leu-rich repeat motif and Pro-rich regions, and implicates putative functions such as methyltransferase or beta lactamase activity. From the Arabidopsis ESTs that were spotted on the array, the DRM1 (thought to be involved in dormancy; Stafstrom et al., 1998) is expressed during germination of the seeds on water but not during osmopriming.
Gene Expression during Slow and Fast Drying
The drying method used to redry the osmoprimed seeds has a major effect on the behavior of the seeds in physiological tests. However, the majority of the genes on the array do not contain significantly different RNA levels between fast-dried and slow-dried osmoprimed seeds. Some general trends can be observed, however. When comparing the RNA levels in osmoprimed dried seeds to that of osmoprimed nondried seeds (common reference), for most genes no significant difference is found, indicating little change occurs during the drying process. For a limited number of genes the RNA levels decline during drying (−2-log ratios compared to the nondried common reference). This would indicate that gene-specific RNA degradation occurs during the drying process.
No genes with increasing RNA levels during fast drying were observed on the array, whereas a small number of genes could be found that had higher RNA levels after slow drying, compared to fast drying, of the osmoprimed seeds. This latter group includes genes for RAB18, Em6, peroxiredoxin, cruciferin, and jacalin lectin family protein (Table IV; Fig. 5, lanes 7–9). Some of these genes are also up-regulated during maturation (Table I). For these genes, the difference in expression could be attributed to a decrease in RNA levels in the fast-dried sample relative to nondried primed seeds, an increase in RNA levels in the slow-dried sample relative to nondried primed seeds, or both. Genes that were higher in fast-dried primed seeds relative to slow-dried primed seeds were also found (Table IV, group 5; Fig. 6A, lanes 7–9). Here the RNA levels had declined during the slow-drying process while remaining unaffected during the fast-drying process. These genes are also highly expressed during germination on water (Table II), and the degradation that occurs during slow drying may contribute to the partial reversal of the physiological state of these seeds to resemble mature seeds.
DISCUSSION
Using a dedicated microarray containing cDNAs isolated from immature (cotyledon stage) or germinating Brassica seeds, gene expression related to seed maturation, germination, osmopriming, and stress tolerance was studied. By selecting gene sets that were representative of the various processes (germination, stress tolerance related), samples could be characterized and grouped accordingly. Interestingly, when including a larger, more random set of genes, samples were clustered differently than when a subset of genes related to relevant processes was used (Fig. 4). This illustrates the usefulness of picking specific gene sets for discriminating samples, something that might not be obvious when all genes (for example on a full genome array) are used for the analysis, since the vast majority of genes on the array might not be involved in the process under study. Genes can be classified into several groups, representing late maturation-related genes, germination-related genes, stress tolerance-related genes, and osmopriming- and drying-related genes.
Genes Expressed during Seed Maturation and Slow Drying Are Correlated to Stress Tolerance
Normally a sample of dry seeds can be heterogeneous, containing seeds of various maturity levels, representing the location on the plant, differences in maturation speed within individual siliques, growth conditions, and the time of harvest. Using chlorophyll content as a measure to differentiate between FM seeds (containing very low chlorophyll levels) and near mature seeds (still containing substantial amounts of chlorophyll), seeds could be sorted for maturity level and analyzed separately. Even though near-mature and FM seeds are visually indistinguishable and originate from the same seed lot, the two seed fractions differed substantially in seed quality (Fig. 1). This is accompanied by differences in RNA levels for several genes (included in Table I). Typical expression patterns of genes from this group include an increase during seed maturation, a decline during seed germination and seed osmopriming, and higher RNA levels in slow-dried than in fast-dried primed seeds.
The mRNA levels from the B. oleracea Em6 gene (homologous to AT2G40170), RAB18 (homologous to AT5G66400), and a hypothetical protein (homologous to AT1G08220) were most strikingly correlated to stress tolerance of the seeds, since they not only increased during seed maturation, but also were differently affected by fast and slow drying. This correlation with longevity was confirmed by RNA blot for Em6 and RAB18 (Fig. 5). Em6 encodes a LEA group 1 protein that accumulates in Arabidopsis seeds late during seed development and is rapidly degraded during seed germination. Interestingly, degradation of Em6 protein was also observed in imbibed dormant Arabidopsis seeds (Bies et al., 1998). BnEm6, a homolog to the AtEm6 gene, has been isolated from B. napus and was shown to have the same expression pattern (Vicient et al., 1998). In our studies, mRNAs for BnEm6 persist in the early phase of germination, as illustrated by the relatively large number of cDNAs encoding this gene found in both the 7-h and the 15-h germination on water cDNA libraries. Contig analysis of these genes reveals the presence of two highly similar variants (>99%) that share more than 95% identity with the AtEm6 gene sequence. With our hybridization conditions both allelic variants behaved as one, with maximum RNA levels found in FM seeds and reduced RNA levels in primed and germinating seeds. Recently, LEA protein classification has been reanalyzed using novel bioinformatics tools (Wise, 2003; Wise and Tunnacliffe, 2004). Based on this analysis, group 1b LEA proteins (to which Em6 belongs) share features with RNA or ATP-binding proteins, DNA gyrases, or molecular chaperones. These features suggest a role for Em6 in protecting DNA integrity during the CD treatments we performed, correlating their presence with improved seed stress tolerance of the slow-dried Brassica seeds. The Arabidopsis RAB18 gene belongs to the group 2 LEA proteins and encodes an abscisic acid (ABA)-inducible dehydrin that is expressed in developing seeds (Lang et al., 1994). In plants the RAB18 protein can accumulate in response to ABA or drought stress and is mainly localized in the nucleus (Nylander et al., 2001). In our study, RAB18 mRNA is degraded during seed germination and osmopriming and reappears after slow drying. It is conceivable that during slow drying the primed seeds are able to perceive the gradual increase in drought stress and respond by reinducing RAB18 gene expression. Additionally, the slightly elevated temperature during slow drying (30°C) may have contributed to reinduction of stress gene expression. During fast drying the moisture content of the seeds drops much more rapidly (data not shown) possibly preventing ABA-mediated induction of RAB18. The hypothetical protein contains a putative mitochondrial-targeting site, but lacks further homology to predicted functional-protein domains. Further analysis of expression and biological function of this gene is needed to understand its role in seed stress tolerance.
Looking at the expression of the genes from Table I in different Arabidopsis tissues, stages, and treatments in the publicly available hybridization data from Genevestigator (https://www.genevestigator.ethz.ch) revealed high expression in mature siliques (seeds), low expression in geminating seeds (mixed seeds from 1–6 d after adding water), and no expression at other stages of plant development for the majority of these genes. Our data provides additional information of the expression levels of these genes at more defined stages of late maturation and early germination. Four of the LEA genes were found to be ABA inducible in plant tissues, while all other genes from Table I increased in expression in seedlings on medium lacking Glc and Suc relative to medium containing Glc and Suc, according to the information in Genevestigator. Only squalene epoxidase, Histone H1, and the sHSP genes were expressed in a select number of additional tissues.
RNA Levels High in Mature Seeds Rapidly Decline during Osmopriming and/or Germination
A number of additional genes with relatively high RNA levels in dry seeds, which declined during osmopriming, germination on water, or both, are also shown in Table I. Several of these genes, such as sHSPs or LEA genes, are known to be expressed during seed maturation and have been proposed to play a role in the acquisition of desiccation tolerance during maturation (Wehmeyer et al., 1996; Cuming, 1999; Raynal et al., 1999). No major differences in RNA levels between high- and low-chlorophyll-containing seeds or between fast- and slow-dried primed seeds were found for these genes, suggesting a limited role for these transcripts in the seed quality differences observed between these seed fractions. It has been known for some time that seed maturation is accompanied by the accumulation of LEA proteins (Galau et al., 1986; Dure et al., 1989). These LEA proteins are often ABA inducible, can also be found in leaves in response to desiccation stress, and are thought to play a role in the survival of the seed after drying and shedding from the mother plant. In total eight different members of the LEA family on the array were preferentially expressed in dry, FM seeds, and their RNA levels rapidly declined during germination and/or osmopriming. Interestingly, individual members of this family behaved differently. Two members were also differentially expressed between fast- and slow-dried primed seeds (see above) suggesting a more general role in seed stress tolerance for these genes. Transcripts for a seed maturation protein belonging to LEA group 5 (Table I) increased during the final stages of seed maturation as well, resulting in a 4-fold difference in RNA levels between low-chlorophyll and high-chlorophyll seeds. Unlike Em6 and RAB18, this gene was not differentially expressed between fast- and slow-dried seeds. Transcripts from the LEA M10 gene were reduced both in germinating and primed seeds, whereas RNA levels of most other LEA genes in Table I did not decrease during osmopriming. It is possible either that the osmotic stress (−1 MPa) encountered by the seeds during osmopriming prevented degradation of these mRNAs or that degradation was accompanied by new, osmotic stress-induced transcription. We were able to differentiate Brassica LEA M10 and LEA M17 homolog RNA levels in our hybridizations, even though these genes are highly similar in Arabidopsis. The two Brassica ESTs that have the LEA M10 and LEA M17 as their closest homolog in Arabidopsis are not similar enough for cross-hybridization on the array (due to a large number of insertions and single nucleotide changes).
Some reports suggested the involvement of sugar and sHSPs in desiccation tolerance and longevity by acting as molecular chaperons (Lee et al., 1995, 1997). The amount of HSP17.6 protein, a seed-expressed, cytoplasmic sHSP, is correlated with both desiccation tolerance and longevity in B. oleracea seeds (Bettey and Finch-Savage, 1998). This gene can enhance osmotolerance of Arabidopsis plants upon overexpression (Sun et al., 2001). In our analysis, transcripts from two sHSP genes were also reduced both in germinating and primed seeds (Table I), including HSP17.6.
The Arabidopsis peroxiredoxin gene (AT1G48130) has previously been shown to be up-regulated during seed maturation and down-regulated during Arabidopsis seed germination (Haslekås et al., 1998). Our experiments confirm this expression pattern in B. oleracea. In primed B. oleracea seeds RNA levels for this gene are intermediate between dry seeds and seeds germinating on water and seem to decrease during fast drying (Table IV). This suggests that in B. oleracea expression of the peroxiredoxin gene is not involved in the reinduction of longevity of primed seeds during slow drying, but its disappearance during fast drying could contribute to the observed quality loss in osmoprimed fast-dried seeds.
Of the storage compound genes only napin differed between high-chlorophyll and low-chlorophyll seeds, indicating a decrease in RNA levels during the final stages of maturation drying. The peak of storage protein gene expression is much earlier during seed development, when cells rapidly expand and accumulate reserves. When screening ESTs from developing Arabidopsis seeds (5–13 DAP), over 50% of cDNA clones represented cruciferin or napin mRNA (White et al., 2000). In this study we looked at three main groups of storage-related proteins and did not differentiate among individual gene family members, which contain enough homology to allow cross-hybridization on the microarray. The three groups encoded, respectively, 2S (napin) storage proteins, 12S (cruciferin) storage proteins, and lipid accumulation-related oleosin. For all three genes, mRNA could still be detected in dry seeds, reflecting stored mRNAs from earlier stages in seed development. Their levels declined rapidly during osmopriming (oleosin, napin) or germination on water (cruciferin). Oleosin is expressed throughout seed maturation, as it is required for lipid body integrity and stability. Oleosin protects the lipids from being degraded prematurely during seed maturation and storage, but during germination it is thought to mediate docking of lipases to allow oil reserve degradation in glyoxysomes (Thompson et al., 1998).
Germination Expression Programs Are Initiated during Osmopriming
Priming is a commonly used technique for improvement of seed quality. Usually, primed seeds are able to germinate faster and more uniformly than unprimed seeds, but depending on the priming conditions, seed species, and initial quality, primed seeds can be poorly storable. In this study we have looked at the effect of an osmopriming protocol that improves germination characteristics and compared the effect of two different drying regimes on longevity and gene expression. This way a number of physiological processes can be studied. In the osmopriming treatment used in this study, seeds are kept on filter paper moistened with PEG solution of high osmolarity (−1 MPa) for 7 d. PEG cannot enter the seeds, but the osmotic pressure it exerts reduces the rate and extent of water uptake by the seed, allowing only partial progression of germination-related events. During this treatment RNA levels of many germination-related genes increase, reflecting the activation of germination-related processes. Genes in this class include a variety of metabolic and cell cycle-related genes, encoding components of the translation machinery (ribosomal subunits and translation initiation factors), and enzymes involved in carbon metabolism, histones, transcription factors, etc. This confirms that during osmopriming, a large number of germination-related molecular events are initiated, consistent with the observation that primed seeds are able to germinate much faster than untreated control seeds. Many of these genes have been previously described to be up-regulated during seed germination and include metabolic enzymes such as S-adenosyl Met synthetase (Gallardo et al., 2002), ribosomal subunits (Toorop et al., 2003), and enzymes involved in sugar metabolism (Gallardo et al., 2001).
However, the high osmolarity of the osmopriming solution prevents completion of germination (radicle protrusion), allowing only early germination-related events to take place. In addition, it is conceivable that the osmotic stress experienced by the seeds during osmopriming results in the induction of stress-related gene expression. This was found during proteome analysis of Arabidopsis seeds (Gallardo et al., 2001), where accumulation of low-Mr HSPs, dehydrins, and catalase was found in osmoprimed seeds. In our study, this was also found for a number of LEA proteins (Table I). Most other stress-related genes that might have been induced by osmopriming treatment were even higher in seeds germinating on water.
Germination Is Accompanied by Huge Shifts in Gene Expression
Upon imbibition on water, the water rapidly enters the cells of the dry seed, and a number of germination-related processes are initiated. Overall RNA levels of a large group of genes after 15 h of germination are quite similar to the RNA levels seen after 7 d of osmopriming (Table II). These genes participate in metabolic processes that are initiated in the seed upon entry of water into the cells. Longer germination periods result in accumulation of higher levels of these gene products, and about 48 h after the start of imbibition radicle protrusion occurs.
In addition to the quantitative differences in gene expression observed (Tables II and III) some genes appear to be preferentially expressed during germination on water and do not accumulate during osmopriming. A number of these encode genes related to stress, wounding, or defense against pathogens. During germination and after radicle protrusion, degradation and physical damage of the endosperm and seed coat occurs by hydrolyzing enzymes and penetration of the radicle, releasing sugars and signal molecules. These in turn may trigger the induction of a number of defense genes that are needed to protect the emerging seedling from pathogen attack. In imbibed tobacco seeds class I β-1,3-glucanases are thought to play a dual role, by hydrolyzing cell walls to allow radicle protrusion and release dormancy and by defending the seed against pathogens (Leubner-Metzger, 2003). In tomato both 1-3-glucanases and endochitinases are expressed during germination, but they do not contribute to endosperm cap weakening, suggesting wound induced expression for seedling defense against pathogens (Morohashi and Matsushima, 2000; Wu and Bradford, 2003). The preferential expression of both 1-3-glucanases and endochitinases upon germination in water but not in osmoprimed seeds, suggests a similar role of these genes during Brassica radicle protrusion.
CONCLUSIONS
This study provides insight into the orchestration of events during early germination processes, prior to radicle protrusion. Through a careful selection of samples differing in physiological parameters, we were able to correlate expression of groups of genes with processes that occur during seed maturation and germination. Very little is known about the molecular events in the first hours of germiantion, which are critical for seedling establishment and survival. Results obtained might provide molecular tools to understand and control the process of priming and elucidate the mechanism by which priming improves seedling performance in the field. This study provides an important first step toward a molecular understanding of the conflicting processes that occur in seeds subjected to changes in water content, where storability, stress tolerance, and preservation compete with germination and growth of a sensitive seedling.
MATERIALS AND METHODS
Seed Samples and Sorting
In the experiments seed lots were used from Brassica oleracea cv Bartolo F1. The seed lot that was sorted for maturity was specifically chosen for its initial poor quality and is not commercially available. In experiments comparing seeds of different maturity, seeds from the poor quality seed lot were sorted for their level of chlorophyll fluorescence using ScanMaster I (Satake, Houston) as described by Jalink et al. (1998). The level of chlorophyll fluorescence is inversely correlated with seed maturity. A good quality seed lot was used for most other experiments.
Germination Test
Germination tests were carried out in 4 replicates of 50 seeds each as recommended by the International Seed Testing Association (ISTA). Seeds were considered germinated when radicles emerged. Seed germination parameters related to germination, such as germination rate (T50) and uniformity (U10–90), were calculated with the software package Seed Calculator2.1 (Plant Research International B.V., Wageningen, The Netherlands). Seedling quality was evaluated according to ISTA (2003). Seedlings were considered as normal when having a healthy root system, shoot axis, and cotelydons. Seedlings with clear deficits in one or more of these structures, for example a blunt root, were considered as abnormal. Seeds without protruded radicle tip were considered as nongerminated.
Priming and Drying Treatments
Seeds were primed in PEG6000 solution at an osmotic potential of −1.0 MPa for 7 d on filter paper in the dark at 20°C. After incubation seeds were washed with running tap water for 5 min to remove the osmotic agent and blotted between filter paper for 15 min to remove excess water on the seed surface. Then seeds were dried to their original moisture content using one of two different methods. In the fast-drying method, seeds were dried in circulating air at 20°C, 32% relative humidity (RH) for 3 d. In the slow-drying method (adapted from Bruggink et al., 1999), seeds were dried in standing air at 30°C, 75% RH for 3 d and dried further in circulating air at 20°C, 32% RH for 3 d. Primed seeds and control seeds were stored dry in hermetically closed tubes at 5°C.
Seed Longevity
Under ambient conditions, even primed B. oleracea seeds can be stored for a considerable time. Therefore, longevity of the seed samples was tested under more severe conditions. In one test seeds were stored at 30°C and 66% RH, a condition that can be met in practice in the tropics. A second longevity test was performed by CD. CD of the seeds was performed by first equilibrating the seeds in a cabinet at 20°C, 85% RH for 3 d; then the seeds were transferred into sealed laminate aluminum bags and kept at 40°C for 0, 1, 2, 3, 4, or 5 d. After the storage or CD, seeds were dried at 20°C, 32% RH for 3 d and kept in hermetically closed tubes at 5°C.
Preparation of the cDNA Microarray
Total RNA was isolated from cotyledon stage (during seed maturation) and germinating (soaked 15 h in water) Brassica napus seeds, and from isolated root tips from germinating (soaked 7 h in water) B. napus seeds using Trizol RNA isolation reagent (Gibco-BRL, Cleveland). One cDNA library (cotelydon stage) was made with the HybriZAP 2.1 kit (Stratagene, La Jolla, CA). Two cDNA libraries were prepared using the Uni-ZAP XR vector (Stratagene). After mass excision of the cDNA-containing plasmids, inserts were PCR amplified from individual bacterial colonies, and clones containing an insert length of 300 bp or more were selected for the chip. Plasmid DNA was isolated from the selected colonies using Multiscreen FB 96-well plates (Millipore, Bedford, MA). Ten-nanogram plasmid DNA was used as template for a large-scale PCR reaction with universal primers containing a C6 amino modification (Isogen Bioscience, Maarssen, The Netherlands) at the 5′ end. PCR products were purified over Multiscreen FB 96 well plates according to the spin protocol provided by the supplier (Millipore) and transferred to 384-well plates for spotting. The samples were arrayed onto CMT-CAPS coated slides (Corning, Corning, NY) using the PixSys 7500 (Biodot) system with four Chipmaker3 pins covering an area of 9.0 × 9.0 mm. The RH in the spotting chamber was held at ±85%. The slides were processed according to the protocol provided by the supplier.
Included on the cDNA microarray were 238 clones of the cotyledon stage seed cDNA library, 599 genes of the 7-h germinating seed root tip library, and 600 genes of the 15-h imbibed seed library. In addition, about 10 germination-related ESTs from tomato (Lycopersicon esculentum) and 30 Arabidopsis (Arabidopsis thaliana) cell cycle or stress-related ESTs were selected from the literature and spotted on the chip. Foreign clones (luciferase) were included as controls for normalization, and yeast (Saccharomyces cerevisiae) genes that lacked cross-hybridization to plant genes were used as negative controls for subtracting the background. Approximately 1,000 clones were sequenced from the 5′ end using the Applied Biosystems (Foster City, CA) big-dye terminator cycle sequence ready-reaction kit.
Labeling and Hybridization
RNA for hybridizations was isolated using a protocol for recalcitrant tissues to remove polysaccharides and other interfering compounds commonly found in dry seeds (Schultz et al., 1994). For labeling, 40 μg total RNA was spiked with luciferase mRNA (1 ng; Promega, Madison, WI) and the mixture was fluorescently labeled by reverse transcription in the presence of 5-(3-aminoallyl)-dUTP (Sigma-Aldrich, St. Louis) using oligo(dT) as a primer (Isogen Bioscience). After preciptiation of the RNA/DNA hybrids and degradation of the RNA, either Cy3 or Cy5 monofunctional dyes (Amersham, Freiburg, Germany) were linked to the aminomodified nucleotides in a separate coupling step, creating fluorescently labeled cDNA. Cy5-labeled target was mixed with Cy3-labeled reference (or vice versa) prior to hybridization. The processed slide was prehybridized in a large volume of hybridization buffer (50% formamide, 5× Denhardt's reagent, 5× SSC, 0.2% SDS, 0.1 mg/mL denatured fish DNA) for 2 h at 42°C to reduce background. Subsequently, the slides were washed and dried in order to mount a cutout frame of 10 × 10 mm (ABgene, Epsom, UK) to the slide. This frame, covered by a plastic coverslip with two small holes enabling easy access, served as a hybridization chamber. After filling the chamber with denatured probe mix in hybridization buffer, the holes were covered and the slide was incubated horizontally at 42°C for 16 h in the dark. Slides were subsequently washed in 1× SSC, 0.1% SDS for 5 min, 0.1× SSC, 0.1% SDS for 5 min, rinsed with 0.1× SSC, and dried. Fluorescence levels for both Cy3 and Cy5 of the individual cDNA spots were determined using a ScanArray3000 laser scanner (Perkin-Elmer, Foster City, CA).
As a common reference osmoprimed seeds (prior to drying) were used for all hybridizations. Nonhybridizing yeast clones were used to calculate background levels for each hybridization. After subtraction of background, data were normalized for relative dye intensity using the relative fluorescence of the luciferase spots.
Separately, normalization was done using an ANOVA approach where gene sample effects can be estimated separately from array effects, dye effects, main effects for samples and genes, and interaction of gene with dye. Also, mixture models were used as an alternative method to identify gene clusters (Yeung et al., 2001).
Northerns
Northern-blot analysis was performed essentially according to standard protocols. Three micrograms total RNA was subjected to gel electrophoresis using glyoxal in a 1.2% agarose gel using 15 mm sodium phosphate buffer, pH 7. Gels were blotted overnight onto Hybond N+ (Amersham) membrane using 7.5 mm NaOH. Hybridization was done overnight in dextranesulfate containing 0.18 mg/mL herring sperm DNA and 32P-labeled probe (PCR fragment labeled using the Megaprime DNA-labeling System kit [Amersham RPN 1604]) at 65°C. The blot was washed using sodium chloride/sodium phosphate/EDTA (SSPE) concentrations ranging from 2× SSPE to 0.1× SSPE in combination with 0.1% SDS. Bands were detected using phosphorimaging screens and were scanned using an FX pro plus molecular imager (Bio-Rad Laboratories, Hercules, CA).
Acknowledgments
We thank Ronny Joosen for help with bioinformatics, and Henk Hilhorst and Wilco Ligterink for critical reading of the manuscript.
This work was supported by the Dutch Ministry of Agriculture, program 241, and by Sumika Agrotech Company (to Y.S.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.051664.
References
- Bettey M, Finch-Savage WE (1998) Stress protein content of mature Brassica seeds and their germination performance. Seed Sci Res 8: 347–355 [Google Scholar]
- Bewley JD, Black M (1994) Seeds: Physiology of Development and Germination, Ed 2. Plenum Press, New York
- Bies N, Aspart L, Carles C, Gallois P, Delseny M (1998) Accumulation and degradation of Em proteins in Arabidopsis thaliana: evidence for post-transcriptional controls. J Exp Bot 49: 1925–1933 [Google Scholar]
- Bino RJ, de Vries JN, Kraak HL, van Pijlen JG (1992) Flow cytometric determination of nuclear replication stages in tomato seeds during priming and germination. Ann Bot (Lond) 69: 231–236 [Google Scholar]
- Bruggink GT, Ooms JJJ, Van der Toorn P (1999) Induction of longevity in primed seeds. Seed Sci Res 9: 49–53 [Google Scholar]
- Buitink J, Hemminga MA, Hoekstra FA (2000) Is there a role for oligosaccharides in seed longevity? An assessment of intracellular glass stability. Plant Physiol 122: 1217–1224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuming AC (1999) LEA proteins. In PR Shewry, R Casey, eds, Seed Proteins. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 753–780
- De Castro RD, van Lammeren AAM, Groot SPC, Bino R, Hilhorst HWM (2000) Cell division and subsequent radicle protrusion in tomato are inhibited by osmotic stress but DNA synthesis and formation of microtubular cytoskeleton are not. Plant Physiol 122: 327–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Castro RD, Zheng X, Bergervoet JHW, De Vos CHR, Bino R (1995) β-Tubulin accumulation and DNA replication in imbibing tomato seeds. Plant Physiol 109: 499–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dure L, Crouch M, Harada J, Ho THD, Mundy J, Quatrano R, Thomas T, Sung ZR (1989) Common amino-acid sequence domains among the lea proteins of higher-plants. Plant Mol Biol 12: 475–486 [DOI] [PubMed] [Google Scholar]
- Galau GA, Hughes DW, Dure L (1986) Abscisic-acid induction of cloned cotton late embryogenesis-abundant (lea) messenger-RNAs. Plant Mol Biol 7: 155–170 [DOI] [PubMed] [Google Scholar]
- Gallardo K, Job C, Groot SPC, Puype M, Demol H, Vandekerckhove J, Job D (2001) Proteomic analysis of Arabidopsis seed germination and priming. Plant Physiol 126: 835–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallardo K, Job C, Groot SP, Puype M, Demol H, Vandekerckhove J, Job D (2002) Importance of methionine biosynthesis for Arabidopsis seed germination and seedling growth. Physiol Plant 116: 238–247 [DOI] [PubMed] [Google Scholar]
- Girke T, Todd J, Ruuska S, White J, Benning C, Ohlrogge J (2000) Microarray analysis of developing Arabidopsis seeds. Plant Physiol 124: 1570–1581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groot SPC, Karssen CM (1987) Gibberellins regulate seed germination in tomato by endosperm weakening: a study with gibberellin-deficient mutants. Planta 171: 525–531 [DOI] [PubMed] [Google Scholar]
- Groot SPC, Soeda Y, Stoopen G, Konings MCJM, van der Geest AHM (2003) Gene expression during loss and regaining of stress tolerance at seed priming and drying. In G Nicolas, KJ Bradford, D Côme, HW Pritchard, eds, The Biology of Seeds: Recent Research Advances. CAB International, Cambridge, MA, pp 279–287
- Haslekås C, Stacy RA, Nygaard V, Culianez-Macia FA, Aalen RB (1998) The expression of a peroxiredoxin antioxidant gene, Atper1, in Arabidopsis thaliana is seed-specific and related to dormancy. Plant Mol Biol 36: 833–845 [DOI] [PubMed] [Google Scholar]
- Heydekker W, Higgins J, Gulliver RL (1973) Accelerated germination by osmotic seed treatment. Nature 246: 42–44 [Google Scholar]
- Holdsworth M, Kurup S, McKibbin R (1999) Molecular and genetic mechanisms regulating the transition from embryo development to germination. Trends Plant Sci 4: 275–280 [Google Scholar]
- Hong TD, Ellis RH (1992) The survival of germinating orthodox seeds after desiccation and hermetic storage. J Exp Bot 43: 239–247 [Google Scholar]
- International Seed Testing Association (2003) International Rules for Seed Testing. The International Seed Testing Association, Bassersdorf, Switzerland
- Jalink H, Van der Schoor R, Frandas A, Van Pijlen JG, Bino R (1998) Chlorophyll fluorescence of Brassica oleracea seeds as a non-destructive marker for seed maturity and seed performance. Seed Sci Res 8: 437–443 [Google Scholar]
- Koornneef M, Bentsink L, Hilhorst H (2002) Seed dormancy and germination. Curr Opin Plant Biol 5: 33–36 [DOI] [PubMed] [Google Scholar]
- Lang V, Mantyla E, Welin B, Sundberg B, Palva ET (1994) Alterations in water status, endogenous abscisic acid content, and expression of RAB18 gene during the development of freezing tolerance in Arabidopsis thaliana. Plant Physiol 104: 1341–1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee G, Pokala N, Vierling E (1995) Structure and in vitro molecular chaperone activity of cytosolic small heat shock proteins from pea. J Biol Chem 270: 10432–10438 [DOI] [PubMed] [Google Scholar]
- Lee G, Roseman A, Saibil H, Vierling E (1997) A small heat shock protein stably binds heat denatured model substrates and can maintain a substrate in a folding-competent state. EMBO J 16: 659–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leubner-Metzger G (2003) Functions and regulation of beta-1,3-glucanases during seed germination, dormancy release and after-ripening. Seed Sci Res 13: 17–34 [Google Scholar]
- Massart DL, Vandeginste BGM, Buydens LMC (1997) In Handbook of Chemometrix and Qualimetrix. Series: Data Handling in Science and Technology, Vol 20A. Elsevier/North-Holland Publishing, Amsterdam, pp 108–113
- McDonald MB (2000) Seed priming. In M Black, JD Bewley, eds, Seed Technology and Its Biological Basis. Sheffield Academic Press, Sheffield, UK, pp 287–325
- Morohashi Y, Matsushima H (2000) Development of beta-1,3-glucanase activity in germinated tomato seeds. J Exp Bot 51: 1381–1387 [PubMed] [Google Scholar]
- Nylander M, Svensson J, Palva ET, Welin BV (2001) Stress-induced accumulation and tissue-specific localization of dehydrins in Arabidopsis thaliana. Plant Mol Biol 45: 263–279 [DOI] [PubMed] [Google Scholar]
- Osborne DJ (1993) Function of DNA synthesis and DNA repair in the survival of embryos during early germination and in dormancy. Seed Sci Res 3: 43–53 [Google Scholar]
- Peng J, Harberd NP (2002) The role of GA-mediated signalling in the control of seed germination. Curr Opin Plant Biol 5: 376–381 [DOI] [PubMed] [Google Scholar]
- Raynal M, Guilleminot J, Gueguen C, Cooke R, Delseny M, Gruber V (1999) Structure, organization and expression of two closely related novel Lea (late-embryogenesis-abundant) genes in Arabidopsis thaliana. Plant Mol Biol 40: 153–165 [DOI] [PubMed] [Google Scholar]
- Ruuska SA, Girke T, Benning C, Ohlrogge JB (2002) Contrapuntal networks of gene expression during Arabidopsis seed filling. Plant Cell 14: 1191–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz DJ, Craig R, Cox-Foster DL, Mumma RO, Medford JI (1994) RNA isolation from recalcitrant plant tissue. Plant Mol Biol Rep 12: 310–316 [Google Scholar]
- Stafstrom JP, Ripley BD, Devitt ML, Drake B (1998) Dormancy-associated gene expression in pea axillary buds: cloning and expression of PsDRM1 and PsDRM2. Planta 205: 547–552 [DOI] [PubMed] [Google Scholar]
- Sun W, Bernard C, van de Cotte B, Van Montagu M, Verbruggen N (2001) At-HSP17.6A, encoding a small heat-shock protein in Arabidopsis, can enhance osmotolerance upon overexpression. Plant J 27: 407–415 [DOI] [PubMed] [Google Scholar]
- Thompson JE, Froese CD, Madey E, Smith MD, Hong Y (1998) Lipid metabolism during plant senescence. Prog Lipid Res 37: 119–141 [DOI] [PubMed] [Google Scholar]
- Toorop PE, Groot SPC, Hilhorst HWM (2003) Expression of a ribosomal protein gene during germination of cabbage (Brassica oleracea f. oleracea) seeds. In G Nicolas, KJ Bradford, D Côme, HW Pritchard, eds, The Biology of Seeds: Recent Research Advances. CAB International, Cambridge MA, pp 191–197
- Vázquez-Ramos JM, Sánchez MdlP (2004) The cell cycle and seed germination. Seed Sci Res 13: 113–130 [Google Scholar]
- Vicient CM, Roscoe TJ, Delseny M (1998) Characterization of an Em-like gene from Brassica napus. J Exp Bot 49: 1061–1062 [Google Scholar]
- Wehmeyer N, Hernandez LD, Finkelstein RR, Vierling E (1996) Synthesis of small heat-shock proteins is part of the developmental program of late seed maturation. Plant Physiol 112: 747–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White JA, Todd J, Newman T, Focks N, Girke T, de Ilarduya OM, Jaworski JG, Ohlrogge JB, Benning C (2000) A new set of Arabidopsis expressed sequence tags from developing seeds: the metabolic pathway from carbohydrates to seed oil. Plant Physiol 124: 1582–1594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise MJ (2003) LEAping to conclusions: a computational reanalysis of late embryo-genesis abundant proteins and their possible roles. BMC Bioinformatics 4: 52–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise MJ, Tunnacliffe A (2004) POPP the question: what do LEA proteins do? Trends Plant Sci 9: 13–17 [DOI] [PubMed] [Google Scholar]
- Wobus U, Weber H (1999) Seed maturation: genetic programmes and control signals. Curr Opin Plant Biol 2: 33–38 [DOI] [PubMed] [Google Scholar]
- Wu CT, Bradford KJ (2003) Class I chitinase and beta-1,3-glucanase are differentially regulated by wounding, methyl jasmonate, ethylene, and gibberellin in tomato seeds and leaves. Plant Physiol 133: 263–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung KY, Fraley C, Murua A, Raftery AE, Ruzzo WL (2001) Model-based clustering and data transformations for gene expression data. Bioinformatics 17: 977–987 [DOI] [PubMed] [Google Scholar]