Abstract
Historically, drugs used in the treatment of cancers also tend to cause damage to healthy cells while affecting cancer cells. Therefore, the identification of novel agents that act specifically against cancer cells remains a high priority in the search for new therapies. In contrast to normal cells, most cancer cells contain multiple centrosomes which are associated with genome instability and tumorigenesis. Cancer cells can avoid multipolar mitosis, which can cause cell death, by clustering the extra centrosomes into two spindle poles, thereby enabling bipolar division. Kinesin-like protein KIFC1 plays a critical role in centrosome clustering in cancer cells, but is not essential for normal cells. Therefore, targeting KIFC1 may provide novel insight into selectively killing of cancer cells. In the present study, we identified a small molecule KIFC1 inhibitor, SR31527, which inhibited microtubule-stimulated KIFC1 ATPase activity with an IC50 value of 6.6 μM. By using bio-layer interferometry technology, we further demonstrated that SR31527 bound directly to KIFC1 with high affinity (Kd = 25.4 nM). Our results from computational modeling and STD-NMR experiment suggested that SR31527 bound to a novel allosteric site of KIFC1 that appears suitable for developing selective inhibitors of KIFC1. Importantly, SR31527 prevented bipolar clustering of extra-centrosomes in triple negative breast cancer (TNBC) cells and significantly reduced TNBC cell colony formation and viability, but was less toxic to normal fibroblasts. Therefore, SR31527 provides a valuable tool for studying the biological function of KIFC1 and serves as a potential lead for the development of novel therapeutic agents for breast cancer treatment.
Keywords: Kinesin, mitosis, breast cancer, inhibitor, allosteric regulation
INTRODUCTION
Inhibition of cell mitosis is a clinically validated strategy for cancer treatment (1). Traditional anti-mitotic drugs such as taxanes and vinca alkaloids inhibit mitotic spindle assembly by targeting tubulin and disrupting microtubule (MT) dynamics (2). However, because MTs also present ubiquitously in normal cells and play very important roles throughout the cell cycle, disruption of MTs results in undesirable side effects, such as peripheral neuropathy, in patients( 2,3).
Kinesins are cytoskeletal motor proteins that utilize the energy from ATP hydrolysis to perform mechanical work along MTs and mediate cellular processes such as cargo transport, spindle and chromosome movement (4). Mitotic kinesins are required for various aspects of mitosis, including bipolar spindle assembly, chromosome alignment, chromosome segregation and cytokinesis (5). Since mitotic kinesins function exclusively during mitosis, compounds specifically inhibit mitotic kinesins could affect only the proliferating cells, and thus are anticipated to be a new type of chemotherapeutic agents with better safety profiles (6). The development of mitotic kinesin inhibitors has undergone significant progress in the past decade. A number of highly selective inhibitors of several mitotic kinesins, including Eg5 and CENP-E, have been identified (7–12), and some of those inhibitors have advanced into clinical trials (13–16). It is very encouraging that neuropathy, which is commonly caused by traditional mitotic inhibitors, was not observed on patients treated with mitotic kinesin inhibitors (5). However, myelosupression is still the primary dose limiting toxicity, and various hematological side effects were observed on patients treated with those mitotic kinesin inhibitors (5) because they non-differentially block the mitosis of both cancer cells and normal cells.
Kinesin like protein (KIFC1), a unique mitotic kinesin, has recently emerged as an new anti-cancer drug target (17). Increased centrosome number, or centrosome amplification, has been reported in a variety of human primary cancers and correlates with aneuploidy, chromosomal instability and tumorigenesis (18–22). In mitosis, supernumerary centrosomes can form multipolar spindles which cause aneuploidy and ultimately cell death. However, cancer cells can avoid multipolar mitosis by clustering the extra centrosomes into two spindle poles thereby enabling bipolar division (23–25). Recent studies indicate that KIFC1 is critical for the centrosome clustering in cancer cells with amplified centrosomes (26–28). Knockdown of KIFC1 induced multipolar spindle mitotic defects in cancer cells containing extra centrosomes and caused cancer cell death (28), but had no effects on normal cells (27,28). Moreover, KIFC1 is required for proper spindle assembly, stable pole-focusing and survival of cancer cells irrespective of normal or supernumerary centrosome number, thus indicating a more general and critical role for KIFC1 in cancer cells (26,27). While the mechanism of how KIFC1 regulates the survival of cancer cells is still under investigation, recent studies clearly implicate KIFC1 as an attractive drug target for the development of cancer cell selective therapeutics.
KIFC1 expression is significantly elevated in a broad panel of cancer tissues (26,27,29). Pannu et al. reported that KIFC1 is overexpressed in human breast cancers, and that KIFC1 overexpression correlates to increased aggressiveness and poorer clinical outcomes in breast cancer patients (26). Recently, we also demonstrated that KIFC1 expression is up-regulated in breast cancer, particularly in triple negative breast cancer (TNBC), and that KIFC1 is highly expressed in human breast cancer cell lines, but is undetectable in primary normal human mammary epithelial cells and weakly expressed in two human fibroblast lines (17). We further showed that KIFC1 silencing significantly reduced breast cancer cell viability (17). In the present studies, we identified a small molecule KIFC1 inhibitor, SR31527, through a high-throughput screening (HTS) campaign. Our results indicated that SR31527 inhibited KIFC1 by binding directly to an allosteric site of KIFC1 without involving MTs. Moreover, SR31527 prevented bipolar clustering of extra-centrosomes in TNBC cells and significantly reduced TNBC cell colony formation and viability, and was less cytotoxic to normal fibroblasts.
MATERIALS AND METHODS
Preparation of active KIFC1 motor protein
To enable high-throughput screening, we established a protocol that allows us to purify a large amount of KIFC1 protein from E. coli. Initially, more than ten constructs encoding human KIFC1 motor domain protein were generated. Among them, two constructs made with pET28 plasmid produced soluble and active KIFC1 proteins in E. coli cells. Protein purification was carried out in two steps with His-tag affinity and gel filtration chromatography. Specifically, cell pellet was resuspended in the lysis buffer (75 mM Tris-HCl pH 8.0, 300 mM NaCl, 5% glycerol, 20 mM imidazole, 0.1% Triton X-100, 0.5 mM TCEP supplemented with a protease-inhibitor cocktail and 1μg /ml Benzonase nuclease) and was lysed by two passages through a French press. Cell debris was removed by centrifugation and the clear supernatant was passed through a Ni-NTA column (HisTrap 5 GE Healthcare) equilibrated with the washing buffer (20 mM Tris-HCl pH 7.5, 300 mM NaCl, 5% glycerol, 20 mM imidazole and 0.5 mM TCEP). The column was then washed extensively with washing buffer to remove non-specific proteins. The bound KIFC1 protein was eluted using a 300 ml linear gradient of 50–400 mM imidazole in an elution buffer (20 mM Tris-HCl pH 7.5, 300 mM NaCl, 5% glycerol, 400 mM imidazole and 0.5 mM TCEP). Eluted fractions were analyzed by SDS-PAGE, and fractions containing KIFC1 were pooled together. Further purification was conducted with a gel-filtration column (Superdex 200 26/60 Amersham Biosciences) and the eluted fractions from the column were analyzed by SDS-PAGE. Fractions containing purified KIFC1 were pooled together and concentrated. With this protocol, we were able to obtain approximately 10–15 mg of KIFC1 protein from one literof the E. coliculture.
High-throughput screening
A biochemical assay measuring MT stimulated ATPase activity of KIFC1 was used for HTS. This assay was based upon the HTS assay of kinesin Eg5 (30), and was run in a 1536-well microtiter plate format. Briefly, 2.5 μl of 15 mM PIPES (pH 7.0) containing 1 mM MgCl2, 6 μg/ml MTs, 20 μM Paclitaxel, 100 μM ATP, 0.02% Tween-20, 2% DMSO, and 35 μg/ml KIFC1 protein were added to each well of 1536-well microtiter plates. The plates were incubated at room temperature for 1 h followed by addition of 1.25 μl ADP HunterTM Plus (DiscoverRx) Reagent A and 2.5 μl Reagent B to detect the ADP production. The plates were further incubated for 0.5 h at room temperature and then bottom read for fluorescence (ex.530/em.590) on the Envision. A pilot screen of 10,000 structurally diverse commercial compounds was performed in a single dose (10 μM) format in duplicates on two separate days and the percent of inhibition data generated on each day was examined for each compound. The values of Pearson’s correlation was 0.93, indicating an excellent correlation. The Z’-value for the campaign was 0.91±0.01; the coefficient of variations (CVs) was 1.36 ± 0.17 for the full reaction and 4.17 ± 1.46 for the background reaction. The hit rate (based on three times the standard deviation (3*δ) plus the average percent of inhibition from all screened compounds) was 1.04%. All of these statistical results are indicative of a robust, reproducible screen ready for HTS. We then utilized this assay and screened additional 20,000 structurally diverse compounds commercially obtained from the Enamine library.
Molecular modeling
Molecular modeling studies were performed on a SGI Altix XE 1200 Linux cluster using the modules of the Schrödinger Suite 2012 (Schrödinger, LLC, New York, NY, 2012). Specifically, SiteMap program was used to identify potential binding pocket(s) by mapping the surface of the KIFC1 crystal structure (PDB ID: 2REP). A pockets with a SiteScore value > 0.80 is considered suitable for thebinding of small molecule compounds. The 3D models of ligands (SR31527, monastrol) were first generated using LigPrep, then docked into the different binding sites (including ATP-binding site, monastrol-binding site, S1 and S2 sites) using Glide program following an Induced-Fit-Docking (IFD) protocol (31) which is capable of sampling dramatic side-chain conformational changes as well as minor changes in protein backbone structure. The default docking parameters were first tested by docking monastrol (an Eg5 inhibitor) into its binding site on Eg5. The docked monastrol-Eg5 model excellently reproduced the crystal structure of monastrol-Eg5 complex (PDB ID: 1Q0B). The same IFD protocol as well as parameters were then employed for the docking studies of SR31527 into the different binding sites on KIFC1. Residues within 5 Å of the docked ligands were allowed to be flexible. The docked results were rankedby the extra -precision (XP) scoring function of Glide .
NMR spectroscopy
The STD-NMR data were collected following established protocols (32,33 ). Samples containing SR31527 and KIFC1 protein at a concentration ratio of 20:1 were prepared in D2O. STD-NMR spectra were recorded with a total of 32 K points, 80 scans, and selective saturation of protein resonances at 0, 0.65, 1.67, and 7.61 ppm (−8.18 ppm for the reference spectra), using a series of SEDUCE pulses (1000 points, 50 ms), for a total saturation time of 10 s (SEDUCE-1 pulse is similar to a Gaussian pulse, and has been used by other laboratories (34)). Reference experiments using the free ligands themselves (i.e. without KIFC1 protein) were performed under the same experimental conditions to verify true ligand binding. No STD signals were present in the difference spectra of the free ligand, indicating that the effects observed in the presence of KIFC1 were due to a true saturation transfer from the protein.
Biolayer interferometry
Biolayer interferometry (BLI) was used to study the kinetics of SR31527 binding to KIFC1 on a ForteBIO Octet (Menlo Park, CA), per manufacturer's instructions and published methods (35,36). Purified His-tagged KIFC1 protein was first bound to HIS2K biosensors coated with penta-His antibody. The biosensors were then transferred into wells containing SR31527 in serial dilutions from 60 μM to 0.74 μM concentrations. Protein-ligand binding was measured by monitoring the changes in the interferometric profile of the wavelength of light passing through the sensor. Following a 300-second incubation, the KIFC1-coated sensor tips were transferred to the kinetics buffer to allow dissociation for 900 seconds. Binding curves were analyzed using the ForteBIO software, which performs a global fit according to the 1:1 Langmuir model to obtain the kinetic rate constants for each set of interaction conditions.
Cell culture
All breast cancer cell lines and lung fibroblast lines were obtained from ATCC, and cultured in DMEM medium containing 10% fetal bovine serum, 2 mM of L-glutamine, 100 units/ml of penicillin, and 100 μg/ml of streptomycin. Cells were grown under standard cell culture conditions at 37 °C in a humidified atmosphere with 5% CO2 . All the cell lines were mycoplasma-negative.
Confocal microscopy
MDA-MB-231, BT549 and MDA-MB-435s cells were grown on glass coverslips. After 24 h treatment of SR31527, the cells were fixed with 4% paraformaldehyde in phosphate-buffered saline for 20 min, and permeabilized with 0.2% Triton X-100 for 10 min at room temperature. The cells were then incubated with anti-α-tubulin-FITC antibody (sigma) and anti-γ-tubulin Cy3 conjugate (sigma) for 45 min, and nuclei were stained with NucRed647 for 10 min. The cells were then examined by a laser-scan confocal microscope (Leica DMI 4000 B). All images were captured with an HCX PL Apo 63x oil immersion objective. Images were processed and analyzed using Leica's LAS Image Analysis software.
Cell viability assay
Cells were seeded into 96-well tissue culture treated microtiter plates at a density of 3000–4000 cells/well. After overnight incubation, the cells were treated with SR31527 for 96 h. Cell viability was measured by the CellTiter-Glo Assay (Promega).
Colony formation assay
Cancer cells were seeded at a density of 500 cells/well into six-well plates. Sixteen hours after the plates were set up, SR31527 was added, and media were replenished every 3 days. After being incubated for 10–14 days, colonies were fixed with 4% formaldehyde, stained with 0.5 mg/mlcrystal violet, and imaged on a FluorChem HD2 Imager System (Alpha Innotech).
RESULTS
High-throughput screening for KIFC1 inhibitors
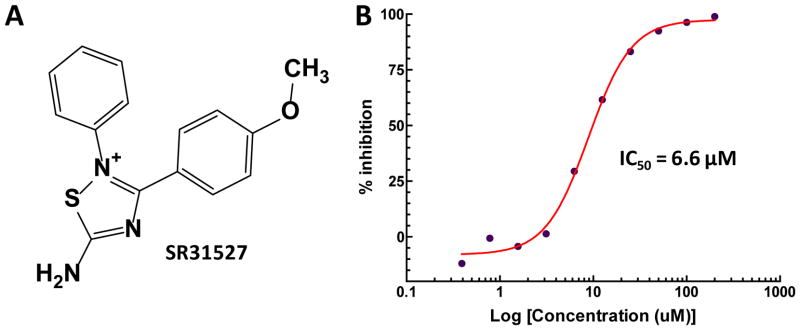
To identify small molecule KIFC1 inhibitors, we established an HTS assay based on an ATPase assay, which detects the MT stimulated enzymatic activity of KIFC1. A diverse library consist of 30,000 structurally representative compounds was first screened in an HTS format. The primary HTS hits were further confirmed in a 10-point 2-fold dilution concentration-response format. Thirty compounds were found to inhibit KIFC1 activity in a concentration-dependent manner (Supplemental Table 1). Among them, SR31527 displayed the most potent inhibitory effect with an IC50 value of 6.6 μM (Figure 1). The rest identified inhibitors are less potent and/or structurally unattractive. For example, while C2 showed a similar IC50 value as SR31527 (Supplemental Table 1), it is a phenanthrene derivative with potential non-selective DNA intercalation properties. In addition, C2 contains a chemically reactive benzoquinone moiety that could potentially react with proteins possessing nucleophilic groups such as thiols. Therefore, we focused our follow-up studies on SR31527.
FIGURE 1.
Structure of SR31527 and its inhibitory effect on KIFC1 function. (A) Chemical structure of SR31527. (B) SR31527 inhibited the MT-stimulated ATPase activity of KIFC1 in a concentration dependent mode with an IC50value of 6.6 μM. The kinetic curve was fitted using GraphPad.
SR31527 binds directly to KIFC1 without involving MT
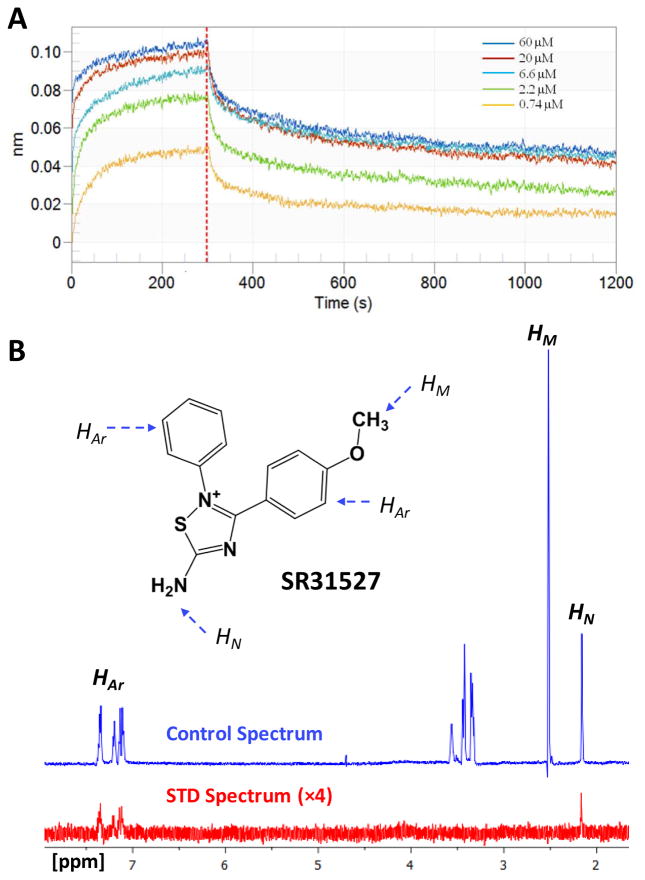
An important advantage of mitotic kinesin inhibitors over traditional antimitotic agents is that they do not interfere with the multi-functional MT, thus may cause fewer side effects. Since our primary assay detects the MT-stimulated ATPase activities, there is a chance that the identified active compounds may inhibit KIFC1 function through a MT related mechanism (37). We therefore utilized Biolayer Interferometry (BLI) to evaluate whether SR31527 can bind directly to KIFC1 without involving MTs. Our BLI results demonstrated direct binding of SR31527 to KIFC1 with a calculated dissociation constant Kd value of 25.4 nM (Figure 2A). We further applied Saturation-Transfer Difference (STD) NMR as a secondary binding assay, which is a sensitive and easily applicable technique that not only detects transient binding, but also provides information regarding which part(s) of a ligand interacts directly with a receptor (32,34,38). The observed STD-NMR spectrum confirmed that SR31527 bound to KIFC1 in the absence of MT, and indicated that the aromatic ring(s) and the amine group of SR31527 interact directly with KIFC1 (Figure 2B).
FIGURE 2.
SR31527 binds directly to KIFC1. (A) The biolayer interferometry (BLI)results confirmed that SR31527 binds KIFC1 (without the existence of microtubules) with a calculated dissociate constant Kd value of 25.4 nM. The experiments were performed and analyzed using the ForteBIO Octet system. (B) The reference 1D 1H NMR spectra of SR31527 in the presence of KIFC1, at 600 MHz and 298 K ( blue-colored) and the corresponding STD-NMR spectrum (×4) (red-colored). SR31527 structure is embedded with its aromatic, methyl and amine hydrogen marked as HAr, HM and HN, respectively, and their corresponding NMR signals assigned.
SR31527 binds to a novel allosteric site of the KIFC1 motor domain
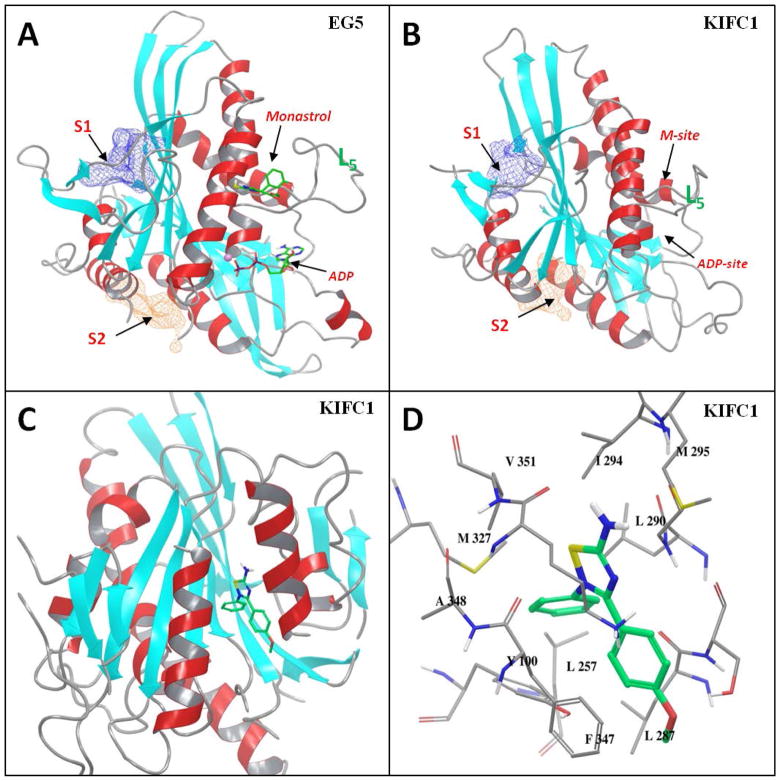
We conducted structural analysis and docking studiesto explore the structural insights ofpotential interactions between SR31527 and KIFC1. We have previously studied kinesin Eg5 through molecular modeling and simulations, and identified several novel allosteric sites in addition to the well-studied monastrol binding-site (M-site) where most known Eg5 inhibitors bind (39). By mapping the surface of the KIFC1 crystal structure, we found that two of the previously identified kinesin allosteric pockets, S1 and S2, also exist on KIFC1 with excellent SiteScore of 1.09 and 0.97, respectively (Figure. 3A & 3B). A Sitescore value of 0.80 has been found to accurately distinguish between drug-binding and non-drug-binding sites (40), suggesting both S1 and S2 are potential binding sites for potent KIFC1 inhibitors. On the other hand, due to the relatively short loop-5 of KIFC1 that forms part of the M-site pocket, the SiteScore of the M-site on KIFC1 is only 0.70 compared to 1.02 on Eg5.
FIGURE 3.
Structural illustration of the binding between SR31527 and KIFC1. (A) The carton presentation of the X-ray structure of the Eg5 motor domain (PDB ID: 1Q0B). The monastrol and ADP-binding sites, the previously identified allosteric S1 and S2 sites, and the loop-5 are marked; (B) The carton representation of the X-ray structure of KIFC1 motor domain (PDB ID: 2REP) with the different sites and the loop-5 marked; (C) The docked KIFC1-SR31527 model with SR31527 bind at the S2 site of KIFC1 (the model was rotated approximately 180 degrees from the orientation of (A)/(B) for better presentation); (D) The close-up view of the KIFC1-SR31527 interaction.
We further performed docking studies to evaluate which site SR31527 may bind to. The SR31527 molecule was docked separately into the different binding pockets of KIFC1, including the M-site, S1, S2 and the ATP binding site (ATP-site). The docked results indicated that SR31527 fitted best at the S2 site with a docking score of −6.5 kcal/mol, comparing to −4.9, −3.1 and −4.8 kcal/mol for the S1, M-site and ATP-site. The S2 site is a cleft-shaped pocket located between the helix α4 and α6 of KIFC1. Visual examination of the docked models further confirmed the structural complementarity between SR31527 and the S2 site: the aromatic rings of SR31527 fit well into the hydrophobic pocket of S2 and interact with Tyr100 and Phe347 through π-π stacking, while the polar amide group points towards the solvent accessible area of the pocket (Figure. 3C & 3D). Interestingly, in our STD-NMR experiments (Figure 2B), the 1D 1H STD spectrum of the SR31527 methyl group was not observable, which suggested that, unlike the other structural elements of the SR31527 molecule, the methyl group was relatively away from the protein, therefore was not significantly affected by the binding. This is consistent with the docked model where the methyl group pointed towards the outside of the S2 pocket and did not form any specific interactions with the protein. Taken together, our results suggest that SR31527 inhibitsKIFC1 by binding directly tothe allosteric S 2site of KIFC1.
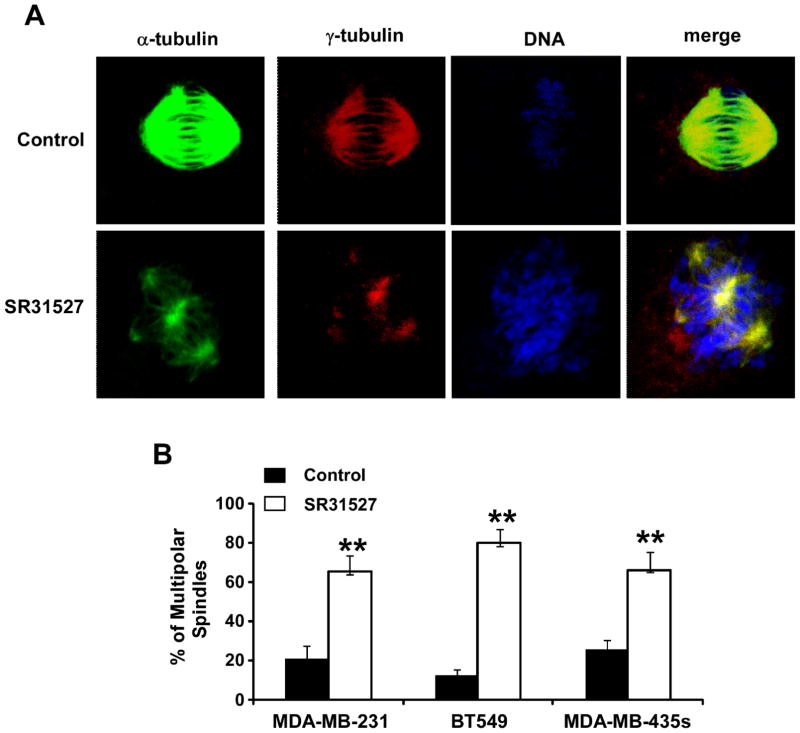
SR31527 triggers multipolar spindle formation in breast cancer cells
KIFC1 is normally a nonessential kinesin motor protein, but plays a critical role in centrosome clustering in cancer cells (27,28). Knockdown of KIFC1 induced multipolar spindle mitotic defects in cancer cell lines containing extra centrosomes, and induced cancer cell death (27,28). Having established that SR31527 binds to KIFC1 and suppresses KIFC1 enzymatic activity, we then examined whether SR31527 induces multipolar spindle formation in breast cancer cells. As shown in Figure 4, SR31527 significantly enhanced the percentage of MDA-MB-231, BT549 and MDA-MB-435s cells containing multipolar spindles, indicating that SR31527 actsas an extra -centrosome de-clustering agentby binding to KIFC1.
FIGURE 4.
SR31527 induced multiple spindle formation in breast cancer cells. (A) BT549 cells were treated with SR31527 at 50 μM. After 24 h incubation, the cells were fixed, permeabilized and immunolabeled for α- and γ-tubulin for the detection of spindles and centrosomes (green and red fluorescent labeling, respectively). DNA was labeled with NucRed647 (blue). (B) MDA-MB-231, BT549 and MDA-MB-435s were treated with SR31527 at 50 μM for 24 h. The cells were then fixed and stained as described above, and the percentage of multipolar spindles was calculated in each cell line treated with DMSO control or SR31527. All the values are the average of triplicate determinations with the s.d. indicated by error bars. **P<0.01 versus corresponding cancer cells treated with DMSO control.
SR31527 decreases the cell viability and colony formation of breast cancer cells and is less cytotoxic to normal human fibroblasts
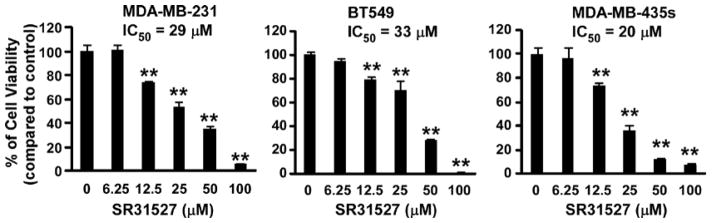
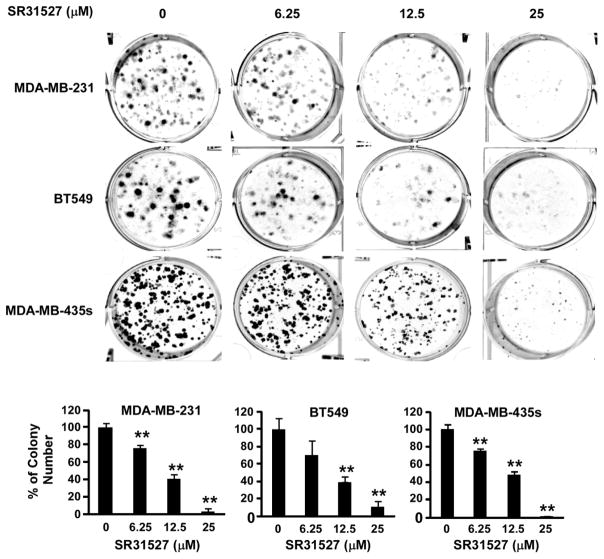
Given that SR31527 can bind to KIFC1, suppress KIFC1 enzymatic activity and induce multiple spindle formation in breast cancer cells, we then examined the effect of SR31527 on cell viability of breast cancer cells. As shown in Figure 5, SR31527 inhibited TNBC cell viability in a concentration dependent manner with IC50 values between 20 and 33 μM in TNBC cell lines MDA-MB-231, BT549 and MDA-MB-435s. To further characterize the anti-cancer activity of SR31527, we performed colony formation assays in TNBC cells. As showed in Figure 6, SR31527 at 6.25 to 25 μM significantly suppressed colony formation in TNBC cells.
FIGURE 5.
SR31527 decreased breast cancer cell viability. Breast cancer MDA-MB-231, BT549 and MDA-MB-435s cells in 96-well plates were treated with SR31527 at the indicated concentrations for 96 h. Cell viability was measured by the CellTiter-Glo assay. All the values are the average of triplicate determinations with the s.d. indicated by error bars. **P<0.01 versus corresponding control value.
FIGURE 6.
SR31527 inhibited breast cancer colony formation. Breast cancer MDA-MB-231, BT549 and MDA-MB-435s cells were treated with SR31527 at the indicated concentrations for 10–12 days. The media were changed every three days. Colonies were fixed with formaldehyde and stained with crystal violet. All the values are the average of triplicate determinations with the s.d. indicated by error bars. **P<0.01 compared to cells treated with DMSO.
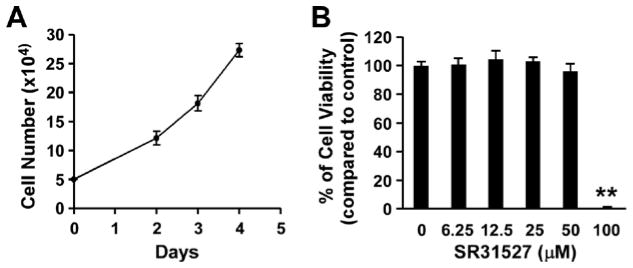
A specific KIFC1 inhibitor should not only display its cytotoxicity against malignant cells, but also be less toxic toward normal cells. In our recent study, we reported that KIFC1 is highly expressed in TNBC cell lines, but is almost undetectable in human lung fibroblast line LL47 (17). We therefore evaluated the effects of SR31527 on LL47 cells. LL47 cells had a moderate growth rate with the doubling time of 41 hours in the logarithmic growth phase (Figure 7A). As shown in Figure 7B, SR31527 at the concentrations between 6.25 and 50 μM had almost no effects on LL47 cell viability, whereas SR31527 at 50 μM was able to kill 65–83% of TNBC cells (Figure 5), suggesting that SR31527 is able to selectively kill malignant cells. However, SR31527 at 100 μM killed almost all of LL47 cells, indicating that SR31527 displaysoff -target cytotoxiceffects at high concentrations.
FIGURE 7.
Effects of SR31527 on LL47 cell viability. (A) LL47 cells were plated in 12-well plates (5 × 104 cells per well), and cells were harvested and counted with trypan blue at 48, 72, and 96 h. All the values are the average of triplicate determinations, and are plotted with s.d. given as error bars. (B) LL47 cells in 96-well plates were treated with SR31527 at the indicated concentrations for 96 h. Cell viability was measured by the CellTiter-Glo assay. All the values are the average of triplicate determinations with the s.d. indicated by error bars. **P<0.01 compared to cells treated with DMSO.
DISCUSSION
Chromosomal instability (CIN) is a hallmark of many tumors and is frequently caused by extra centrosomes that transiently disrupt the normal bipolar spindle geometry needed for accurate chromosome segregation (18,19,41,42). It has been demonstrated that KIFC1 is required for the survival of cancer cells with multiple centrosomes, and more importantly, it is nonessential for normal cells. Therefore, specific inhibition of KIFC1 could be a promising strategy to selectively kill cancer cells without damaging normal healthy cells. In the present study, we identified a small molecule KIFC1 inhibitor, SR31527, which binds directly to KIFC1 with an IC50 value of 6.6 μM against MT-stimulated KIFC1 ATPase activity. Moreover, SR31527 prevented bipolar clustering of extra-centrosomes in TNBC cells and significantly reduced TNBC cell viability with IC50 values between 20 and 33 μM in TNBC cell lines MDA-MB-231, BT549 and MDA-MB-435s. Importantly, SR31527 displayed no cytotoxic effects on LL47 fibroblasts at the concentration up to 50 μM, indicating that SR31527 shows good selectivityagainst cancer cells.
KIFC1 has emerged as an anti-cancer drug target in recent years. To date, only two groups have reported specific KIFC1 inhibitors in the literature (37,43,44). Scientists at AstraZeneca identified a small molecule KIFC1 inhibitor, AZ82, which inhibited MT-stimulated KIFC1 ATPase activity with an IC50 of 0.3 μM and triggered multipolar spindle formation and mitotic catastrophe in cells with amplified centrosomes. However, a nonspecific cytotoxic effect of AZ82 was observed at 4 μM, which prevented further studies regarding whether it can selectively kill cancer cells with amplified centrosomes. Additional studies indicated that AZ82 bound specifically to KIFC1/MT complex but did not interact directly with KIFC1 or MT when they were not associated (37). Another KIFC1 inhibitor (CW069) recently reported by Watts and coworkers was computationally designed based on the inhibitors of kinesin Eg5 (43). CW069 bound specifically to KIFC1 and inhibited its ATPase activity with an IC50 value of 75 μM. It increased multipolar spindle formation in neuroblastoma N1E-115 cells, but had no effects on bipolar spindle formation in normal human dermal fibroblasts (NHDF). CW069 was also more potent at decreasing cell viability of NIE-115 cells (IC50 = 86 μM) than the NHDF cells (IC50 187 μM) (43). In the present study, we demonstrated that SR31527 binds directly to KIFC1 without interacting with MT. Moreover, SR31527 exhibits high selectivity for cytotoxicity toward TNBC cells compared with normal fibroblasts. All together, these findings suggest that by exclusively binding to KIFC1, KIFC1 inhibitors could selectively kill cancer cells over normal cells. However, a recent study demonstrated that KIFC1 is essential for bipolar spindle formation and genomic stability in the primary human fibroblast line IMR-90 (45). Moreover, KIFC1 is able to promote breast cancer cell proliferation and enhance cell cycle kinetics through G2 and M-phases via centrosome clustering-independent mechanisms in breast cancer cells (26). Therefore, future studies are required to address whether there are any other cell cycle abnormalities in normal cells and cancer cells treated with SR31527.
Our computer modeling and NMR results indicated that SR31527 bound to a novel allosteric S2 site on KIFC1. We further attempted mutagenesis studies of the S2 site residues, but failed to obtain active mutant KIFC1 proteins (data not shown), suggesting residues at the S2 site are important for the structure and function of KIFC1. The allosteric binding of SR31527 is different from AZ82 and CW069, both putatively bind to the M-site of KIFC1. This allosteric S2 site was originally found by us on kinesin Eg5 based on computational analysis of molecular dynamics simulation results (39), and was later confirmed by the crystal structure of an Eg5-inhibitor complex (46). Since all kinesin proteins contain a structurally similar motor domain, it is not very surprising that such a similar S2 site was also found on KIFC1. The structural features of kinesin proteins are highly conserved, although KIFC1 is the only kinesin which displays a pivotal role in cancer cell centrosome clustering, spindle assembly and survival (27,28). Therefore, identifying selective inhibitors of KIFC1 is critical for developing cancer cell-selective agents. One well-known strategy for achieving selectivity is to target a protein’s allosteric binding sites (47–51). While targeting the allosteric M-site for selective Eg5 inhibitors has been successful (11), the M-Site is quite specific for the Eg5 motor due to its unique long loop-5 which forms the lid of the pocket. Therefore, a similar strategy may not work well for other kinesins with short loop-5, such as KIFC1. The S2 site is mainly formed by two structurally conserved helices and has the structural properties that are suitable for the tight binding of small molecule ligands, therefore, it offers an opportunity to identify selective KIFC1 inhibitors. SR31527 thus provides a valuable tool to study the allosteric regulation of KIFC1 and serves as a potential lead for the development of a novel therapeutic agent for cancer treatment. Further structural biology studies, such as through X-ray crystallography, shall provide additional structural and biological details of the allosteric KIFC1-inhibitor interactions for drug discovery purposes.
Summary of statement.
KIFC1 is nonessential for normal cells, but critical for the survival of cancer cells. In this study, we identified a new KIFC1 inhibitor which significantly reduced the viability of TNBC cells, but was less toxic to normal fibroblasts.
Acknowledgments
We thank Dr. N. Rama Krishna and Dr. Ronald Shin of the UAB High Field NMR Facility for their supports on the collection of NMR spectrum.
Funding information This work was partially supported by Southern Research Internal Fund, Alabama Innovation Fund and grants from the National Institutes of Health R01CA124531 and R21CA182056
Footnotes
Declarations of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Author contribution statement WZ coordinated the study, performed the computational modeling and wrote the paper. YW and RL designed and purified the protein. LZ, IP, RB, LW and LR performed the HTS studies. JM, SA, CA and MS contributed their medicinal chemistry expertise to the study and the manuscript. RB and VG conducted the binding assays studies. WL, BX and YL were responsible for the cell based assays and related writings.
References
- 1.Nagle A, Hur W, Gray NS. Antimitotic agents of natural origin. Curr Drug Targets. 2006;7:305–326. doi: 10.2174/138945006776054933. [DOI] [PubMed] [Google Scholar]
- 2.Hagiwara H, Sunada Y. Mechanism of taxane neurotoxicity. Breast cancer. 2004;11:82–85. doi: 10.1007/BF02968008. [DOI] [PubMed] [Google Scholar]
- 3.Quasthoff S, Hartung HP. Chemotherapy-induced peripheral neuropathy. Journal of neurology. 2002;249:9–17. doi: 10.1007/pl00007853. [DOI] [PubMed] [Google Scholar]
- 4.Miki H, Okada Y, Hirokawa N. Analysis of the kinesin superfamily: insights into structure and function. Trends in Cell Biology. 2005;15:467–476. doi: 10.1016/j.tcb.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 5.Huszar D, Theoclitou ME, Skolnik J, Herbst R. Kinesin motor proteins as targets for cancer therapy. Cancer and Metastasis Reviews. 2009;28:197–208. doi: 10.1007/s10555-009-9185-8. [DOI] [PubMed] [Google Scholar]
- 6.Sarli V, Giannis A. Inhibitors of mitotic kinesins: next-generation antimitotics. ChemMedChem. 2006;1:293–298. doi: 10.1002/cmdc.200500045. [DOI] [PubMed] [Google Scholar]
- 7.Jiang C, You QD, Li ZY, Guo QL. Kinesin spindle protein inhibitors as anticancer agents. Expert Opinion on Therapeutic Patents. 2006;16:1517–1532. [Google Scholar]
- 8.Bergnes G, Brejc K, Belmont L. Mitotic kinesins: Prospects for antimitotic drug discovery. Current Topics in Medicinal Chemistry. 2005;5:127–145. doi: 10.2174/1568026053507697. [DOI] [PubMed] [Google Scholar]
- 9.Coleman PJ, Fraley ME. Inhibitors of the mitotic kinesin spindle protein. Expert Opinion on Therapeutic Patents. 2004;14:1659–1667. [Google Scholar]
- 10.Wood KW, Lad L, Luo L, Qian X, Knight SD, Nevins N, Brejc K, Sutton D, Gilmartin AG, Chua PR, Desai R, Schauer SP, McNulty DE, Annan RS, Belmont LD, Garcia C, Lee Y, Diamond MA, Faucette LF, Giardiniere M, Zhang S, Sun CM, Vidal JD, Lichtsteiner S, Cornwell WD, Greshock JD, Wooster RF, Finer JT, Copeland RA, Huang PS, Morgans DJ, Jr, Dhanak D, Bergnes G, Sakowicz R, Jackson JR. Antitumor activity of an allosteric inhibitor of centromere-associated protein-E. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:5839–5844. doi: 10.1073/pnas.0915068107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.El-Nassan HB. Advances in the discovery of kinesin spindle protein (Eg5) inhibitors as antitumor agents. European journal of medicinal chemistry. 2013;62:614–631. doi: 10.1016/j.ejmech.2013.01.031. [DOI] [PubMed] [Google Scholar]
- 12.Knight SD, Parrish CA. Recent progress in the identification and clinical evaluation of inhibitors of the mitotic kinesin KSP. Current Topics in Medicinal Chemistry. 2008;8:888–904. doi: 10.2174/156802608784911626. [DOI] [PubMed] [Google Scholar]
- 13.Gerecitano JF, Stephenson JJ, Lewis NL, Osmukhina A, Li J, Wu K, You Z, Huszar D, Skolnik JM, Schwartz GK. A Phase I trial of the kinesin spindle protein (Eg5) inhibitor AZD4877 in patients with solid and lymphoid malignancies. Investigational new drugs. 2013;31:355–362. doi: 10.1007/s10637-012-9821-y. [DOI] [PubMed] [Google Scholar]
- 14.Holen K, DiPaola R, Liu G, Tan AR, Wilding G, Hsu K, Agrawal N, Chen C, Xue L, Rosenberg E, Stein M. A phase I trial of MK-0731, a kinesin spindle protein (KSP) inhibitor, in patients with solid tumors. Investigational new drugs. 2012;30:1088–1095. doi: 10.1007/s10637-011-9653-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee RT, Beekman KE, Hussain M, Davis NB, Clark JI, Thomas SP, Nichols KF, Stadler WM. A University of Chicago consortium phase II trial of SB–715992 in advanced renal cell cancer. Clinical genitourinary cancer. 2008;6:21–24. doi: 10.3816/CGC.2008.n.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Woessner R, Tunquist B, Lemieux C, Chlipala E, Jackinsky S, Dewolf W, Jr, Voegtli W, Cox A, Rana S, Lee P, Walker D. ARRY-520, a novel KSP inhibitor with potent activity in hematological and taxane-resistant tumor models. Anticancer research. 2009;29:4373–4380. [PubMed] [Google Scholar]
- 17.Li Y, Lu W, Chen D, Boohaker RJ, Zhai L, Padmalayam I, Wennerberg K, Xu B, Zhang W. KIFC1 is a novel potential therapeutic target for breast cancer. Cancer biology & therapy. 2015;16:1316–1322. doi: 10.1080/15384047.2015.1070980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lingle WL, Barrett SL, Negron VC, D'Assoro AB, Boeneman K, Liu W, Whitehead CM, Reynolds C, Salisbury JL. Centrosome amplification drives chromosomal instability in breast tumor development. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:1978–1983. doi: 10.1073/pnas.032479999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.D'Assoro AB, Lingle WL, Salisbury JL. Centrosome amplification and the development of cancer. Oncogene. 2002;21:6146–6153. doi: 10.1038/sj.onc.1205772. [DOI] [PubMed] [Google Scholar]
- 20.Brinkley BR. Managing the centrosome numbers game: from chaos to stability in cancer cell division. Trends Cell Biol. 2001;11:18–21. doi: 10.1016/s0962-8924(00)01872-9. [DOI] [PubMed] [Google Scholar]
- 21.Acilan C, Saunders WS. A tale of too many centrosomes. Cell. 2008;134:572–575. doi: 10.1016/j.cell.2008.08.007. [DOI] [PubMed] [Google Scholar]
- 22.Boland CR, Komarova NL, Goel A. Chromosomal instability and cancer: not just one CINgle mechanism. Gut. 2009;58:163–164. doi: 10.1136/gut.2008.160143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Godinho S, Kwon M, Pellman D. Centrosomes and cancer: how cancer cells divide with too many centrosomes. Cancer and Metastasis Reviews. 2009;28:85–98. doi: 10.1007/s10555-008-9163-6. [DOI] [PubMed] [Google Scholar]
- 24.Gergely F, Basto R. Multiple centrosomes: together they stand, divided they fall. Genes & Development. 2008;22:2291–2296. doi: 10.1101/gad.1715208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Quintyne NJ, Reing JE, Hoffelder DR, Gollin SM, Saunders WS. Spindle multipolarity is prevented by centrosomal clustering. Science. 2005;307:127–129. doi: 10.1126/science.1104905. [DOI] [PubMed] [Google Scholar]
- 26.Pannu V, Rida PC, Ogden A, Turaga RC, Donthamsetty S, Bowen NJ, Rudd K, Gupta MV, Reid MD, Cantuaria G, Walczak CE, Aneja R. HSET overexpression fuels tumor progression via centrosome clustering-independent mechanisms in breast cancer patients. Oncotarget. 2015;6:6076–6091. doi: 10.18632/oncotarget.3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kleylein-Sohn J, Pollinger B, Ohmer M, Hofmann F, Nigg EA, Hemmings BA, Wartmann M. Acentrosomal spindle organization renders cancer cells dependent on the kinesin HSET. J Cell Sci. 2012;125:5391–5402. doi: 10.1242/jcs.107474. [DOI] [PubMed] [Google Scholar]
- 28.Kwon M, Godinho SA, Chandhok NS, Ganem NJ, Azioune A, Thery M, Pellman D. Mechanisms to suppress multipolar divisions in cancer cells with extra centrosomes. Genes & Development. 2008;22:2189–2203. doi: 10.1101/gad.1700908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pawar S, Donthamsetty S, Pannu V, Rida P, Ogden A, Bowen N, Osan R, Cantuaria G, Aneja R. KIFCI, a novel putative prognostic biomarker for ovarian adenocarcinomas: delineating protein interaction networks and signaling circuitries. Journal of ovarian research. 2014;7:53. doi: 10.1186/1757-2215-7-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang B, Senator D, Wilson CJ, Ng SC. Development of a high-throughput robotic fluorescence-based assay for HsEg5 inhibitor screening. Anal Biochem. 2005;345:326–335. doi: 10.1016/j.ab.2005.07.020. [DOI] [PubMed] [Google Scholar]
- 31.Sherman W, Day T, Jacobson MP, Friesner RA, Farid R. Novel procedure for modeling ligand/receptor induced fit effects. Journal of Medicinal Chemistry. 2006;49:534–553. doi: 10.1021/jm050540c. [DOI] [PubMed] [Google Scholar]
- 32.Jayalakshmi V, Krishna NR. CORCEMA refinement of the bound ligand conformation within the protein binding pocket in reversibly forming weak complexes using STD-NMR intensities. Journal of Magnetic Resonance. 2004;168:36–45. doi: 10.1016/j.jmr.2004.01.017. [DOI] [PubMed] [Google Scholar]
- 33.Krishna NR, Jayalakshmi V. Complete relaxation and conformational exchange matrix analysis of STD-NMR spectra of ligand-receptor complexes. Progress in Nuclear Magnetic Resonance Spectroscopy. 2006;49:1–25. [Google Scholar]
- 34.Peng JW, Lepre CA, Fejzo J, Abdul-Manan N, Moore JM. Nuclear magnetic resonance-based approaches for lead generation in drug discovery. Methods Enzymol. 2001;338:202–230. doi: 10.1016/s0076-6879(02)38221-1. [DOI] [PubMed] [Google Scholar]
- 35.Do T, Ho F, Heidecker B, Witte K, Chang L, Lerner L. A rapid method for determining dynamic binding capacity of resins for the purification of proteins. Protein expression and purification. 2008;60:147–150. doi: 10.1016/j.pep.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 36.Abdiche Y, Malashock D, Pinkerton A, Pons J. Determining kinetics and affinities of protein interactions using a parallel real-time label-free biosensor, the Octet. Anal Biochem. 2008;377:209–217. doi: 10.1016/j.ab.2008.03.035. [DOI] [PubMed] [Google Scholar]
- 37.Wu J, Mikule K, Wang W, Su N, Petteruti P, Gharahdaghi F, Code E, Zhu X, Jacques K, Lai Z, Yang B, Lamb ML, Chuaqui C, Keen N, Chen H. Discovery and mechanistic study of a small molecule inhibitor for motor protein KIFC1. ACS chemical biology. 2013;8:2201–2208. doi: 10.1021/cb400186w. [DOI] [PubMed] [Google Scholar]
- 38.Jayalakshmi V, Krishna NR. Complete relaxation and conformational exchange matrix (CORCEMA) analysis of intermolecular saturation transfer effects in reversibly forming ligand-receptor complexes. Journal of Magnetic Resonance. 2002;155:106–118. doi: 10.1006/jmre.2001.2499. [DOI] [PubMed] [Google Scholar]
- 39.Zhang W. Exploring the intermediate states of ADP-ATP exchange: a simulation study on Eg5. J Phys Chem B. 2011;115:784–795. doi: 10.1021/jp107255t. [DOI] [PubMed] [Google Scholar]
- 40.Sheth PR, Shipps GW, Seghezzi W, Smith CK, Chuang CC, Sanden D, Basso AD, Vilenchik L, Gray K, Annis DA, Nickbarg E, Ma Y, Lahue B, Herbst R, Le HV. Novel Benzimidazole inhibitors Bind to a Unique Site in the Kinesin Spindle Protein Motor Domain. Biochemistry. 2010;49:8350–8358. doi: 10.1021/bi1005283. [DOI] [PubMed] [Google Scholar]
- 41.Ganem NJ, Godinho SA, Pellman D. A mechanism linking extra centrosomes to chromosomal instability. Nature. 2009;460:278–U146. doi: 10.1038/nature08136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sluder G, Nordberg JJ. The good, the bad and the ugly: the practical consequences of centrosome amplification. Curr Opin Cell Biol. 2004;16:49–54. doi: 10.1016/j.ceb.2003.11.006. [DOI] [PubMed] [Google Scholar]
- 43.Watts CA, Richards FM, Bender A, Bond PJ, Korb O, Kern O, Riddick M, Owen P, Myers RM, Raff J, Gergely F, Jodrell DI, Ley SV. Design, synthesis, and biological evaluation of an allosteric inhibitor of HSET that targets cancer cells with supernumerary centrosomes. Chemistry & biology. 2013;20:1399–1410. doi: 10.1016/j.chembiol.2013.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang B, Lamb ML, Zhang T, Hennessy EJ, Grewal G, Sha L, Zambrowski M, Block MH, Dowling JE, Su N, Wu J, Deegan T, Mikule K, Wang W, Kaspera R, Chuaqui C, Chen H. Discovery of Potent KIFC1 Inhibitors Using a Method of Integrated High-Throughput Synthesis and Screening. J Med Chem. 2014;57:9958–9970. doi: 10.1021/jm501179r. [DOI] [PubMed] [Google Scholar]
- 45.Kim N, Song K. KIFC1 is essential for bipolar spindle formation and genomic stability in the primary human fibroblast IMR-90 cell. Cell structure and function. 2013;38:21–30. doi: 10.1247/csf.12014. [DOI] [PubMed] [Google Scholar]
- 46.Ulaganathan V, Talapatra SK, Rath O, Pannifer A, Hackney DD, Kozielski F. Structural insights into a unique inhibitor binding pocket in kinesin spindle protein. J Am Chem Soc. 2013;135:2263–2272. doi: 10.1021/ja310377d. [DOI] [PubMed] [Google Scholar]
- 47.Changeux JP. 50th anniversary of the word "Allosteric". Protein Science. 2011;20:1119–1124. doi: 10.1002/pro.658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ozbabacan SEA, Gursoy A, Keskin O, Nussinov R. Conformational ensembles, signal transduction and residue hot spots: Application to drug discovery. Current Opinion in Drug Discovery & Development. 2010;13:527–537. [PubMed] [Google Scholar]
- 49.Eglen RM, Reisine T. Human kinome drug discovery and the emerging importance of atypical allosteric inhibitors. Expert Opinion on Drug Discovery. 2010;5:277–290. doi: 10.1517/17460441003636820. [DOI] [PubMed] [Google Scholar]
- 50.Fenton AW. Allostery: an illustrated definition for the 'second secret of life'. Trends Biochem Sci. 2008;33:420–425. doi: 10.1016/j.tibs.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lindsley JE, Rutter J. Whence cometh the allosterome? Proceedings of the National Academy of Sciences of the United States of America. 2006;103:10533–10535. doi: 10.1073/pnas.0604452103. [DOI] [PMC free article] [PubMed] [Google Scholar]