Abstract
The widely known vocabulary gap between health consumers and healthcare professionals hinders information seeking and health dialogue of consumers on end-user health applications. The Open Access and Collaborative Consumer Health Vocabulary (OAC CHV), which contains health-related terms used by lay consumers, has been created to bridge such a gap. Specifically, the OAC CHV facilitates consumers’ health information retrieval by enabling consumer-facing health applications to translate between professional language and consumer friendly language. To keep up with the constantly evolving medical knowledge and language use, new terms need to be identified and added to the OAC CHV. User-generated content on social media, including social question and answer (social Q&A) sites, afford us an enormous opportunity in mining consumer health terms. Existing methods of identifying new consumer terms from text typically use ad-hoc lexical syntactic patterns and human review. Our study extends an existing method by extracting n-grams from a social Q&A textual corpus and representing them with a rich set of contextual and syntactic features. Using K-means clustering, our method, simiTerm, was able to identify terms that are both contextually and syntactically similar to the existing OAC CHV terms. We tested our method on social Q&A corpora on two disease domains: diabetes and cancer. Our method outperformed three baseline ranking methods. A post-hoc qualitative evaluation by human experts further validated that our method can effectively identify meaningful new consumer terms on social Q&A.
Keywords: Controlled vocabularies, Consumer health vocabulary, Consumer health information, Social Q&A, Ontology enrichment
Graphical abstract
1. Introduction
Over the past two decades, a variety of controlled vocabularies and domain ontologies in biomedicine have been established to encode biomedical entities, terms, and their relationships [1]. Well-curated controlled vocabularies lay a solid foundation in various healthcare information systems such as electronic health records (EHRs) and clinical decision support systems [2]. For example, the International Classification of Diseases 9th Revision, Clinical Modification (ICD-9-CM) is widely used to encode the diagnoses and procedures in healthcare administrative documents for billing purposes [3]. SNOMED CT is recommended for encoding problem lists in EHRs by the Meaningful Use Stage 2 of the Health Information Technology for Economic and Clinical Health (HITECH) Act [4]. RxNORM is a reference terminology that normalizes names of all clinical drugs available on the U.S. market with links to many of the drug vocabularies commonly used in pharmacy management [5]. Importantly, the Unified Medical Language System (UMLS), developed and maintained by the U.S. National Library of Medicine (NLM), has integrated more than 190 source vocabularies into its Metathesaurus (META). The UMLS has a high-level semantic network of 127 semantic types which represent the broad semantics of the concepts in the health domain [1]. The META’s source vocabularies include not only professional terminologies such as the ICD, SNOMED CT, and RxNORM, but also vocabularies used by healthcare consumers such as the Open Access and Collaborative Consumer Health Vocabulary (OAC CHV, “CHV” in short) [6]. The CHV was created to complement the existing scope of the UMLS, and to assist the needs for developing consumer-facing health applications. According to a recent Pew Research survey, 72% of the Internet users seek for health information online [7]. With the advent of Web 2.0, user-generated and experience-based health information emerges as collective wisdoms and important knowledge assets [8]. It is therefore important to understand the medical terms adopted by the health consumers when they describe their health issues on these platforms. These terms can thus be used to enrich the CHV and further strengthen its role in building consumer-oriented health applications, such as a consumer-friendly clinical trial search engine.
In contrast to medical terminologies that are primarily used by health professionals (e.g., SNOMED CT), the CHV represents lay people’s health-related terms and expressions that aim at bridging the vocabulary gap between consumers and professionals, and at facilitating information seeking needs of health consumers [6, 9, 10]. Plovnick and Zeng [11] showed that consumers’ search queries, when reformulated with professional terms, can yield better retrival accuracy of health information. As the UMLS pulls together terms of the same meaning from different sources into a unified concept, it can facilitate query reformulation by mapping consumer terms to their corresponding professional terms. As in UMLS 2015AA version, 53.8% of the concepts in the CHV are covered in SNOMED CT. Nevertheless, controlled vocabularies need to be constantly updated with new entities to ensure domain completeness as the medical knowledge evolves [12]. As new health-related terms are consistently emerging and being adopted by health information consumers, the CHV should continue adding these new terms to satisfy their information and communication needs. The development of professional vocabularies (e.g., SNOMED CT) receives suffiicient support from teams of domain experts as well as professional organizations (e.g., SNOMED International and NLM). In contrast, the CHV employs an open-access and collaborative approach [13] to identify or extract lay people’ health-related terms from a variety of consumer-generated text corpora on various platforms such as PatientsLikeMe [14], MedHelp [15, 16], MedLinePlus [17, 18], and Wikipedia [19].
The goal of our study is to develop a framework to identify new consumer health terms that are syntactically and contextually similar to existing CHV terms from Yahoo! Answers, a popular US-based social question and answer (Q&A) platform. Social Q&A is an online community-based question and answer service that allows its users to post their questions and answer others’ questions on a wide range of topics in everyday life. Yahoo! Answers also has a comprehensive list of topic categories devoted to health-related Q&As, which generate rich text corpora with consumer health terms.
Recently, Chandar et al. [20] introduced a similarity-based method to identify potential new SNOMED CT terms in a text corpus of clinical trial eligibility criteria. We extend this similarity-based technique using additional linguistic and contextual features used in previous CHV studies (e.g., [18]) with a set of text mining techniques that includes term extraction, normalization, syntactic parsing, and clustering. Specifically, we propose a method and test it on the social Q&A data from Yahoo! Answers to identify new consumer health terms that can be potentially added to the CHV. We then assess the validity of our method through both a quantitative evaluation and an expert review. Our findings systematically addressed the following research questions:
RQ1: Do the existing CHV terms in the social Q&A text corpus exhibit good clustering characteristics based on syntactic and contextual features?
RQ2: Can the proposed similarity-based technique identify consumer health terms that are syntactically and contextually similar to existing CHV terms in the text corpus?
Health-related terms used by consumers could vary across disease domains. Thus, the current study focuses on analyzing Q&As related to diabetes and cancer. As a common chronic condition in the United States, diabetes can lead to an array of complications such as hypertension, stroke, and kidney disease [21]. Cancer, on the other hand, is the second leading cause of death in the United States, and has a major impact on society across the world [22]. Also a previous study found that the diabetes-related questions and answers posted on Yahoo! Answers has a reasonably good coverage of UMLS concepts [23]. In short, findings from our study will provide a foundation for developing automated and domain-agnostic methods to identify new consumer health terms from consumer-generated textual corpora.
2. Related Work
2.1. Corpus-Based Ontology Learning and Term Identification
Domain-expert-driven ontology (or taxonomy) development involves iterative discussion and reconciliation, which is labor intensive and time consuming. On the other hand, the vast amount of data on the web present ample opportunities for semi-automated learning to ease the burden of ontology curators. Existing ontology learning methods can be broadly categorized as learning from structured data and learning from unstructured data. Unstructured data represents the majority of the user-generated content on today’s social networking platforms, online communities, and social Q&A sites, where consumers increasingly discuss and share their health issues [24]. Thus, we surveyed the ontology learning methods on unstructured data, which primarily include symbolic, statistical, and hybrid approaches. The symbolic approach uses linguistic patterns to identify entities and their relations; this approach usually favors accuracy over recall [19]. The statistical approach uses the frequency of noun or noun phrases in documents retrieved from the web to discover concepts [25]. In the biomedical domain, due to the semantic complexity of the biomedical text, biomedical ontology learning often employs a hybrid of statistical and symbolic methods [26]. For example, Lossio-Ventura et al. leveraged both linguistic and statistical approaches to identify biomedical entities from PubMed abstracts [27]. They further developed a supervised method that uses a set of statistical, lexical, and semantic features to predict the relations between the entities based on a manually annotated dataset. However, creating gold standard labeled datasets is laborious and subjective. Hoxha et al. [28] recently developed Ontofier, an unsupervised ontology learning framework that uses the agglomerative hierarchical clustering to learn domain taxonomies. They used agglomerative hierarchical clustering to produce a dendrogram (i.e., a tree diagram) that can be pruned to be a taxonomy with parent-child relations. They applied the framework on free-text clinical trial eligibility criteria and MEDLINE abstracts to build domain taxonomies of UMLS concepts. The limitation of their work is that only UMLS concepts were used to build the taxonomy and that the framework has only been tested on biomedical text corpora. Since the CHV does not have hierarchical relationships, we focused on methods that can effectively identify meaningful terms in text.
2.2. CHV Development from Consumer-Generated Text Corpora
Over the past few years, online health platforms have become increasingly popular for lay people to share experience, express concerns, and seek health information. These platforms have attracted more and more attention of researchers as a fertile ground to mine consumer terms [14–17, 19]. Zeng et al. [18] proposed a general term identification method for CHV development that incorporates collaborative human review and automated term recognition. They identified over 700 consumer terms from MedLinePlus query log files [29] using logistic regression and human review [18]. They also provided review criteria for determining whether a term is a valid CHV term, which we adapted in our study. Keselman et al. [17] mapped the consumer health terms extracted from MedLinePlus query logs to the UMLS concepts, and found that non-mapping concepts constituted a small proportion of consumer health terms, which may affect the process of building CHV. Later, Doing-Harris & Zeng-Treitler [14] created a computer-assistant update (CAU) system to identify new candidate CHV terms from text messages posted on a health-related social networking site, PatientsLikeMe.com. Recently, MacLean and Heer [15] trained a conditional random field (CRF) classifier on crowd-labeled datasets of discussion posts on MedHelp and user comments on CureTogether to identify consumer health terms. To simultaneously extract consumer and professional term pairs, Vydiswaren et al. [19] leveraged pre-defined lexical patterns such as “A, also known as B” to automatically extract the synonymous pairs of terms A and B from Wikipedia, and labeled them as either consumer or professional terms. In another study, over 120,000 discussion messages on MedHelp were examined to identify consumer expressions that co-occurred with pre-selected terms associated with adverse drug reactions [16]. These studies often require labor-intensive manual reviews by domain experts or the crowd, and/or rely on ad hoc lexical syntactic patterns, limiting their scalability.
2.3. Similarity-Based Term Identification
Chandar et al. [20] tested the feasibility of a similarity-based approach to recommend concepts mined from a text corpus to a standard terminology. In their study [20], n-grams, extracted from a text corpus of over 180,000 clinical trial summaries on ClinicalTrials.gov, were represented by a set of features that characterize their syntactic and contextual information. In Chandar et al.’s study, the n-grams that can be covered by SNOMED CT were clustered into groups based on their linguistic, syntactic, and contextual similarity, while the n-grams that were not covered by SNOMED CT were ranked based on their distance to the nearest cluster. Each n-gram was represented with: (1) capitalization features that capture the capitalization information of the n-gram, (2) syntactic features, i.e., Part-of-Speech (POS) tags of the n-gram, (3) prefixes and suffixes of the n-gram, and (4) the syntactic features, prefixes, suffixes of the tokens that co-occur with the n-gram. The advantage of this approach is that rather than giving a binary decision on the inclusion of the new terms, it ranks the candidate terms based on their similarities to existing terms, thereby allowing terminology designers to choose the number of terms to review based on their affordable effort. In this work, we adapted Chandar’s method [20] to identify new consumer health terms from social Q&A data by adding a set of new features such as the C-value [30]. To match terms against the established UMLS vocabularies, we employed a fuzzy matching method that allows minor lexical variants to improve the recall of matching terms. We also strengthened the validation method. Specifically, instead of using the UMLS terms in the text corpus as the validation data in a single test, we created random samples of the CHV terms for training and testing in multiple tests. We employed a multidimensional validation method that uses the numbers of clusters and the top percentage of the ranking results as variables to test the validity and the robustness of our method on multiple datasets.
3. Methods
3.1. Data Collection
Yahoo! Answers is a social Q&A site, where people seek information through raising questions and receiving answers from others who are willing to share their information, knowledge, experiences, and opinions on a wide range of topics. A major motivation for consumers to post questions on a social Q&A site is that their searches on web search engines with short queries often fail to retrieve useful information for their specific problems [31]. Social Q&A sites such as Yahoo! Answers allow consumers to ask questions in full sentences using natural language. Such questions also tend to contain more context information, and more closely resemble open-domain language than texts written by professionals [32]. Meanwhile, the answers provided on the site are also mainly written in lay language. Therefore, the textual data on social Q&A sites can be a good source for mining consumer terms. As one question may receive more than one answer, we only used the questions and the “best answers” of the questions (as voted by users) to ensure the quality of the text corpus.
Using the application program interface (API) of Yahoo! Answers, we collected a total of 58,422 questions and their corresponding best answers (one for each question) in the diabetes category, and 81,433 questions and answers in the cancer category. Park et al. [23] recently assessed the UMLS terminology coverage in the same dataset but did not find a significant difference between the questions and the answers with respect to the frequently-used terms and semantic types. Therefore, we combined each question with its corresponding best answer as one document in the corpus.
3.2. A Similarity-Based N-gram Model
The key concepts in this paper and their definitions are described in Table 1.
Table 1.
The key concepts in this paper and their definitions
| Concept | Definition |
|---|---|
| Target vocabulary | The vocabulary to which we would like to suggest new terms. In this study, our target vocabulary is the CHV. |
| N-gram | A contiguous sequence of n words in a sentence. In this study, we included up to 5-grams, since n-grams with n between 1 to 5 can cover over 99% of the terms of interest [33]. |
| Seed term | An n-gram extracted from the text corpus that can be found in the target vocabulary (i.e., CHV). |
| Candidate term | An n-gram that is not covered by the target vocabulary but could be potentially added to the target vocabulary. In order to qualify as a candidate term, an n- gram may be subject to some constraints, e.g., occurring more than 5 times in the corpus. |
| Term context | Either the entire sentence that contains the term, or a window of 10 words before or after the term in its sentence. |
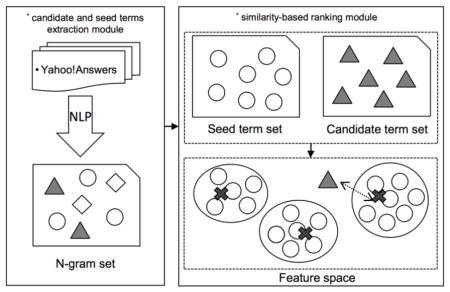
Essentially, our method, simiTerm, consists of two major modules (Figure 1): (1) a candidate and seed terms extraction module; and (2) a similarity-based ranking module. In the first module, we applied natural language processing (NLP) techniques (e.g., sentence splitting, tokenization, Part-of-Speech tagging, fuzzy matching) to extract terms from the textual corpora, and constructed syntactic and contextual features for each term. We identified terms that can be matched to existing CHV terms as seed terms; and the rest as candidate terms. In the second module, we grouped the seed terms using the K-means clustering, and ranked the candidate terms based on their distance to the center of the nearest seed term cluster.
Figure 1.

The conceptual process of consumer term recommendation method simiTerm.
3.3. Data Processing and Analysis Workflow
Considering the huge volume of social Q&A datasets, we implemented our method on the Apache Spark platform, a widely used distributed computing framework [34]. Figure 2 shows the workflow of simiTerm.
Figure 2.
The workflow of data processing of simiTerm.
3.3.1. Step 1: Candidate and Seed Terms Extraction
We adopted the OpenNLP library [35], which has good performance for general purpose NLP tasks [36]. We first split the documents into sentences and extracted tokens from each sentence. To normalize lexical variations of a token (e.g., ‘calorie’ and its plural form ‘calories’), we used the LVG (Lexical Variant Generation) tool [37] to convert each token to its basic form. Then, we obtained a stream of tokens for the sentence in which the term appears. The n-grams were then extracted by iterating over the contiguous tokens (up to 5-grams were extracted). A stop words list can effectively exclude meaningless n-grams such as prepositions ‘of’, ‘on’, or the article ‘a’, ‘an’ and ‘the’. We excluded n-grams beginning or ending with a stop word or a digit. We also excluded symbols. We considered n-grams with the same string but different POS tags as different n-grams. We also excluded n-grams with term frequency less than 5 in the whole dataset.
Next, we obtained a set of n-grams from the text corpus. To determine whether an n-gram is a seed term or not, we employed a fuzzy matching approach [38] to identify n-grams that exist in the CHV. We generated the basic form of each existing CHV term using same techniques described above. If an n-gram’s basic form is the same as any CHV term’s basic form, the n-gram is recognized as a seed term; otherwise it is a candidate term. Figure S1 in the Supplementary Material illustrates an example of the fuzzy matching process. We use the notation “UMLS w/o CHV” to indicate that a term is in the UMLS but not in CHV. Only CHV terms were considered as seed terms. The non-CHV terms, including UMLS w/o CHV terms and terms that are not in UMLS, were considered as candidate terms.
3.3.2. Step 2: Feature Vector Construction
We used a rich set of features to represent a term extracted from the textual corpus, including both linguistic and contextual features that have been shown to be effective for identifying new CHV terms [14–17, 19]. The linguistic features included POS tags, prefix/suffix, syntactic patterns (e.g., term “insulin receptor” matches the pattern “Noun+Noun”), capitalization status, and C-value [39]. We also used standard features, including term frequency (TF), document frequency (DF), and a number of task-specific features such as similarity to UMLS terms, similarity to CHV terms, and the UMLS semantic type (if any). Different from the linguistic features that characterize the term itself, the contextual features characterize the words in the context of the current term, including their semantic types and their POS tags. We describe the details of each feature below.
Term frequency (TF)
The number of occurrences of a term in the whole text corpus.
Document frequency (DF)
The number of documents that a term occurs in a text corpus. Both term frequency and document frequency indicate how often a term is used in a corpus. However, the document frequency emphasizes on how widely a term is used across documents. For example, common terms such as ‘diabetes’ are likely to occur more often across different documents than rare terms such as ‘Lantus.’
C-value
It combines linguistic and statistical information of a term [39]. C-value takes into account the length of the term and whether the term is nested by other terms or not. The C-value of term a is calculated by
| (1) |
where |a| is the number of words in a. tf(a) is the term frequency of term a. b is a term that contains term a. tf(b) is term frequency of term b. Ta is the set of terms that contain term a. P(Ta) is the number of terms in Ta.
Similarity to seed/UMLS terms
The similarity between a term and a existing CHV term based on the Levenshtein distance (i.e., edit distance) [40], which is the minimum number of single-character edits required to change one term into the other term. The similarity between term t and s is defined by
| (2) |
where s is the seed term that matches t. L-dist(t,s) is the Levenshtein distance (case ignored) between t and s. |t| and |s| are the number of letters t and s contain, respectively. Since we ignored the case when calculating the Levenshtein distance, if t and s are different in case, we applied 10% penalty on their similarity, considering that the difference in case status is less important than the difference in letters. Note that we also obtained the similarity between UMLS w/o CHV terms and n-grams with the same method, which is noted as UMLS similarity.
Containing a seed/UMLS term
This Boolean feature indicates whether a term contains a seed/UMLS term or not. For example, the candidate term “blood pressure test” contains the CHV term “blood pressure.” It is thus more likely to be a CHV term than those that do not contain any CHV terms. The value of this feature is True if a term contains an existing UMLS/CHV term, otherwise False.
Syntactic patterns
We first unified similar POS tags as one tag. For example, there are multiple tags for noun annotated by OpenNLP: NN (singular or mass noun), NNS (plural noun), NNP (singular proper noun), and NNPS (plural proper noun). We unified all these noun tags into ‘N, noun’. We then used three dichotomous features to represent the syntactic structure of a noun phrase: (1) Noun+Noun, two or more contiguous nouns; (2) (Adjective|Noun)+Noun, adjective followed by a noun; (3) ((Adjective|Noun)+|((Adjective|Noun)*(Noun Preposition)?)(Adjective|Noun)*)Noun, a noun phrase including a preposition. Each syntactic pattern is treated as a sub-feature. If a term matches one of the patterns above, the value of the corresponding sub-feature is True, otherwise False. Note that POS tagging is done on the entire sentence that contains the term of interest.
Capitalization patterns
Often if a letter in a term is capitalized, the term may be a special term (e.g., a drug name). We used three dichotomous features to represent this information: first letter of the first word capitalized, first letter of each word capitalized, and all the letters of all the words capitalized. If a term matches one of the capitalization patterns, the value of corresponding feature will be True, otherwise False.
Prefixes/suffixes
In English, some prefixes and suffixes of a word have special meanings. For example, un-, dis- and anti- are commonly used as negative prefixes; –able and -ative are adjective suffixes; -iasis and -iatric are medicine related suffixes. We obtained a list of prefixes [41] and suffixes [42] provided by the NLM. Every prefix or suffix in the lists is considered as a sub-feature. Note that we matched only the longest prefix or suffix. If a term contains any of the prefixes or suffixes in one of its words, the corresponding sub-feature will be marked as True, otherwise False.
POS context
We assume that two terms occurring in the similar syntactic context are more similar than those occurring in different syntactic contexts. We used the feature POS context to encode such information. We collected the POS tags of the words near a candidate term (the words within a window of 10 in the same sentence). Every POS tag is a sub-feature, the value of which is based on the occurrence of the respective POS tag in the contexts of the term of all the sentences that the term occurs. The formula for calculating the value of a POS context sub-feature is:
| (3) |
where tf(t) is the term frequency of term t, type is a POS tag, count(type) is the number of words with the POS tag in the context of term t, and contexts are all the contexts of all the sentences in which term t occurs. Table S1 in the Supplementary Material shows the number of occurrences of the POS tags in the contexts of the two terms ‘sugar level’ and ‘glucose level.’
Seed term context
This feature is constructed based on the observation that people tend to use domain-specific words when describing their problems in a certain disease domain. For example, people may use multiple diabetes-related CHV terms such as ‘sugar blood,’ ‘HbA1c,’ ‘glucose,’ ‘diet’ to describe their diabetes issue in a sentence. If a non-CHV term such as ‘calorie intake’ is frequently used along with a few CHV terms, it is more likely to be consumer health terms. To incorporate this assumption in our model, we added two sub-features: (1) stickiness to seed terms: the average number of CHV terms that co-occur with the candidate term in a sentence, and (2) distance to the nearest seed terms: the average number of words between the nearest seed term and a candidate term in a sentence.
Semantic context
The feature semantic context is based on the assumption that if the semantic context of an n-gram is similar to that of a CHV term, the n-gram is similar to the CHV term. According to a recent work of ours [23], we used the 12 most frequent semantic types to construct 12 semantic context sub-features that can cover more than 80% of the UMLS terms in our dataset: “Amino Acid, Peptide, or Protein,” “Body Part, Organ, or Organ Component,” “Disease or Syndrome,” “Finding,” “Medical Device,” “Organic Chemical,” “Pharmacologic Substance,” “Sign or Symptom,” “Therapeutic or Preventive Procedure.” “Finding,” “Pharmacologic Substance,” and “Disease or Syndrome.” The method for calculating these sub-features is similar to that for the feature POS context features. We give a concrete example of the semantic context feature in Table S2 in the Supplementary Material.
Our method can measure the similarity between the candidate terms and the seed terms based on their feature vectors. For the features with Boolean values, we converted True to 1 and False to 0. In addition, we rescaled the values of all the features into the range [0, 1]. For value x in feature f,
| (4) |
where x is the original value, x′ is the normalized value, min(f) is the minimum value of the feature f, max(f) is the maximum value of feature f. We alleviated the effect of the outliers by employing the z-score (also known as “standard score”), which is defined by
| (5) |
where x is the feature value, μ is the mean and σ is the standard deviation of the vector. If the z-score of an element x is greater 2.0 (i.e., it is greater than 97.5% of all the elements), we replaced it with (μ + 2.0 * σ).
3.3.3. Step 3: Ranking Similarity of the Candidate Terms to the Seed Terms
Before we devise an effective method for identifying consumer terms from text corpus, it is important to have a glance at the structural characteristics of the data. To visualize the data, we used the Principal Component Analysis (PCA) [43] to reduce the dimensionality of the data to three. To obtain similarities of the candidate terms to the seed terms, we first used K-means clustering [44] to group the seed terms, and then calculated the similarity of the candidate terms to these seed term clusters.
Step 3.1: Clustering Seed Terms with K-means Clustering
K-means clustering partitions terms into k clusters based on their distance to each other in the feature vector space [44]. In simiTerm, we employed K-means++ to obtain an initial set of centers that is provably close to the optimum solution [45]. As K-means clustering may not converge, we set the maximum number of iterations of each experiment to be 1,000. We calculated the average size of the clusters and then excluded the clusters with a size below 10% of the average size. The terms in these clusters were assigned to the nearest cluster in the rest of clusters.
K-means clustering requires a user-specified parameter: k, the number of clusters. We used the evaluation results based on a held-out set of seed terms to find the optimal k when the margin of performance increase (i.e., recall, precision, and F-score) between two consecutive k values is insignificant.
Step 3.2: Calculating the Similarity of a Candidate Term to the Clusters
For each candidate term, we used the Euclidean distance [46] between the candidate term and its nearest cluster center as the similarity score of the candidate term.
Step 3.3: Ranking the Candidate Terms
We consider the average TF of the terms in a cluster as the importance of the cluster. Thus, we first ranked the candidate terms by the average TF of the clusters to which they belong, and then by their similarity scores. Note that in order to evaluate the similarity score based purely on the feature vector representation of the terms, we ranked the terms only by similarity score in the quantitative evaluation.
3.3.4. Evaluation Approaches
Our evaluation approaches consist of a quantitative evaluation and a qualitative evaluation. In the quantitative evaluation, we employed repeated random sub-sampling validation [47]. Our assumption is that existing CHV terms are contextual and syntactically similar in the text corpus. We randomly picked 70% of the seed terms as training data, and the other 30% of the seed terms with all the other candidate terms as validation data (see Figure 3). We clustered the training data with K-means clustering and assessed if the seed terms in the validation data can be ranked higher by our method than three baseline methods: random ranking, TF-based ranking, and C-value based ranking. We chose TF and C-value as baselines because prevoius studies on the development of CHV have used TF and/or C-value to determine the validiy of a term to be CHV term [18]. We computed three quality measures including recall, precision, and F-score, which are calculated using following formulae:
| (6) |
| (7) |
| (8) |
where n is the top n terms in the ranked list of the validation data. Top n terms in the ranked list are predicted to be true (positive), while others are predicted to be false (negative). Seed terms in the top n terms were considered as true positives, while non-seed-terms in the top n terms were considered as false positives. We chose n from 0 to two times of the number of seed terms in the validation data. Specifically, we calculated the quality measures at every 5% increment of the number of the seed terms in the validation data. The coefficient 0.5 in F-score indicates our emphasis on precision over recall [48, 49]. We repeated this process (random split, clustering, and ranking) for 10 times, and calculated the average value of each quality measure.
Figure 3.
Random split of the terms in the quantitative evaluation.
To assess the perceived quality of our method, we performed a manual review of a sample of recommended terms. Raters reviewed a sample of candidate terms suggested by our algorithm to assess whether these terms should be added into CHV, following the CHV development guideline [18]. Inter-rater agreement was assessed.
4. Results
4.1. Basic Characteristics of the Datasets
From the diabetes Q&A dataset, a total of 28,780 n-grams were extracted. Out of the 28,780 unique n-grams extracted, 7,374, (25.62%) n-grams are CHV terms, while only 2,465 (8.56%) n-grams were identified as UMLS w/o CHV terms. From the cancer Q&A dataset, a total of 36,537 unique n-grams were extracted. Out of the 36,537 n-grams extracted, 8,879 (24.3%) n-grams are CHV terms, 2,895 (7.92%) n-grams were UMLS w/o CHV terms. Table S3 in the Supplementary Material gives the details of the n-grams extracted from the diabetes Q&A dataset. We also plotted the trend of TF for different types of n-grams from diabetes Q&A dataset ordered by TF in decreasing order in Figure S2 in the Supplementary Material.
For every n-gram, we constructed a feature vector with 313 features. Figure S3 in the Supplementary Material illustrates the 3D scatter plot of the data after the number of dimensions of the feature matrix of 7,374 seed term was reduced to three by PCA. As shown in Figure S3, the seed terms represented by the feature vectors can be clustered with clear boundaries.
4.2. Quantitative Evaluation Results on the Diabetes Q&A Dataset
We performed the quantitative evaluation of simiTerm on diabetes and cancer Q&A datasets separately. For the diabetes Q&A dataset, when the TF threshold “5” was used, there were 7,374 seed terms. As 30% of them were in the validation dataset (see Figure 3), there were 2,212 (7,374*30%) seed terms and a total of 23,618 (28,780 – 7,374*70%) terms in validation dataset. The random ranking baseline is a randomly shuffled list of terms in the validation dataset. Therefore, the precision of this baseline is 9.78% (2,212/22,618). For the TF-based and C-value-based ranking, we first ranked all the terms in the validation dataset by TF and C-value respectively, and then calculated the the recall, precision and F-score for every 5% increment of the number of seed terms in the validation dataset. We tested our method with k (number of clusters) from 5 to 200 with an increment of 5 (one grey line for each k in Figure 4). K-means clustering may produce different results with the same data and parameters due to the randomly initialized center points, we therefore ran K-means clustering algorithm 10 times for every k, calculated the recall, precision and F-score of each experiment, and took the average of these measures as the final result. Figure 4 (a), (b), (c) show that our method outperforms all three baselines. TF-based ranking is the best baseline, while the random ranking is the worst baseline. We obtained the best F-score at the threshold about top 50% of the total number of seed terms in the validation dataset.
Figure 4.
Evaluation results of simiTerm on the diabetes Q&A dataset with three baselines: random-order-based ranking, TF-based ranking, and C-value-based ranking for three quality measures with two TF threshold values: (a) recall, TF > 5; (b) precision, TF > 5; (c) F-score, TF >5, (d) recall, TF > 100; (e) precision, TF > 100; (f) F-score, TF > 100. The bright grey colors represent smaller k values while dark grey colors repersent larger k values.
We further used a higher TF threshold “100” to test the robustness of simiTerm. As such, a total of 3,541 n-grams from diabetes Q&A dataset were included, in which 1,895 (53.52%) were identified as seed terms. Similarly, we analyzed the precision, recall, and F-score of simiTerm and three baseline ranking methods. As shown in Figure 4 (d), (e) and (f), simiTerm also outperformed all three baselines in all the experiments. Table 2 shows the three quality measures of simiTerm (k = 150, TF threshold = 5 or 100) comparing with the TF-based ranking baseline for the diabetes Q&A dataset. These results demonstrated that: 1) using a higher TF value as a filtering criterion can retain candidate terms of higher quality; and 2) simiTerm consistently outperformed the baseline methods (thus robust) with candidate terms of different quality settings.
Table 2.
The recall, precision and F-score of simiTerm (TF threshold: 100 or 5) and TF-based ranking (k: the number of cluster, which is 150) in the diabetes Q&A dataset.
| % of total seed terms | 50 | 75 | 100 | 125 | 150 | 175 | 200 | |
|---|---|---|---|---|---|---|---|---|
| Recall (%) | k=150, TF threshold = 100 | 31.4 | 41.5 | 49.1 | 57.5 | 69.3 | 79.0 | 83.8 |
| k=150, TF threshold = 5 | 22.4 | 27.7 | 31.4 | 34.3 | 36.9 | 39.4 | 42.4 | |
| TF baseline | 23.5 | 30.9 | 37.9 | 46.3 | 50.1 | 54.6 | 61.1 | |
| Precision (%) | k=150, TF threshold = 100 | 62.8 | 55.3 | 49.1 | 46.0 | 46.2 | 45.1 | 41.9 |
| k=150, TF threshold = 5 | 44.9 | 36.9 | 31.4 | 27.5 | 24.6 | 22.5 | 21.2 | |
| TF baseline | 46.9 | 41.2 | 37.9 | 37.0 | 33.4 | 31.2 | 30.5 | |
| F-score | k=150, TF threshold = 100 | .522 | .520 | .491 | .479 | .496 | .493 | .466 |
| K=150, TF threshold = 5 | .37 | .35 | .31 | .29 | .27 | .25 | .23 | |
| TF baseline | .391 | .387 | .379 | .386 | .360 | .341 | .339 | |
In addition to the three baselines (i.e., TF-based ranking, C-value-based ranking, and random order), we also assessed if our selected features outperformed the Bag-of-Words (BoW) features using diabetes Q&A dataset. We aggregated all the sentences in which a term T occurs as a document D. All the terms with TF > 100 that co-occur with T in D formed the vector representation of T. We applied PCA to perform dimension reduction, keeping 95% of the variance among all the features. We then applied the same clustering algorithm on the terms represented with the BoW features. As shown in Figure S4 in the supplementary material, the clustering algorithm with BoW features performed worse than the TF and the C-value baselines and similar to the random-order baseline. Further, we did not consider a classification approach (e.g., Support Vector Machine and Random Forest), since it gives binary decisions and cannot rank the terms based on their similarities to the existing terms.
4.3. Quantitative Evaluation Results on the Cancer Q&A Dataset
We also performed the quantitative evaluation on the cancer Q&A dataset. As shown in Figure 5, simiTerm also outperformed three baselines across all of our experiments. These results demonstrated the robustness of simiTerm on text corpora on a different disease domain.
Figure 5.
Evaluation results of simiTerm on the cancer Q&A dataset: (a) recall, TF > 100; (b) precision, TF > 100; (c) F-score, TF >100 in cancer Q&A dataset.
4.4. Manual Review of the Recommended Diabetes-Related Consumer Health Terms
We first examined the characteristics of the clusters (k=150, TF threshold =100) generated by simiTerm on the diabetes Q&A dataset. For unigrams, prefixes and suffixes had great influence on the clustering results. For example, as shown in Table 3, all terms in Cluster 99 have suffix ‘–ment’, while all terms in Cluster 106 have prefix ‘hypo-.’ We further found that simiTerm can effectively group terms with the similar meaning into the same cluster. For example, the terms in Cluster 113 are related to glucose, e.g., ‘glucose meter,’ and ‘glucose tablet.’ The terms in Cluster 8 are all related to blood sugar. Even though we did not emphasize the semantics of the terms in our set of features, our method could roughly cluster terms into semantically uniform groups.
Table 3.
Example terms in the clusters of diabetes-related terms when k = 150, TF threshold = 100 were used in simiTerm. (K: cluster index; TF: term frequency; N: noun; A: adjective; V: verb).
| K | Type | Term (POS tag) | TF |
|---|---|---|---|
| 99 | chv | equipment (N) | 169 |
| 99 | chv | government (N) | 326 |
| 99 | chv | experiment (N) | 215 |
| 99 | chv | environment (N) | 230 |
| 99 | chv | adjustment (N) | 219 |
| 99 | chv | management (N) | 540 |
| 99 | umls | improvement (N) | 281 |
| 99 | umls | statement (N) | 187 |
| 99 | chv | element (N) | 115 |
| 99 | umls | assignment (N) | 110 |
| 106 | chv | hypoglycaemia (N) | 197 |
| 106 | chv | hypoglycemia (N) | 5330 |
| 106 | chv | hyperglycemia (N) | 968 |
| 106 | chv | hypothyroidism (N) | 1000 |
| 106 | other | hypo (N) | 1107 |
| 113 | chv | blood glucose level (NNN) | 3419 |
| 113 | umls | glucose meter (NN) | 1096 |
| 113 | chv | glucose control (NN) | 247 |
| 113 | chv | blood glucose (NN) | 7993 |
| 113 | chv | blood glucose meter (NNN) | 404 |
| 113 | chv | glucose level (NN) | 7809 |
| 113 | chv | glucose metabolism (NN) | 157 |
| 113 | other | glucose tablet (NN) | 247 |
| 113 | other | control glucose level (VNN) | 672 |
| 113 | other | raise blood glucose (VNN) | 105 |
| 8 | chv | high blood sugar level (ANNN) | 334 |
| 8 | chv | low blood sugar level (ANNN) | 193 |
| 8 | other | good blood sugar (ANN) | 119 |
| 8 | other | high sugar level (ANN) | 248 |
| 8 | chv | elevate blood sugar (ANN) | 123 |
In the manual review, two researchers JB and JH reviewed a sample of the top 500 diabetes-related candidate terms suggested by simiTerm to assess whether these terms should be added into the CHV. The 500 terms consisted of 100 terms in each of the 1-gram, 2-gram, 3-gram, 4-gram, and 5-gram category. An example sentence that contains each term was also presented to the raters. JH has extensive research experience in health communication and computer-mediated communication. JB has extensive research experience in social media analysis and biomedical ontologies. The manual review process started with a discussion of the review criteria for being a valid consumer term (e.g., noun phrases with independent semantics) [18], as well as independent assessments of whether a term: 1—“should not be added to CHV,” 2—“possibly can be added to CHV,” or 3—“should be added to CHV.” Note that one of the review criteria in [18] is “N-grams representing existing UMLS medical concepts are considered to be CHV terms, but CHV terms may represent non-UMLS concepts.” Thus, we also provided the souce information of the term to the raters. The inter-rater reliability, i.e., Cohen’s kappa, of the two raters on the 500 terms achieved .57, indicating “substantial” agreement [50]. Most of the disagreements are due to the fact that some of the CHV criteria in the development guideline are subjective [18]. For example, one of the review criteria is “CHV terms should be specific to the medical domain, for example, ‘Google’ and ‘Yahoo’ are general words, not CHV terms.” However, the terms ‘government’ and ‘management’, which are not strictly medical terms, are both CHV terms. Overall, our rater agreement level can be considered good and acceptable.
For those terms that received different ratings from the two raters, we recoded the terms that received “1” and “2” as “1”, “2” and “3” as “3”. In summary, 325 (65%) terms should not be added to CHV, 5 (1%) terms could possibly be added to CHV, and 170 (34%) terms should be added to CHV. Figure 6 illustrates the results of the manual review stratified by term length. In particular, 3-grams in the sample contained more meaningful terms than terms of other length. Among the 170 terms that should be added to CHV, 131 (77%) were not covered by the UMLS. Table 4 lists the top 10 terms that “should be added to CHV” ranked by their similarities to existing CHV terms. N-grams such as ‘calorie intake,’ ‘WebMD,’ which were not frequent in the dataset but relevant to diabetes were ranked high. Interestingly, ‘Yahoo,’ ‘Google,’ ‘YouTube,’ ‘WebMD’ were closest to the same cluster that contains the CHV term ‘Amazon.’ We were also able to identify important diabetes-related lay terms, such as ‘Mendosa’ (the lay term of ‘Mendosa’s Glycemic Index Diet’), ‘med’ (the lay term of ‘medication’ or ‘medicine’), and ‘carb intake’ (the lay term of ‘carbohydrate intake’).
Figure 6.
Manual review results of top 500 suggested terms after conflict resolution.
Table 4.
Top 10 new diabetes-related consumer terms that were suggested by simiTerm to be added to CHV.
| Rank | Term | Already in the UMLS? | Term Frequency | Document Frequency | Example CHV Terms in the Cluster | Example Sentence |
|---|---|---|---|---|---|---|
| 1 | room | Yes | 1,000 | 833 | home, hospital, work, site | I know for a fact that if I get blood draws done more than a week before my appointment, my doctor will not review them until I am in the examining room with her. |
| 2 | lab | Yes | 1,550 | 1,141 | level, blood, test, weight | If blood draw it needs to go to the labs to be processed and takes a bit of time. |
| 3 | calorie intake | Yes | 119 | 103 | food intake, carbohydrate intake, sugar intake | It does sound like it is your calorie intake, but that is not directly fixed by drinking or eating sugar, you may need to increase how much you eat prior to a workout. |
| 4 | carb | Yes | 12,532 | 5,731 | food, water, sugar, carbohydrate | You need to stick to low Glycemic Index carbs. |
| 5 | issue | Yes | 2,913 | 2,407 | life, day, year, hour, month | That is a normal blood sugar, but there are other causes that explain your symptoms, such as an issue with the pituitary gland or diabetes inspidus, which is different from diabetes mellitus and has nothing to do with blood sugar |
| 6 | spike | Yes | 1,328 | 1,018 | change, sign, stress, energy | It is normal for your blood sugar to spike after a meal. |
| 7 | article | Yes | 816 | 680 | change, sign, stress, energy | One night I was reading a magazine in bed and it was an article about diabetes. |
| 8 | range | Yes | 5,298 | 3,636 | life, day, year, hour, month | The best range is 70 to 99 when waking up and 70 to 140 after meals. |
| 9 | WebMD | No | 223 | 191 | Amazon, www | WebMD might be a good place to start. |
| 10 | bloodstream | Yes | 1,001 | 740 | medicine, drug, pill, urine | The endocrine system is a system of glands, each of which secretes a type of hormone directly into the bloodstream to regulate the body. |
5. Discussion
Existing term identification methods often rely on ad hoc pre-defined lexical syntactic patterns. In this study, we automated part of the CHV development process by applying a similarity-based text mining technique to identify terms on social Q&A that were syntactically and contextually similar to the existing CHV terms. Even though CHV can cover over a quarter of the extracted n-grams, there is still great potential to identify meaningful terms from UMLS w/o CHV and from the terms that cannot be covered by UMLS. The results indicated that most of the top ranked recommended terms are UMLS terms, confirming that consumers do frequently use UMLS terms that are not covered by CHV. Even though user-generated content on social media often contain sentences with grammatical errors and typos, our method simiTerm can still yield significant improvement over the baselines. Specifically, the fuzzy matching employed in simiTerm may have alleviated the problem of typos by using the root forms of the terms.
As is presented in Figure 4, our method with a higher TF threshold values (i.e., 100) yielded better performance than a low TF threshold (i.e., 5). It may be because higher TF threshold can filter out those n-grams that are less frequently used by consumers and are thus more likely to be noisy terms. Therefore, in the manual evaluation, we applied TF threshold 100. From the manual review of the suggested terms, we found that some of the CHV review criteria of Zeng et al. [18] such as “CHV terms should be specific to the medical domain” are vague, leading to a certain level of disagreement among coders. This is not uncommon in manual annotation tasks and warrants further efforts in building a more objective CHV development guideline. We thus only consider the manual review as a way to assess the general appropriateness of the terms recommended by simiTerm. The final decision for including a new term in a controlled vocabulary should be made by terminology curators. As our method can rank the candidate terms based on their similarities to existing CHV terms instead of giving a binary decision for each term, the curators can decide on the number of terms for manual review based on the amount of effort they would like to make. Further, using Apache Spark, our method can scale up to large corpora. With a modular design, other researchers can modify individual modules to better meet the needs of their own tasks. To allow the broad community to contribute to the problem, we made our tool simiTerm publicly available in GitHub (https://github.com/henryhezhe2003/simiTerm).
It is also our hope that researchers can leverage the CHV to develop innovate user-friendly health applications that benefit ordinary health consumers more broadly. For example, clinical trial recruitment is a critical issue in clinical research. The lack of enrollment or slow accrual is the major reason for withdrawing or suspending clinical trials. The clinical trial registry ClinicalTrials.gov was created by the NLM to disseminate the clinical trial opportunities and results. However, the trial descriptions in ClinicalTrials.gov are extremely difficult to read for ordinary people [51]. Existing clinical trial portals that use data from ClinicalTrials.gov, such as Antidote and Dory/TrialX, tend to improve trial search with locations or domain-specific questions. However, they still use the same trial descriptions provided by ClinicalTrials.gov without simplification. CHV can be utilized to simplify medical text such as trial descriptions and eligibility criteria by replacing professional jargons with lay terms. A consumer-friendly clinical trial search engine with simplified trial description may improve the understandability of trials and have an impact on trial recruitment. To achieve this goal, the CHV should be consistently expanded with new consumer-friendly terms. Our method automates the process of identifying these potential new consumer terms from social media, recommends top ranked new terms according to their similarities to existing CHV terms, and thus reduces the amount of manual work required.
5.1. Limitations and Future Work
Some limitations should be noted. First, social Q&A could be a good venue to observe its users’ natural expression of their health conditions but may not be generalizable to that of non-users and to other online information sources. In future work, we will apply simiTerm to different textual datasets to identify new consumer health terms. Second, when applying the clustering method, ideally, one should use the optimal set of features. However, it is challenging to select an optimal set of features due to the complex nature of our task and the poor quality of the social media dataset. Our features were selected empirically, which may contain inherent biases and correlations. In future work, sophisticated feature selection methods such as regularized trees could be exploited to alleviate this limitation.
6. Conclusions
In this study, we represented n-grams extracted from the social Q&A textual corpora with a set of linguistic and contextual features. We identified n-grams that are covered by the CHV. Based on these features, we generated a ranked list of new terms that are syntactically and contextually similar to the existing CHV terms on social Q&A. Our similarity-based method outperformed TF-based and C-value-based ranking baselines in identifying new CHV terms. According to the post-hoc qualitative evaluation by human experts, our method was shown to effectively identify useful consumer health terms from social questions and answers in Yahoo! Answers.
Supplementary Material
Highlights.
We devise a similarity-based method to identify consumer terms from social media text
Rich syntactic and contextual features are used to represent terms
Our method outperforms three term ranking baselines
Our study confirms the value of social media for mining consumer terms
Acknowledgments
We would like to thank Dr. Warren Allen for providing the computing resource for this work.
Funding
This work was supported by the start-up fund of Florida State University and an Amazon Web Services Education and Research Grant Award (PI: He). The work was partially supported by National Center for Advancing Translational Sciences under the Clinical and Translational Science Award UL1TR001427 (PI: Nelson & Shenkman). The content is solely the responsibility of the authors and does not represent the official view of the National Institutes of Health.
Footnotes
Conflict of Interest
None.
Contribution
ZH conceptualized and designed the study. SO collected and provided the social Q&A data from Yahoo! Answers. ZC implemented the method. ZH and ZC performed the data analysis and drafted the initial version. JB and JH manually reviewed the sample of 500 terms. ZH and JB extensively revised the manuscript iteratively for important intellectual content. All authors contributed to the methodology development, and results interpretation. All authors edited the paper significantly, and gave final approval for the version to be published. ZH takes primary responsibility for the research reported here.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bodenreider O. Biomedical ontologies in action: role in knowledge management, data integration and decision support. Yearb Med Inform. 2008:67–79. [PMC free article] [PubMed] [Google Scholar]
- 2.Cimino JJ. High-quality, standard, controlled healthcare terminologies come of age. Methods Inf Med. 2011;50(2):101–4. [PMC free article] [PubMed] [Google Scholar]
- 3.Finnegan R. ICD-9-CM coding for physician billing. J Am Med Rec Assoc. 1989;60(2):22–3. [PubMed] [Google Scholar]
- 4.Agrawal A, He Z, Perl Y, Wei D, Halper M, Elhanan G, et al. The readiness of SNOMED problem list concepts for meaningful use of electronic health records. Artif Intell Med. 2013;58(2):73–80. doi: 10.1016/j.artmed.2013.03.008. [DOI] [PubMed] [Google Scholar]
- 5.Bennett CC. Utilizing RxNorm to support practical computing applications: capturing medication history in live electronic health records. J Biomed Inform. 2012;45(4):634–41. doi: 10.1016/j.jbi.2012.02.011. [DOI] [PubMed] [Google Scholar]
- 6.Zeng QT, Tse T. Exploring and developing consumer health vocabularies. J Am Med Inform Assoc. 2006;13(1):24–9. doi: 10.1197/jamia.M1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pew. Health Fact Sheet. 2016 Oct 31; Available from: http://www.pewinternet.org/fact-sheets/health-fact-sheet/
- 8.Metzger MJ, Flanagin AJ. Using Web 2.0 technologies to enhance evidence-based medical information. J Health Commun. 2011;16(Suppl 1):45–58. doi: 10.1080/10810730.2011.589881. [DOI] [PubMed] [Google Scholar]
- 9.Lewis D, Brennan P, McCray A, Tuttle MBJ. If We Build It, They Will Come: Standardized Consumer Vocabularies. Studies in Health Technology and Informatics. 2001;84:1530. [Google Scholar]
- 10.Smith C, Stavri Z. Consumer health vocabulary. In: Lewis D, Eysenbach G, Kukafka R, Stavri Z, Jimison H, editors. Consumer health informatics. 2005. [Google Scholar]
- 11.Plovnick RM, Zeng QT. Reformulation of consumer health queries with professional terminology: a pilot study. J Med Internet Res. 2004;6(3):e27. doi: 10.2196/jmir.6.3.e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cimino JJ. Desiderata for controlled medical vocabularies in the twenty-first century. Methods Inf Med. 1998;37(4–5):394–403. [PMC free article] [PubMed] [Google Scholar]
- 13.CHV Wiki Page. February 20, 2016 Available from: http://consumerhealthvocab.chpc.utah.edu/CHVwiki/
- 14.Doing-Harris KM, Zeng-Treitler Q. Computer-assisted update of a consumer health vocabulary through mining of social network data. J Med Internet Res. 2011;13(2):e37. doi: 10.2196/jmir.1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.MacLean DL, Heer J. Identifying medical terms in patient-authored text: a crowdsourcing-based approach. J Am Med Inform Assoc. 2013;20(6):1120–7. doi: 10.1136/amiajnl-2012-001110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiang L, Yang C, editors. Using co-occurrence analysis to expand consumer health vocabularies from social media data. Healthcare Informatics (ICHI), 2013 IEEE International Conference on; Philadelphia, PA: IEEE; 2013. [Google Scholar]
- 17.Keselman A, Smith CA, Divita G, Kim H, Browne AC, Leroy G, et al. Consumer health concepts that do not map to the UMLS: where do they fit? J Am Med Inform Assoc. 2008;15(4):496–505. doi: 10.1197/jamia.M2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zeng QT, Tse T, Divita G, Keselman A, Crowell J, Browne AC, et al. Term identification methods for consumer health vocabulary development. J Med Internet Res. 2007;9(1):e4. doi: 10.2196/jmir.9.1.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vydiswaran VG, Mei Q, Hanauer DA, Zheng K. Mining consumer health vocabulary from community-generated text. AMIA Annu Symp Proc. 2014;2014:1150–9. [PMC free article] [PubMed] [Google Scholar]
- 20.Chandar P, Yaman A, Hoxha J, He Z, Weng C. Similarity-based recommendation of new concepts to a terminology. AMIA Annu Symp Proc. 2015;2015:386–95. [PMC free article] [PubMed] [Google Scholar]
- 21.ADA. Diabetes complications. 2015 April 25, 2016 Available from: http://www.diabetes.org/living-with-diabetes/complications/
- 22.CDC. Statistics for Different Kinds of Cancer. November 1, 2016 Available from: https://www.cdc.gov/cancer/dcpc/data/types.htm.
- 23.Park M, He Z, Chen Z, Oh S, Bian J. Consumers’ use of UMLS concepts on social media: diabetes-related textual data analysis in blog and social Q&A sites. JMIR Diabetes. 2016;4(4):e41. doi: 10.2196/medinform.5748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oh S. The characteristics and motivations of health answerers for sharing information, knowledge, and experiences in online environments. Journal of the American Society for Information Science and Technology. 2012;63(3):543–57. [Google Scholar]
- 25.Sanchez D, Moreno A. Recent Advances in Artificial Intelligence Research and Development. Vol. 113. IOS Press; 2004. Creating ontologies from Web documents; pp. 11–8. [Google Scholar]
- 26.Sabou M, Wroe C, Goble C, Mishne G, editors. Learning Domain Ontologies for Web Service Descriptions: an Experiment in Bioinformatics. Proceedings of the 14th International World Wide Web Conference; 2005; Chiba, Japan. [Google Scholar]
- 27.Lossio-Ventura JA, Hogan WR, Modave F, Hicks A, Guo Y, He Z, et al., editors. Towards Building an Obesity-Cancer Knowledge Base: Biomedical Entity Identification and Relation Detection. Proceedings of the 2016 IEEE International Conference on Bioinformatics and Biomedicine; 2016; Shenzhen, China: IEEE; pp. 1081–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hoxha J, Jiang G, Weng C. Automated learning of domain taxonomies from text using background knowledge. J Biomed Inform. 2016;63:295–306. doi: 10.1016/j.jbi.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.National Library of Medicine. MedlinePlus. Bethesda, MD: Available from: http://www.medlineplus.gov. [Google Scholar]
- 30.Frantzi KT, Ananiadou S, Mima H. Automatic recognition of multi-word terms: the C-value/NC-value method. Int J on Digital Libraries. 2000;3(2):115–30. [Google Scholar]
- 31.Shah C, Oh J, Oh S. Exploring characteristics and effects of user participation in online social Q&A sites. First Monday. 2008;13(9) [Google Scholar]
- 32.Roberts K, Demner-Fushman D. Interactive use of online health resources: a comparison of consumer and professional questions. J Am Med Inform Assoc. 2016;23(4):802–11. doi: 10.1093/jamia/ocw024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.The SPECIALIST Lexicon. February 20, 2016 Available from: https://lsg3.nlm.nih.gov/LexSysGroup/Projects/lexicon/current/docs/designDoc/UDF/wordStats/index.html.
- 34.Zaharia M, Chowdhury M, Tathagata D, Ankur D, Justin M, Murphy M, et al. Resilient distributed datasets: A fault-tolerant abstraction for in-memory cluster computing. Proceedings of the 9th USENIX conference on Networked Systems Design and Implementation; 2012; USENIX Association; [Google Scholar]
- 35.Baldridge J. The openNLP project. 2005 January 18, 2016 Available from: http://opennlp.apache.org/index.
- 36.Buyko E, Wermter J, Poprat M, Hahn U, editors. Automatically adapting an NLP core engine to the biology domain. Proceedings of the ISMB 2006 Joint Linking Literature, Information and Knowledge for Biology and the 9th Bio-Ontologies Meeting; 2006 August 6–10; Fortaleza, Brazi. 2006. [Google Scholar]
- 37.McCray AT, Srinivasan S, Browne AC. Lexical methods for managing variation in biomedical terminologies. Proc Annu Symp Comput Appl Med Care. 1994:235–9. [PMC free article] [PubMed] [Google Scholar]
- 38.Pal S. Dictionary Based Annotation at Scale with Spark SolrTextTagger and OpenNLP. Mendeley Data. 2015 [Google Scholar]
- 39.Frantzi KT, Ananiadou S, Tsujii J, editors. The C-value/NC-value Method of Automatic Recognition for Multi-Word Terms. ECDL ‘98 Proceedings of the Second European Conference on Research and Advanced Technology for Digital Libraries; 1998. [Google Scholar]
- 40.Levenshtein VI. Binary codes capable of correcting deletions, insertions, and reversals. Soviet Physics Doklady. 1966;10:707. [Google Scholar]
- 41.NLM. Prefix List - 2014. 2014 January 2016 Available from: https://lsg2.nlm.nih.gov/LexSysGroup/Projects/lvg/2014/docs/designDoc/UDF/derivations/prefixList.html.
- 42.NLM. Derivational Suffix List. 2015 January 2016 Available from: https://lexsrv2.nlm.nih.gov/LexSysGroup/Projects/lvg/2015/docs/designDoc/UDF/derivations/suffixList.html.
- 43.Abdi H, Williams LJ. Principal component analysis. Wiley Interdisciplinary Reviews: Computational Statistics. 2010:433–59. [Google Scholar]
- 44.Hartigan JA, Wong MA. Algorithm AS 136: A K-Means Clustering Algorithm. Journal of the Royal Statistical Society, Series C. 1979;28(1):100–8. [Google Scholar]
- 45.Bahmani B, Moseley B, Vattani A, Kumar R, Vassilvitskii S. Scalable k-means++ Proceedings of the VLDB Endowment. 2012;5(7):622–33. [Google Scholar]
- 46.Deza E, Deza MM. Encyclopedia of Distances. Springer; 2009. [Google Scholar]
- 47.Kohavi R, editor. A study of cross-validation and bootstrap for accuracy estimation and model selection. Proceedings of the 14th international joint conference on Artificial intelligence; 1995. [Google Scholar]
- 48.Van Rijsbergen CJ. Information Retrieval. 2. Butterworth; 1979. [Google Scholar]
- 49.F1 Score. 2016 April 8, 2016 Available from: https://en.wikipedia.org/wiki/F1_score.
- 50.Landis L, Koch G. The measurement of observer agreement for categorical data. Biometrics. 1977;33(1):159–74. [PubMed] [Google Scholar]
- 51.Wu DT, Hanauer DA, Mei Q, Clark PM, An LC, Proulx J, et al. Assessing the readability of ClinicalTrials.gov. J Am Med Inform Assoc. 2016;23(2):269–75. doi: 10.1093/jamia/ocv062. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.







