Abstract
The midbody is a microtubule-rich structure that forms between dividing cells during final stages of cytokinesis. Previously thought to be a transient structure, midbodies are now suggested to have additional roles beyond regulating cytokinesis. While the role midbodies play during abscission are now well established, their function in regulating polarity and cell signaling are only beginning to be understood. Due to the newly found interest in the structure and functions of midbodies, new techniques must be developed in order to further the study of this once-thought transient structure. Here, we describe several approaches used to explore post-mitotic roles of the midbodies.
Keywords: cytokinesis, abscission, midbody, epithelial cell polarity, endosomes
INTRODUCTION
The midbody (MB) is a subcellular structure that forms during cell division and was initially described over 100 years ago by Walther Flemming. The MB forms during the ingression of the cleavage furrow as the central spindle microtubules are compacted and cross-linked within a thin intracellular bridge connecting two daughter cells. It is now well established that the MBs play key roles in orchestrating cytokinesis by recruiting various mitotic kinases, such as Aurora B and Plk1, as well as Rab11/FIP3-containing furrow endosomes, membrane severing ESCRT complex and the microtubule severing enzyme spastin, all of which are responsible for the mediating abscission during late stages of cytokinesis (Schiel et al. 2012; D. Yang, et al 2008; Elia et al. 2011)
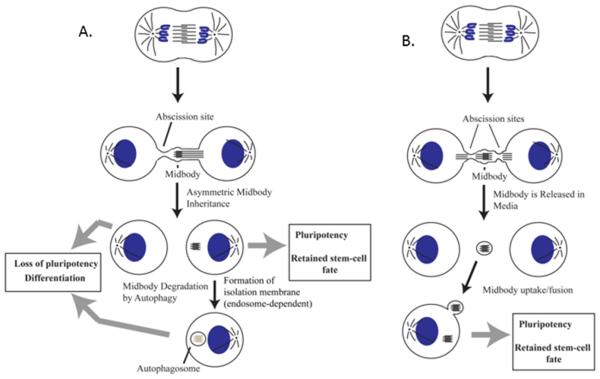
Recently, it has become apparent that in addition to regulating cytokinesis, MBs also have non-mitotic roles. Several studies have highlighted that the MB can be asymmetrically inherited during cell division, which may confer properties (such as a polarity cue or a signaling platform) to the cell which inherited it (Figure 1A) (Ettinger et al. 2011; Kuo et al 2012; Pollarolo et al. 2011; Singh & Pohl 2014; Wilcock et al. 2007). Conversely, other studies have revealed that symmetric abscission leads to a release of the MB to the extracellular space (Figure 1B) (Elia et al., 2011). Interestingly, some recent studies suggest that these extracellular MBs may be engulfed by surrounding cells, although the functional significance of this MB uptake remains unclear. It was proposed that MB uptake process relies on the enrichment of phosphatidylserines in the outer leaflet of the midbody membrane (Chai et al., 2012), a process that appears to be similar to an actin-dependent phagocytosis-like uptake of apoptotic cells (Crowell, Gaffuri, Gayraud-morel, & Tajbakhsh, 2014).
Figure 1. Midbodies can either be asymmetrically inherited or released into the extracellular space.
(A) Model detailing the possible fates of an asymmetrically inherited midbody. Upon asymmetric inheritance, the midbody can remain as a post-mitotic midbody in the cell and potentially influence pluripotency, or can be degraded via autophagy.
(B) Model detailing the possible fate of a midbody following symmetric division, where it is released into extracellular space and can be potentially uptaken by surrounding cells.
In the past few years, the MB has been shown to help define the polarity of daughter cells in vivo and in vitro. It was found that during spinal cord neurogenesis in chick embryos the MB is inherited and displaced towards the apical surface of dividing neural progenitors. Additionally, upon relocation to the apical surface, the MB positions itself at the site of forming neurites (Wilcock et al., 2007). In the fly notum, polarized accumulation of post-mitotic furrow markers at the midbody (such as RhoA and Aurora A) also preceded neurite or ‘sprout’ outgrowth in newly-born neurons (Pollarolo et al., 2011). Finally, during the early development of Caenorhabditis elegans, the MB was shown to function as a polarity cue which is necessary for directing mitotic spindle orientation for the following cell division (Singh & Pohl, 2014). Similarly, using 3D mammalian epithelial cultures, we have recently demonstrated that the formation of the apical membrane initiation site (AMIS) is guided by the MB, thus allowing for Rab11/FIP5 endosomes to deliver apical cargo during lumenogenesis (Li, Mangan, Cicchini, Margolis, & Prekeris, 2014a).
Numerous recent studies have now begun to recognize the potential roles for MBs during stem cell maintenance and or carcinogenesis (Dionne, Wang, & Prekeris, 2015). It has been shown that cells that have differentiated in vitro from human embryonic stem cells (hESCs) have fewer MBs as compared to hESCs (Kuo et al. 2012) Additionally, MBs accumulate in the basal compartment of seminiferous tubules in testes, where germline cells reside (Kuo et al. 2012). Somewhat controversial to this, another phenomenon that has been observed is the capacity for different cell types to release MBs at different frequencies. For example, proliferating cells (such as stem cells) tend to release MBs at a higher rate, while cancer cells appear to retain them (Ettinger et al., 2011). Furthermore, it is known that post-mitotic intracellular MBs can be degraded via selective macroautophagy. This autophagic degradation of MBs is necessary to prevent their accumulation, and it was suggested that cancer cells may have a decreased capacity of inducing post-mitotic MB degradation (Kuo et al. 2012).
Despite accumulating evidence that MBs influence cell polarity and differentiation, many questions remain. How cells regulate MB uptake and degradation is unknown. What determines whether cells undergo asymmetric cytokinesis and which cell inherits the MB also remains unclear. Finally, despite many correlative studies, there is no clearly defined signaling pathway(s) that is dependent on MB accumulation and can regulate cell 'stemness' and proliferation. Here we describe a few newly developed techniques and approaches that lead to the beginning of defining the post-mitotic MB functions and regulation.
MIDBODY AND EPITHELIAL CELL POLARITY
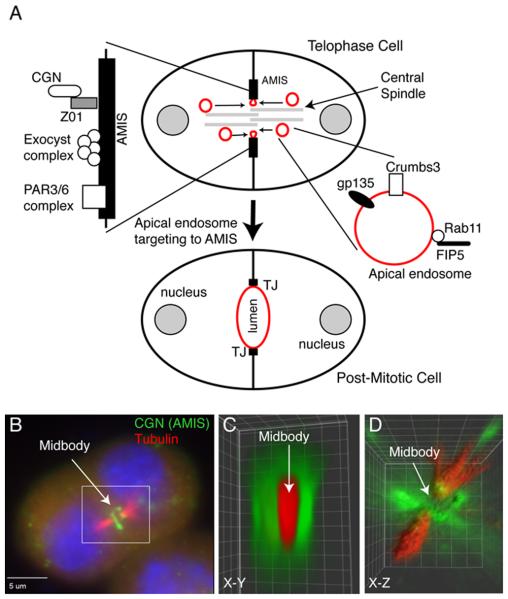
Epithelial tissues are composed of polarized cells, which function as selectively permeable barriers. The plasma membrane of epithelial cells is divided into apical and basolateral domains, and specialized protein complexes between adjacent cells, such as the tight junctions (TJs), maintain the separation of apical and basolateral plasma membrane. Additionally, epithelial cells coordinate their polarization with neighboring cells within 3D space to form an apical lumen, a key step in the establishment of renal and gut architecture, and thereby function (Blasky, Mangan, & Prekeris, 2015). Despite the importance of lumenogenesis for epithelia function, the mechanisms governing this process remain to be fully understood. One of the models of de novo lumen formation proposes that upon first cell division, Rab11/FIP5 protein complex-containing apical endosomes are transported to the site of the forming lumen, where they fuse with a specialized apical PM site known as the AMIS to initiate single lumen formation (Fig. 2A). These Rab11-endosomes were shown to contain gp135 (apical glycoprotein), Crumbs3 (apical CRB polarity complex), TUBA (GEF for Cdc42), myosin-Vb, Sec15 (Exocyst subunit), and Rab8/Rabin8. The delivery of these apical cargo proteins to the site of the forming lumen is required for polarized epithelial cyst formation in vitro (Blasky et al., 2015).
Figure 2. Midbody-dependent recruitment of apical plasma membrane proteins determines the site of nascent apical lumen formation.
(A) Schematic model depicting the role of midbody during lumen formation. Red marks apical endosomes and apical plasma membrane. AMIS stands for apical membrane initiation site.
(B-D) MDCK epithelial cells grown in Matrigel/Collagen matrix were fixed and immunostained with anti-cingulin (CGN; AMIS marker, antibody generated in the Prekeris lab) and anti-acetylated microtubule (central spindle marker) antibodies. Panel B show a single image plane. Panels C and D show 3D rendering based on the entire Z-stack (reproduced from [(Li, Mangan, et al., 2014a)] with permission from EMBO Reports).
Recent work has demonstrated that midbody formation during telophase plays a major role in epithelial polarization. It was shown that during fly development midbodies associates with TJs, thus providing polarization cues for newly formed daughter cells. Similarly, we have shown that during apical lumen formation in vitro, the midbody serves as the first “symmetry breaking” event that establishes the apical lumen site, and that the lumen formation is initiated by AMIS assembly around the midbody (Li, Mangan et al. 2014; Li et al. 2014). Thus, it is becoming clear that the midbody regulates spatiotemporal properties of AMIS/TJ formation. What remains unknown is how and why the AMIS forms around the midbody and how the midbody-associated AMIS mediates targeting of apical endosomes during epithelia morphogenesis.
Analyzing the role of MB during epithelia lumenogenesis
Overview
Epithelial cell interactions that create apical lumens occur in 3 dimensions (3D) but most of our information about lumen formation is derived from the use of 2 dimensional (2D) models, leaving significant gaps in our understanding of the cellular and molecular mechanisms that drive lumen morphogenesis. Recently, 3D epithelial cultures have emerged as an in vitro technique that allows dissection of the molecular mechanisms governing lumenogenesis. The major strength of these 3D assays is the ability to perform high-resolution imaging during different stages of epithelia polarization while retaining the in vivo-like complexity of lumen formation in a 3D extracellular matrix. Here, we will describe the 3D in vitro lumenogenesis assays using the Madin-Darby canine kidney (MDCK) cell line. Also, similar assays can be performed using other spontaneously polarizing epithelial cell lines, such as Caco-2 (human intestinal epithelial cells), MCF10A (human mammary epithelial cells) or WIF2B (human hepatocyte cell line).
Procedure
a) Embedding MDCK cells in 3D matrix
Carry out steps 1-8 in a tissue culture hood.
Plate MDCK cells on 10 cm tissue culture plate and let grow for 24 hours at 37°C. The plated cells should not be fully confluent since cells need to be in growth phase. Typically we want to have cells at 50-70% confluency after 24 hour incubation. Thus, we usually plate cells at 30% confluency.
A couple hours before plating cells, thaw Matrigel (Corning, #354230) on ice. It is very important to keep Matrigel cold even while thawing, since it rapidly solidifies at room temperature. While Matrigel alone is sufficient to grow polarized MDCK cysts, Matrigel is a very soft extracellular matrix. Thus, in some cases it may be beneficial to supplement it with purified collagen I. We typically add collagen I to a final 3 mg/ml concentration to increase Matrigel stiffness.
Aspirate media and rinse cells with 10 mL PBS. Add 2 mL 0.25% Trypsin-EDTA and let sit at 37°C for 10-15 minutes. MDCK cells are usually difficult to dislodge. Thus, if needed, they can be incubated for 20-25 minutes.
Dislodge cells by gently tapping at the side of 10 cm dish. Harvest cells by adding 8 mL MDCK media (Dulbeco’s modified Eagle media supplemented with 10% Fetal Bovine Serum) to plate and transferring cells to 15 ml tube. Sediment cells by centrifugation at 1000 x rpm for 3 minutes.
Aspirate media and resuspend cells in 1 mL of MDCK media by pipetting up and down 50-100 times using 1 mL blue pipette tip. Pipetting up and down repeatedly helps to separate cells and is crucial for this assay to embed individual cells in Matrigel. Embedding cell clumps will lead to formation of cell aggregates with multiple lumens. We found that pipetting cells 40 times with a 1 mL pipette usually gives a maximum number of individual cells. However, this number may vary, thus it is advisable to initially pipette cells varying number of times, while monitoring the efficiency of cell separation.
Count cells to determine number of cells/mL and add 20,000 cells to 25 μL MDCK media in 1.5 mL tube. The optimum number of cells to plate may vary. Plating too many cells may cause clumping of dividing cells and overlap of fluorescence, making it difficult to distinguish single cells and observe them over time. Plating too few cells will make it more difficult to find single cells that are entering metaphase. We recommend testing a range of 10,000-50,000 cells to find the optimum number of cells for each assay.
Make a 75% Matrigel/Collagen solution by adding 75 μL Matrigel/Collagen to the 25 μL cell solution from step 6. Gently mix by pipetting up and down. Immediately plate the cell/Matrigel mix by placing a drop onto the center of either a 8-chamber dish (immunofluorescent microscopy) or a 5 cm gridded glass-bottom dish (time-lapse microscopy). This step needs to be done quickly, since the Matrigel-cell mixture solidifies readily at room temperature.
Place cells in the incubator and let solidify in the tissue culture incubator at 37°C for 30 minutes. Add serum-supplemented media.
Incubate the cells for 12-18 hours. If grown in Matrigel, the majority of MDCK cells will undergo first cell division within 24 hours of embedding. Thus, cells analyzed 12-18 hours post-embedding will either just have completed first cell division or will be at different stages of cytokinesis. Since AMIS is usually recruited to the midbody at late telophase, at 12-18 hours of incubation post-embedding will enhance the chance to find cells at the initial stages of AMIS recruitment of lumenogenesis.
b) Analyzing the lumenogenesis by immunofluorescence microscopy
Gently aspirate media and fix cells with 3% paraformaldehyde for 30 minutes.
Permeabilize cells with 0.5% Triton X-100 for 10 minutes, followed by 2×20 minute quenching with 100 mM Glycine in phosphate buffered saline (PBS).
Block non-specific binding by incubating cells in immunofluorescence buffer (PBS with 0.05% Triton X-100, 0.1% bovine serum albumin, 10% FBS, and 10 mM NaN3) for 4 hours at room temperature.
Block non-specific mice IgG by incubating cells in immunofluorescence buffer with 20 mg/ml of goat anti-mouse F(ab)2 fragments (Jackson ImmunoResearch, #115-006-006) for 40 minutes. This is a key step if mice primary antibodies will be used for immunofluorescence, since Matrigel is rich in mice IgG. This block then needs to be followed with 3×20 minutes washes with immunofluorescence buffer to remove any non-bound anti-mouse F(ab)2 fragments.
Incubate with primary antibodies diluted in immunofluorescence buffer overnight. To visualize midbody and central spindle, we typically use mice anti-acetylated tubulin antibodies (Sigma #T7451). To visualize AMIS, any previously published anti-ZO1, anti-occludin or anti-cingulin antibodies can be used. The AMIS also contain other membrane transport and polarity proteins, such as Par3/6 complex, the Exocyst complex, Syntaxin 3, Slp4a (Blasky et al., 2015). However all these proteins, while somewhat enriched at the AMIS, are also present at the basolateral plasma membrane. As a result, staining using these markers leads to difficulties identifying the AMIS, especially at the early stages of its formation.
Incubate 3×20 minutes with immunofluorescence buffer to remove non-bound primary antibodies, followed by incubation with appropriate fluorescent tag-conjugated secondary antibodies (in immunofluorescence buffer) for 60 minutes. Wash again 3×20 minutes with immunofluorescence buffer.
Aspirate liquid, remove chamber walls and dry slide for at least 1.5 hours before mounting in any suitable mounting medium, such as Vectashield.
c) Analyzing the lumenogenesis by time-lapse microscopy
Mount dish on fluorescent microscope and use a gridded chart to label the location of several individual cells, especially those that may be in metaphase (starting division). After every time point, cells can be returned to incubator. The grid coordinates will help to find cells for following time points. This is a preferred method when long (2-8 hour) time-lapses are used, since the entire time-lapse set will take 1-4 days. If short time lapses (under 1 hour) are used, a fluorescent microscope equipped with environmental control chamber needs to be used. In that case, several cells can be picked (if microscope has X-Y motorized stage) and imaged without moving cells back to incubator.
Adjust focus to set top and bottom of each cell and take initial 0.2 μm-step z-stack images. In order to be sure to include the whole cell in the range of imaging from top to bottom, it is best to set the focus a little beyond the point when the cell first begins to blur. It is crucial for the experiment to pick cells that are fully suspended within the matrix and do not touch the bottom of the dish. Any contact with glass-bottom will set a competing polarity axis and usually leads to disorganized and multi-luminal cysts.
Repeat imaging according to time frame for desired observation. This will result in taking a mini-z-stack for every time point. Generally, avoid taking more than 50 time-lapse z-stacks. Taking fewer time-points or fewer images in each mini-z-stack will decrease photo-damage to cells. Excessive imaging can be a problem, since mitotic cells are especially sensitive to photo-damage. Shorter exposures will also reduce photo-bleaching.
Taking mini-z-stacks at every time point allows the use of post-acquisition image analysis to generate three-dimensional images of MDCK cells at each time point during lumen formation. Alternatively, individual images that best represent the lumen formation dynamics can be selected and displayed/analyzed for every time point.
Data Interpretation
Immunofluorescence microscopy (IFM) and time-lapse analysis are two great complementary approaches to analyze AMIS formation around midbody during lumenogenesis. IFM allows analysis of the presence of various polarity and protein transport regulators during initial AMIS recruitment and formation. However, to truly understand spatiotemporal dynamics of AMIS formation and lumen initiation, IFM findings need to be confirmed by time-lapse microscopy. Since at 12-18 hours post-embedding, cells will be at different stages of division and lumenogenesis, it can sometimes be challenging to determine the order and timing of protein recruitment to the midbody and AMIS. This issue is complicated due to the fact that cells at late telophase look very similar to cells that already completed first division and have established a nascent lumen. Thus, arbitrarily chosen fixed cell images may not always represent a real order of lumen formation. Consequently, time-lapse analysis is the only approach that can fully define the spatiotemporal dynamics and regulation of nascent lumen formation.
ANALYSIS OF MIDBODY UPTAKE BY INTERPHASE CELLS
Now that it has come to light that midbodies can either be asymmetrically inherited by one daughter cell or can be uptaken from the extracellular environment, it is important to develop techniques that allow further examination of the potential post-mitotic midbody functions. Recent work using C. elegans suggests that MBs may be engulfed by surrounding cells due to the presence of phosphatidylserine on the outer leaflet of the plasma membrane remnant surrounding the midbody. Phosphatidylserines may act similarly to phagocytic "eat me" signal for cells to uptake the MB through an actin-dependent mechanism. Several techniques, including MB purification and MB uptake analysis, recently emerged to allow initial analysis of the machinery and functional consequences of MB accumulation in post-mitotic cells.
(A) Midbody purification
Overview
This protocol has been modified to get the best yield of MBs using HeLa cells stably expressing MKLP1-GFP (Kuo et al, 2012). Based on recent studies (Ettinger et al., 2011), it is likely that this protocol needs to be adjusted to different certain cell types, as cell lines can release midbodies at different frequencies. Additionally, it is possible that midbodies derived from different cells can contain different proteins and lipids, thus, the sucrose gradient centrifugation conditions (described below) may need to be adjusted for each cell type.
Procedure
Grow ten 15cm dishes of HeLa cells to sub-confluency (~70%) in Dulbecco’s modified Eagle media supplemented with 10% Fetal Bovine Serum.
Once 70% sub-confluency is reached, collect conditioned media (referred to as CM) from all ten plates and place into 50 mL Corning conical tubes.
Centrifuge CM at 300 x g for 5 minutes at 4°C. Remove the supernatants and place into fresh 50 mL conical tubes, and perform another 300 x g centrifugation for 5 minutes at 4°C. This centrifugation step will remove majority of intact cells or cell debris.
Remove the supernatants and place into fresh 50 mL conical tube. Centrifuge the CM at 10,000 x g for 30 minutes at 4°C.
Discard the supernatants. Combine the pellets and resuspend in 1 mL sterile phosphate-buffered saline (PBS). Place into a fresh 1.5 mL microcentrifuge tube.
Centrifuge the combined pellets at 10,000 x g for 30 minutes at 4°C. Discard the supernatant.
Resuspend the pellet in nuclease free water, 6mM MgCl2, 250 mM sucrose, and 250mM NaCl to a final volume of 200 uL.
Prepare a step-density gradient in a 1.3 mL ultracentrifuge tube consisting of 400 uL of 1M and 2M sucrose placed on top of 300 uL of a 40% glycerol cushion. For our sucrose gradients, we typically use a buffer that consists of 20 mM HEPES, pH7.4, 6 mM MgCl2, 1mM Dithiothreitol, and 100 mM NaCl.
Layer the resuspended pellet fraction on top of the step-gradient. Ultracentrifuge using a TLS-55 rotor by spinning at 3000 x g for 20 minutes at 4°C.
Immediately following centrifugation collect the visible opaque band at 2M sucrose/glycerol interphase and place into a fresh 1.5 mL microcentrifuge tube.
Centrifuge at 10,000 x g for 30 minutes at 4°C. Discard the supernatant, and resuspend the pellet in 20-40 uL of sterile PBS. Here, the pellet may not be visible, so it is important to keep track of which side of the tube the pellet may be on. Proceed immediately to MB uptake experiment.
(B) Midbody uptake analysis
Overview
Here, we describe the protocol and methods for analysis to assess MB uptake by HeLa cells. As stated before, it is likely that MBs isolated from different cell types may have distinct composition and properties. Additionally, various cell lines may have different capacities to uptake MBs. Both of these compounding factors may lead to different results when isolating and ‘feeding’ MBs to various cell types. We will use the term ‘feeding’ to describe the process in which we physically pipette MBs onto the cell population.
Procedure
One day prior to MB ‘feeding’, plate HeLa cells on collagen-coated 18mm coverslips placed in 12 well dishes. These cells should be grown to sub-confluency (~70%).
Immediately prior to MB ‘feeding’, remove media from wells and add 300 uL of fresh media. From the midbody isolation protocol, pipette 10-20 uL of isolated midbodies onto HeLa cells.
Centrifuge the plate at 1,050 rpm for 45 minutes to assist the MBs in sinking to the cells. Add 200 uL fresh media to each well following centrifugation. Incubate cells for 1-2 hours. From our experience cells need a couple of hours to internalize the MBs.
Wash cells with PBS and fix for 30 minutes in 4% paraformaldehyde.
Fix and immunostain cells using a standard immunofluorescence protocol.
Collect images using a 63x or 100x oil objectives and a fluorescent microscope. To ensure non-bias in collecting images, random sets of cells were imaged without first examining if those sets of cells had midbodies in the viewing field. Since MBs can be present at various sub-cellular locations, z-mini-stack (1 μm step size) needs to be imaged for every cell analyzed.
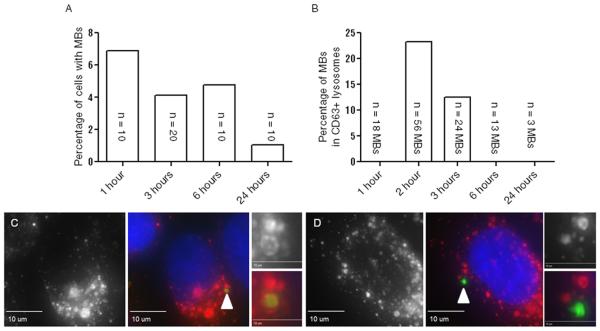
To assess the efficiency of MB uptake, all individual z-planes should be examined. Since our HeLa cells stably express a known MB marker MKLP1-GFP, the uptaken midbodies can be readily identified. Alternatively, MBs can marked using antibodies that recognize endogenous MB-resident proteins, such as Cep55, Cyk4, or MKLP1. The number of MBs internalized versus the number of cells in the viewing field can be directly compared to achieve a ‘MB accumulation frequency’ at different time points (Figure 3). If needed, cells can be co-stained for other various cellular markers. For example, co-staining with anti-CD63 (lysosomal marker, gift from Dr. Andrew Peden) antibody will allow to determine whether the midbody is present in cytosol or is targeted to lysosomal degradative pathway (Figure 3).
Figure 3. Quantification of midbody uptake.
(A) Time course of MB feeding. The percentage on the y-axis represents the number of MBs per nuclei in a random viewing field of cells. N represents a number of MBs counted.
(B) Number of MBs present in CD63-positive lysosomes. Colocalization of MKLP1 and CD63 was scored to identify MBs undergoing autophagy. The percentage on the y-axis represents the number of CD63-positive MBs per the total number of midbodies counted for that time-point.
(C) Representative image of MB present in CD63-positive lysosome. Left panel shows CD63 (red) only. Middle panel shows merged image of CD63 (red) MKLP1 (green) and DNA (blue). White arrowhead points to engulfed midbody. Right panel shows inset images from middle panel. Right top panel shows CD63 (red) only. Right bottom shows merged image of CD63 (red) and MKLP1 (green).
(D) Representative image of MB within a cell but not within a CD63-positive lysosome. Left panel shows CD63 (red) only. Middle panel shows merged image of CD63 (red) MKLP1 (green) and DNA (blue).White arrowhead points to midbody. Right panel shows inset images from middle panel. Right top panel shows CD63 (red) only. Right bottom shows merged image of CD63 (red) and MKLP1 (green).
Data Interpretation
It is becoming clear that MBs are engulfed via phagocytosis-like machinery. As a result, it is possible that some of the MBs are quickly routed to the degradative lysosomal pathway and do not participate in signaling. Consistent with this, we do see that a fraction of internalized MBs co-stain with lysosomal markers, such as CD63, during first 2 hours of post-feeding (Figure 3C). Furthermore, as shown in Figure 3A, the number of MBs remaining in cells 24 hours post-feeding is markedly decreased compared to first hour, indicating that they are likely targeted to the lysosomes for degradation. Interestingly, after 3 hours of post-feeding, the majority of intracellular MBs did not colocalize with CD63-positive lysosomes, potentially indicating that these MBs have evaded phagocytic degradation (Fig. 3B), although the machinery governing this process remains completely unknown. These evidence suggest that MBs can be inherited from the extracellular environment and retained in cells, without being degraded, for at least 24 hours post-internalization. What remains to be determined is whether these MBs actually function as a signaling platform and whether they affect the properties and proliferation of cells that retain and accumulate post-mitotic MBs.
SUMMARY
MBs have recently emerged as putative regulators of several post-mitotic cell functions and properties, including cell polarization and proliferation. Furthermore, recent data indicate that MBs can also be uptaken from the extracellular milieu, thus potentially serving as platforms for parallel signal transfer between cells. Further studies, however, will be needed to further understand the regulation and functional significance of MB inheritance, uptake and accumulation. Here, we have introduced several new techniques that will allow researchers to start investigating post-mitotic MB functions.
ACKNOWLEDGMENTS
We are grateful to Abitha Jacob and Dr. Alexander Blasky for critical reading of the manuscript. We thank Dr. Andrew Peden, University of Sheffield, for the CD63 antibody. I apologize to all colleagues whose work could not be cited due to space limitations. Work in Rytis Prekeris laboratory is supported by NIH (DK064380) and Cancer League of Colorado.
REFERENCES
- Blasky AJ, Mangan A, Prekeris R. Polarized Protein Transport and Lumen Formation During Epithelial Tissue Morphogenesis. Annual Review of Cell and Developmental Biology. 2015;31:575–591. doi: 10.1146/annurev-cellbio-100814-125323. doi:10.1146/annurev-cellbio-100814-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai Y, Tian D, Yang Y, Feng G, Cheng Z, Li W, Ou G. Apoptotic regulators promote cytokinetic midbody degradation in C. Elegans. Journal of Cell Biology. 2012;199(7):1047–1055. doi: 10.1083/jcb.201209050. doi:10.1083/jcb.201209050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowell EF, Gaffuri A, Gayraud-Morel B, Tajbakhsh S. Engulfment of the midbody remnant after cytokinesis in mammalian cells. Journal of Cell Science. 2014;127:3840–3851. doi: 10.1242/jcs.154732. doi:10.1242/jcs.154732. [DOI] [PubMed] [Google Scholar]
- Yang D, Rismanchi N, Renvoise B, Lippincott-Scwartz J, Blackstone C, Hurley J. Structural basis for midbody targeting of spastin by the ESCRT-III protein CHMP1B. Nature Structural Molecular Biology. 2008;15(12):1278–1286. doi: 10.1038/nsmb.1512. doi:10.1038/nsmb.1512.Structural. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dionne LK, Wang XJ, Prekeris R. Midbody: From cellular junk to regulator of cell polarity and cell fate. Current Opinion in Cell Biology. 2015;35:51–58. doi: 10.1016/j.ceb.2015.04.010. doi:10.1016/j.ceb.2015.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elia N, Sougrat R, Spurlin TA, Hurley JH, Lippincott-schwartz J. Dynamics of endosomal sorting complex required for transport ( ESCRT ) machinery during cytokinesis and its role in abscission. PNAS. 2011;108(12):4846–4851. doi: 10.1073/pnas.1102714108. doi:10.1073/pnas.1102714108/-/DCSupplemental.www.pnas.org/cgi/doi/10.1073/pnas.1102714108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ettinger AW, Wilsch-Bräuninger M, Marzesco A-M, Bickle M, Lohmann A, Maliga Z, Karbanová J, Corbeil D, Hyman A, Huttner WB. Proliferating versus differentiating stem and cancer cells exhibit distinct midbody-release behaviour. Nature Communications. 2011;2:503. doi: 10.1038/ncomms1511. doi:10.1038/ncomms1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Kuehn EW, Prekeris R. Kinesin-2 mediates apical endosome transport during epithelial lumen formation. Cellular Logistics. 2014;4:e28928. doi: 10.4161/cl.28928. doi:10.4161/cl.28928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Mangan A, Cicchini L, Margolis B, Prekeris R. FIP5 phosphorylation during mitosis regulates apical trafficking and lumenogenesis. EMBO Reports. 2014a;15(4):428–437. doi: 10.1002/embr.201338128. doi:10.1002/embr.201338128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollarolo G, Schulz JG, Munck S, Dotti CG. Cytokinesis remnants define first neuronal asymmetry in vivo. Nature Neuroscience. 2011;14(12):1525–1533. doi: 10.1038/nn.2976. doi:10.1038/nn.2976. [DOI] [PubMed] [Google Scholar]
- Schiel JA, Simon GC, Zaharris C, Weisz J, Castle D, Wu CC, Prekeris R. FIP3-endosome-dependent formation of the secondary ingression mediates ESCRT-III recruitment during cytokinesis. Nature Cell Biology. 2012;14:1068–78. doi: 10.1038/ncb2577. doi:10.1038/ncb2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh D, Pohl C. Coupling of Rotational Cortical Flow, Asymmetric Midbody Positioning, and Spindle Rotation Mediates Dorsoventral Axis Formation in C. elegans. Developmental Cell. 2014;28(3):253–267. doi: 10.1016/j.devcel.2014.01.002. doi:10.1016/j.devcel.2014.01.002. [DOI] [PubMed] [Google Scholar]
- Kuo T-C, Chen C-T, Baron D, Onder T, Loewer S, Almeida S, Weismann C, Xu P, Houghton J-M, Gao F-B, Daley G. Midbody accumulation through evasion of autophagy contributes to cellular reprogramming and tumorigenicity. 2012;127(10):358–366. doi: 10.1038/ncb2332. S. D. doi:10.1016/j.jsbmb.2011.07.002.Identification. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilcock AC, Swedlow JR, Storey KG. Mitotic spindle orientation distinguishes stem cell and terminal modes of neuron production in the early spinal cord. Development (Cambridge, England) 2007;134:1943–54. doi: 10.1242/dev.002519. doi:10.1242/dev.002519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Mangan A, Cicchini L, Margolis B, Prekeris R. FIP5 phosphorylation during mitosis regulates apical trafficking and lumenogenesis EMBO Reports. 2014;15(4):428–437. doi: 10.1002/embr.201338128. http://dx.doi.org/10.1002/embr.201338128. [DOI] [PMC free article] [PubMed] [Google Scholar]