Abstract
Dictyostelids are free-living phagocytes that feed on bacteria in diverse habitats. When bacterial prey is in short supply or depleted, they undergo multicellular development culminating in the formation of dormant spores. In this work, we tested isolates representing four dictyostelid species from two genera (Dictyostelium and Polysphondylium) for the potential to feed on biofilms preformed on glass and polycarbonate surfaces. The abilities of dictyostelids were monitored for three hallmarks of activity: 1) spore germination on biofilms, 2) predation on biofilm enmeshed bacteria by phagocytic cells and 3) characteristic stages of multicellular development (streaming and fructification). We found that all dictyostelid isolates tested could feed on biofilm enmeshed bacteria produced by human and plant pathogens: Klebsiella oxytoca, Pseudomonas aeruginosa, Pseudomonas syringae, Erwinia amylovora 1189 (biofilm former) and E. amylovora 1189 Δams (biofilm deficient mutant). However, when dictyostelids were fed planktonic E. amylovora Δams the bacterial cells exhibited an increased susceptibility to predation by one of the two dictyostelid strains they were tested against. Taken together, the qualitative and quantitative data presented here suggest that dictyostelids have preferences in bacterial prey which affects their efficiency of feeding on bacterial biofilms.
Keywords: Dictyostelium, Polysphondylium, Erwinia amylovora, Pseudomonas aeruginosa, Pseudomonas syringae, myxamoebae, phagocyte
Introduction
Generally, bacterial cells are not planktonic in the environment (Brock 1967; Henrici 1933). Instead bacteria attach to solid surfaces, natural or man-made, and become part of dynamic and structurally heterogeneous communities (Branda et al. 2005; Costerton et al. 1978; Fux et al. 2005; Kumar and Anand 1998; Rodríguez and Bishop 2007; Romero and Kolter 2011). These communities are enmeshed in extracellular polymeric substances (EPS) called matrices (reviewed in Branda et al. 2005; Burmolle et al. 2010; Fux et al. 2003; Hall-Stoodley et al. 2004; More et al. 2014; Richards and Melander 2009). The complex interactions within biofilm communities help protect resident cells from antibacterial compounds (Anderl et al. 2000; Donlan 2002; Donlan and Costerton 2002; Walters et al. 2003) and phagocytic immune defenses (Bjarnsholt et al. 2005; Jesaitis et al. 2003; Mittal et al. 2006; Thurlow et al. 2011; Walker et al. 2005). As a result, surface grown bacteria can be difficult or even impossible to eradicate.
Organisms that prey on bacteria inevitably encounter surfaces covered by biofilms in the diverse aquatic and terrestrial environments they live in. For example, protozoa occur widely in close association with natural biofilm communities where they feed on biofilm-enmeshed bacteria, disrupt biofilm structure and cause biofilm sloughing (Huws et al. 2005; Jackson and Jones 1991; Pedersen 1982; Weitere et al. 2005). These observations suggest that research on bacterial predators could inform fundamentally new strategies to controlled removal of undesired biofilms.
These observations prompted us to investigate dictyostelids (Brefeld 1869; Raper 1935), which are members of a single clade within the supergroup of Amoebozoa (Heidel et al. 2011; Schaap 2011; Schaap et al. 2006). When presented with bacterial lawns, dictyostelids feed, grow and divide in the form of solitary myxamoebae. However, starvation triggers a transition from the solitary form to a multicellular assemblage comprised of non-feeding cells, which undergo complex development culminating in the production of spore-laden sori (Konijn et al. 1967; Raper 1935, 1937; Raper and Rahn 1984) (Fig. 1). Spores can be lyophilized and maintained in a dormant state for decades (Cotter and Raper 1968; Cotter et al. 1992; Raper and Rahn 1984; van Es et al. 1996).
Figure 1.
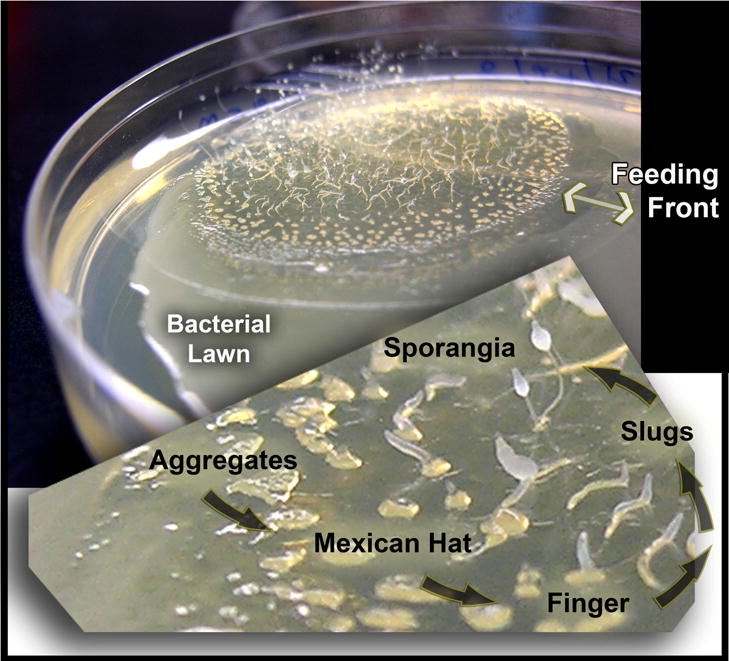
The multicellular development of dictyostelids. Dictyostelids emerge from spores as motile phagocytic myxamoebae and feed on bacterial lawns. Starvation triggers a developmental program, which can be exhibited as morphologically staged rings. Myxamoebae nearest the pristine bacterial lawn (the feeding front) have not entered the developmental pathway. Loose aggregations of myxamoebae proceed to tighter formations including “Mexican hats”. Eventually slugs form, migrate, and produce fruiting bodies. Similar but not identical patterns are common to all dictyostelid species of group 4.
Dictyostelids are abundant in temperate woodland soils but they have also been identified in diverse habitats that include caves, agricultural land, prairies, marshes, sandy mesas, tundra, and tropical forest (e.g., Cavender 1973, 2013; Cavender et al. 2002; Landolt et al. 2008; Romeralo et al. 2010, 2011b; Swanson et al. 2001, 2002; Vadell et al. 2011). In most cases they are found in association with decaying leaves and organic waste (solid surfaces). Given their global distribution, it is not surprising that dictyostelids can feed on a wide range of bacteria (Raper 1937; Raper and Smith 1939). Food “preferences” and competitive advantages amongst dictyostelid isolates have been observed (Eisenberg et al. 1989; Horn 1971; Ketcham et al. 1988; Ketcham and Eisenberg 1989). Experiments profiling gene expression during D. discoideum feeding on Gram positive or Gram negative bacteria suggest prey and predators communicate with each other using chemical cues (Carilla-Latorre et al. 2008; Nasser et al. 2013). The known diversity of dictyostelid social interactions, food preference and corresponding transcriptional alterations augur well for the prospect of identifying dictyostelid strains that use different strategies to prey on biofilm enmeshed bacteria.
The ability of D. discoideum to interact with bacterial biofilm communities has been observed but remains largely unexplored (Yang et al. 2011, 2012). In this study, we tested the abilities of four species of naturally occurring dictyostelid isolates to grow on static bacterial biofilms established on glass and polycarbonate surfaces. We utilized multiple species of bacteria with well-documented propensities for forming biofilms. Specifically, we tested predator-prey interactions between dictyostelids and Klebsiella oxytoca (Tang et al. 2009), Pseudomonas aeruginosa (Mann and Wozniak 2012) Pseudomonas syringiae (Laue et al. 2006) and Erwinia amylovora (Koczan et al. 2009, 2011). Our observations presented here support the hypothesis that biofilm grazing is a common property among dictyostelid myxamoebae. Moreover, data collected from our static biofilm assays on glass and polycarbonate surfaces provide evidence that dictyostelid isolates use diverse strategies to consume biofilm-enmeshed bacterial cells.
Results
Characteristics of dictyostelids used in this study
Biofilms provide a protective matrix that enables some bacteria to survive or even kill grazing protozoans while their planktonic counterparts are eliminated (Matz et al. 2005, 2008). As soil dwelling bacterivores, we hypothesized that dictyostelid myxamoebae may possess the ability to feed on bacterial biofilms. To begin investigating this possibility, we generated working stocks by allowing lyophilized spores of five dictyostelid strains to germinate. All dictyostelid strains were able to grow on lawns of K. oxytoca as described (Methods) and demonstrated morphological and behavioral characteristics appropriate for their taxonomic designations (Raper and Rahn 1984; Swanson et al. 2002). These hallmarks derive from the life cycle of the organism, which includes a solitary myxamoebae stage and progressive stages of aggregation and multicellular development (Fig. 1).
Nearly all known species of dictyostelids have been subdivided into four major groups based on phylogenetic analysis of their 18S ribosomal RNA sequences (Schaap et al. 2006). Our selections of D. giganteum and D. discoideum represent major subclades of group 4 (4A and 4B, respectively) whereas D. aureostipes is specific to group 1 (Schaap 2011; Schaap et al. 2006). The single Polysphondylium isolate chosen for study, a P. pallidum strain (El Salvador), falls into Group 2B. Each species identity was confirmed by Sanger sequencing of the 18S rDNA locus.
To determine if germination efficiencies of dictyostelids differ significantly we tested several variables that are known to affect spore germination efficiencies differently in multiple dictyostelid species (Cotter et al. 2000; Cotter and Raper 1968). There is evidence that some germinating spores can secrete discadenine (inhibitor) that rapidly diffuses, thereby preventing germination of nearby spores (Cohen and Ceccarin 1967; Nomura 1977). We therefore undertook standard quantitative assays to examine spore germination frequencies in the presence of bacteria (K. oxytoca) (Cavender and Raper 1965a, b). We found that the efficiencies of germination for the four naturally occurring dictyostelid strains and the axenic mutant, Dd-AX3, fell within a range of less than one order of magnitude (Supplementary Material Table S1). Thus, in subsequent experiments, we standardized spore counts to account for these differences.
K. oxytoca forms multilayer biofilms on glass surfaces
Analysis of the spatial organization of bacterial populations by Confocal Laser Scanning Microscopy (CLSM) has been a useful tool for the observation of dynamic changes within biofilms (Lawrence and Neu 1999). We used CLSM to characterize the cell assemblages formed by K. oxytoca on a glass surface. For this experiment, a bacterial community was grown using a fluorescent derivative of M5al, strain KOF001 (Table 1 and Methods). Z-stacks of the biofilm at 600x magnification were taken at 2 μm Z distance intervals using a Nikon A1R instrument with two excitation lasers (488 nm and 561 nm). The resulting images were assembled and color coded according to depth to generate the three dimensional representation of structure (Supplementary Material Fig. S1). We found that the KOF001 cells produced a multilayered bacterial assemblage on the glass surface. Thickness measurements of 50 – 75 μm were recorded near the edges of the assemblage while the depths of the interior exceeded the limits of detection (>125 μm).
Table 1.
Strains used in this study
| Isolate | Species | Location & relevant information |
|---|---|---|
| Dictyostelids: | ||
| Dd-AX3 | D. discoideum | Little Butts Gap, NC, USA. Axenic derivative of NC-4 (Raper and Rahn, 1984), called A3 in original citation (Loomis, 1971). See also (http://dictybase.org). axenic strains history. |
| Dg-WS-142 | D. giganteum | Madison, WI, USA (1954). Previously described (Raper and Rahn, 1984; Weber and Raper, 1971). Lyophilized 8/16/71. |
| Da-WS-309 | D. aureostipes | Dayton, OH, USA (1956). Isolated from the National Cash Register Woods. Deciduous woods with poplar, oak, maple and cherry. Lyophilized 12/22/70. |
| Dd-WS-647 | D. discoideum | St. Tammany Parish, LA, USA (1976). Collected by Ann C. Worley from soil and highly decomposed leaf litter, approx. 20 yards from a creek; Honey Island Swamp near Pearl River. Bottom lands covered w/tupelo & cyprus. Many hardwood shrubs and small trees. Lyophilized 8/29/79. |
| Pp-ES | P. pallidum | San Salvador, El Salvador (Raper and Rahn, 1984). Lyophilized 4/57. |
|
Bacteria: | ||
| S17.1 | E. coli | Chromosomally modified donor used for conjugal transfer of plasmid DNA (Simon et al., 1983). |
| M5al | K. oxytoca | Suspected biofilm-forming strain based on studies of other K. oxytoca strains (Bao et al., 2013; Hamilton and Wilson, 1955; Pengra and Wilson, 1958; Yoch and Pengra, 1966). Biofilm formation by M5al established – This study. |
| KOF001 | K. oxytoca | M5al harboring plasmid pFL300 and expressing d- Tomato red fluorescent protein. This study. |
| PAO1 | P. aeruginosa | Biofilm-forming pathogen of humans (Banin et al., 2005; Diggle et al., 2006). |
| 207.2 | P. syringae | Biofilm-forming pathogen of beans. Unpublished. Provided by Dr. Patricia McManus, Department of Plant Pathology, University of Wisconsin-Madison. |
| Ea 1189 | E. amylovora | Biofilm-forming pathogen of rosaceous species such as apple and pear (Burse et al., 2004; Koczan et al., 2009). |
| Ea 1189 Δams | E. amylovora | Biofilm-deficient (in vitro and in planta) nonpathogenic mutant of strain Ea 1189 (Koczan et al., 2009; Zhao et al., 2009). |
CLSM 3 dimensional imaging of myxamoebae movements in K. oxytoca biofilm
Next, we modified our method to detect myxamoebal movement in 3 dimensional space. Dictyostelid strain Dg-WS-142 was selected for the experiment due to its robust antibacterial activity. Here, non-fluorescent cells of Dg-WS-142 (2.5 × 104 myxamoebae) were introduced onto a coverslip biofilm comprised of fluorescent KOF001 bacteria and time lapse images from only the top and bottom Z stacks were captured. (https://media.bact.wisc.edu/filutowicz/Protist-SuppMaterial-Sanders2016.html).
Further analysis of the time lapse images demonstrated the adherence of sessile fluorescent bacteria to glass, reinforcing the evidence of bacterial population formation generated in the original CSLM experiment (Supplementary Material Fig. S1). The architecture of the population is comprised of a largely sessile central mass of multiple layers of cells, within which reside “pools” of free living bacteria (highlighted in the video). These pools may correspond to “water channels” described for K. pneumoniae by other investigators (Singla et al. 2014). Additionally, our observation of amoeboid movement of non-fluorescent myxamoebae over the time course suggests that a biofilm of KOF001 might not be an impenetrable barrier. Dg-WS-142 appears able to freely migrate throughout and submerge within the biofilm-enmeshed bacterial cells. At several points (both positional and temporal) myxamoebae disappear from view and seemingly reappear at a nearby location. This “burrowing” behavior was evident both at the basal population layer in close proximity to the glass surface (bottom Z stack) which is assumed to have a low oxygen tension (de Beer et al. 1994; Stewart 2003) and in the upper portion of the biofilm (top Z stack) which is assumed to be oxygenated (de Beer et al. 1994; Stewart 2003).
Comparative analysis of dictyostelids feeding on K. oxytoca biofilms established on glass surface
To evaluate the relative robustness of predatory activity, we monitored the feeding and multicellular development of five dictyostelid isolates when added to biofilms of K. oxytoca. In a typical experiment, 2.5 × 104 myxamobae were added to a coverslip biofilm, then the sessile bacterial prey and moving predatory myxamoebae were monitored using time-lapse microscopy. (https://media.bact.wisc.edu/filutowicz/Protist-SuppMaterial-Sanders2016.html). Snapshots and descriptions of the most striking observations are presented in Figure 2. For all dictyostelids examined, myxamoebae fed on bacteria adhered to glass surfaces and each dictyostelid strain has been ranked based on its biofilm erosion speed (Fig. 2). Of particular note, in the upper panel of the Figure 2, strains Dd-AX3 and Dg-WS-142 demonstrated nearly complete biofilm erosion in less than 2 days. We also observed variation in the amounts of the prey that remained unconsumed after myxamoebae streaming and multicellular development.
Figure 2.
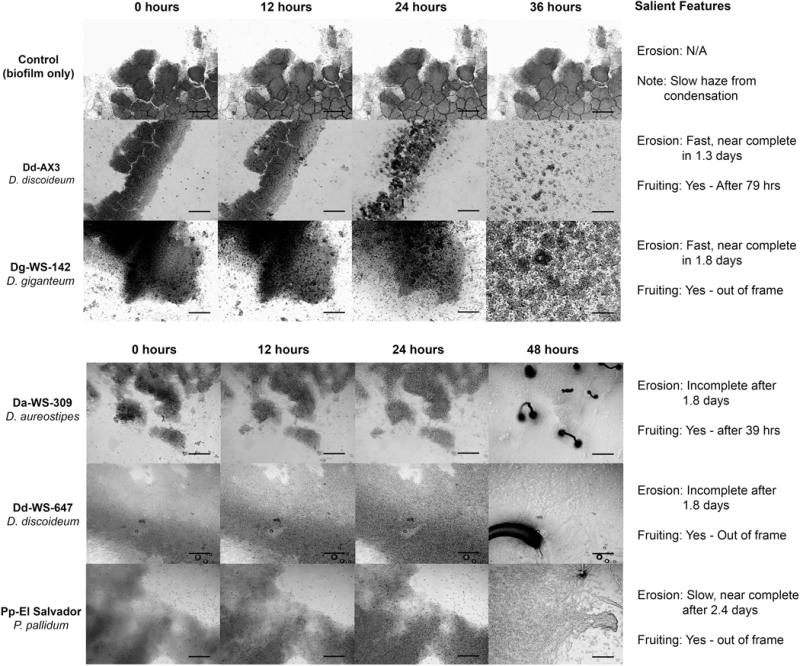
Microscopic evidence (light microscopy) for biofilm destruction by dictyostelids. A chronological sequence of micrographs taken at 40× (total magnification) ranging from time 0 to 36 hrs (or 48 hrs) after dictyostelid myxamoebae deployment onto K. oxytoca biofilms. Each sequence shows the dynamic changes that occur when a biofilm is under attack by the myxamoebae of a dicytostelid species. Differences in the completeness of biofilm breakdown (erosion) and the speed at which it occurs are noted for each dictyostelid species (“Salient Features” column). Multicellular differentiation events are noted (“Fruiting”) and all scalebars are set at 200 μm.
In follow up recordings, bacterial consumption by myxamoebae was monitored at high magnification (320×) to gain more insight on how individual predator cells approach biofilm enmeshed prey colonizing glass surfaces (Table 2). Qualitative analyses of the time-lapse images suggest that different species of dictyostelids employ different tactics to attack communities of bacteria. For example, the cells of strain Dg-WS-142 appeared to be unique in separating chunks of 10–30 K. oxytoca cells from the main biofilm body (https://media.bact.wisc.edu/filutowicz/Protist-SuppMaterial-Sanders2016.html). This process seems to involve individual dictyostelid cells crawling into or under the biofilm as they tear it into much smaller pieces leading to the release of bacterial cell agglomerates. In contrast, Pp-ES myxamoebae appear to access prey at the biofilm’s edges without dislodging large bacterial assemblages.
Table 2.
Time lapse microscopy at 320 × magnification
| Isolate | Movement on/in biofilm | Apparent biofilm consumption strategy |
|---|---|---|
| Pp -ES | Possible 3 dimensional movement. | Cooperative - Myxamoebae jointly attack large chunks of biofilm-emeshed bacterial cells. |
| Dg-WS-142 | 3 dimensional movement throughout the biofilm | Cooperative - Similar to Pp-ES but higher efficiency. Myxamoebae collectively attack large clusters of attached biofilm sloughing it to smaller edible cell clusters. |
Time lapse videos can be viewed at https://media.bact.wisc.edu/filutowicz/Protist-SuppMaterial-Sanders2016.html).
CLSM and Scanning Electron Microscopy (SEM) provide evidence for K. oxytoca biofilm formation on PC filters
Bacteria of various species form biofilms on PC filters, laying the foundation for a useful tool to quantify the susceptibility of bacteria to antibiotics (Anderl et al. 2000; Merritt et al. 2005; Singh et al. 2010; Walters et al. 2003). PC filters were used to investigate predator/prey interactions because filter based assays allow quantitative determination of the consumption of bacteria by myxamoebae as a function of time. Before these quantitative assays were performed, however, we established colonies of K. oxytoca on PC filters, and subjected the filters to CLSM imaging as we did for biofilms preformed on glass coverslips. The CLSM data demonstrated that bacterial assemblages exhibited an average thickness of approximately 125 μm after 2 days of incubation (Fig. 3A). This is consistent with previous studies of the biofilm thickness in K. pneumoniae (Singla et al. 2014).
Figure 3. K. oxytoca biofilm degradation by different dictyostelids.
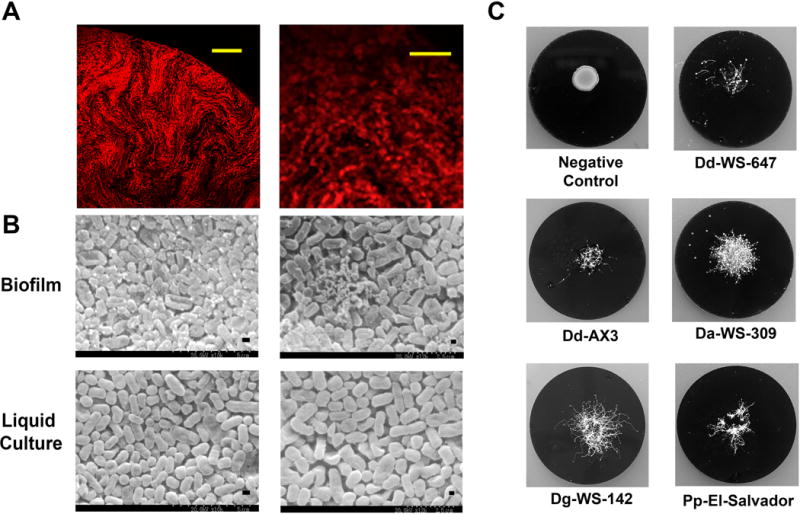
(A) Left: Confocal micrograph (600X magnification) of a sodium chloride washed KOF001 biofilm assembled on PC filter. Right: A digitally zoomed image showing the arrangement of cells. Scale bars are provided (10 μm and 25 μm). (B) Biofilm and planktonic cells (liquid culture) of KOF001 visualized by SEM imaging. Scale bars are provided (2.5μm and 5 μm). (C) Representative images of filter grown biofilms after 6 days consumption by dictyostelids. The images detail biofilm destruction and completion of the full developmental cycle for all five dictyostelid strains examined.
One of the limitations of CLSM is that it can detect bacterial cell arrangements and motion but not extracellular polymeric substance (EPS), which is not fluorescent. For that reason we carried out Scanning Electron Microscopy (SEM) on colonies of KOF001 pre grown on PC filters resting on TSAgar and on TSBroth grown KOF001 with the latter cells existing as planktonic cells (Figs 3B, 4B). We noted K. oxytoca inocula incubated on solid surfaces displayed both bacterial cells and EPS (Fig. 3B, biofilm) whereas cells grown in liquid culture did not have visible EPS (Fig. 3B, liquid culture). We conclude that KOF001 is similar to other Klebsiella strains in that it possesses the ability to form biofilm on PC filters, and our methods are suitable for establishing biofilm communities on glass and polycarbonate surfaces.
Figure 4.
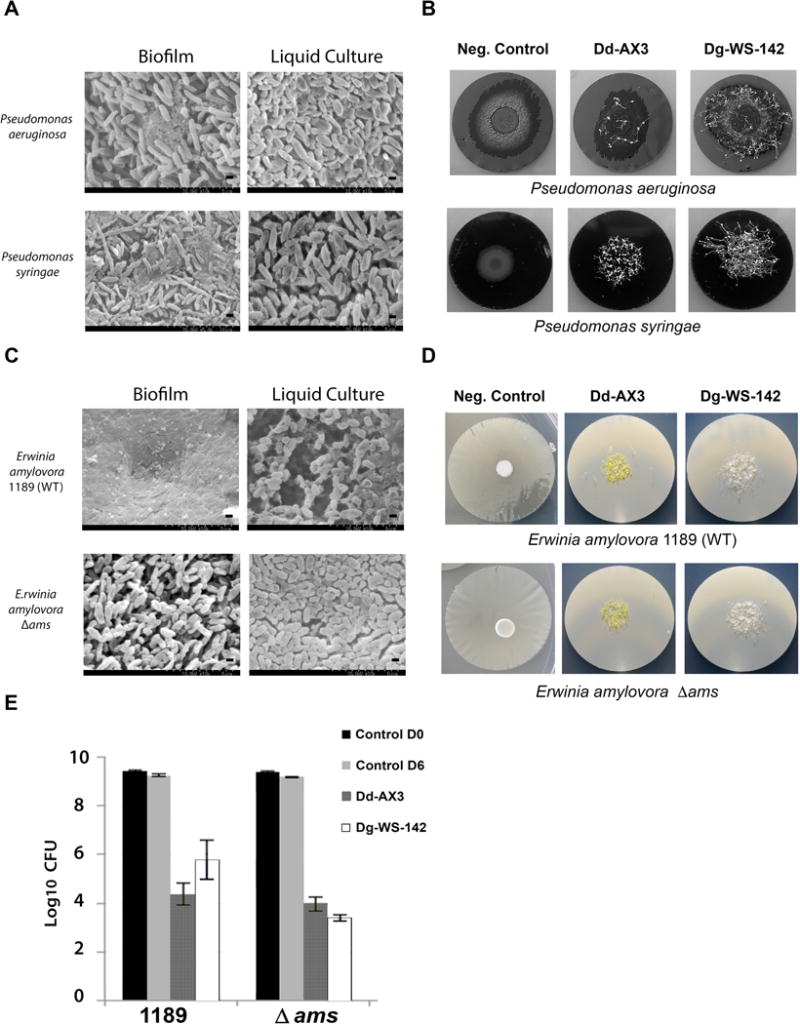
Anti-biofilm activities of dictyostelid myxamoebae against other Gram negative bacterial pathogens of plants and animals. (A) SEM images showing composition differences of” Biofilm” and planktonic (“Liquid Culture”) specimens of P. aeruginosa and P. syringae. Scale bars are provided (2.5μm and 5 μm). (B) Images of P. aeruginosa and P. syringae biofilms grown on PC membranes 7 days after inoculation with 2.5 × 104 dictyostelid spores. Negative Control biofilms were not inoculated with dictyostelids. Images show biofilm destruction and the formation of multicellular fruiting bodies (fluffy patches). (C) SEM images of E. amylovora 1189 and E. amylovora 1189 Δ ams “Biofilm” and planktonic “Liquid Culture” specimens. (D) Corresponding macroscale images of E. amylovora 1189 and E. amylovora 1189 Δ ams on PC membranes 5 days after inoculation with 2.5 × 104 spores of Dd-AX3 or Dg-WS-142. (E) Quantification of surviving bacteria after 5 days’ treatment with 2.5 × 104 spores (inoculum size) of Dd-AX3 or Dg-WS-142. Averages of two experiments, each experiment was performed with duplicate PC filters.
Dictyostelids consume K. oxytoca biofilms on microporous PC filters
Having confirmed the ability of K. oxytoca to form biofilms on PC filters we surveyed a panel of dictyostelid strains (five) from four species for characteristic behaviors when presented with PC-filter biofilms (Fig. 3C). Specifically, we assessed: 1) the ability of dictyostelid spores to germinate, 2) the growth of myxamoebae in size and number with the concomitant decrease of live bacterial prey, and 3) multicellular development. These long-term (7-day) quantitative assays for biofilm destruction were conducted in triplicate with three PC filters for each strain and the numbers of surviving bacteria were quantified as described (Materials and Methods). As shown in Table 3, dictyostelid deployment had a significant effect on bacterial cell survival in all cases, reducing the numbers of viable cells by a minimum of 2 log10 in comparison to the untreated control. Dictyostelid strains Dd-AX3 and Dg-WS-142 exhibited the highest levels of predatory activity (>4 log10 reduction of viable bacteria).
Table 3.
Percent survival of K. oxytoca after 7 days incubation at 23°C with various dictyostelid strains.
| Strain | Dictyostelid Species | Bacterial % Survival | Standard Error | p-value |
|---|---|---|---|---|
| Control | K. oxytoca biofilm (no dictyostelids) | 100 | 10.9 | ---------- |
| Dg-WS-142 | D. giganteum | 0.0150 | 0.00770 | 1.61×10 −5 |
| Dd-AX3 | D. discoideum | 0.0391 | 0.00860 | 1.61×10 −5 |
| Dd-WS-647 | D. discoideum | 0.178 | 0.0515 | 1.62×10 −5 |
| Pp-ES | P. pallidum | 0.441 | 0.0674 | 1.66×10 −5 |
| Da-WS-309 | D. aureostipes | 2.54 | 0.690 | 1.87×10 −5 |
Data collected after 7 days incubation at 23°C
Statistical analysis: Two sample T-test (treated vs. control)
Dictyostelids feed with varying efficiency on biofilms of different species of Gram negative bacteria
To further explore the diversity of biofilm destruction capabilities of dictyostelids, we performed the PC filter assays using three additional species of bacteria: P. aeruginosa (human/animal pathogen (Costerton et al. 1999; Lyczak et al. 2000), P. syringae and E. amylovora (plant pathogens, (Billing 1974; Koczan et al. 2009, 2011; McManus and Jones 1994; Vanneste 2000). For these assays, we narrowed our focus to the two dictyostelid strains that were most efficient in destroying biofilms of K. oxytoca in our previous experiments (Dd-AX3 and Dg-WS-142) (Table 3). SEM analysis revealed major architectural differences between the biofilm enmeshed bacterial populations and bacteria grown in liquid cultures then concentrated and transferred to PC filters less than one hour prior to SEM imaging (Methods). This was true when either P. aeruginosa or P. syringiae were examined. EPS and bacterial cells are visible in the images of the biofilms grown on surface of PC for extended periods of time and at different temperatures (described in Methods) but in samples of bacteria grown in liquid cultures only rod like cells were visible (Fig. 4A). The results using the two other bacterial species (Fig. 4A, B) were similar to our data with K. oxytoca (Fig. 3C). That is, the treatment of the other bacterial species with dictyostelids resulted in the loss of identifiable biofilm and the appearance of sori (Fig. 4B).
To determine if EPS production altered bacterial consumption by dictyostelids we utilized isogenic strains of E. amylovora, one capable of forming biofilms (1189) and the other biofilm-deficient (1189 Δams). E. amylovora 1189 forms biofilms on solid surfaces in vitro and in the vascular systems of apples and pears. In sharp contrast, the Δams mutant does not form biofilms neither in vitro nor in planta (Koczan et al., 2009). Specifically, E. amylovora 1189 Δams lacks a 12-gene operon that encodes proteins required for the biosynthesis of amylovoran, a heteropolymer composed of branched repeating unit of galactose, glucose and pyruvate residues that constitutes an adherence matrix necessary for biofilm formation (Nimtz et al. 1996; Zhao et al. 2009). SEM imaging revealed significant differences between these bacterial strains. Individual rod-shaped bacterial cells are readily distinguishable in liquid media and PC-filter grown cells of E. amylovora 1189 Δams. Specifically, cells of 1189 grown on a PC filter are markedly different in appearance in that an EPS is visible but individual cells are not. The biofilm deficient Δams mutant lacks a strongly defined EPS and appears to instead produce colonies made of disconnected “free cells“ (Fig. 4C). Amylovoran is considered a pathogenicity factor because mutants that are deficient in its production are avirulent (Bellemann and Geider 1992). The structural differences between bacterial communities in the wildtype and Δams mutant E. amylovora appeared not to be inhibitory to Dd-AX3 predation. Bacterial consumption of both Erwinia strains were similar at the end of the 5-day experiment (CFUs reduction of ~ 5 log10) (Fig. 4D, E). A much greater difference in predation efficiency was seen for Dg-WS-142, which consumed the cells of the biofilm forming E. amylovora 1189 less robustly (CFUs reduced approximately 4 log10) in comparison to the Δams mutant (CFUs reduced approximately 6 log10) (Fig. 4E).
Discussion
The ability of bacteria to form biofilms is a powerful survival tool that can disrupt or maintain the health of many ecosystems. A variety of physical and biological forces can act as an auto-regulating system to keep biofilm proliferation in balance. Bacteria themselves produce compounds that break down biofilms, presumably for autoregulation, competition or both (Donlan 2002; Hall-Stoodley et al. 2004; Kaplan 2010; Kolodkin-Gal et al. 2010; Lee et al. 2007; Parsek and Greenberg 2005; Richards and Melander 2009; Romero and Kolter 2011). Moreover, many organisms rely on bacteria as a partial or sole nutrient source. Thus, one might expect some bacteriovores to have evolved a means to access and consume bacteria protected by EPS matrices. Indeed, some non-sporulating amoebae and myxobacteria are known to exhibit anti-biofilm properties (Berleman et al. 2008; Jackson and Jones 1991; Matz et al. 2005; Weitere et al. 2005). Contrary to the commonly held notion that biofilms are protected against predation by protozoa, Acanthamoeba castellanii clearly has the capacity to graze on mixed biofilm communities and to become integrally associated with them, and the ciliate Colpoda maupasi can reduce biofilm thickness by up to 60% (Huws et al. 2005).
Studies on the feeding behavior of D. discoideum and its prey preferences (presented to the predator as lawns) indicate that these myxamoebae can feed on a variety of benign and pathogenic species of bacteria, both Gram negative and Gram positive (Depraitere and Darmon 1978; Raper and Smith 1939). In this study we examined four naturally occurring isolates (and the mutant Dd-AX3) of dictyostelid myxamoebae for their abilities to feed on biofilm enmeshed cells. Spores from all isolates were able to germinate in the presence of bacterial biofilms leading to reductions in the number of viable bacteria; although not all dictyostelids performed these functions with equal efficiency. Furthermore all examined dictyostelid strains appeared to undergo typical aggregation and multicellular behavior when feeding on biofilm enmeshed bacteria. Although bacterial prey deprivation is a key trigger for multicellular development, one of our observations was noteworthy. All examined dictyostelid strains entered the social stage despite having not exhausted the food supply in the biofilm as determined by macroscopic observation, light microscopy and quantitative analysis of bacterial cell number (Table 3, Fig. 4). Although we do not know what prevents the examined dictyostelid strains to consume bacteria to completion, other investigators reported that one-third of dictyostelids collected in wild habitats engage in husbandry of bacteria. Instead of consuming all bacteria, those “farmers” stop feeding early and incorporate bacteria into their fruiting bodies (Brock et al. 2011). It remains to be determined whether the five strains examined in this work are farmers or are not.
Neither the mechanisms used by myxamoebae to phagocytose biofilm-enmeshed bacteria nor the reasons for the apparent isolate specific variations were explored in this work. The chemical nature of EPS is diverse and varies in terms of carbohydrates, proteins, nucleic acids, lipids. EPS also varies in concentration and form (Branda et al. 2005; Flemming and Wingender 2010; More et al. 2014). Thus, it would not be surprising if the differences in the chemistry and scaffolding of the EPS matrices produced by the bacteria we examined (K. oxytoca (Tang et al. 2009) P. aeruginosa (Ude et al. 2006), P. syringae (Laue et al. 2006) and E. amylovora (Koczan et al. 2009; Nimtz et al. 1996) were critical in determining the susceptibility of biofilms to predation by dictyostelids. Our experiments on dictyostelid biofilm degradation investigate this notion by testing bacterial cell survival after dictyostelid feeding on one strain competent in biofilm formation (E. amylovora 1189) and another that is unable to form biofilms due to inability to produce amylovoran (E. amylovora 1189 Δams). We conclude that the lack of biofilm formation by E. amylovora 1189 Δams had a relatively small effect on its susceptibility to predation between dictyostelid predators. Specifically, biofilm formation provided a slightly protective effect against one dictyostelid strain (Dg-WS-142) but had minimal effect on susceptibility when a different predator was employed (Dd-AX3).
The underlying mechanisms responsible for the differences in feeding behaviors among dictyostelids remains to be determined. We suspect that genetic background differences between the dictyostelid strains examined may be key. One possible mechanism of dictyostelid feeding behavior diversity would be if dictyostelids utilized secreted products that facilitate the breakdown of biofilms with different chemical/structural components. One group of the candidate products consists of proteins that are known to be secreted by developing Dictyostelium cells in large numbers (Bakthavatsalam and Gomer 2010). Gene ontology analysis suggests that many of the 349 secreted proteins are involved in protein and carbohydrate metabolism, and proteolysis (Bakthavatsalam and Gomer 2010). Other variables that likely influence the predator/prey relationship include temperature and the shear forces of finger-like pseudopods myxamoebae utilize to capture bacterial prey. Further studies of Dictyostelid predatory activities upon EPS-enmeshed and planktonic bacteria will shed light on the ecological roles of this group of Amoebozoa and their astonishing success in the diverse terrestrial habitats they have been able to successfully inhabit.
Methods
Strains and plasmids
Dictyostelids from two genera (Dictyostelium and Polysphondylium) and bacterial strains used in this work are listed in Table 1. The mutant Dictyostelium discoideum strain Dd-AX-3 was included because it is a broadly studied model organism with the ability to be grown axenically (without bacteria) (Loomis 1971; Sussman and Sussman 1967). M5al is a strain of K. oxytoca, commonly used in our lab to propagate diverse dictyostelids. Strain KOF001, a fluorescent derivative of M5al, was generated using recombinant DNA from 3 parental plasmids and conjugation techniques. First, a DNA fragment from pFL129 (Wild et al. 2004) was isolated that contained a γ origin of replication and a marker for resistance to tetracycline (tet). This fragment was then ligated to a pC9 (McGhee and Jones 2000) DNA fragment, resulting in a dual origin plasmid, pFL299. Second, pFL299 was linearized, and then mixed with a partial digest of plasmid p67T1 (Singer et al. 2010). Ligated products were transformed into S17.1 and a clone bearing pFL300 was identified based on phenotype, molecular weight, and restriction mapping. Significant features of pFL300 include 1) the gene for d-Tomato red fluorescent protein (rfp), 2) the origins of replication (oriV) and conjugal transfer (oriT) from RSF1010 (Meyer 2009), 3) a second origin of replication (narrow host-range) from pC9 (derived from pEA29), and 4) the aforementioned tet-resistance marker. Following conjugation to the intrinsically ampicillin-resistant recipient M5al (via filter mating), KOF001 was selected for based on genetic markers conferring antibiotic (tetracycline and ampicillin) resistance.
18S rDNA sequencing
Methods were based on previous work (Romeralo et al. 2011a; Vadell et al. 2011). 18S rDNA was sequenced via the Sanger sequencing method (Sanger and Coulson 1975). Clustal W2 nucleotide alignments (Larkin et al. 2007) and BLAST (Boratyn et al. 2013) confirmed the taxonomic assignment of each species of dictyostelid used in this study.
Media, growth conditions
SM/2 (supplemented with 0.5% D-glucose) and a medium for the propagation of axenic D. discoideum have been described (Fey et al. 2007; Raper and Rahn 1984; Watts and Ashworth 1970). Final concentrations of antibiotics (when used) were as follows: 50 μg/ml ampicillin and 15 μg/ml tetracycline. Solid media contained agar at concentration of 1.5%, suspended in SM/2 or water (“water agar”). Monocultures of K. oxytoca and P. aeruginosa were grown at 37 °C; P. syringae, E. amylovora and dictyostelids were grown at 23 °C. Biofilms were formed on glass coverslips (Fisherbrand, 12-554A), and 25 mm diameter polycarbonate (PC) filters of 0.2 μm pore size. Black (Millipore, GTBPO1300) and white (GE, K02BP02500) microporous membrane filters were used interchangeably for qualitative and quantitative experiments and to aid in imaging. Our procedure for the revitalization of spore stocks made use of published observations and methods (Bonner 2006; Fey et al. 2007) (http://dictybase.org). Spores were harvested from mature sori under a dissecting microscope. The collected spores were suspended in SM/2 medium containing 20% DMSO and stored at −80°C. Spore germination efficiencies were determined using standard quantitative methods developed by K. Raper’s laboratory (Cavender and Raper 1965a, b; Cotter and Raper 1968).
Establishing biofilms on coverslips and polycarbonate membranes
The procedure for generating K. oxytoca biofilms on glass coverslips relied on a custom-built polypropylene platform that holds coverslips immersed in sterilized medium. Schematics of the platform are provided in Supplementary Material Figure S2; treatment and use of the platform are described in the figure legend. Coverslips (30 mm × 22 mm) were immersed in K. oxytoca grown in liquid culture with mixing. After 48 hours of incubation, biofilms consistently formed at the air-medium interface. Biofilm containing coverslips were rinsed three times by immersion in water followed by mild shaking to dislodge planktonic bacteria and then moved to a humidified Petri plate growth chamber (Supplementary Material Fig. S3) for microscopic imaging. For experiments using microporous PC filters as structural supports, our method for establishing biofilms was derived from previously published work (Anderl et al. 2000; Merritt et al. 2005; Singh et al. 2010), PC filters were laid atop solid growth medium (SM/2, LB or TSA) and 5 – 10 μl of overnight bacterial culture was spotted on the upper surface of the filter. Tryptic Soy broth (TSB) and LB broth media were also used where indicated. After establishing the biofilms, PC filters were transferred to SM/2 or water agar plates.
Time lapse microscopy of K. oxytoca biofilms
Dictyostelid myxamoebae were harvested from SM/2 agar plates after co-culture with M5al had produced “feeding fronts”, i.e., a predator/prey interface where advancing undifferentiated myxamoebae are present in high numbers (see Fig. 1). Using a sterilized microbiological loop, myxamoebae were scraped from the feeding front, transferred to 1 ml of sterile 0.9% sodium chloride and quantified using a hemocytometer (Fey et al. 2007). A coverslip biofilm was then transferred to water agar plate and inoculated with 2.5 × 104 myxamoebae (in 30 μl). The inoculated coverslip was placed in a humidified Petri plate growth chamber (Supplementary Material Fig. S3) and sealed with Parafilm (Bemis, PM992) to reduce biofilm drying. Samples were then transferred to the stage of a Celestron LCD microscope for time lapse imaging at room temperature.
To visualize phagocytic feeding on biofilms, images at 40× magnification (total) were captured at 13-minute intervals and concatenated using FrameByFrame 1.1 software (Philip Brendel ©2009; http://sourceforge.net/projects/framebyframe) to produce time-lapse .mov files (5 frames/second playback). Similarly prepared samples were also examined in high magnification studies (320×) using a light microscope (Leitz Labovert) equipped with an eyepiece camera (Celestron). Micrographs were taken at 30-second intervals for 8 hours and processed using FrameByFrame (.mov files, 5 frames/second).
Confocal Laser Scanning Microscopy
ImageJ software (imagej.nih.gov/ij/download/) was used with the Nikon ND2 reader plugin(http://rsbweb.nih.gov/ij/plugins/nd2-reader.html) to reassemble the files from NIS elements (Nikon, https://www.microscopyu.com) and produce .mov format videos. Time-lapse micrographs were as follows; 2 frames per second, 3 seconds per frame, 14 seconds overall with 90 seconds of real time video.
Scanning Electron Microscopy of static biofilms and Planktonic (“Liquid Culture” grown) specimens
Colonies grown upon PC filters (or concentrated planktonic bacteria) were fixed overnight at 4°C in 0.1 M phosphate buffer (pH 7.4) containing 1.5% glutaraldehyde and 1% tannic acid. Dehydration of the fixed samples proceeded through a series of 15-min. ethanol treatments using the following percentages: 30, 50, 70, 80, 90, 95, 100. A molecular sieve type 4A was adopted to trap all the remaining water in the last treatment with ethanol. After ethanol dehydration, a critical point drying process was applied. Individual samples were deposited into separate sample holders and placed within the chamber of a critical point dryer (Tousimis Samdri-780A) filled with type 4A molecular sieve dried ethanol. A cylinder containing liquid CO2 and an internal siphon tube was used to exchange ethanol with liquid CO2. Samples were soaked at least 4 times for 10 minutes during the process to ensure the exchange. Dried samples were placed onto double-sided, sticky, carbon film (12.7 mm) and adhered onto the aluminum SEM pin stub specimen mount (12.7mm). Specimens adhered onto the pin stubs were placed into an Auto Conductavac IV (See Vac Inc.) with a gold sputter target. The sputter coater consisted of a layer of gold (~5nm) to create a conductive layer at the specimen surface for electron microscopy imaging. Images were taken by using a Hitachi S-3200N SEM, with an accelerating voltage at 5–10kv.
Surveys of the destruction of biofilms grown on PC membranes
For macroscopic observations of predator/prey interactions, 103 bacterial cells (in 5 μl) were applied to the surface of a PC filter and incubated at 37 °C for 48 hours (P. aeruginosa and K. oxytoca) or at 25 °C for 48 hours (P. syringiae and E. amylovora) until the biofilm reached ~109 CFU. PC biofilm assemblages were transferred to water agar plates, or SM/2 agar plates. The biofilms were inoculated with 1 × 104 spores (in 10 μl overlay) per biofilm assemblage and Parafilm-wrapped plates were incubated for anywhere from 4 to 7 days at 23 °C with lids facing down. Planktonic (“Liquid Culture”) specimens were prepared as follows. 1 ml aliquots of late log liquid cultures (0.8 at OD650) grown in LB medium at 28 °C (E. amylovora) or in TSB medium at 28°C (P. syringae) or TSB at 37 °C (P. aeruginosa, and K. oxytoca) were centrifuged for 5 min. at 6,000 rpm, cell pellet was washed with 5 ml of 0.9% NaCl, the suspension was centrifuged for 5 min at 6,000 rpm, resuspended in 50μl 0.9%NaCl and transferred onto surface of PC filter resting on water agar. After 1 hour incubation (absorbtion of NaCl solution by agar), samples were fixed for SEM.
Quantitative biofilm disruption assays
PC filters with pre-grown biofilm (109 cells) were placed on SM2 agar and incubated at 23 °C. Biofilms were inoculated with bacteria-free dictyostelid spores (104). Following incubation for specific times membranes were transferred to 15-ml conical tubes (PC) containing 2 ml 0.9 (% w/v) sodium chloride. Capped tubes were vortexed vigorously and the dispersion of biofilm enmeshed cells to individual bacterial cells was monitored microscopically before samples were diluted and plated. All samples for each time point were normalized to the average CFU of the bacterial control (BC) biofilms at the start of the experiment, i.e., T=0. Again, data are expressed as a function of % survival, allowing comparisons to be made across independent experiments. Percent survival of bacterial cells in a given sample was calculated using the formula: DS/BC × 100 where DS is the dictyostelid-treated sample and BC is the bacteria-only control. For each dictyostelid strain, statistical analysis was performed as follows: Normalized sample data was averaged across all experiments (triplicate technical replicates, duplicate for experiments with E. amylovora in two separate experiments) and the standard error (SE) was calculated using the formula SE = S/√n where n is the number of observations of the sample (n=6) and S is sample standard deviation. Finally, a two sample t-test was performed to determine if the mean surviving CFU differed, significantly, in the treated samples vs. non-treated controls.
Supplementary Material
Acknowledgments
We thank Drs. Patricia McManus, Gary Roberts, George Sundin and Douglas Weibel for the bacterial strains. This work was supported by National Institutes of Health grant R21 AI096070, United States Department of Agriculture project WIS01412 and State Economic Engagement and Development Research Program 101-PRJ89MN awarded to MF laboratory.
Abbreviations
- BLAST
basic local alignment search tool
- CFU
colony forming units
- CLSM
confocal laser scanning microscopy
- DMSO
dimethyl sulfoxide
- NaCl
sodium chloride
- PC
Poly-Carbonate
- rDNA
DNA locus encoding ribosomal RNA
- SEM
scanning electron microscopy
- SM/2
slime mold medium [½ conc.]
- Dd
Dictyostelium discoideum
- Dg
Dictyostelium giganteum
- Da
Dictyostelium aureostipes
- Pp
Polysphondylium pallidum
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Monitoring Editor: Sandra L. Baldauf
Disclosure Statement
Dr. Marcin Filutowicz is required by the UW-Madison Conflict of Interest Committee to disclose a financial interest in AmebaGone, Inc., a Madison based firm that he founded.
References
- Anderl JN, Franklin MJ, Stewart PS. Role of antibiotic penetration limitation in Klebsiella pneumoniae biofilm resistance to ampicillin and ciprofloxacin. Antimicrob Agents Chemother. 2000;44:1818–1824. doi: 10.1128/aac.44.7.1818-1824.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakthavatsalam D, Gomer RH. The secreted proteome profile of developing Dictyostelium discoideum cells. Proteomics. 2010;10:2556–2559. doi: 10.1002/pmic.200900516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banin E, Vasil ML, Greenberg EP. Iron and Pseudomonas aeruginosa biofilm formation. Proc Natl Acad Sci USA. 2005;102:11076–11081. doi: 10.1073/pnas.0504266102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao G, Zhang Y, Du C, Chen Z, Li Y, Cao Z, Ma Y. Genome Sequence of Klebsiella oxytoca M5al, a Promising Strain for Nitrogen Fixation and Chemical Production. Genome Announc. 2013;1 doi: 10.1128/genomeA.00074-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellemann P, Geider K. Localization of transposon insertions in pathogenicity mutants of Erwinia amylovora and their biochemical characterization. J Gen Microbiol. 1992;138:931–940. doi: 10.1099/00221287-138-5-931. [DOI] [PubMed] [Google Scholar]
- Berleman JE, Scott J, Chumley T, Kirby JR. Predataxis behavior in Myxococcus xanthus. Proc Natl Acad Sci USA. 2008;105:17127–17132. doi: 10.1073/pnas.0804387105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billing E. The effect of temperature on the growth of the fireblight pathogen, Erwinia amylovora. J Appl Bacteriol. 1974;37:643–648. doi: 10.1111/j.1365-2672.1974.tb00488.x. [DOI] [PubMed] [Google Scholar]
- Bjarnsholt T, Jensen PO, Burmolle M, Hentzer M, Haagensen JA, Hougen HP, Calum H, Madsen KG, Moser C, Molin S, Hoiby N, Givskov M. Pseudomonas aeruginosa tolerance to tobramycin, hydrogen peroxide and polymorphonuclear leukocytes is quorum-sensing dependent. Microbiology. 2005;151:373–383. doi: 10.1099/mic.0.27463-0. [DOI] [PubMed] [Google Scholar]
- Bonner JT. Migration in Dictyostelium polycephalum. Mycologia. 2006;98:260–264. doi: 10.3852/mycologia.98.2.260. [DOI] [PubMed] [Google Scholar]
- Boratyn GM, Camacho C, Cooper PS, Coulouris G, Fong A, Ma N, Madden TL, Matten WT, McGinnis SD, Merezhuk Y, Raytselis Y, Sayers EW, Tao T, Ye J, Zaretskaya I. BLAST: a more efficient report with usability improvements. Nucleic Acids Res. 2013;41:W29–W33. doi: 10.1093/nar/gkt282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branda SS, Vik S, Friedman L, Kolter R. Biofilms: the matrix revisited. Trends Microbiol. 2005;13:20–26. doi: 10.1016/j.tim.2004.11.006. [DOI] [PubMed] [Google Scholar]
- Brefeld O. Dictyostelium mucoroides. Ein neuer Organismus aus der und der Verwandtschaft der Myxomyceten. Abh Seckenberg Naturforsch Ges. 1869;7:85–107. [Google Scholar]
- Brock DA, Douglas TE, Queller DC, Strassmann JE. Primitive agriculture in a social amoeba. Nature. 2011;469:393–396. doi: 10.1038/nature09668. [DOI] [PubMed] [Google Scholar]
- Brock TD. Life at high temperatures. Evolutionary, ecological, and biochemical significance of organisms living in hot springs is discussed. Science. 1967;158:1012–1019. doi: 10.1126/science.158.3804.1012. [DOI] [PubMed] [Google Scholar]
- Burmolle M, Thomsen TR, Fazli M, Dige I, Christensen L, Homoe P, Tvede M, Nyvad B, Tolker-Nielsen T, Givskov M, Moser C, Kirketerp-Moller K, Johansen HK, Hoiby N, Jensen PO, Sorensen SJ, Bjarnsholt T. Biofilms in chronic infections - a matter of opportunity - monospecies biofilms in multispecies infections. FEMS Pathog Dis. 2010;59:324–336. doi: 10.1111/j.1574-695X.2010.00714.x. [DOI] [PubMed] [Google Scholar]
- Burse A, Weingart H, Ullrich MS. NorM, an Erwinia amylovora multidrug efflux pump involved in in vitro competition with other epiphytic bacteria. Appl Environ Microbiol. 2004;70:693–703. doi: 10.1128/AEM.70.2.693-703.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carilla-Latorre S, Calvo-Garrido J, Bloomfield G, Skelton J, Kay RR, Ivens A, Martinez JL, Escalante R. Dictyostelium transcriptional responses to Pseudomonas aeruginosa: common and specific effects from PAO1 and PA14 strains. BMC Microbiol. 2008;8:109. doi: 10.1186/1471-2180-8-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavender JC. Geographical distribution of Acrasieae. Mycologia. 1973;65:1044–1054. [Google Scholar]
- Cavender JC. A Global Overview of Dictyostelid Ecology with Special Emphasis in North American Forests. In: Romeralo M, Baldauf S, Escalante R, editors. Dictyostelids Evolution, Genomics and Cell Biology. Springer; Dordrecht: 2013. pp. 149–166. [Google Scholar]
- Cavender JC, Raper KB. The Acrasieae in nature. I. Isolation. Am J Bot. 1965a;52:294–296. [PubMed] [Google Scholar]
- Cavender JC, Raper KB. The Acrasieae in nature. II. Forest soil as a primary habitat. Am J Bot. 1965b;52:297–302. [PubMed] [Google Scholar]
- Cavender JC, Stephenson SL, Landolt JC, Vadell EM. Dictyostelid cellular slime moulds in the forests of New Zealand. New Zeal J Bot. 2002;40:235–264. [Google Scholar]
- Cohen A, Ceccarin C. Inhibition of spore germination in cellular slime moulds. Ann Bot. 1967;31:479–487. [Google Scholar]
- Costerton JW, Geesey GG, Cheng KJ. How bacteria stick. Sci Am. 1978;238:86–95. doi: 10.1038/scientificamerican0178-86. [DOI] [PubMed] [Google Scholar]
- Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- Cotter DA, Raper KB. Properties of germinating spores of Dictyostelium discoideum. J Bacteriol. 1968;96:1680–1689. doi: 10.1128/jb.96.5.1680-1689.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotter DA, Sands TW, Virdy KJ, North MJ, Klein G, Satre M. Patterning of development in Dictyostelium discoideum: factors regulating growth, differentiation, spore dormancy, and germination. Biochem Cell Biol. 1992;70:892–919. doi: 10.1139/o92-137. [DOI] [PubMed] [Google Scholar]
- Cotter DA, Mahadeo DC, Cervi DN, Kishi Y, Gale K, Sands T, Sameshima M. Environmental regulation of pathways controlling sporulation, dormancy and germination utilizes bacterial-like signaling complexes in Dictyostelium discoideum. Protist. 2000;151:111–126. doi: 10.1078/1434-4610-00012. [DOI] [PubMed] [Google Scholar]
- de Beer D, Stoodley P, Roe F, Lewandowski Z. Effects of biofilm structures on oxygen distribution and mass transport. Biotechnol Bioeng. 1994;43:1131–1138. doi: 10.1002/bit.260431118. [DOI] [PubMed] [Google Scholar]
- Depraitere C, Darmon M. Growth of “Dictyostelium discoideum” on different species of bacteria (author’s transl) Ann Microbiol (Paris) 1978;129 B:451–461. [PubMed] [Google Scholar]
- Diggle SP, Stacey RE, Dodd C, Camara M, Williams P, Winzer K. The galactophilic lectin, LecA, contributes to biofilm development in Pseudomonas aeruginosa. Environ Microbiol. 2006;8:1095–1104. doi: 10.1111/j.1462-2920.2006.001001.x. [DOI] [PubMed] [Google Scholar]
- Donlan RM. Biofilms: microbial life on surfaces. Emerg Infect Dis. 2002;8:881–890. doi: 10.3201/eid0809.020063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donlan RM, Costerton JW. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin Microbiol Rev. 2002;15:167–193. doi: 10.1128/CMR.15.2.167-193.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg RM, Hurd LE, Ketcham RB. The cellular slime-mold guild and Its bacterial prey - growth-rate variation at the interspecific and intraspecific levels. Oecologia. 1989;79:458–462. doi: 10.1007/BF00378661. [DOI] [PubMed] [Google Scholar]
- Fey P, Kowal AS, Gaudet P, Pilcher KE, Chisholm RL. Protocols for growth and development of Dictyostelium discoideum. Nat Protoc. 2007;2:1307–1316. doi: 10.1038/nprot.2007.178. [DOI] [PubMed] [Google Scholar]
- Flemming HC, Wingender J. The biofilm matrix. Nat Rev Microbiol. 2010;8:623–633. doi: 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- Fux CA, Costerton JW, Stewart PS, Stoodley P. Survival strategies of infectious biofilms. Trends Microbiol. 2005;13:34–40. doi: 10.1016/j.tim.2004.11.010. [DOI] [PubMed] [Google Scholar]
- Fux CA, Stoodley P, Hall-Stoodley L, Costerton JW. Bacterial biofilms: A diagnostic and therapeutic challenge. Expert Rev Anti-infect Ther. 2003;1:667–683. doi: 10.1586/14787210.1.4.667. [DOI] [PubMed] [Google Scholar]
- Hall-Stoodley L, Costerton JW, Stoodley P. Bacterial biofilms: from the natural environment to infectious diseases. Nature Rev Microbiol. 2004;2:95–108. doi: 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- Hamilton, Wilson . Nitrogen Fixation in Aerobacter aerogenes. In: Toivonen NJ, editor. Biochemistry of Nitrogen A Collection of Papers on Biochemistry of Nitrogen and Related Subjects, Dedicated to Artturi Ilmari Virtanen on the Occasion of his 60th Birthday, Jan 15 1955. Suomalainen Tiedeakatemia; Helsinki: 1955. p. 535. [Google Scholar]
- Heidel AJ, Lawal HM, Felder M, Schilde C, Helps NR, Tunggal B, Rivero F, John U, Schleicher M, Eichinger L, Platzer M, Noegel AA, Schaap P, Glöckner G. Phylogeny-wide analysis of social amoeba genomes highlights ancient origins for complex intercellular communication. Genome Res. 2011;21:1882–1891. doi: 10.1101/gr.121137.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrici AT. Studies of freshwater bacteria: I. A direct microscopic technique. J Bacteriol. 1933;25:277–287. doi: 10.1128/jb.25.3.277-287.1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn E. Food competition among the cellular slime molds (Acrasieae) Ecology. 1971;52:475–484. [Google Scholar]
- Huws SA, McBain AJ, Gilbert P. Protozoan grazing and its impact upon population dynamics in biofilm communities. J Appl Microbiol. 2005;98:238–244. doi: 10.1111/j.1365-2672.2004.02449.x. [DOI] [PubMed] [Google Scholar]
- Jackson SM, Jones EBG. Interactions within biofilms: disruption of biofilm structure by protozoa. Kieler Meeresforsch. 1991;8:264–268. [Google Scholar]
- Jesaitis AJ, Franklin MJ, Berglund D, Sasaki M, Lord CI, Bleazard JB, Duffy JE, Beyenal H, Lewandowski Z. Compromised host defense on Pseudomonas aeruginosa biofilms: characterization of neutrophil and biofilm interactions. J Immunol. 2003;171:4329–4339. doi: 10.4049/jimmunol.171.8.4329. [DOI] [PubMed] [Google Scholar]
- Kaplan JB. Biofilm dispersal: mechanisms, clinical implications, and potential therapeutic uses. J Dent Res. 2010;89:205–218. doi: 10.1177/0022034509359403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketcham RB, Eisenberg RM. Clonal diversity in populations of Polysphondylium pallidum, a cellular slime mold. Ecology. 1989;70:1425–1433. [Google Scholar]
- Ketcham RB, Don RL, Shenk MA, Eisenberg RM. Do interactions of cellular slime mold species regulate their densities in soil? Ecology. 1988;69:193–199. [Google Scholar]
- Koczan JM, Lenneman BR, McGrath MJ, Sundin GW. Cell surface attachment structures contribute to biofilm formation and xylem colonization by Erwinia amylovora. Appl Environ Microbiol. 2011;77:7031–7039. doi: 10.1128/AEM.05138-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koczan JM, McGrath MJ, Zhao Y, Sundin GW. Contribution of Erwinia amylovora exopolysaccharides amylovoran and levan to biofilm formation: implications in pathogenicity. Phytopathology. 2009;99:1237–1244. doi: 10.1094/PHYTO-99-11-1237. [DOI] [PubMed] [Google Scholar]
- Kolodkin-Gal I, Romero D, Cao S, Clardy J, Kolter R, Losick R. D-amino acids trigger biofilm disassembly. Science. 2010;328:627–629. doi: 10.1126/science.1188628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konijn TM, Van De Meene JG, Bonner JT, Barkley DS. The acrasin activity of adenosine-3′,5′-cyclic phosphate. Proc Natl Acad Sci USA. 1967;58:1152–1154. doi: 10.1073/pnas.58.3.1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar CG, Anand SK. Significance of microbial biofilms in food industry: a review. Int J Food Microbiol. 1998;42:9–27. doi: 10.1016/s0168-1605(98)00060-9. [DOI] [PubMed] [Google Scholar]
- Landolt JC, Cavender JC, Stephenson SL, Vadell EM. New species of dictyostelid cellular slime moulds from Australia. Aust Syst Bot. 2008;21:50–66. [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Laue H, Schenk A, Li H, Lambertsen L, Neu TR, Molin S, Ullrich MS. Contribution of alginate and levan production to biofilm formation by Pseudomonas syringae. Microbiology. 2006;152:2909–2918. doi: 10.1099/mic.0.28875-0. [DOI] [PubMed] [Google Scholar]
- Lawrence JR, Neu TR. Confocal laser scanning microscopy for analysis of microbial biofilms. Methods Enzymol. 1999;310:131–144. doi: 10.1016/s0076-6879(99)10011-9. [DOI] [PubMed] [Google Scholar]
- Lee J, Jayaraman A, Wood TK. Indole is an inter-species biofilm signal mediated by SdiA. BMC Microbiol. 2007;7:42. doi: 10.1186/1471-2180-7-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loomis WF., Jr Sensitivity of Dictyostelium discoideum to nucleic acid analogues. Exp Cell Res. 1971;64:484–486. doi: 10.1016/0014-4827(71)90107-8. [DOI] [PubMed] [Google Scholar]
- Lyczak JB, Cannon CL, Pier GB. Establishment of Pseudomonas aeruginosa infection: lessons from a versatile opportunist. Microbes Infect. 2000;2:1051–1060. doi: 10.1016/s1286-4579(00)01259-4. [DOI] [PubMed] [Google Scholar]
- Mann EE, Wozniak DJ. Pseudomonas biofilm matrix composition and niche biology. FEMS Microbiol Rev. 2012;36:893–916. doi: 10.1111/j.1574-6976.2011.00322.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matz C, McDougald D, Moreno AM, Yung PY, Yildiz FH, Kjelleberg S. Biofilm formation and phenotypic variation enhance predation-driven persistence of Vibrio cholerae. Proc Nat Acad Sci of USA. 2005;102:16819–16824. doi: 10.1073/pnas.0505350102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matz C, Moreno AM, Alhede M, Manefield M, Hauser AR, Givskov M, Kjelleberg S. Pseudomonas aeruginosa uses type III secretion system to kill biofilm-associated amoebae. ISME J. 2008;2:843–852. doi: 10.1038/ismej.2008.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGhee GC, Jones AL. Complete nucleotide sequence of ubiquitous plasmid pEA29 from Erwinia amylovora strain Ea88: gene organization and intraspecies variation. Appl Environ Microbiol. 2000;66:4897–4907. doi: 10.1128/aem.66.11.4897-4907.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McManus PS, Jones AL. Epidemiology and genetic analysis of streptomycin-resistant Erwinia amylovora from Michigan and evaluation of oxytetracycline for control. Phytopathology. 1994;84:627–633. [Google Scholar]
- Merritt JH, Kadouri DE, O’Toole GA. Growing and analyzing static biofilms. Curr Protoc Microbiol. 2005;22:1B.1.1–1B.1.18. doi: 10.1002/9780471729259.mc01b01s00. B:1B.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer R. Replication and conjugative mobilization of broad host-range IncQ plasmids. Plasmid. 2009;62:57–70. doi: 10.1016/j.plasmid.2009.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittal R, Sharma S, Chhibber S, Harjai K. Effect of macrophage secretory products on elaboration of virulence factors by planktonic and biofilm cells of Pseudomonas aeruginosa. Comp Immunol Microbiol Infect Dis. 2006;29:12–26. doi: 10.1016/j.cimid.2005.11.002. [DOI] [PubMed] [Google Scholar]
- More TT, Yadav JS, Yan S, Tyagi RD, Surampalli RY. Extracellular polymeric substances of bacteria and their potential environmental applications. J Environ Manag. 2014;144:1–25. doi: 10.1016/j.jenvman.2014.05.010. [DOI] [PubMed] [Google Scholar]
- Nasser W, Santhanam B, Miranda ER, Parikh A, Juneja K, Rot G, Dinh C, Chen R, Zupan B, Shaulsky G, Kuspa A. Bacterial discrimination by dictyostelid amoebae reveals the complexity of ancient interspecies interactions. Curr Biol. 2013;23:862–872. doi: 10.1016/j.cub.2013.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimtz M, Mort A, Domke T, Wray V, Zhang Y, Qiu F, Coplin D, Geider K. Structure of amylovoran, the capsular exopolysaccharide from the fire blight pathogen Erwinia amylovora. Carbohydr Res. 1996;287:59–76. doi: 10.1016/0008-6215(96)00070-5. [DOI] [PubMed] [Google Scholar]
- Nomura TTY, Abe H, Uchiyama M. Cytokinin activity of discadenine: A spore germination inhibitor of Dictyostelium discoideum. Phytochemistry. 1977;16:1819–1820. [Google Scholar]
- Parsek MR, Greenberg EP. Sociomicrobiology: the connections between quorum sensing and biofilms. Trends Microbiol. 2005;13:27–33. doi: 10.1016/j.tim.2004.11.007. [DOI] [PubMed] [Google Scholar]
- Pedersen K. Factors regulating microbial biofilm development in a system with slowly flowing seawater. Applied Environ Microbiol. 1982;44:1196–1204. doi: 10.1128/aem.44.5.1196-1204.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pengra RM, Wilson PW. Physiology of nitrogen fixation by Aerobacter aerogenes. J Bacteriol. 1958;75:21–25. doi: 10.1128/jb.75.1.21-25.1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raper KB. Dictyostelium discoideum, a new species of slime mold from decaying forest leaves. J Agr Res. 1935;50:135–147. [Google Scholar]
- Raper KB. Growth and development of Dictyostelium discoideum with bacterial associates. J Agr Res. 1937;55:289–316. [Google Scholar]
- Raper KB. Developmental patterns in simple slime molds. Growth. 1941;5:41–76. [Google Scholar]
- Raper KB, Rahn AW. The Dictyostelids. Princeton University Press; Princeton, NJ: 1984. [Google Scholar]
- Raper KB, Smith NR. The growth of Dictyostelium discoideum upon pathogenic bacteria. J Bacteriol. 1939;38:431–445. doi: 10.1128/jb.38.4.431-445.1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards JJ, Melander C. Controlling bacterial biofilms. Chembiochem: a Eur J Chem Biol. 2009;10:2287–2294. doi: 10.1002/cbic.200900317. [DOI] [PubMed] [Google Scholar]
- Rodríguez S, Bishop P. Three-dimensional quantification of soil biofilms using image analysis. Environ Eng Sci. 2007;24:96–103. [Google Scholar]
- Romeralo M, Moya-Larano J, Lado C. Social amoebae: environmental factors influencing their distribution and diversity across south-western. Eur Microb Ecol. 2011b;61:154–165. doi: 10.1007/s00248-010-9715-5. [DOI] [PubMed] [Google Scholar]
- Romeralo M, Cavender JC, Landolt JC, Stephenson SL, Baldauf SL. An expanded phylogeny of social amoebas (Dictyostelia) shows increasing diversity and new morphological patterns. BMC Evol Biol. 2011a;11:84. doi: 10.1186/1471-2148-11-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romeralo M, Landolt JC, Cavender JC, Laursen GA, Baldauf SL. Two new species of dictyostelid cellular slime molds from Alaska. Mycologia. 2010;102:588–595. doi: 10.3852/09-107. [DOI] [PubMed] [Google Scholar]
- Romero D, Kolter R. Will biofilm disassembly agents make it to market? Trends Microbiol. 2011;19:304–306. doi: 10.1016/j.tim.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F, Coulson AR. A rapid method for determining sequences in DNA by primed synthesis with DNA polymerase. J Mol Biol. 1975;94:441–448. doi: 10.1016/0022-2836(75)90213-2. [DOI] [PubMed] [Google Scholar]
- Schaap P. Evolutionary crossroads in developmental biology: Dictyostelium discoideum. Development. 2011;138:387–396. doi: 10.1242/dev.048934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaap P, Winckler T, Nelson M, Alvarez-Curto E, Elgie B, Hagiwara H, Cavender J, Milano-Curto A, Rozen DE, Dingermann T, Mutzel R, Baldauf SL. Molecular phylogeny and evolution of morphology in the social amoebas. Science. 2006;314:661–663. doi: 10.1126/science.1130670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon R, Priefer U, Puhler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- Singer JT, Phennicie RT, Sullivan MJ, Porter LA, Shaffer VJ, Kim CH. Broad-host-range plasmids for red fluorescent protein labeling of gram-negative bacteria for use in the zebrafish model system. Appl Environ Microbiol. 2010;76:3467–3474. doi: 10.1128/AEM.01679-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R, Ray P, Das A, Sharma M. Penetration of antibiotics through Staphylococcus aureus and Staphylococcus epidermidis biofilms. Journal of Antimicrob Chemother. 2010;65:1955–1958. doi: 10.1093/jac/dkq257. [DOI] [PubMed] [Google Scholar]
- Singla S, Harjai K, Chhibber S. Artificial Klebsiella pneumoniae biofilm model mimicking in vivo system: altered morphological characteristics and antibiotic resistance. J Antibiot (Tokyo) 2014;67:305–309. doi: 10.1038/ja.2013.139. [DOI] [PubMed] [Google Scholar]
- Stewart PS. Diffusion in biofilms. J Bacteriol. 2003;185:1485–1491. doi: 10.1128/JB.185.5.1485-1491.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussman R, Sussman M. Cultivation of Dictyostelium discoideum in axenic medium. Biochem Biophys Res Commun. 1967;29:53–55. doi: 10.1016/0006-291x(67)90539-6. [DOI] [PubMed] [Google Scholar]
- Swanson A, Spiegel F, Cavender J. Taxonomy, slime molds, and the questions we ask. Mycologia. 2002;94:968–969. [PubMed] [Google Scholar]
- Swanson A, Vadell E, Cavender J. Global distribution of forest soil dictyostelids. J Biogeogr. 2001;26:133–148. [Google Scholar]
- Tang X, Flint SH, Bennett RJ, Brooks JD, Morton RH. Biofilm growth of individual and dual strains of Klebsiella oxytoca from the dairy industry on ultrafiltration membranes. J Ind Microbiol Biotechnol. 2009;36:1491–1497. doi: 10.1007/s10295-009-0637-5. [DOI] [PubMed] [Google Scholar]
- Thurlow LR, Hanke ML, Fritz T, Angle A, Aldrich A, Williams SH, Engebretsen IL, Bayles KW, Horswill AR, Kielian T. Staphylococcus aureus biofilms prevent macrophage phagocytosis and attenuate inflammation in vivo. J Immunol. 2011;186:6585–6596. doi: 10.4049/jimmunol.1002794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ude S, Arnold DL, Moon CD, Timms-Wilson T, Spiers AJ. Biofilm formation and cellulose expression among diverse environmental Pseudomonas isolates. Environ Microbiol. 2006;8:1997–2011. doi: 10.1111/j.1462-2920.2006.01080.x. [DOI] [PubMed] [Google Scholar]
- Vadell EM, Cavender JC, Romeralo M, Edwards SM, Stephenson SL, Baldauf SL. New species of dictyostelids from Patagonia and Tierra del Fuego, Argentina. Mycologia. 2011;103:101–117. doi: 10.3852/09-301. [DOI] [PubMed] [Google Scholar]
- van Es S, Virdy KJ, Pitt GS, Meima M, Sands TW, Devreotes PN, Cotter DA, Schaap P. Adenylyl cyclase G, an osmosensor controlling germination of Dictyostelium spores. J Biol Chem. 1996;271:23623–23625. doi: 10.1074/jbc.271.39.23623. [DOI] [PubMed] [Google Scholar]
- Vanneste JL. Erwinia amylovora. CABI Pub.; Wallingford, Oxon, UK; New York, NY: 2000. Fire Blight: the Disease and its Causative Agent. [Google Scholar]
- Walker TS, Tomlin KL, Worthen GS, Poch KR, Lieber JG, Saavedra MT, Fessler MB, Malcolm KC, Vasil ML, Nick JA. Enhanced Pseudomonas aeruginosa biofilm development mediated by human neutrophils. Infect Immun. 2005;73:3693–3701. doi: 10.1128/IAI.73.6.3693-3701.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters MC, 3rd, Roe F, Bugnicourt A, Franklin MJ, Stewart PS. Contributions of antibiotic penetration, oxygen limitation, and low metabolic activity to tolerance of Pseudomonas aeruginosa biofilms to ciprofloxacin and tobramycin. Antimicrob Agents Chemother. 2003;47:317–323. doi: 10.1128/AAC.47.1.317-323.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts DJ, Ashworth JM. Growth of myxameobae of the cellular slime mould Dictyostelium discoideum in axenic culture. Biochem J. 1970;119:171–174. doi: 10.1042/bj1190171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber AT, Raper KB. Induction of fruiting in two aggregateless mutants of Dictyostelium discoideum. Dev Biol. 1971;26:606–615. doi: 10.1016/0012-1606(71)90143-6. [DOI] [PubMed] [Google Scholar]
- Weitere M, Bergfeld T, Rice SA, Matz C, Kjelleberg S. Grazing resistance of Pseudomonas aeruginosa biofilms depends on type of protective mechanism, developmental stage and protozoan feeding mode. Environ Microbiol. 2005;7:1593–1601. doi: 10.1111/j.1462-2920.2005.00851.x. [DOI] [PubMed] [Google Scholar]
- Wild J, Czyz A, Rakowski SA, Filutowicz M. γ origin plasmids of R6K lineage replicate in diverse genera of Gram-negative bacteria. Annals Microbiol. 2004;54:471–480. [Google Scholar]
- Yang L, Liu Y, Markussen T, Hoiby N, Tolker-Nielsen T, Molin S. Pattern differentiation in co-culture biofilms formed by Staphylococcus aureus and Pseudomonas aeruginosa. FEMS Immunol Med Microbiol. 2011;62:339–347. doi: 10.1111/j.1574-695X.2011.00820.x. [DOI] [PubMed] [Google Scholar]
- Yang L, Hengzhuang W, Wu H, Damkiaer S, Jochumsen N, Song Z, Givskov M, Hoiby N, Molin S. Polysaccharides serve as scaffold of biofilms formed by mucoid Pseudomonas aeruginosa. FEMS Immunol Med Microbiol. 2012;65:366–376. doi: 10.1111/j.1574-695X.2012.00936.x. [DOI] [PubMed] [Google Scholar]
- Yoch DC, Pengra RM. Effect of amino acids on the nitrogenase system of Klebsiella pneumoniae. J Bacteriol. 1966;92:618–622. doi: 10.1128/jb.92.3.618-622.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Sundin GW, Wang D. Construction and analysis of pathogenicity island deletion mutants of Erwinia amylovora. Can J Microbiol. 2009;55:457–464. doi: 10.1139/w08-147. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


