Abstract
Reflux esophagitis damages the squamous epithelium that normally lines the esophagus, and promotes replacement of the damaged squamous lining by the intestinal metaplasia of Barrett’s esophagus, the precursor of esophageal adenocarcinoma. Thus, to prevent the development of Barrett’s metaplasia and esophageal adenocarcinoma, the pathogenesis of reflux esophagitis must be understood. We have reported that reflux esophagitis, both in a rat model and in humans, develops as a cytokine-mediated inflammatory injury (i.e. cytokine sizzle), not as a caustic chemical injury (i.e. acid burn), as traditionally has been assumed. Moreover, reflux induces activation of hypoxia inducible factor (HIF)-2α which enhances the transcriptional activity of nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) causing increases in pro-inflammatory cytokines and in migration of T lymphocytes, an underlying molecular mechanism for this cytokine-mediated injury. In some individuals, reflux esophagitis heals with Barrett’s metaplasia. A number of possibilities exist for the origin of the progenitor cells that give rise to this intestinal metaplasia including those of the esophagus, the proximal stomach or the bone marrow. However, intestinal cells are not normally found in the esophagus, the stomach, or the bone marrow. Thus, the development of Barrett’s intestinal metaplasia must involve some molecular reprogramming of key developmental transcription factors within the progenitor cell, a process termed transcommitment, which may be initiated by the noxious components of the gastric refluxate. This review will highlight recent studies on the pathogenesis of reflux esophagitis and on reflux-related molecular reprogramming of esophageal squamous epithelial cells in the pathogenesis of Barrett’s metaplasia.
Keywords: Barrett’s esophagus, cytokine, NF-κB, CDX2, squamous cells
Introduction
Gastroesophageal reflux disease (GERD) is widely regarded as the main cause of esophageal inflammation because the reflux of acid, bile salts, and other noxious agents contained in refluxed gastric juice result in reflux esophagitis [1]. Complications of reflux esophagitis include esophageal ulceration, stricture formation, and the development of Barrett’s esophagus, a condition which predisposes to esophageal adenocarcinoma [2]. In the United States, GERD is extremely common with over 20% of adult Americans have heartburn and/or regurgitation at least once per week. In Japan, the prevalence of GERD symptoms has been increasing over the past two decades [3]. During the 1990s, 10.3% of Japanese patients being seen for routine follow-up had GERD symptoms whereas the rate of these symptoms increased to 18.9% during 2000–2010 [3]. Barrett’s esophagus is one of the serious complication of reflux esophagus because of its increased risk of progression to esophageal adenocarcinoma. In the United States, approximately 5.6% of adults have long-segment (≥ 3 cm of columnar mucosa with goblet cells) Barrett’s esophagus and 10–15% have shorter segments (< 3 cm) of disease [2,4]. In addition, the frequency of esophageal adenocarcinoma has increased by more than 7 fold in the past four decades in the United States [2]. Within Japan, estimates on the prevalence of short-segment Barrett’s esophagus rage from 1.2– 59% and for long-segment from 0.2–1.4% [3]. Although Barrett’s esophagus appears to be increasing in Japan, esophageal adenocarcinoma still accounts for <5% of esophageal cancer cases in this country [3]. In order to make advances into preventing Barrett’s esophagus and the other serious esophageal complications of reflux esophagitis, a better understanding of the pathogenesis of reflux esophagitis and its contribution to the development of Barrett’s esophagus is essential. This review will focus on current research concepts supported by recently published key studies on the development of reflux esophagitis and Barrett’s esophagus.
Pathogenesis of Reflux Esophagitis: Acid Burn or Cytokine Sizzle
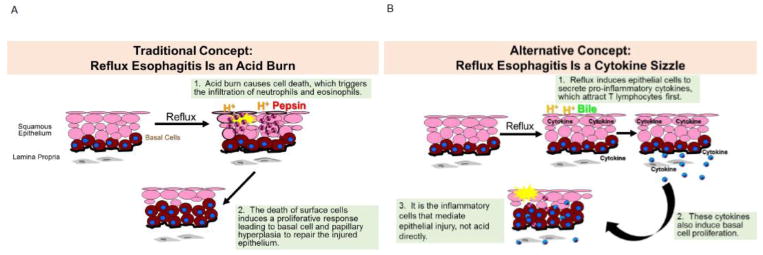
In 1935, gastroenterologist Asher Winkelstein reported in the Journal of the American Medical Association, patients who had heartburn and other esophageal symptoms associated with endoscopic and histologic signs of inflammation in the distal esophagus. Winkelstein went on to propose that these patients had “peptic esophagitis…resulting from the irritant action on the mucosa of free hydrochloric acid and pepsin.” For more than 80 years, this traditional concept that reflux esophagitis results from an acid-peptic “burn” has been a widely held belief which has for the most part gone unchallenged. In this model, reflux esophagitis is thought to start when refluxed acid and pepsin damage the proteins of the junctional complexes that bind cells together to make the epithelium impermeable to water, hydrogen ions, and other solutes [5]. However, when these junctional proteins are damaged, the epithelium becomes permeable, and allows acid to enter and attack the epithelial cells. This acid burn causes the death of surface epithelial cells, which triggers the infiltration of neutrophils and eosinophils and induces proliferation of esophageal basal cells, efforts that aid in repairing the injured epithelium (Figure 1A) [6].
Figure 1.
Concepts on the Pathogenesis of Reflux Esophagitis. (A) The traditional concept has been that reflux esophagitis results from a caustic (acid) burn. When esophageal squamous epithelium is exposed to reflux, acid and pepsin are thought to damage the junctions between the cells, making the epithelium permeable and allowing acid to seep into the epithelium and injure the epithelial cells. This acid burn causes cell death, which triggers the infiltration of neutrophils and eosinophils into the epithelium. The death of surface cells is also assumed to induce a proliferative response leading to basal cell and papillary hyperplasia to repair the injured epithelium. (B) The alternative concept that we propose is that reflux esophagitis develops as a cytokine-mediated inflammatory injury (i.e. cytokine sizzle). In this model, the reflux of acid and bile salts doesn’t destroy epithelial cells directly, but rather induces them to secrete pro-inflammatory cytokines, which attract T lymphocytes first. These cytokines also induce basal cell proliferation. Ultimately, it is inflammatory cells that mediate the epithelial injury, not the direct acid burn.
In 2008, our group began using a rat model in which reflux esophagitis was induced by creating a surgical esophagoduodenostomy. We noted that erosive esophagitis took weeks to develop after the surgical induction of reflux in our animal model. Esophageal injury due to an acid burn should develop rapidly, and we were puzzled by the long delay between the onset of reflux and the appearance of esophagitis. Using this animal model, we studied the histologic events of reflux esophagitis beginning at post-operative day 3 and 7 and then every week after out to post-operative week 8 with comparison to sham-operated control animals [7]. On postoperative day 3, we found esophageal inflammation most prominent in the submucosa, an esophageal mucosa that was intact, and the inflammatory cell infiltrate in the submucosa was exclusively lymphocytes [7]. By post-operative week 1, the lymphocytic-predominant inflammation reached the lamina propria and by post-operative week 3, the epithelial layer was inflamed [7]. Using immunostaining for CD3, a T cell marker and CD20, a B cell marker, we found that the infiltrating lymphocytes were CD3+ and CD20−, demonstrating that they were T lymphocytes [7]. Basal cell proliferation (i.e. hyperplasia) began at post-operative week 1 and peaked in degree by week 4, but it wasn’t until week 4 that we began to see death of surface epithelial cells (i.e. erosions) [7]. In contrast to the acid burn model, we found in this animal model that inflammation did not start in the mucosa, and the first inflammatory cells were lymphocytes, not neutrophils or eosinophils. We also found that basal cell hyperplasia occurred while the surface epithelial cells were still intact so this hyperplasia was not due to the death of surface cells [7].
Since our initial observation was the infiltration of a lymphocytic infiltrate, we postulated that the reflux of acid and bile induces esophageal epithelial cells to secrete pro-inflammatory cytokines. Using cultures of esophageal squamous cells derived from patients with GERD, we found that cells secreted interleukin (IL)-8 and IL-1β, potent pro-inflammatory cytokines, when they were exposed to acidic bile salts, and that secretion of IL-8 induced the migration of lymphocytes and neutrophils [7]. Expression of IL-8 by reflux-stimulated esophageal squamous cells was also observed in our animal model in vivo [7]. Based on these findings, we proposed an alternative concept for the pathogenesis of reflux esophagitis in which reflux esophagitis begins as a cytokine-mediated injury (i.e. cytokine sizzle) rather than a caustic chemical injury [7]. In this model, the reflux of acid and bile doesn’t destroy epithelial cells directly, but rather induces them to secrete pro-inflammatory cytokines. These cytokines attract lymphocytes first, rather than neutrophils or eosinophils, and they induce the basal cell and papillary proliferation characteristic of GERD [5]. We postulate that ultimately, it is inflammatory cells that mediate the epithelial injury through a cytokine sizzle, rather than the direct caustic effects of an acid burn (Figure 1B).
Reflux Esophagitis in Humans: A Likely Result of the Cytokine Sizzle
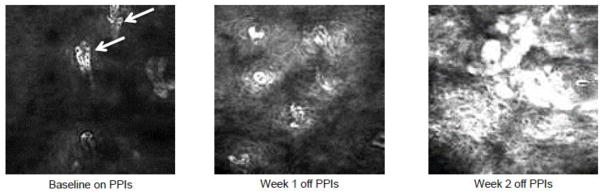
Our alternative concept on the pathogenesis of reflux esophagitis was based on rat and cell lines studies, and it was not clear if this model is applicable to humans. Validation that acute reflux esophagitis in humans is cytokine-mediated (i.e. not primarily an acid burn) could have important implications for the prevention and treatment of GERD, but the logistics of conducting such a validation study are challenging. For example, GERD patients typically have years of symptoms before seeking medical attention [8], and physicians rarely, if ever, see patients with “acute” GERD. Thus, the early histologic changes of reflux esophagitis had not been evaluated prospectively in humans. It has been known for decades that severe, erosive reflux esophagitis healed by proton pump inhibitor (PPI) therapy will return in most cases within 6 to 12 months after PPIs are stopped, although the rapidity with which erosive esophagitis redevelops has not been clear [9,10]. So, we induced acute reflux esophagitis by temporarily interrupting PPI therapy in patients with severe GERD [11]. Using an endoscopy database, we identified 12 patients with Los Angeles (LA) Grade C reflux esophagitis, and treated them with PPIs twice daily for at least one month [11]. While patients were taking their PPIs, we formed endoscopy using high definition white light and confocal laser endomicroscopy (CLE) with biopsy of the distal esophagus, and we stopped the PPIs. At one and two week, we repeated the endoscopy and CLE with biopsy; at the end of week 2, we restarted the patients on their PPIs [11]. Within 2 weeks after stopping PPIs, all 12 patients developed endoscopic reflux esophagitis with 5 patients developing LA Grade C in this short time period. At 1 and 2 weeks after stopping PPIs, significant increases were found in lymphocytes infiltrating the epithelium; neutrophils and eosinophils were few in number. Immunostaining for CD3 and CD20 demonstrated that the lymphocytes were almost exclusively CD3+ T cells, similar to our rat studies [11]. CLE imaging demonstrated significant increases in intercellular space width in the proximal and distal esophagus (i.e. dilation of intercellular space) and in capillary width within 2 weeks after stopping PPIs [11]. We also observed by CLE that the widened intercellular spaces contained increases in fluorescein, the intravenous contrast agent given to patients to enhance identification of cells and capillaries (Figure 2). In the acid burn model for the pathogenesis of reflux esophagitis, dilation of intercellular spaces (DISs), a characteristic GERD feature, is thought to result from acid-induced damage of the proteins of the junctional complexes causing increases in epithelial permeability which allow water to enter from the luminal surface and expand the intercellular spaces [12]. However, our observation that blood-borne fluorescein increases in the intercellular spaces suggests that perhaps DISs result from reflux-induced inflammation increasing vascular permeability which allows for the leakage of fluid out of the blood vessels and into the intercellular spaces causing their expansion. Overall, our findings in GERD patients with acute reflux esophagitis induced by interrupting PPI therapy for 2 weeks are consistent with our earlier findings in our rat studies suggesting that the pathogenesis of reflux esophagitis may be mediated by the cytokine sizzle rather than the acid burn (Figure 1B).
Figure 2.
Confocal Laser Endomicroscopy (CLE) Images Of Acute Reflux Esophagitis. Representative images from the distal esophagus of an individual study subject at baseline on proton pump inhibitors (PPIs), week 1 and week 2 off of PPIs. White arrows indicate fluorescein within the intraepithelial capillaries at baseline on PPIs. By 1 and 2 weeks off PPIs, the fluorescein has leaked from the blood vessels into the intercellular spaces enhancing the identification of the individual squamous cells. Images courtesy of Dr. Kerry B. Dunbar
Hypoxia-Inducible Factor (HIF)-2α: Initiator of the Cytokine Sizzle
Inflammed tissues, such as reflux esophagitis, often are hypoxic, and hypoxia induces the expression of hypoxia-inducible factors (HIFs). HIFs are heterodimeric transcription factors that have HIF-α subunits (either HIF-1α or HIF-2α), which are oxygen regulated and a HIF-1β subunit, which is constitutively expressed [13]. HIFs play a key role in enabling cells to respond to hypoxia stress and in mediating inflammatory processes [13–16]. Under normoxic conditions, HIFs are inactive because the HIF-a subunits are degraded by proteasomes. In the setting of hypoxia, however, proteosomal degradation is inhibited, the HIF-a subunits are stabilized, allowing for them to accumulate within the cell. The HIF-a subunit then binds to the HIF-1B subunit, they translocate to nucleus, and induce the transcription of target genes that contain hypoxia responsive elements (HREs) [14,17,18].
In a mouse model of colonic inflammation, Shah et al. found that colonic inflammation developed in pattern very similar to our rat model of esophagitis, with inflammation starting in the submucosa that subsequently progressed to the mucosal surface associated with an increase in proliferation and an increase in expression of pro-inflammatory cytokines [19]. Subsequent studies by this group demonstrated that it was HIF-2α, and not HIF-1α, that mediated the colonic inflammation in this mouse model [20]. In addition to hypoxia, HIF can be induced by the production of reactive oxygen species (ROS) and we have shown that human esophageal squamous cell in culture exposed to acid and bile salts increase the intracellular production of ROS [21]. Therefore, we reasoned that refluxed acid and bile salts may cause esophageal squamous epithelium to produce ROS which activate HIF-2α to induce the expression of pro-inflammatory mediators. Indeed, using our cultured esophageal squamous cells, we found that exposure to acidic bile salts increased ROS production, increased HIF-2α expression and activity, and increased mRNA expression of pro-inflammatory molecules including T lymphocyte attracting chemokines [22]. Moreover, we found that the nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) pathway was a major effector of the HIF-2α-mediated esophageal epithelial cell inflammatory response to acidic bile salts [22]. These findings in esophageal squamous cells in culture suggests that HIF-2α might play a role in inducing acute reflux esophagitis in our patients during the 2 week interruption of their PPI therapy [11].
Using esophageal biopsies from our patients, we found no change in squamous epithelial cell immunostaining for HIF-1α from baseline to 2 weeks after stopping PPIs. In contrast, we observed an increase in epithelial cell cytoplasmic and nuclear HIF-2α staining form baseline to 1 and 2 weeks after stopping PPIs [22]. Using an index called an H score [23,24], we quantitated the HIF-1α and HIF-2α staining in all 12 patients. There was no significant change in squamous epithelial cells staining for HIF-1α from baseline to 2 weeks after stopping PPIs. In contrast, HIF-2α squamous epithelial cell staining significantly increased at 1 weeks after stopping PPIs and remained elevated at week 2 [22]. In esophageal biopsies from these same 12 patients, we also found increases in mRNA expression of the pro-inflammatory mediators including IL-8 and IL-1β. To determine if changes in HIF-2α protein expression were associated with changes in mRNA expression of the pro-inflammatory mediators, we computed eta2 values, which are used for non-linear correlations [25]. Values of 0.14 or greater generally are interpreted as indicating a large association [25]. At 1 week, we found large associations between the HIF-2α H score and mRNA expression levels of IL-1β and by week 2, large associations were found with IL-8 [22]. We next sought to determine whether HIF-2α enhanced NF-κB/p65 activity in human esophageal biopsies. Immunostaining for the active, phosphorylated form of p65 in esophageal biopsies demonstrated significant increases at week 1, which remained elevated at week 2 after stopping PPIs [22]. Non-linear correlations between H scores of HIF-2α and phosphorylated p65 demonstrated large associations between these proteins at 1 and 2 weeks after stopping PPIs [22]. Furthermore, non-linear correlations demonstrated large associations between phosphorylated p65 and IL-8 and IL-1β at week 2 off PPI therapy [22]. Thus our in vitro and in vivo findings have elucidated molecular mechanisms whereby the reflux of acid and bile salts causes esophagitis through cytokine-mediated mechanisms triggered by HIF-2α.
Barrett’s Esophagus: A Serious Complication of Reflux Esophagitis
Reflux esophagitis can lead to Barrett’s esophagus, which develops through metaplasia. Metaplasia occurs when one adult tissue type replaces another, usually as a response to tissue damage and regeneration from chronic inflammation [26–28]. Metaplasia might represent a protective adaptation to chronic injury, but metaplasia also can predispose to malignancy for reasons that are not clear. [29] In the esophagus, chronic inflammation due to reflux esophagitis damages the squamous epithelium and allows for its replacement by an abnormal columnar epithelium (specialized intestinal metaplasia) comprising a mixture of gastric and intestinal cell phenotypes. [2,30] This metaplastic epithelium is called Barrett’s esophagus and is a major risk factor for esophageal adenocarcinoma [31,32].
The pathogenesis of this disorder remains poorly understood. A topic of intense research interest is the identity of the cell of origin for the specialized intestinal metaplasia of Barrett’s esophagus. A number of potential sources have been proposed including the esophagus, the proximal stomach or the bone marrow. However, intestinal cell are not normally found in the esophagus, the stomach or the bone marrow. Thus, it would seem that Barrett’s metaplasia results from a process called cellular reprogramming in which the expression of key developmental transcription factors is altered in a way that changes the cell’s phenotypic commitment [27]. It is generally accepted that GERD is the condition that induces the cellular reprogramming, but the identity of the progenitor cells whose reprogramming gives rise to Barrett’s metaplasia remains unclear [27]. We will review some key studies addressing the mature esophageal squamous epithelial cell and the esophageal squamous epithelial progenitor cell as the potential origin of Barrett’s metaplasia and the effects of noxious components found in gastroesophageal reflux on esophageal progenitor molecular reprogramming. However, these studies do not refute the alternative possibility that a columnar progenitor cell (in the gastric cardia or at the gastroesophageal junction) also might be a precursor of Barrett’s metaplasia nor are these possible origins mutually exclusive.
Origin of Barrett’s Esophagus: Fully, Differentiated Esophageal Squamous Epithelial Cell
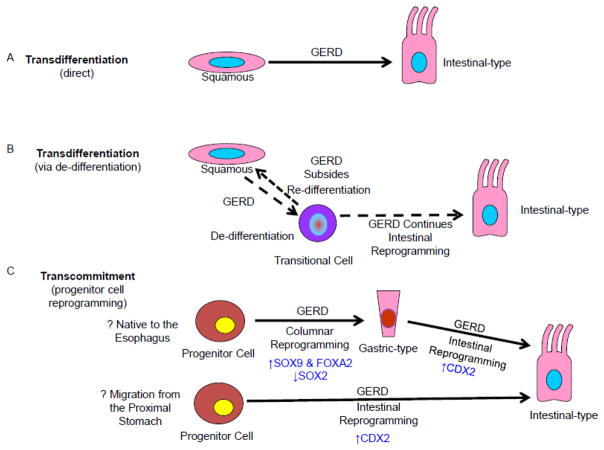
Barrett’s metaplasia may result from transdifferentiation, the process in which one fully differentiated cell type (i.e. squamous) changes directly into another (i.e. intestinal) [28]. Direct transdifferentiation is a molecular reprogramming event that does not require the cell to divide in order to change its phenotype (Figure 3A) [28]. Explants of mouse embryonic columnar-lined esophagus grown in vitro have been shown to lose columnar cell markers and gain squamous cell markers, with a subset of cells simultaneously expressing both types of markers [33]. This switch in marker expression can occur without accompanying changes in cell death or proliferation, suggesting that one cell type can convert directly into another without any intermediary cell divisions [33,34]. It is also possible that the cells with features of both cell types (transitional cells) represent de-differentiated cells that can re-program into the new cell type through a series of intervening cell divisions (Figure 3B)[28,35].
Figure 3.
Conceptual Overview of GERD-Induced Cellular Reprogramming in the Pathogenesis of Barrett’s Metaplasia. Potential pathways for the origin of Barrett’s metaplasia include (A) Direct transdifferentiation is the process in which an individual, fully differentiated cell (i.e. squamous) change directly into another type of fully differentiated cell (i.e. intestinal-type cell) in the setting of GERD. (B) Transdifferentiation in the setting of GERD may result in de-differentiated cells with features of both squamous and intestinal cell types (transitional cells). If GERD subsides, this transitional cell can re-differentiate into a squamous cell. If GERD continues, this transitional cell can reprogram into the new intestinal-type cell through a series of intervening cell divisions. (C) Transcommitment is the process in which immature progenitor cells are reprogrammed in the setting of GERD to give rise to the gastric and intestinal cell types that comprise Barrett’s metaplasia. Progenitor cells that are native to the esophagus undergo reprogramming to columnar gastric-type cells. Some of these gastric-type columnar cells undergo further reprogramming into intestinal-type cells. Progenitor cells may migrate from the proximal stomach into the esophagus, but some of these gastric progenitor cells would still have to undergo reprogramming into intestinal-type cells. Some of the transcription factors that have been implicated in these GERD-induced reprogramming processes are indicated in blue.
Some studies on patients with Barrett’s esophagus have suggested that a transdifferentiation process might underlie the pathogenesis of the esophageal metaplasia. Biopsy specimens taken at the squamo-columnar junction (SCJ) in Barrett’s patients can show a “multilayered epithelium” with a basal layer of squamous cells covered by a superficial layer of columnar cells [36]. Immunocytochemical staining of this multilayered epithelium demonstrates that some cells display both squamous and columnar cell features [36], and scanning electron microscopy has demonstrated a “distinctive cell” at the squamocolumnar junction with ultrastructural characteristics of both squamous and columnar cells [37].
Origin of Barrett’s Esophagus: Esophageal Squamous Epithelial Progenitor Cell
Despite this indirect evidence for transdifferentiation, it seems unlikely that the variety of gastric and intestinal cell types that comprise Barrett’s metaplasia develop solely through the transdifferentiation of mature esophageal squamous cells [38]. It is more likely that a metaplasia, in which one tissue type converts into another, arises from undifferentiated progenitor cells that have the capacity to produce and maintain multiple cell type [28]. In the setting of GERD-induced tissue damage, immature progenitor cells are molecularly reprogrammed to express columnar rather than squamous developmental transcription factors, thereby differentiating into the multiple columnar cell types of Barrett’s metaplasia. This molecular reprogramming process has been called transcommitment, and the responsible progenitor cells might be native to the esophagus, or they might migrate into the esophagus from the proximal stomach when the squamous epithelium is damage by GERD (Figure 3C). In support of a molecular reprogramming process of squamous esophageal progenitor cells, immortalized esophageal squamous cell lines and tissues exposed to acid and bile salts in vitro or GERD in vivo increase their expression of the columnar transcription factors SOX9 and forkhead box protein A2 (FOXA2), which are targets of the Hedgehog pathway, and of the intestinal transcription factor caudal-related homeobox transcription factor 2 (CDX2), a target of the (NF-κB) pathway (Figure 3C) [39–43]. In addition, esophageal squamous cells exposed to nitric oxide, another noxious component of reflux, in vitro or in vivo decrease their expression of sex determining region Y-box 2 (SOX2), a transcription factor that promotes stratified squamous epithelia development, through inhibition of protein kinase B (PKB or Akt) pathway signaling (Figure 3C) [44,45].
GERD-Induced Columnar Transcription Factors SOX9 and FOXA2: Targets of the Hedgehog Pathway
SOX9 and FOXA2 are transcription factors that characterize columnar cells and both are targets of the Hedgehog (Hh) pathway. Hh ligands bind to their transmembrane receptor called patched (PTCH) to activate pathway signaling. In the absence of ligand binding, PTCH inhibits smoothened (Smo), a protein that transduces the signal downstream. Following the binding of Hh to PTCH, Smo is released from PTCH inhibition and activates Gli transcription factors to regulate downstream target genes [40]. Wang et al. demonstrated that esophageal squamous cell lines and squamous tissues exposed to acid and bile salts in vitro or to gastroesophageal reflux in vivo exhibit Hedgehog pathway signaling. Following exposure to acid and bile salts, esophageal squamous epithelial cells secrete the hedgehog ligand, sonic hedgehog. The secretion by the epithelial cells of this ligand binds the PTCH receptor located on stromal fibroblasts leading to the secretion of bone morphogenic protein 4 (BMP4). BMP4 then binds to its receptors, the BMP type I receptors located on the esophageal squamous cells and this epithelial-mesenchymal Hh signaling causes the squamous cells to produce the transcription factor SOX-9 [39]. Furthermore, esophageal squamous cells expressing plasmids containing sonic hedgehog, Gli1, or a constitutively active BMP receptor type IA induced the columnar cell transcription factor FOXA2 [40]. Expression of SOX-9 and FOXA2 induces genes that influence columnar and goblet cell differentiation such as cytokeratin 8 and mucin 2 (MUC2) [39,40]. Thus reflux of acid and bile salts could initiate reprogramming of progenitors in the esophagus by activation of Hedgehog signaling and upregulation of SOX9 and FOXA2.
The GERD-Induced Intestinal Transcription Factor CDX2: Target of the NF-κB pathway
CDX2 is a key developmental transcription factor that directs formation of intestinal epithelium and is a target of the NF-κB pathway [46,47]. In fact, two putative NF-κB binding sites have been identified in the CDX2 promoter [48]. Uninflamed esophageal squamous epithelium does not express active NF-κB, but expression of the activated form of this transcription factor has been found in the esophageal epithelium as reflux-induced inflammation ensues [22,49]. In previous studies, we demonstrated that acid and bile salt exposure induces NF-κB signaling in esophageal squamous cells in culture [22,43,50]. In the pancreas, furthermore, NF-κB signaling has been shown to play a key role in the molecular reprogramming process that underlies pancreatic acinar-to-ductal metaplasia [51].
CDX2 expression is frequently found in biopsy specimens of Barrett’s metaplasia, which is not surprising since intestinal-type columnar cells are characteristic of this esophageal metaplasia [52–55]. Like NF-κB, CDX2 expression has been found in biopsy specimens of esophageal squamous epithelium inflamed by GERD, but not in uninflamed esophageal epithelium [55]. In animal models of reflux esophagitis, the reflux-damaged esophageal epithelium increases Cdx2 expression before the development of a Barrett’s-like metaplasia [44,56,57]. In cultured esophageal squamous cells from rats and some human subjects, acid and bile salt exposures have also been shown to increase activity of the Cdx2 promoter and increase Cdx2 mRNA expression [58–61]. Tamagawa et al. demonstrated that bile salts increase CDX2 expression, which in turn decreases HES1 and increases atonal homolog 1 (ATOH1) expression, targets that reflect decreases in Notch pathway signaling [62,63]. Moreover, the combination of increased CDX2 and decreased Notch signaling led to increases in MUC2 and delta-like 1 (Dll1), genes that further promote goblet cell differentiation [63]. These studies suggest that the reflux of acid and bile salts could initiate reprogramming of progenitors in the esophagus by activation of NF-κB signaling and upregulation of CDX2.
GERD-Suppressed Stratified Squamous Epithelial Transcription Factor SOX2: A Target of the Akt Pathway
The majority of studies on the molecular events underlying the intestinal metaplasia of Barrett’s esophagus have focused primarily on the upregulation of genes involved in columnar and intestinal differentiation such as SOX9 and CDX2 [39,43]. However, it seems equally plausible that the molecular reprogramming of squamous-to-columnar metaplasia also involves the downregulation of genes that regulate squamous differentiation such as SOX2 and the isoforms of tumor protein p63 (p63) [64,65]. Furthermore, noxious components of gastroesophageal reflux, other than acid and bile salts, like nitric oxide (NO) may play a role in the reprogramming process.
Iijima et al. found that high concentrations of NO can be generated in the esophageal lumen during episodes of gastroesophageal reflux [66]. Dietary nitrate is commonly found in green, leafy vegetables. When these foods are ingested, dietary nitrate is absorbed and secreted into the saliva. Oral bacteria then reduce the nitrate to nitrite, which is then swallowed. When nitrite comes in contact with refluxed gastric acid, NO is rapidly generated. This NO can react with oxygen to form highly toxic reactive nitrogen species that result in damage to the tissue [67]. In GERD patients with and without Barrett’s esophagus, NO generated from dietary nitrate has been shown to reach genotoxic concentrations at the gastroesophageal junction [68]. Moreover, Endo et al. found that dietary supplementation with nitrates accelerated the development of metaplasia in a rat model of reflux esophagitis [69]. However, very little had been known regarding the mechanisms whereby exposure of the esophagus to NO, generated from dietary nitrate, might facilitate the development of Barrett’s metaplasia.
Using esophageal squamous cells in culture, Asanuma et al. found that exposure to the small molecule NO donor, NOC9, profoundly reduced SOX2 mRNA expression compared to cells exposed to acid and bile salts [70]. NOC9 exposure caused S-nitrosylation of Akt, which blocked its phosphorylation, and interfered with its downstream signaling leading to reductions in SOX2 mRNA and protein [45]. Moreover, the generation of NO by NOC9 decreased the expression of the TA and ΔNP isoforms of p63, another transcription factor that promotes stratified squamous epithelia, and increased the expression of CDX2 [65,71]. Using tissue specimens from rats with surgically-induced reflux esophagitis fed postoperative diets with and without NO-supplementation, the investigators found diminished staining for SOX2 in the squamous-lined distal esophagus of rats fed an NO-supplemented diet compared to rats feed a normal diet [45]. These findings suggest that the generation of NO by gastroesophageal reflux could initiate reprogramming of progenitors in the esophagus by inhibiting Akt signaling causing reduction in SOX2, by decreasing the TA and ΔNP isoforms of p63, and by upregulating CDX2, events that might lead to the development of the intestinal metaplasia of Barrett’s esophagus.
Conclusions
Data from a rat model and humans suggest that reflux esophagitis develops as a cytokine-mediated inflammatory injury (i.e. sizzle), not as a caustic chemical injury (i.e. acid burn), as has traditionally been assumed. Molecular mechanisms elucidated in esophageal squamous cell lines demonstrate that acid and bile salts, the major components of gastroesophageal reflux, induce HIF-2α which enhances NF-κB transcriptional activity resulting in the production of pro-inflammatory molecules including chemokines that attract T lymphocytes. Esophageal biopsies of patients with acute reflux esophagitis at 1 and 2 weeks after stopping PPIs demonstrate large associations between HIF-2α production, NF-κB activation, and pro-inflammatory mediator expression in support of such a mechanism. The past few years have seen an explosion of research into the origin of Barrett’s esophagus, and controversy currently exits as to whether GERD-induced molecular reprogramming of progenitors that are native to the esophagus is involved. Investigations into this issue have uncovered the role of signaling pathways like Hedgehog, NF-κB, and Akt and transcription factors like SOX9, FOXA2, CDX2, and SOX2 that conceivably could induce squamous-to-columnar molecular reprogramming. Thus, new insights into understanding the pathogenesis of reflux esophagitis and reflux-related reprogramming of native esophageal progenitors have highlighted potential molecular pathways and molecules for future targeted therapies to prevent the development of Barrett’s esophagus.
Acknowledgments
This work was supported by the National Institutes of Health Grants R01-DK063621 and R01-DK103598.
Footnotes
Compliance with ethical standards
Conflict of interest The author declares that she has no conflict of interest.
References
- 1.Spechler SJ. Carcinogenesis at the gastroesophageal junction: Free radicals at the frontier. Gastroenterology. 2002;122:1518–1520. doi: 10.1053/gast.2002.33368. [DOI] [PubMed] [Google Scholar]
- 2.Spechler SJ, Souza RF. Barrett’s esophagus. The New England journal of medicine. 2014;371:836–845. doi: 10.1056/NEJMra1314704. [DOI] [PubMed] [Google Scholar]
- 3.Iwakiri K, Kinoshita Y, Habu Y, Oshima T, Manabe N, Fujiwara Y, Nagahara A, Kawamura O, Iwakiri R, Ozawa S, Ashida K, Ohara S, Kashiwagi H, Adachi K, Higuchi K, Miwa H, Fujimoto K, Kusano M, Hoshihara Y, Kawano T, Haruma K, Hongo M, Sugano K, Watanabe M, Shimosegawa T. Evidence-based clinical practice guidelines for gastroesophageal reflux disease 2015. Journal of gastroenterology. 2016;51:751–767. doi: 10.1007/s00535-016-1227-8. [DOI] [PubMed] [Google Scholar]
- 4.Spechler SJ. Clinical practice. Barrett’s esophagus The New England journal of medicine. 2002;346:836–842. doi: 10.1056/NEJMcp012118. [DOI] [PubMed] [Google Scholar]
- 5.Ismail-Beigi F, Horton PF, Pope CE., 2nd Histological consequences of gastroesophageal reflux in man. Gastroenterology. 1970;58:163–174. [PubMed] [Google Scholar]
- 6.Orlando RC. Pathophysiology of gastroesophageal reflux disease. Journal of clinical gastroenterology. 2008;42:584–588. doi: 10.1097/MCG.0b013e31815d0628. [DOI] [PubMed] [Google Scholar]
- 7.Souza RF, Huo X, Mittal V, Schuler CM, Carmack SW, Zhang HY, Zhang X, Yu C, Hormi-Carver K, Genta RM, Spechler SJ. Gastroesophageal reflux might cause esophagitis through a cytokine-mediated mechanism rather than caustic acid injury. Gastroenterology. 2009;137:1776–1784. doi: 10.1053/j.gastro.2009.07.055. [DOI] [PubMed] [Google Scholar]
- 8.Lundell LR, Dent J, Bennett JR, Blum AL, Armstrong D, Galmiche JP, Johnson F, Hongo M, Richter JE, Spechler SJ, Tytgat GN, Wallin L. Endoscopic assessment of oesophagitis: Clinical and functional correlates and further validation of the los angeles classification. Gut. 1999;45:172–180. doi: 10.1136/gut.45.2.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hetzel DJ, Dent J, Reed WD, Narielvala FM, Mackinnon M, McCarthy JH, Mitchell B, Beveridge BR, Laurence BH, Gibson GG, et al. Healing and relapse of severe peptic esophagitis after treatment with omeprazole. Gastroenterology. 1988;95:903–912. doi: 10.1016/0016-5085(88)90162-x. [DOI] [PubMed] [Google Scholar]
- 10.Chiba N. Proton pump inhibitors in acute healing and maintenance of erosive or worse esophagitis: A systematic overview. Canadian journal of gastroenterology = Journal canadien de gastroenterologie. 1997;11(Suppl B):66b–73b. [PubMed] [Google Scholar]
- 11.Dunbar KB, Agoston AT, Odze RD, Huo X, Pham TH, Cipher DJ, Castell DO, Genta RM, Souza RF, Spechler SJ. Association of acute gastroesophageal reflux disease with esophageal histologic changes. JAMA: the journal of the American Medical Association. 2016;315:2104–2112. doi: 10.1001/jama.2016.5657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tobey NA, Gambling TM, Vanegas XC, Carson JL, Orlando RC. Physicochemical basis for dilated intercellular spaces in non-erosive acid-damaged rabbit esophageal epithelium. Diseases of the esophagus: official journal of the International Society for Diseases of the Esophagus/ISDE. 2008;21:757–764. doi: 10.1111/j.1442-2050.2008.00841.x. [DOI] [PubMed] [Google Scholar]
- 13.Loboda A, Jozkowicz A, Dulak J. Hif-1 and hif-2 transcription factors--similar but not identical. Molecules and cells. 2010;29:435–442. doi: 10.1007/s10059-010-0067-2. [DOI] [PubMed] [Google Scholar]
- 14.Taylor CT. Interdependent roles for hypoxia inducible factor and nuclear factor-kappab in hypoxic inflammation. The Journal of physiology. 2008;586:4055–4059. doi: 10.1113/jphysiol.2008.157669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eltzschig HK, Carmeliet P. Hypoxia and inflammation. The New England journal of medicine. 2011;364:656–665. doi: 10.1056/NEJMra0910283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zinkernagel AS, Johnson RS, Nizet V. Hypoxia inducible factor (hif) function in innate immunity and infection. Journal of molecular medicine (Berlin, Germany) 2007;85:1339–1346. doi: 10.1007/s00109-007-0282-2. [DOI] [PubMed] [Google Scholar]
- 17.Scholz CC, Taylor CT. Hydroxylase-dependent regulation of the nf-kappab pathway. Biological chemistry. 2013;394:479–493. doi: 10.1515/hsz-2012-0338. [DOI] [PubMed] [Google Scholar]
- 18.Haddad JJ, Harb HL. Cytokines and the regulation of hypoxia-inducible factor (hif)-1alpha. International immunopharmacology. 2005;5:461–483. doi: 10.1016/j.intimp.2004.11.009. [DOI] [PubMed] [Google Scholar]
- 19.Shah YM, Ito S, Morimura K, Chen C, Yim SH, Haase VH, Gonzalez FJ. Hypoxia-inducible factor augments experimental colitis through an mif-dependent inflammatory signaling cascade. Gastroenterology. 2008;134:2036–2048. 2048.e2031–2033. doi: 10.1053/j.gastro.2008.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xue X, Ramakrishnan S, Anderson E, Taylor M, Zimmermann EM, Spence JR, Huang S, Greenson JK, Shah YM. Endothelial pas domain protein 1 activates the inflammatory response in the intestinal epithelium to promote colitis in mice. Gastroenterology. 2013;145:831–841. doi: 10.1053/j.gastro.2013.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feagins LA, Zhang HY, Zhang X, Hormi-Carver K, Thomas T, Terada LS, Spechler SJ, Souza RF. Mechanisms of oxidant production in esophageal squamous cell and barrett’s cell lines. American journal of physiology Gastrointestinal and liver physiology. 2008;294:G411–417. doi: 10.1152/ajpgi.00373.2007. [DOI] [PubMed] [Google Scholar]
- 22.Huo X, Agoston A, Dunbar KB, Cipher DJ, Zhang X, Yu C, Cheng E, Zhang Q, Pham TH, Tambar UK, Bruick RK, Wang DH, Odze RD, Spechler SJ, Souza RF. Hypoxia-inducible factor 2a plays a role in mediating oesophagitis in gastro-oesophageal reflux disease. Gut. 2016 doi: 10.1136/gutjnl-2016-312595. Epub. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hirsch FR, Varella-Garcia M, Bunn PA, Jr, Di Maria MV, Veve R, Bremmes RM, Baron AE, Zeng C, Franklin WA. Epidermal growth factor receptor in non-small-cell lung carcinomas: Correlation between gene copy number and protein expression and impact on prognosis. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2003;21:3798–3807. doi: 10.1200/JCO.2003.11.069. [DOI] [PubMed] [Google Scholar]
- 24.John T, Liu G, Tsao MS. Overview of molecular testing in non-small-cell lung cancer: Mutational analysis, gene copy number, protein expression and other biomarkers of egfr for the prediction of response to tyrosine kinase inhibitors. Oncogene. 2009;28(Suppl 1):S14–23. doi: 10.1038/onc.2009.197. [DOI] [PubMed] [Google Scholar]
- 25.Cohen J. Statistical power analysis for the behavioral sciences. 2. New Jersey: Lawrence Erlbaum Associates; 1988. [Google Scholar]
- 26.Spechler SJ. Intestinal metaplasia at the gastroesophageal junction. Gastroenterology. 2004;126:567–575. doi: 10.1053/j.gastro.2003.11.061. [DOI] [PubMed] [Google Scholar]
- 27.Burke ZD, Tosh D. Barrett’s metaplasia as a paradigm for understanding the development of cancer. Current opinion in genetics & development. 2012;22:494–499. doi: 10.1016/j.gde.2012.08.001. [DOI] [PubMed] [Google Scholar]
- 28.Slack JM. Metaplasia and transdifferentiation: From pure biology to the clinic. Nature reviews Molecular cell biology. 2007;8:369–378. doi: 10.1038/nrm2146. [DOI] [PubMed] [Google Scholar]
- 29.Tosh D, Slack JM. How cells change their phenotype. Nature reviews Molecular cell biology. 2002;3:187–194. doi: 10.1038/nrm761. [DOI] [PubMed] [Google Scholar]
- 30.McDonald SA, Lavery D, Wright NA, Jansen M. Barrett oesophagus: Lessons on its origins from the lesion itself. Nature reviews Gastroenterology & hepatology. 2015;12:50–60. doi: 10.1038/nrgastro.2014.181. [DOI] [PubMed] [Google Scholar]
- 31.Spechler SJ. Barrett esophagus and risk of esophageal cancer: A clinical review. JAMA the journal of the American Medical Association. 2013;310:627–636. doi: 10.1001/jama.2013.226450. [DOI] [PubMed] [Google Scholar]
- 32.Thrift AP, Pandeya N, Whiteman DC. Current status and future perspectives on the etiology of esophageal adenocarcinoma. Frontiers in oncology. 2012;2:11. doi: 10.3389/fonc.2012.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu WY, Slack JM, Tosh D. Conversion of columnar to stratified squamous epithelium in the developing mouse oesophagus. Developmental biology. 2005;284:157–170. doi: 10.1016/j.ydbio.2005.04.042. [DOI] [PubMed] [Google Scholar]
- 34.Beresford WA. Direct transdifferentiation: Can cells change their phenotype without dividing? Cell differentiation and development: the official journal of the International Society of Developmental Biologists. 1990;29:81–93. doi: 10.1016/0922-3371(90)90026-s. [DOI] [PubMed] [Google Scholar]
- 35.Eberhard D, Tosh D. Transdifferentiation and metaplasia as a paradigm for understanding development and disease. Cellular and molecular life sciences: CMLS. 2008;65:33–40. doi: 10.1007/s00018-007-7428-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boch JA, Shields HM, Antonioli DA, Zwas F, Sawhney RA, Trier JS. Distribution of cytokeratin markers in barrett’s specialized columnar epithelium. Gastroenterology. 1997;112:760–765. doi: 10.1053/gast.1997.v112.pm9041237. [DOI] [PubMed] [Google Scholar]
- 37.Shields HM, Zwas F, Antonioli DA, Doos WG, Kim S, Spechler SJ. Detection by scanning electron microscopy of a distinctive esophageal surface cell at the junction of squamous and barrett’s epithelium. Digestive diseases and sciences. 1993;38:97–108. doi: 10.1007/BF01296780. [DOI] [PubMed] [Google Scholar]
- 38.Corbett JL, Tosh D. Conversion of one cell type into another: Implications for understanding organ development, pathogenesis of cancer and generating cells for therapy. Biochemical Society transactions. 2014;42:609–616. doi: 10.1042/BST20140058. [DOI] [PubMed] [Google Scholar]
- 39.Wang DH, Clemons NJ, Miyashita T, Dupuy AJ, Zhang W, Szczepny A, Corcoran-Schwartz IM, Wilburn DL, Montgomery EA, Wang JS, Jenkins NA, Copeland NA, Harmon JW, Phillips WA, Watkins DN. Aberrant epithelial-mesenchymal hedgehog signaling characterizes barrett’s metaplasia. Gastroenterology. 2010;138:1810–1822. doi: 10.1053/j.gastro.2010.01.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang DH, Tiwari A, Kim ME, Clemons NJ, Regmi NL, Hodges WA, Berman DM, Montgomery EA, Watkins DN, Zhang X, Zhang Q, Jie C, Spechler SJ, Souza RF. Hedgehog signaling regulates foxa2 in esophageal embryogenesis and barrett’s metaplasia. The Journal of clinical investigation. 2014;124:3767–3780. doi: 10.1172/JCI66603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mari L, Milano F, Parikh K, Straub D, Everts V, Hoeben KK, Fockens P, Buttar NS, Krishnadath KK. A psmad/cdx2 complex is essential for the intestinalization of epithelial metaplasia. Cell reports. 2014;7:1197–1210. doi: 10.1016/j.celrep.2014.03.074. [DOI] [PubMed] [Google Scholar]
- 42.Milano F, van Baal JW, Buttar NS, Rygiel AM, de Kort F, DeMars CJ, Rosmolen WD, Bergman JJ, VAM J, Wang KK, Peppelenbosch MP, Krishnadath KK. Bone morphogenetic protein 4 expressed in esophagitis induces a columnar phenotype in esophageal squamous cells. Gastroenterology. 2007;132:2412–2421. doi: 10.1053/j.gastro.2007.03.026. [DOI] [PubMed] [Google Scholar]
- 43.Huo X, Zhang HY, Zhang XI, Lynch JP, Strauch ED, Wang JY, Melton SD, Genta RM, Wang DH, Spechler SJ, Souza RF. Acid and bile salt-induced cdx2 expression differs in esophageal squamous cells from patients with and without barrett’s esophagus. Gastroenterology. 2010;139:194–203. e191. doi: 10.1053/j.gastro.2010.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tatsuta T, Mukaisho K, Sugihara H, Miwa K, Tani T, Hattori T. Expression of cdx2 in early grcl of barrett’s esophagus induced in rats by duodenal reflux. Digestive diseases and sciences. 2005;50:425–431. doi: 10.1007/s10620-005-2452-9. [DOI] [PubMed] [Google Scholar]
- 45.Asanuma K, Huo X, Agoston A, Zhang X, Yu C, Cheng E, Zhang Q, Dunbar KB, Pham TH, Wang DH, Iijima K, Shimosegawa T, Odze RD, Spechler SJ, Souza RF. In oesophageal squamous cells, nitric oxide causes s-nitrosylation of akt and blocks sox2 (sex determining region y-box 2) expression. Gut. 2015 doi: 10.1136/gutjnl-2015-309272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guo RJ, Suh ER, Lynch JP. The role of cdx proteins in intestinal development and cancer. Cancer biology & therapy. 2004;3:593–601. doi: 10.4161/cbt.3.7.913. [DOI] [PubMed] [Google Scholar]
- 47.Beck F. The role of cdx genes in the mammalian gut. Gut. 2004;53:1394–1396. doi: 10.1136/gut.2003.038240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim S, Domon-Dell C, Wang Q, Chung DH, Di Cristofano A, Pandolfi PP, Freund JN, Evers BM. Pten and tnf-alpha regulation of the intestinal-specific cdx-2 homeobox gene through a pi3k, pkb/akt, and nf-kappab-dependent pathway. Gastroenterology. 2002;123:1163–1178. doi: 10.1053/gast.2002.36043. [DOI] [PubMed] [Google Scholar]
- 49.O’Riordan JM, Abdel-latif MM, Ravi N, McNamara D, Byrne PJ, McDonald GS, Keeling PW, Kelleher D, Reynolds JV. Proinflammatory cytokine and nuclear factor kappa-b expression along the inflammation-metaplasia-dysplasia-adenocarcinoma sequence in the esophagus. The American journal of gastroenterology. 2005;100:1257–1264. doi: 10.1111/j.1572-0241.2005.41338.x. [DOI] [PubMed] [Google Scholar]
- 50.Huo X, Zhang X, Yu C, Zhang Q, Cheng E, Wang DH, Pham TH, Spechler SJ, Souza RF. In oesophageal squamous cells exposed to acidic bile salt medium, omeprazole inhibits il-8 expression through effects on nuclear factor-kappab and activator protein-1. Gut. 2014;63:1042–1052. doi: 10.1136/gutjnl-2013-305533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liou GY, Doppler H, Necela B, Krishna M, Crawford HC, Raimondo M, Storz P. Macrophage-secreted cytokines drive pancreatic acinar-to-ductal metaplasia through nf-kappab and mmps. The Journal of cell biology. 2013;202:563–577. doi: 10.1083/jcb.201301001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vallbohmer D, DeMeester SR, Peters JH, Oh DS, Kuramochi H, Shimizu D, Hagen JA, Danenberg KD, Danenberg PV, DeMeester TR, Chandrasoma PT. Cdx-2 expression in squamous and metaplastic columnar epithelia of the esophagus. Diseases of the esophagus: official journal of the International Society for Diseases of the Esophagus/ISDE. 2006;19:260–266. doi: 10.1111/j.1442-2050.2006.00586.x. [DOI] [PubMed] [Google Scholar]
- 53.Phillips RW, Frierson HF, Jr, Moskaluk CA. Cdx2 as a marker of epithelial intestinal differentiation in the esophagus. The American journal of surgical pathology. 2003;27:1442–1447. doi: 10.1097/00000478-200311000-00006. [DOI] [PubMed] [Google Scholar]
- 54.Groisman GM, Amar M, Meir A. Expression of the intestinal marker cdx2 in the columnar-lined esophagus with and without intestinal (barrett’s) metaplasia. Modern pathology: an official journal of the United States and Canadian Academy of Pathology, Inc. 2004;17:1282–1288. doi: 10.1038/modpathol.3800182. [DOI] [PubMed] [Google Scholar]
- 55.Eda A, Osawa H, Satoh K, Yanaka I, Kihira K, Ishino Y, Mutoh H, Sugano K. Aberrant expression of cdx2 in barrett’s epithelium and inflammatory esophageal mucosa. Journal of gastroenterology. 2003;38:14–22. doi: 10.1007/s005350300001. [DOI] [PubMed] [Google Scholar]
- 56.Pera M, Pera M, de Bolos C, Brito MJ, Palacin A, Grande L, Cardesa A, Poulsom R. Duodenal-content reflux into the esophagus leads to expression of cdx2 and muc2 in areas of squamous epithelium in rats. Journal of gastrointestinal surgery: official journal of the Society for Surgery of the Alimentary Tract. 2007;11:869–874. doi: 10.1007/s11605-007-0162-7. [DOI] [PubMed] [Google Scholar]
- 57.Ingravallo G, Dall’Olmo L, Segat D, Fassan M, Mescoli C, Dazzo E, Castoro C, Polimeno L, Rizzetto C, Baroni MD, Zaninotto G, Ancona E, Rugge M. Cdx2 hox gene product in a rat model of esophageal cancer. Journal of experimental & clinical cancer research: CR. 2009;28:108. doi: 10.1186/1756-9966-28-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu T, Zhang X, So CK, Wang S, Wang P, Yan L, Myers R, Chen Z, Patterson AP, Yang CS, Chen X. Regulation of cdx2 expression by promoter methylation, and effects of cdx2 transfection on morphology and gene expression of human esophageal epithelial cells. Carcinogenesis. 2007;28:488–496. doi: 10.1093/carcin/bgl176. [DOI] [PubMed] [Google Scholar]
- 59.Kazumori H, Ishihara S, Rumi MA, Kadowaki Y, Kinoshita Y. Bile acids directly augment caudal related homeobox gene cdx2 expression in oesophageal keratinocytes in barrett’s epithelium. Gut. 2006;55:16–25. doi: 10.1136/gut.2005.066209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Marchetti M, Caliot E, Pringault E. Chronic acid exposure leads to activation of the cdx2 intestinal homeobox gene in a long-term culture of mouse esophageal keratinocytes. Journal of cell science. 2003;116:1429–1436. doi: 10.1242/jcs.00338. [DOI] [PubMed] [Google Scholar]
- 61.Hu Y, Williams VA, Gellersen O, Jones C, Watson TJ, Peters JH. The pathogenesis of barrett’s esophagus: Secondary bile acids upregulate intestinal differentiation factor cdx2 expression in esophageal cells. Journal of gastrointestinal surgery: official journal of the Society for Surgery of the Alimentary Tract. 2007;11:827–834. doi: 10.1007/s11605-007-0174-3. [DOI] [PubMed] [Google Scholar]
- 62.Tamagawa Y, Ishimura N, Uno G, Yuki T, Kazumori H, Ishihara S, Amano Y, Kinoshita Y. Notch signaling pathway and cdx2 expression in the development of barrett’s esophagus. Laboratory investigation; a journal of technical methods and pathology. 2012;92:896–909. doi: 10.1038/labinvest.2012.56. [DOI] [PubMed] [Google Scholar]
- 63.Tamagawa Y, Ishimura N, Uno G, Aimi M, Oshima N, Yuki T, Sato S, Ishihara S, Kinoshita Y. Bile acids induce delta-like 1 expression via cdx2-dependent pathway in the development of barrett’s esophagus. Laboratory investigation; a journal of technical methods and pathology. 2016;96:325–337. doi: 10.1038/labinvest.2015.137. [DOI] [PubMed] [Google Scholar]
- 64.Que J, Okubo T, Goldenring JR, Nam KT, Kurotani R, Morrisey EE, Taranova O, Pevny LH, Hogan BL. Multiple dose-dependent roles for sox2 in the patterning and differentiation of anterior foregut endoderm. Development (Cambridge, England) 2007;134:2521–2531. doi: 10.1242/dev.003855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Watanabe H, Ma Q, Peng S, Adelmant G, Swain D, Song W, Fox C, Francis JM, Pedamallu CS, DeLuca DS, Brooks AN, Wang S, Que J, Rustgi AK, Wong KK, Ligon KL, Liu XS, Marto JA, Meyerson M, Bass AJ. Sox2 and p63 colocalize at genetic loci in squamous cell carcinomas. The Journal of clinical investigation. 2014;124:1636–1645. doi: 10.1172/JCI71545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Iijima K, Henry E, Moriya A, Wirz A, Kelman AW, McColl KE. Dietary nitrate generates potentially mutagenic concentrations of nitric oxide at the gastroesophageal junction. Gastroenterology. 2002;122:1248–1257. doi: 10.1053/gast.2002.32963. [DOI] [PubMed] [Google Scholar]
- 67.Jaiswal M, LaRusso NF, Gores GJ. Nitric oxide in gastrointestinal epithelial cell carcinogenesis: Linking inflammation to oncogenesis. American journal of physiology Gastrointestinal and liver physiology. 2001;281:G626–634. doi: 10.1152/ajpgi.2001.281.3.G626. [DOI] [PubMed] [Google Scholar]
- 68.Suzuki H, Iijima K, Scobie G, Fyfe V, McColl KE. Nitrate and nitrosative chemistry within barrett’s oesophagus during acid reflux. Gut. 2005;54:1527–1535. doi: 10.1136/gut.2005.066043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Endo H, Iijima K, Asanuma K, Ara N, Ito H, Asano N, Uno K, Koike T, Imatani A, Shimosegawa T. Exogenous luminal nitric oxide exposure accelerates columnar transformation of rat esophagus. International journal of cancer Journal international du cancer. 2010;127:2009–2019. doi: 10.1002/ijc.25227. [DOI] [PubMed] [Google Scholar]
- 70.Asanuma K, Huo X, Agoston A, Zhang X, Yu C, Cheng E, Zhang Q, Dunbar KB, Pham TH, Wang DH, Iijima K, Shimosegawa T, Odze RD, Spechler SJ, Souza RF. In oesophageal squamous cells, nitric oxide causes s-nitrosylation of akt and blocks sox2 (sex determining region y-box 2) expression. Gut. 2016;65:1416–1426. doi: 10.1136/gutjnl-2015-309272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ishiyama F, Iijima K, Asanuma K, Ara N, Yoshitake J, Abe Y, Koike T, Imatani A, Ohara S, Shimosegawa T. Exogenous luminal nitric oxide exacerbates esophagus tissue damage in a reflux esophagitis model of rats. Scandinavian journal of gastroenterology. 2009;44:527–537. doi: 10.1080/00365520802699260. [DOI] [PubMed] [Google Scholar]