Abstract
Transcripts from the late promoter of human papillomavirus type 16 (HPV16) are upregulated upon host cell differentiation. Differentiation-dependent transcript regulation is thought to sequester viral antigens in the uppermost epithelial layers, facilitating immune evasion. The mechanisms regulating late promoter upregulation during differentiation are poorly characterized. We show that the late promoter is upregulated at the transcriptional level and that the viral enhancer stimulates promoter activity. Using kinase inhibition and chromatin immunoprecipitation analysis, we show evidence for differentiation-dependent enhancement of transcript elongation. Three factors that promote transcript elongation, cyclin dependent kinase 9 (CDK9), CDK8 (a subunit of the Mediator complex), and bromodomain containing protein 4 (Brd4) are recruited to viral genomes upon differentiation, and each plays a role in promoter activity. These results shed light on the transcriptional processes utilized by HPV16 for proper regulation of gene expression during the viral life cycle.
Keywords: papillomavirus, gene expression, late promoter, CDK9, CDK8, P-TEFb, Mediator, Pol II carboxy terminal domain, Brd4, differentiation, transcriptional elongation
Introduction
Human papillomaviruses (HPVs) are small DNA viruses that infect keratinocytes of stratified squamous epithelia and are linked to a range of cancers, including cervical and an increasing subset of oropharyngeal cancers (Doorbar et al., 2012; Forman et al., 2012). The HPV life cycle is organized around the ability of the virus to persist and produce viral progeny for months or years in the face of constant cellular turnover and immune surveillance (Bodily and Laimins, 2011; Westrich et al., 2016). Most infections are cleared within two years (Moscicki et al., 2012; Woodman et al., 2001), but in a minority of cases, infections can last for decades. The ability of high risk HPVs to persist for long periods allows proliferating infected cells to accumulate genetic changes needed for cancer development, and thus long-term persistence is the primary risk factor for the development of cervical cancer (Moscicki et al., 2012). The pattern of viral gene expression during the HPV life cycle is a key strategy that contributes to viral persistence (Bodily and Laimins, 2011). Upon infection, a pool of cells containing the HPV genome is established in keratinocytes of the basal epithelial layer, where viral copy number and gene expression are maintained at low levels. As host cells detach from the basement membrane and pass through the stages of squamous differentiation, viral gene expression and replication are activated (Bodily and Laimins, 2011). The viral late promoter, or p670 (Figure 1a), drives expression of transcripts encoding the antigenic late proteins, and these transcripts are found at high levels only upon cellular differentiation (Grassmann et al., 1996; Hummel et al., 1992; Ruesch and Laimins, 1998). The late promoter also regulates viral intermediate genes (including E1, E2, E1^E4, and E5), which are involved in amplification of the viral genome (Bedell et al., 1991; Fehrmann et al., 2003; Peh et al., 2004; Wilson et al., 2005). Thus the late promoter facilitates viral persistence by targeting high level genome replication and capsid protein synthesis to the differentiated strata in the host epithelium, shielding the virus from immune-mediated clearance (Frazer, 2009; Gielen et al., 1988; Nardelli et al., 2002).
Figure 1.
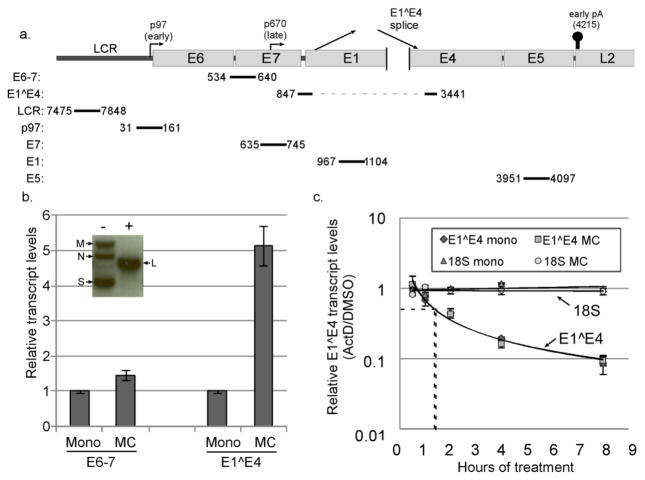
The HPV16 late promoter is transcriptionally upregulated. a. Schematic diagram of the HPV16 genome showing the location of the long control region (LCR), early and late promoters, early polyadenylation signal, and the position of qPCR amplicons examined in this study. b. Inset: Southern blot analysis of DNA from HPV16 cells digested with a restriction enzyme that does not cut the viral genome (HindIII, -) or which cuts the genome once (BamHI, +), showing the supercoiled (S), nicked circular (N), multimeric (M), and linear bands (L) indicative of episomal maintenance of the HPV16 genome. Graph: Cells containing episomal HPV16 were cultured in either monolayer (mono) or methylcellulose (MC) for 24 hours. Total RNAs were isolated and subjected to RT-qPCR analysis using E6–7 or E1^E4 primers. Ct values were normalized to cyclophilin A (Cyc) as an internal control and then to the monolayer sample for each experiment. Values represent the means of 10 (E6–7) or 15 (E1^E4) independent experiments and bars represent ± one standard error of the mean. c. Cells grown in either monolayer or MC were treated with DMSO or actinomycin D (ActD, 5μg/μl) for the indicated time. Total RNAs were subjected to RT-qPCR for the E1^E4 spliced product or the 18S rRNA. Ct values were normalized to Cyc, and the corresponding DMSO control in monolayer or MC was set to 1. Dotted lines represent the t1/2 for each decay curve. Bars represent ± one standard error of the mean.
Although factors that regulate the viral early promoter have been studied in great detail (Bernard, 2013), the differentiation-specific cellular factors that are responsible for upregulation of HPV late promoter activity remain unclear. One reason that late promoter regulation is poorly understood is that the promoter is imbedded within the E7 open reading frame in the viral genome (see Figure 1a). Consequently, genetic analysis of the late promoter in the context of the complete HPV genome has the potential to disrupt the essential functions of E7. Differentiation-dependent late promoter activation also depends on the virus being present in the cell as an episome (Frattini et al., 1996). Consequently, the tools available for analysis of late promoter regulation are quite restricted. Despite these challenges, previous work has shed light on late promoter regulation in high-risk HPVs. Viral genome amplification is not required for promoter activation (Bodily and Meyers, 2005; Spink and Laimins, 2005), making HPV late promoters different from late promoters of other viruses which depend on genome replication for their activities. Changes in overall chromatin structure and histone modifications are found in the promoter region upon differentiation (del Mar Pena and Laimins, 2001; Wooldridge and Laimins, 2008), although the functional significance of these changes is not clear. Studies using luciferase reporters have suggested that loss of transcriptional repressors such as CDP and YY1 (Ai et al., 2000; Ai et al., 1999; Kukimoto and Kanda, 2001; Sato et al., 2007) or binding of transcriptional activators such as hSkn-1a or C/EBPβ (Kukimoto and Kanda, 2001; Kukimoto et al., 2006) over the course of differentiation are important for regulating promoter activity. The viral enhancer found in the upstream regulatory region (URR; also called the long control region, LCR) can augment late promoter reporter activity in HPV31 (Bodily and Meyers, 2005). The viral protein E7 can activate the late promoter, although factors that promote cell cycle progression have an inhibitory effect (Bodily et al., 2013). Chromatin immunoprecipitation (ChIP) studies of the late promoter in the context of complete, episomal viral genomes have confirmed that a variety of transcription factors associate with the late promoter in both differentiated and undifferentiated conditions, including c-Myb, C/EBPα, C/EBPβ, NFAT, YY1, NF1, Oct-1, c-Jun, and Sp1 (Carson and Khan, 2006; Wooldridge and Laimins, 2008). Of these, only C/EBPβ has so far been shown to be functionally relevant in the context of infected cells (Gunasekharan et al., 2012), but how these various factors cooperate to regulate the transcriptional process remains unknown.
RNA polymerase II- (Pol II)-mediated transcription is regulated at many levels, including recruitment of the polymerase complex to the promoter, transcriptional initiation, polymerase pausing, and transcript elongation (Heidemann et al., 2013; Li et al., 2007). A crucial aspect of understanding transcriptional regulation is sorting out which step or steps in the transcriptional cycle control the overall activity of a given promoter. Following the binding of gene-specific transcription factors to the promoter and modification of the surrounding chromatin, the multi-subunit Mediator complex is recruited. Mediator coordinates recruitment of Pol II and the general transcription factors to the promoter to form the pre-initiation complex (PIC). Mediator also facilitates transcript initiation by Pol II. Thus, Mediator translates the binding of gene-specific transcription factors into transcriptional activity by Pol II (Allen and Taatjes, 2015; Malik and Roeder, 2010; Taatjes, 2010). At the majority of promoters following initiation, pausing factors arrest the polymerase complex 30–50 base pairs downstream of the transcriptional start site (Bataille et al., 2012; Hargreaves et al., 2009; Lenasi and Barboric, 2010; Rahl et al., 2010). Elongation factors are then recruited to the Pol II complex, which promote continued transcript synthesis until termination. Pol II can then be recruited again to the promoter to repeat the process. Each of these stages in the transcription cycle can be a point of regulation and are often associated with phosphorylation of the heptad repeats in the Pol II carboxy-terminal domain (CTD), which has been shown to be mediated by transcriptional cyclin dependent kinases (CDKs)(Adelman and Lis, 2012; Kohoutek and Blazek, 2012).
Transcriptional CDKs play critical roles in regulating Pol II activity and other aspects of transcriptional regulation. Transcription factor IIH (TFIIH), a general transcription factor and component of the PIC, includes CDK7/cyclin H as components and depends strongly on Mediator for its recruitment and activity (Boeing et al., 2010). CDK7 can phosphorylate serine 5 (Ser5) of the CTD heptad repeats of Pol II, which promotes transcriptional initiation (Heidemann et al., 2013; Kohoutek and Blazek, 2012; Lenasi and Barboric, 2010). Following initiation, positive transcription elongation factor b (P-TEFb), an elongation factor which contains CDK9/cyclin T, is recruited to the paused Pol II complex (Malik and Roeder, 2010). CDK9 phosphorylates serine 2 (Ser2) of the Pol II CTD heptad repeat motif, thus promoting the transition to transcription elongation (Kohoutek and Blazek, 2012; Lenasi and Barboric, 2010; Malumbres and Barbacid, 2005; Rahl et al., 2010). In addition to its role in transcriptional elongation, phosphorylation of the CTD by CDK9 stimulates assembly of RNA processing factors, facilitates alternative splicing, and regulates mRNA transport, thus functionally linking transcription with RNA processing (Lenasi and Barboric, 2010).
Several factors are known to facilitate recruitment of P-TEFb to the Pol II complex, including bromodomain protein 4 (Brd4), which binds to acetylated chromatin and helps P-TEFb associate with Mediator to promote recruitment (Donner et al., 2010; Dooley et al., 2016; Loven et al., 2013; Ren et al., 2016; Yang et al., 2005). Brd4 is one of several members of the bromodomain and extra-terminal domain (BET) family of transcriptional regulatory proteins, but Brd4 is the family member most associated with HPV (reviewed in (Iftner et al., 2016)). Brd4 binds to and regulates the HPV early promoter (Schweiger et al., 2006; Wu et al., 2006; Wu et al., 2016; Yan et al., 2010) and viral genome replication (Wang et al., 2013; Wu et al., 2016; You et al., 2005). The Mediator kinase subcomplex (CDK8/cyclin C) can cooperate with Brd4 to promote recruitment and activation of P-TEFb/CDK9 to promote transcriptional elongation (Allen and Taatjes, 2015; Donner et al., 2010; Galbraith et al., 2013; Hargreaves et al., 2009; Jang et al., 2005; Schroder et al., 2012; Yang et al., 2005). In this way, CDK8 and Brd4 positively regulate the synthesis of full length, mature transcripts. Importantly, CDK8 association with Mediator is incompatible with PIC formation but promotes transcriptional elongation (Allen and Taatjes, 2015). Thus the CDK8/kinase module serves as a molecular switch to help transition from initiation to the elongation phase of transcription.
We began this study by reasoning that transcription factors that regulate the late promoter would ultimately feed into the central CDK- and Mediator-dependent steps of transcription (Kohoutek and Blazek, 2012; Malumbres and Barbacid, 2005). We wished to determine the pattern of binding and activity of these transcriptional regulatory factors at the HPV16 late promoter, enhancer, and downstream sequences during differentiation-dependent promoter upregulation in order to deduce which step(s) of the transcriptional cycle are most important for activation. Our findings support a model that the differentiation-dependent activation of the HPV16 late promoter is not regulated at the level of transcriptional initiation or PIC formation. Instead, recruitment of the CDK8 module of the Mediator complex, followed by association of CDK9, is the critical step in late promoter activation. Additionally, BET family proteins contribute to differentiation-dependent association of CDK8 with the viral genome. These findings suggest that HPV has evolved to use regulated transcriptional elongation as a means of controlling viral late gene expression.
Materials and methods
Plasmids and drugs
The late promoter reporter (pGL2-16 late pro) was described previously (Bodily et al., 2011a). pGL2 -LCR/Late was created by PCR using the primers 16LCR 5′ and 16Late pro 3′ (Supplementary Table 1) with pUCHPV16 as a template and cloning the fragment into the HindIII/KpnI site of pGL2b. pGL2 LCR was created similarly using the primers16LCR5′ and 16 early 3′. pGL2 -LCR/Late TATA and pGL2 LCR/TATA were created using site-directed mutagenesis with the QuickChange II XL Site Directed Mutagenesis kit (Agilent) using p97TATA 5′ and p97TATA 3′ primers listed in Supplementary Table 1. The HPV31 late promoter reporter was described previously (Spink and Laimins, 2005). JQ1 (Filippakopoulos et al., 2010) was a generous gift from James Bradner (Dana Farber Cancer Institute, Harvard Medical School). Flavopiridol and actinomycin D were obtained from Sigma. Senexin A was obtained from Tocris. All drugs were dissolved in DMSO.
Cell culture, transfection, and CRISPR
Human foreskin keratinocytes (HFKs) were isolated from neonatal foreskins and HFKs containing HPV16 genomes (HPV16 cells) were created by transfection and selection as previously described (Bodily et al., 2011a; Wilson and Laimins, 2005). Cells containing HPV31 genomes were created similarly (Wilson and Laimins, 2005). HFKs and keratinocyte-derived cell lines were cultivated in E medium with 5% fetal bovine serum in the presence of mitomycin C-treated NIH-3T3 J2 fibroblast feeders (Wilson and Laimins, 2005). Differentiation was induced by suspending cells in E medium containing 1.6% methylcellulose (MC) for 24 hrs, followed by washing with phosphate buffered saline (PBS)(Wilson and Laimins, 2005). Drugs were added to MC or to monolayer control media prior to adding the cells. Cell lines derived from at least three donors were used in each experiment. Episomal maintenance of the viral DNA was confirmed by Southern blotting as described (Fehrmann et al., 2003).
For CDK8 knockdown experiments, three donor backgrounds of HPV16 cells were infected with either control LentiCRISPR lentivirus particles or particles targeting CDK8 (Sigma) along with polybrene (2ug/ml). Feeder fibroblasts were added 6 hrs after infection followed by puromycin (5ug/ml) selection. Following selection, cells were seeded in monolayer or methylcellulose for 24 hrs and analyzed for CDK8 protein levels using western blotting. Cell lines with less than 50% CDK8 were analyzed for HPV transcript and genome levels.
Luciferase assays
Keratinocytes transfected with reporter plasmids overnight using polyethyleneimine (PEI; Polysciences) were trypsinized, and then divided, one half into monolayer and one half into MC culture (Bodily et al., 2011b). A plasmid expressing Renilla luciferase was cotransfected as an internal control (Bodily et al., 2013). The total level of DNA in each transfection was kept constant in each sample by addition of empty vector DNA. Following 24 hrs of incubation, lysates were prepared and assayed for luciferase activity using the Dual-Luciferase® Reporter Assay System (Promega) according to manufacturer’s instructions.
RNA extraction, qPCR, and Western blotting
Total RNA was isolated using RNA-STAT 60™ (TelTest, Inc), digested with RNAs-free DNAse (Promega), phenol-chloroform extracted, and reverse transcribed using qScript (Quanta) as described previously (Bodily et al., 2013). Specificity for RT-qPCR for RNA rather than contaminating DNA is shown in Supplementary Figure 1. Quantitative PCR was performed using the PerfeCTa® SYBR® Green SuperMix ROX (Quanta) on an Applied Biosystems StepOne Plus™ real time PCR machine using the primers listed in Supplementary Table 1. Cyclophilin A (Cyc) was used as an internal housekeeping control because its levels are stable throughout keratinocyte differentiation (Steele et al., 2002). Where appropriate, curves were fit using Microsoft Excel. Western blotting was performed as reported previously (Bodily et al., 2011a), except that blocking and antibody dilution were performed using Odyssey Blocking Buffer (Li-Cor). Antibodies are listed in Supplementary Table 2. Blots were imaged using near-IR secondary antibodies on a Li-Cor Odyssey Infrared Imager. Band intensities were measured using Image J.
Chromatin immunoprecipitation
Cells grown in monolayer culture were trypsinized and resuspended in 10 ml E medium. Cells grown in MC were washed with PBS and resuspended in 10 ml E medium. Formaldehyde was added to a final concentration of 1% and incubated for 15 minutes at room temperature with rocking. 1 ml 1.25M glycine was added and incubated for an additional 5 minutes at room temperature. Cells were then washed three times with ice cold PBS containing protease inhibitors and resuspended in 1x Lysis Buffer (Cell Signaling) at a final cell density of 10 million cells/ml. Cells were sonicated briefly and then treated with micrococcal nuclease (final concentration 60 U/μl, New England Biolabs) for 1 hr on ice. EDTA was added to a final concentration of 50 mM and debris was removed by centrifugation. Chromatin was diluted in ND buffer (20 mM Tris pH 8, 137 mM NaCl, 1% NP-40, 10% glycerol, and 2 mM EDTA) containing protease inhibitors and pre-cleared with blocked Protein G Dynabeads (Invitrogen). Following pre-clearing, antibodies (Supplementary Table 2) were added and incubated overnight at 4°C with rotation. Pol II ChIPs included the addition of anti-IgY following one hour of incubation with anti-Pol II antibodies. Pre-blocked Dynabeads were then added and incubated at 4°C with rotation for 1–3 hrs. Beads were washed twice with ND buffer containing protease inhibitors, twice with ND buffer containing 0.3 M NaCl, twice with LiCl buffer (250 mM LiCl, 0.5% NP-40, 0.5% sodium deoxycholate), once with TE buffer containing 0.2% triton X-100, and once with TE buffer alone. Beads were resuspended in TE buffer containing 0.3% SDS, 200 mM NaCl, and 0.5 mg/ml proteinase K (Sigma) and incubated for 2 hrs at 45°C followed by 65°C overnight. Supernatants were removed and beads were washed with TE buffer containing 500 mM NaCl. DNA was purified using the PCR Clean-up DNA Purification Kit (MoBio). Immunoprecipitated DNA fragments were subjected to qPCR as described above using the primers listed in Supplementary Table 1. Significance was calculated using Welch’s unequal variances t test.
Results
We sought in our studies to determine what core transcriptional regulatory factors could be responsible for upregulation of transcripts derived from the HPV16 late promoter upon differentiation (Figure 1a). Because the differentiation-dependent increase in transcripts from the late promoter is only seen when mucosal HPVs are episomal (Frattini et al., 1996), human foreskin keratinocytes (HFKs) harboring episomally replicating HPV16 genomes (HPV16 cells) were used in our studies. These cell lines were created by transfecting the HPV16 genome into primary foreskin keratinocytes, followed by drug selection and outgrowth of immortalized clones (Bodily et al., 2011a; Wilson and Laimins, 2005). HPV16 cells derived from at least three different donors were used in each of our experiments to account for donor-to-donor variability. Episomal status of the viral genome in these cells was confirmed regularly by Southern blot analysis (Figure 1b, inset). HPV16 cells grown in monolayer culture are maintained in an undifferentiated state. Differentiation of HPV-containing keratinocytes can be achieved in several ways. In these studies we take advantage of the fact that cells suspended in methylcellulose (MC) for 24–48 hours undergo sufficient cellular differentiation to support increased levels of intermediate and late viral transcripts (Bodily et al., 2013; Ruesch et al., 1998; Wilson and Laimins, 2005)(Figure 1b, see Supplementary Figure 3a for cellular differentiation markers). HPV transcripts are heterogeneous, possessing many alternatively spliced forms; but the E1^E4 splice is found in the majority of transcripts originating from the late promoter (Ozbun and Meyers, 1999; Van Doorslaer et al., 2013). E1^E4-containing transcripts can also originate from the early promoter, but since the early promoter-derived transcripts (E6–E7) are not substantially increased upon differentiation in our system (Figure 1b), increased levels E1^E4-containing transcripts upon differentiation are derived primarily from the late promoter. We also observe an increase in L1/L2 transcripts upon culture in MC (not shown), but L1/L2 transcripts are under additional levels of post-transcriptional control, including the need to bypass the early polyadenylation signal (Schwartz, 2013; Terhune et al., 2001). L1/L2 levels are therefore less suitable as a measure of late promoter activation. Thus we use an increase in the levels of the E1^E4 spliced product as determined by RT-qPCR as a measure of late promoter upregulation upon differentiation.
The HPV16 late promoter is transcriptionally rather than post-transcriptionally regulated
At the outset of our studies it was important to resolve an important issue regarding HPV late promoter activity. Although the HPV late promoters are frequently referred to as “differentiation inducible,” it had not been demonstrated whether the increase in transcripts originating from the native viral late promoter upon differentiation is due to increased synthesis (transcriptional activation) or decreased degradation (transcript stabilization). Given the extensive post-transcriptional processing signals present in HPV transcripts (see (Schwartz, 2013) for a review), it is possible that differentiation-dependent transcript stabilization rather than increased transcription rate is responsible for the abundance of intermediate and late transcripts in differentiated cells. Before proceeding with further studies of late promoter regulation, we considered it important to clearly resolve whether transcript stabilization or transcriptional activation is responsible for the increase in late promoter-derived transcripts upon differentiation.
To measure transcript stability, HPV16 cells were grown in either monolayer or MC culture for 22 hours. The mRNA synthesis inhibitor actinomycin D (ActD) was then added to the cultures for various lengths of time before harvest, for a total time of 30 hours in MC culture. DMSO-treated cultures were used as controls. Because ActD prevents the synthesis of new mRNAs, the decline in mRNA levels over time represents a measure of the rate of mRNA decay (Jeon and Lambert, 1995). RNAs were harvested and the levels of E1^E4 transcripts were measured by RT-qPCR. DMSO controls were set to 1 in both monolayer and MC culture. As seen in Figure 1c, E1^E4 transcripts declined over time upon ActD treatment in both monolayer and MC. The decay curves of RNAs from cells grown in monolayer and those from MC were superimposable, each with a half-life of around 84 minutes. By contrast, 18S rRNA and Cyc levels were stable throughout the duration of this experiment (Figure 1c and not shown). These data show that E1^E4 spliced transcripts are equally labile in both differentiated and undifferentiated cells. Therefore, the increase in late promoter-derived transcripts upon differentiation cannot be accounted for by changes in transcript stability and must therefore be due to increased transcript synthesis.
The LCR contributes to late promoter activity
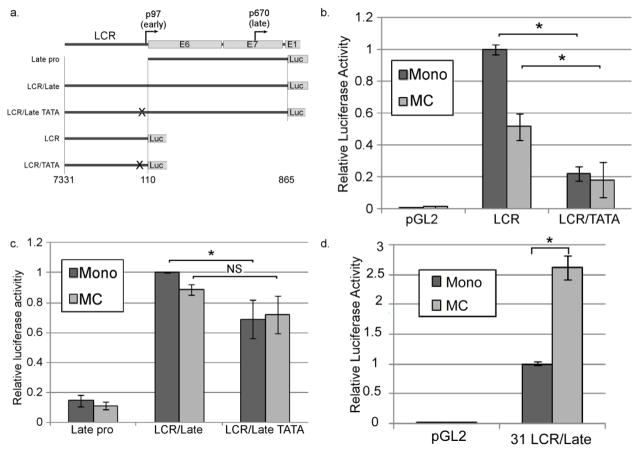
Previous work using luciferase reporters derived from HPV31 showed that the enhancer elements in the viral LCR are necessary for differentiation-dependent late promoter upregulation (Bodily and Meyers, 2005). We tested whether the LCR also influences late transcription in HPV16. Because the LCR is also important for the activity of the early promoter and contains the viral origin of replication, we could not test the role of the LCR in the context of the complete viral genome. Instead, we cloned fragments of the HPV16 genome containing either the late promoter region alone (downstream of the early promoter, Late Pro) or the late promoter region together with the early promoter and LCR (LCR/Late; Figure 2a). Luciferase was placed in the position of the E1 ORF. An early promoter reporter was also made by placing luciferase in the position of E6 to test the activity of the LCR alone. Because the early promoter is contained within the LCR/Late reporter, a version of each reporter plasmid was made containing a mutation in the early promoter TATA box to reduce the contribution of the early promoter to luciferase expression. These plasmids (LCR, LCR/TATA, LCR/Late, LCR/Late TATA) were transfected into HPV16 cells, which were then cultured in either monolayer or MC for 24 hours, followed by luciferase assay. HPV16 cells were used for these experiments to ensure that any virally encoded or induced factors needed for promoter regulation would be present (Bodily et al., 2013). Mutation of the TATA box in the context of the early promoter/LCR reporter (LCR/TATA) significantly reduced early promoter activity as expected (Figure 2b). Analysis of the late promoter reporters (Figure 2c) allows us to make several conclusions. 1) Inclusion of the LCR resulted in substantially higher levels of luciferase activity than the Late Pro construct alone. This suggested that the enhancer elements in the LCR contribute strongly to late promoter activity. 2) Mutation of the early TATA box resulted in a modest but significant decrease in luciferase activity in monolayer, indicating that the early promoter does contribute to transcription as far downstream as the E1 ORF, as predicted by transcript mapping (Ozbun and Meyers, 1998; Van Doorslaer et al., 2013). 3) Despite loss of the early TATA box in the LCR/Late TATA construct, high levels of luciferase activity were still seen indicating that the majority of the activity of the LCR/Late construct was attributable to the late promoter. 4) Luciferase levels were not increased when cells were grown in MC, in contrast to reporters containing the analogous region of HPV31, which are upregulated upon growth in MC (Figure 2d)(Bodily and Meyers, 2005). These observations suggest that the LCR contributes to overall late promoter activity, but what is needed for differentiation induced activation of the late promoter is not captured by these reporter constructs, and thus the upstream cis elements may not be sufficient to mediate promoter upregulation.
Figure 2.
The LCR contributes to late promoter activity. a. The portions of the viral genome included in the reporter plasmids. b and c. HPV16-containing cells were transfected with the indicated reporter and cultivated in either monolayer or MC for 24 hrs. Luciferase activities in lysates were measured and normalized to Renilla luciferase as an internal control. d. HPV31-containing cells were transfected with a reporter consisting of the HPV31 LCR and late promoter sequence (analogous to LCR/Late) and analyzed as in b and c. Values represent the means of 4 (b), 9 (c), or 6 (d) experiments and bars represent ± one standard error of the mean.
Processive elongation rather than recruitment of Pol II to the late promoter correlates with late promoter activation
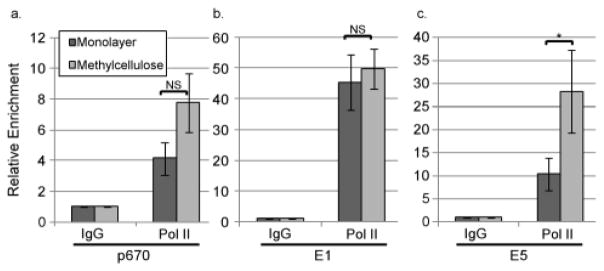
The observation that promoter activity does not increase from our reporters upon differentiation suggests that some aspect of the transcription cycle other than promoter activity itself may be regulated upon differentiation. Pol II recruitment, initiation, pausing, and elongation can all serve as points of regulation for transcription, so we sought to better understand whether and how the association of Pol II with promoter and downstream sequences changes during differentiation. HPV16 cells were grown in monolayer culture, which represents the inactive state of the promoter, or MC culture in which the promoter is activated. Following 24 hours of culture, chromatin was crosslinked and harvested, and chromatin immunoprecipitation (ChIP) was performed using antibodies specific for Pol II. Following ChIP, qPCR was performed to measure the levels of viral DNA recovered in the ChIP, which serves as an indicator of the amount of Pol II associated with viral DNA. qPCR signals were normalized to the levels of input DNA to eliminate the effect of any changes in viral genome copy number during differentiation. The mean increase in Pol II binding upon differentiation to the region surrounding the late promoter start site (p670, 635–745) on a per genome basis approached but did not achieve significance (p=0.06)(Figure 3a). In some experiments, we observed that Pol II recruitment did increase at the late promoter upon differentiation. In other experiments, we did not observe increased Pol II binding, even in samples in which late promoter-derived transcripts were upregulated. These observations suggest that the increased level of promoter activity during differentiation may not result from increased recruitment of the polymerase to the promoter region.
Figure 3.
Pol II occupancy at the late promoter does not significantly increase upon differentiation but does increase near the polyadenylation site. Chromatin immunoprecipitation (ChIP) was performed using non-specific IgG or antibodies specific for Pol II on chromatin from HPV16 cells grown in monolayer or MC for 24 hours. qPCR was performed on the ChIP products using primers targeting (a) the late promoter/p670 region, (b) the E1 open reading frame, or (c) the E5 open reading frame. Ct values were normalized first to input and then to the IgG control for each experiment. Values represent means of 5 (a), 3 (b), or 4 (c) experiments and bars represent ± one standard error of the mean. *=p<0.05, NS=p>0.05.
Most cellular promoters are pre-populated with Pol II that does not engage in productive transcription but is paused at a promoter proximal site until a signal is received to recruit P-TEFb and trigger transcript elongation (Hargreaves et al., 2009). If the late promoter is regulated at the level of pause release, we expected that regions downstream of the promoter would show a differentiation-dependent increase in the levels of Pol II present. We used ChIP to examine the levels of Pol II in two regions downstream of the promoter. The E1 region is located about 300 nucleotides downstream of the late promoter start site and would thus be downstream of a promoter proximal pause site. If Pol II promoter-proximal pausing is released in response to differentiation, we would expect to see increased levels of Pol II in the E1 region. However, the occupancy of E1 by Pol II did not change (Figure 3b), indicating that Pol II transits the E1 ORF at similar levels regardless of differentiation. The E5 ORF is located immediately before the early polyadenylation signal, and would thus only be occupied by Pol II that had efficiently elongated over 3200 base pairs and was approaching termination. Most transcripts originating from the late promoter in cells grown in MC utilize the early polyadenylation signal. (Ozbun and Meyers, 1998; Terhune et al., 2001; Van Doorslaer et al., 2013). When we examined the E5 region, we found that occupancy by Pol II is increased significantly upon differentiation (Figure 3c). These results suggest that elongation, and specifically the ability of Pol II to processively elongate transcripts through the length of the entire early region, is the step in transcript synthesis that is differentiation-dependent.
CDK9 is recruited downstream of the late promoter upon differentiation
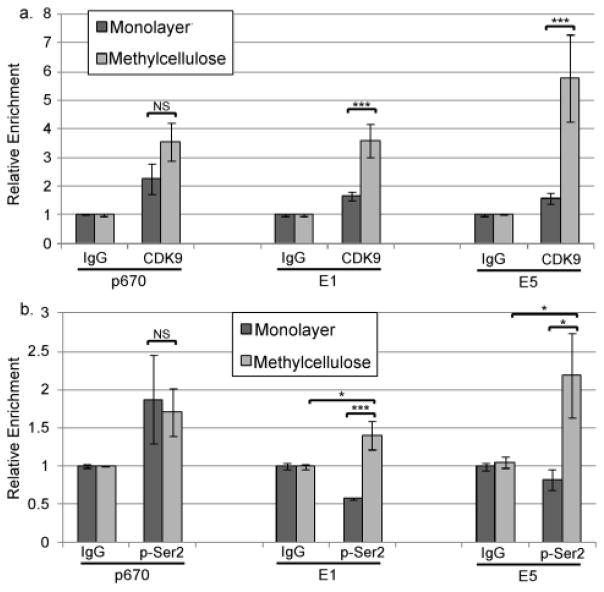
To further investigate whether elongation is critical for late promoter activation, we examined P-TEFb, a critical component of the Pol II elongation complex (Kohoutek and Blazek, 2012; Lenasi and Barboric, 2010; Malumbres and Barbacid, 2005; Rahl et al., 2010). P-TEFb is recruited to paused Pol II complexes by Brd4 and/or Mediator and the CDK9 subunit phosphorylates Ser2 of the CTD, leading to pause release and elongation. P-TEFb continues to be associated with elongating Pol II, phosphorylating additional Ser2 repeats as the complex moves along the length of the gene body up to the termination signal (Kohoutek and Blazek, 2012; Lenasi and Barboric, 2010). Therefore if transcriptional elongation is the step in mRNA synthesis that is regulated by differentiation, we would expect to see an increase in P-TEFb occupancy in cells grown in MC. ChIP using antibodies specific for CDK9 showed that, like Pol II, the level of CDK9 present around the promoter start site p670 tended to increase upon differentiation but the increase did not achieve statistical significance. In contrast, the occupancy of the E1 and E5 regions by CDK9 was significantly increased in differentiating cells (Figure 4a), suggesting that differentiation facilitates association of P-TEFb with Pol II complexes downstream of the late promoter. Although total recovery using pSer2-specific antibodies in our ChIP assays was not high, we could detect a significant accumulation of pSer2-phosphorylated Pol II upon differentiation at sites downstream of the promoter (Figure 4b), suggesting that CDK9 recruited to the Pol II complexes is active.
Figure 4.
CDK9 recruitment is upregulated at downstream sites upon differentiation. ChIP was performed using non-specific IgG or antibodies specific for (a) CDK9 or (b) Ser2-phosphorylated Pol II using chromatin from HPV16 cells grown in monolayer or MC for 24 hours. qPCR was performed on the ChIP products using primers targeting p670, E1, or E5 regions. Ct values were normalized first to input and then to the IgG control for each experiment. Values represent means of 3 experiments and bars represent ± one standard error of the mean. *=p<0.05, ***=p<0.01, NS=p>0.05.
CDK9 activity is required for late promoter upregulation
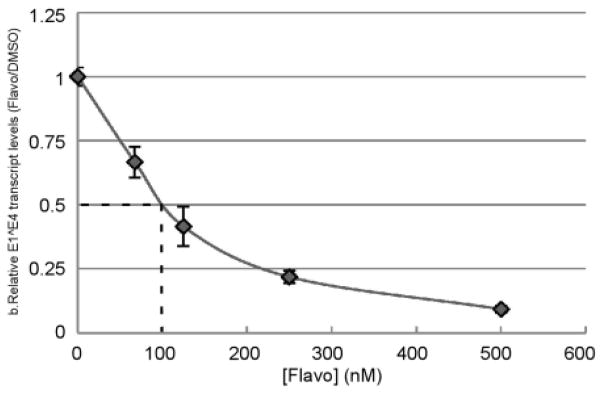
To investigate the functional significance of CDK9 for late promoter activity, we treated HPV16 cells with flavopiridol, a CDK inhibitor specific for CDK9 (Senderowicz, 2003). As shown in Figure 5, fold induction of late transcripts upon differentiation was inhibited by flavopiridol in a dose-dependent manner with an IC50 of around 100 nM, which is within the concentration range that specifically inhibits transcriptional elongation (Donner et al., 2010; Patel et al., 2013). We also attempted shRNA-mediated knockdown of CDK9, but were unsuccessful in obtaining stable clones (not shown). We do not know whether flavo directly affects recruitment of CDK9 to the viral genome, but in combination with the ChIP data, these results suggest that transcript elongation is a critical component of late viral gene regulation and that recruitment and activity of CDK9 are critical for the induction of late transcripts during differentiation.
Figure 5.
Late transcription requires CDK9 activity. Cells containing episomal HPV16 were cultured for 24 hrs in monolayer or MC in the presence of various concentrations of flavopiridol (flavo), and total RNAs were subjected to RT-qPCR analysis using primers specific for the E1^E4 spliced mRNA. Fold induction of E1^E4 transcripts upon differentiation in the DMSO-treated sample within each experiment was set to 1. The dotted line represents the IC50. Values represent the average of 6 experiments and bars represent ± one standard error of the mean. ***=P<0.01, NS=p>0.05.
CDK8 associates with the viral genome upon differentiation
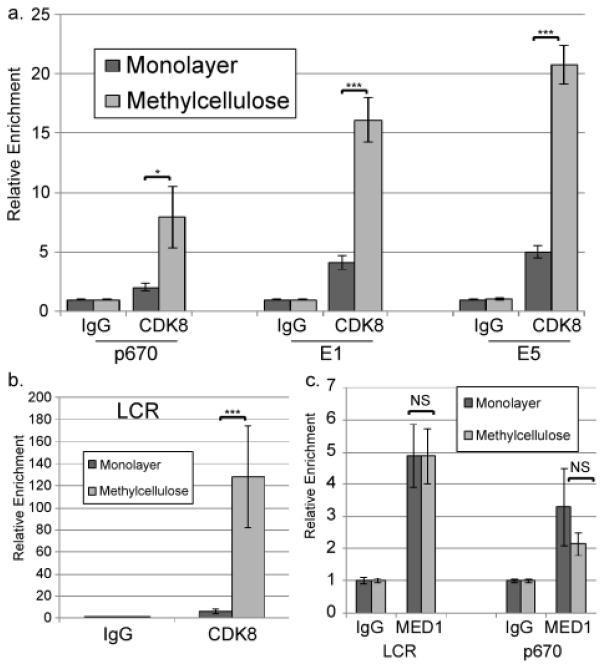
CDK8, a component of the kinase module of the Mediator complex (Allen and Taatjes, 2015; Donner et al., 2007a; Malik and Roeder, 2010), is one of several factors that can promote recruitment of CDK9/P-TEFb to facilitate transcriptional elongation. To understand how CDK9 is recruited to the viral genome, we investigated whether CDK8 is associated with episomal HPV genomes and how this association changes upon differentiation. ChIP analysis determined that CDK8 could be detected at the late promoter/p670 in undifferentiated cells, but it is substantially increased in cells grown in MC (Figure 6a). Others have found that CDK8 can be observed throughout transcribed cellular gene bodies, suggesting that it can be part of elongating transcriptional complexes (Donner et al., 2007a). Similarly, we observed CDK8 association with downstream sites upon differentiation, consistent with the possibility that CDK8 may accompany the Pol II holoenzyme during transcription of the HPV genome (Figure 6a). Mediator functions in part by serving as a bridge between specific DNA binding transcription factors and general transcriptional machinery, including the Pol II complex and P-TEFb (Allen and Taatjes, 2015). As part of that function, Mediator plays a role in creating and maintaining DNA loops that allow association of enhancer elements with promoters (Allen and Taatjes, 2015). Given the result above that the viral LCR acts to increase the activity of the late promoter (Figure 2b), we tested whether CDK8 might associate with the LCR during differentiation upstream of late promoter activation. ChIP analysis showed that CDK8 is found at the enhancer region in the LCR, and that this association increases dramatically upon differentiation (Figure 6b). Thus association of CDK8 with the viral genome correlates with late promoter activation.
Figure 6.
CDK8 is recruited to the viral genome upon differentiation. ChIP was performed using non-specific IgG or antibodies specific for CDK8 (a, b) or MED1 (c) on chromatin from HPV16 cells grown in monolayer or MC for 24 hours. qPCR was performed on the ChIP products using primers specific for the p670 region (a, c), the E1 region (a), the E5 region (a), or the central LCR (b, c). Ct values were normalized first to input and then to the IgG control for each experiment. Values represent means of 3 (a), 6 (b), or 3 (c) experiments and bars represent ± one standard error of the mean. *=p<0.05, ***=p<0.01, NS=p>0.05.
CDK8 is a component of the kinase module of Mediator, which reversibly associates with the larger Mediator complex at various stages of transcription (Allen and Taatjes, 2015). The kinase module is not associated with Mediator during the assembly of the PIC or initiation of transcription, but it is recruited to facilitate the transition from initiation to elongation (Allen and Taatjes, 2015). We sought to determine whether the recruitment of CDK8 is associated with recruitment of the larger Mediator complex or whether CDK8 is recruited separately. ChIP using antibodies against the core Mediator component MED1 showed that Mediator is associated with the LCR, the late promoter, and the early promoter in HPV16 cells grown in monolayer culture (Figure 6c and Supplementary Figure 2a). However, unlike CDK8, the levels of MED1 binding did not increase upon differentiation at any of these sites. These results suggest that, like Pol II, Mediator is already present on the viral genome, but that differentiation-dependent recruitment of the CDK8/kinase module is associated with transcriptional activation of the late promoter.
CDK8 is functionally important for late promoter activity
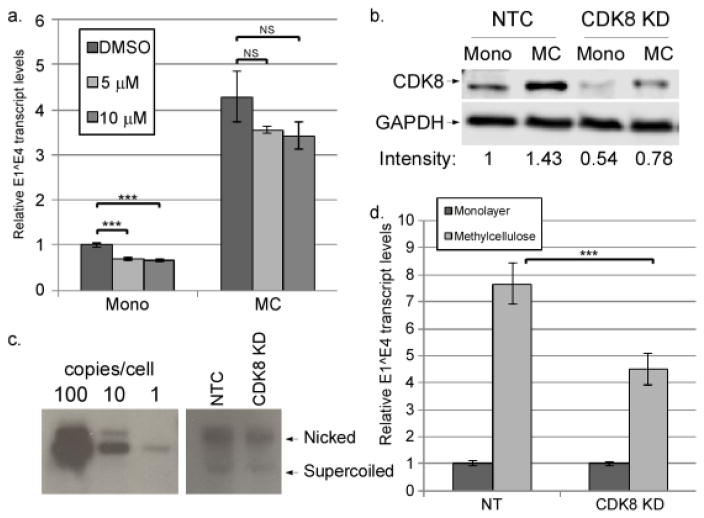
Data so far indicated that CDK8 occupancy at both the late promoter and the LCR enhancer correlates strongly with late promoter activation. The mechanisms by which CDK8 serves to recruit P-TEFb to target genes are not clear. Previous studies show that shRNA-mediated knockdown of CDK8 results in a failure to recruit P-TEFb (Galbraith et al., 2013), but it is not known whether kinase activity of CDK8 is involved or whether P-TEFb recruitment occurs through protein-protein interactions. To determine whether the kinase activity of CDK8 is important for HPV16 late promoter activation, we treated HPV16 cells in monolayer or MC culture with various concentrations of senexin A, a CDK8 kinase inhibitor (Porter et al., 2012), and measured the upregulation of E1^E4 transcripts upon culture in MC. We found that senexin A had a statistically significant but modest suppressive effect on E1^E4 transcripts in monolayer (which primarily originate from the early promoter), but no effect in MC, in which E1^E4 transcripts are primarily made from the late promoter (Figure 7a). This suggests that differentiation-dependent late promoter activation is independent of CDK8-mediated phosphorylation. Western blotting against the CDK8 phosphorylation site in STAT1, Ser-727 (Bancerek et al., 2013; Poss et al., 2016) confirmed that senexin A can inhibit CDK8 in our system (Supplementary Figure 2b)
Figure 7.
CDK8 contributes to late promoter activation, but not through its kinase activity. a. HPV16 cells were cultured for 24 hrs in monolayer or MC in the presence of the indicated concentration of senexin A or DMSO. Total RNAs were isolated and subjected to RT-qPCR analysis using primers specific for the E1^E4 spliced mRNA. Ct values were normalized to Cyc and then to the DMSO control. b and c. CDK8 was knocked down in HPV16-containing cells using CRISPR technology. b. A representative western blot of CDK8 levels in lysates from HPV16 cells infected with CDK8-CRISPR lentiviruses (CDK8 KD) or non-target control lentivirus (NTC) grown in monolayer (mono) or methylcellulose (MC). Relative CDK8 band intensities are indicated below the blots. c. A representative Southern blot showing HPV16 genome copy number in NTC and CDK8 KD cells. Copy number controls corresponding to 100, 10, or 1 viral genome copy per cell are shown. d. Total RNAs were isolated from non-target control or CDK8 KD cells and levels of E1^E4 transcripts were measured using RT-qPCR as in (a). Values represent the means of 4 (a) or 5 (d) experiments and bars represent ± 1 standard error of the mean. ***=P<0.01, NS=p>0.05.
Previous studies have shown that knockdown of CDK8 affects a different spectrum of genes than inhibition of kinase activity, suggesting that CDK8 plays a structural role that is independent of kinase activity (Poss et al., 2016). CDK8 has been successfully knocked down by others, although with some negative effects on cellular growth and viability (Galbraith et al., 2010; Poss et al., 2016). Using CRISPR technology, we were able to obtain a partial knockdown of CDK8 in HPV16-containing keratinocytes in both monolayer and MC (Figure 7b). Cell growth was not significantly altered (not shown), and the copy number of HPV16 episomal genomes was not noticeably different in CDK8 knockdown cells as measured by Southern blot (Figure 7c) or quantitative PCR (not shown). Importantly, the partial knockdown of CDK8 resulted in a corresponding reduction in E1^E4 levels upon late promoter activation in cells grown in MC (Figure 7d). These results indicate that CDK8 is important for late promoter activation, likely through mechanisms other than its kinase activity.
BET family proteins are necessary for late promoter activation
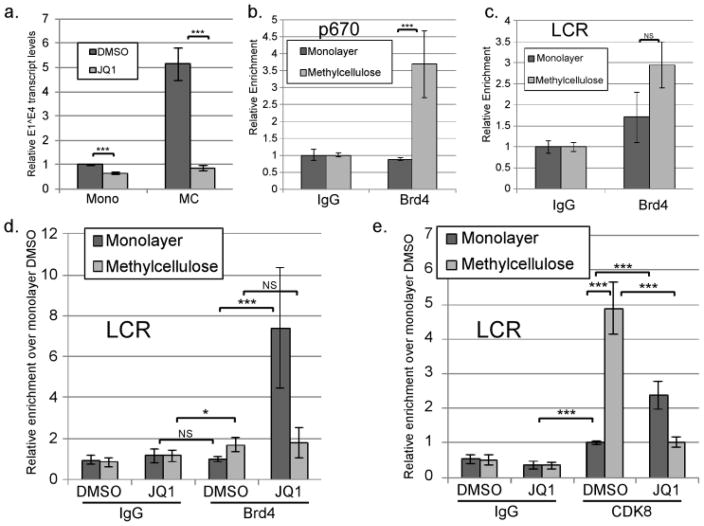
Brd4 is another cellular factor that can recruit CDK9/P-TEFb to promoters, sometimes in cooperation with CDK8 (Hargreaves et al., 2009; Kohoutek, 2009; Patel et al., 2013; Yan et al., 2010). Brd4 is important for regulation of the HPV early promoter and viral DNA replication (Iftner et al., 2016; Schweiger et al., 2006; Wang et al., 2013; Yan et al., 2010). To determine whether Brd4 or another BET family protein has a functional role in late promoter activation, we cultured cells containing episomal HPV16 genomes in monolayer or MC in the presence or absence of JQ1, which interacts with the bromodomains of BET family proteins and prevents binding to acetylated chromatin (Filippakopoulos et al., 2010). Treatment with JQ1 completely blocked late promoter activation in MC at the relatively low concentration of 250 nM (Figure 8a). JQ1 at this concentration did not inhibit early promoter activity as measured by E6–E7 transcript levels (Supplementary Figure 2c). The modest reduction in E1^E4 levels in monolayer may be due do inhibition of basal or residual late promoter activity in monolayer. JQ1 at this concentration also did not have an effect on cell growth or survival (not shown).
Figure 8.
CDK8 binding to the viral genome depends on BET family proteins and correlates with Brd4 association. HPV16 cells were grown in monolayer or MC for 24 hours in the presence of DMSO or 250 nM JQ1. a. Total RNAs were isolated and subjected to RT-qPCR analysis using primers specific for the E1^E4 spliced mRNA. Ct values were normalized to Cyc and then to the corresponding DMSO control. Values represent the means of 9–11 experiments. ChIP was performed using non-specific IgG or antibodies specific for Brd4 (b-d) or CDK8 (e). qPCR was performed on the ChIP products using primers specific for p670/late promoter (b) or the central LCR (c–e). Ct values were normalized to the input and IgG control and then to the monolayer DMSO sample for each experiment. Values represent means of 3 experiments and bars represent ± one standard error of the mean. *=p<0.05, ***=p<0.01, NS=p>0.05.
Although JQ1 can inhibit other BET family proteins, we chose to focus on Brd4 because of its involvement in elongation and its known role in HPV transcriptional regulation (reviewed in Iftner et al., 2016). To determine whether Brd4 might directly regulate the late promoter, ChIP analysis was performed. We were unable to detect Brd4 binding to the viral genome at the late promoter in undifferentiated cells, but binding was detected in cells differentiating in MC (Figure 8b). Brd4 is also known to bind to enhancer elements, including the LCR enhancer elements of integrated HPV16 genomes (Di Micco et al., 2014; Dooley et al., 2016; Nagarajan et al., 2014). Given its role in regulating the early promoter (Jha et al., 2010; Wang et al., 2013; Yan et al., 2010) and the late promoter (Figure 8a), we were surprised to find Brd4 was only weakly detectible at the LCR and that the trend toward increased binding to the LCR upon differentiation was not significant (Figure 8c). Therefore even though Brd4 and CDK8 both associate with the enhancer elements in the LCR, only CDK8 binding increases appreciably upon differentiation. Interestingly, ChIP analysis showed that Brd4 binding increased significantly upon differentiation at the early promoter, p97 (Supplementary Figure 2d), although early promoter activity was not strongly increased by differentiation (Figure 1b).
Next we performed ChIP analysis targeting Brd4 binding in cells treated with DMSO or JQ1 in either monolayer or MC. Again, Brd4 was not detectible on the viral LCR in monolayer but was detectible at low levels upon differentiation (Figure 8d). Upon treatment with JQ1, however, association of Brd4 with the LCR in monolayer culture unexpectedly and dramatically increased. We interpret this result to mean that, because JQ1 disrupts association of Brd4’s bromodomains with acetylated chromatin, Brd4 becomes displaced from the host chromosomes making it more mobile in the nucleus (Korb et al., 2015), and therefore available for binding to the viral genome. A similar observation was made in the context of HSV1 infection, in which treatment with JQ1 caused an increased association of Brd4 with HSV1 promoters (Ren et al., 2016). Because of the presence of JQ1, increased binding of Brd4 to the viral genome must not be mediated through the bromodomains but instead through one of the other domains of the protein. JQ1-induced Brd4 association in monolayer cultures did not result in a corresponding increase in transcriptional activity of either the early or late promoter (Figure 8a and Supplementary Figure 2e). In contrast to the effect in monolayer, Brd4 binding to the viral genome did not increase upon treatment with JQ1 in MC culture (Figure 8d). This finding indicates that the mechanism of association between Brd4 and the LCR differs between monolayer and MC.
Inhibition of bromodomains impacts recruitment of CDK8 to the viral genome
Because JQ1 displaces Brd4 from binding to acetylated chromatin, and JQ1 prevented late promoter upregulation (Figure 8a), our data suggest that binding of Brd4 (or perhaps another BET family protein) to acetylated chromatin may be required for promoter activation during differentiation. Brd4 can be found in some variations of the Mediator complex and can cooperate with CDK8 in recruiting elongation factors to promoters (Jiang et al., 1998; Wu and Chiang, 2007). To further examine the role of Brd4 in late promoter activation, we tested whether JQ1 treatment could affect CDK8 recruitment to the genome. We reasoned that Brd4 may associate with the viral genome upon differentiation and recruit CDK8/Mediator, which then promotes transcriptional elongation through protein-protein interactions. If so, then treatment with JQ1 should reduce or eliminate CDK8 recruitment to the genome in differentiated cells. As we had observed before, CDK8 binding to the LCR was robust and increased substantially upon differentiation in cells treated with DMSO. Significantly, binding of CDK8 to the viral genome in MC was dramatically reduced in the presence of JQ1, suggesting that Brd4 or another BET family protein is necessary to recruit CDK8 to the viral genome in differentiating cells (Figure 8e). Brd4 could recruit CDK8 directly, but we also observed that JQ1 was able to efficiently inhibit cellular differentiation, as measured by the induction of the differentiation-dependent genes K10, TGM1 (Supplementary Figure 3a), FLN, and INV (not shown). Thus JQ1 may prevent differentiation-dependent CDK8 recruitment to the viral genome by disrupting overall differentiation of the cells. However, like Brd4, CDK8 is recruited to the viral genome upon treatment of cells in monolayer with JQ1 (Figure 8e). Because JQ1 specifically targets bromodomain-containing proteins such as Brd4, and CDK8 does not contain a bromodomain, Brd4 is likely recruiting CDK8 to the viral genome directly. Association of CDK8 with the E5 region of the genome followed a very similar pattern to that seen at the LCR: differentiation-dependent increase in association blocked by JQ1 treatment, and an increase in binding in monolayer culture induced by JQ1 (Supplementary Figure 3b). From these data we conclude that CDK8 is recruited to the genome in a BET protein-dependent manner.
Discussion
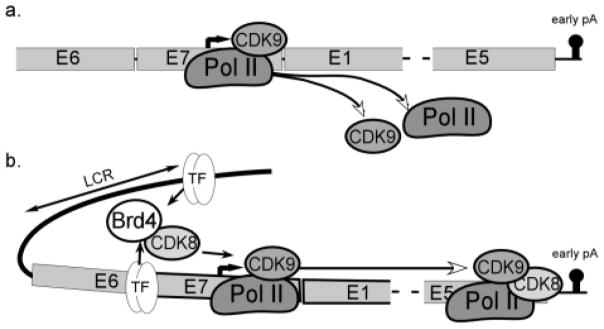
In this study, we have examined how core transcriptional regulatory complexes change during differentiation-dependent HPV16 late promoter activation. These data suggest that the primary mode of late promoter upregulation is through transcript elongation. We found that Pol II is present at the late promoter and at proximal downstream sites in undifferentiated cells. CDK9 is critical to late transcript upregulation, and recruitment of CDK9 to promoter-distal sites is associated with a differentiation-dependent increase in Pol II occupancy at the 3′ end of the early region, just before the early polyadenylation signal. CDK8, which can recruit CDK9 to promoters, binds to the genome in a JQ1-sensitive manner upon differentiation and increases late promoter activation. These observations together suggest a model (Figure 9) in which Pol II is able to initiate transcription and elongate at least as far as the E1 region in undifferentiated cells but is unable to elongate efficiently through the early region because low levels of CDK9-containing elongation complexes associate with the viral genome. Upon differentiation, transcription factors in either the LCR or the late promoter recruit Brd4, which in turn recruits CDK8 and brings the LCR into contact with the late promoter, facilitating association of CDK9 with Pol II complexes and transcript elongation.
Figure 9.
a. Under undifferentiated (monolayer) conditions, Pol II and CDK9 are present at the late promoter and initiate transcription, but elongating complexes do not continue to the end of the early region. b. Under differentiating conditions, the viral DNA loops to bring the LCR enhancer elements into association with the late promoter. Transcription factors binding to the enhancer and/or promoter recruit Brd4 or another BET family member, which then recruits CDK8. Either together or individually, CDK8 facilitates the processivity of elongating Pol II/CDK9 complexes to achieve complete transcription of the early region.
To our knowledge this is the first report examining the role of transcriptional elongation in the control of differentiation-inducible HPV gene expression. The presence of elongation complexes along genes is a strong predictor of the level of gene expression, and much of the regulation of eukaryotic transcription occurs following the initiation step during the transition to elongating polymerase (Adelman and Lis, 2012; Allen and Taatjes, 2015; Mayer et al., 2010). Although P-TEFb may be considered part of the general transcriptional machinery, its levels and recruitment can be regulated in a signal- and differentiation-dependent manner; thus P-TEFb is essential to the proper regulation of many mammalian genes (Fujita et al., 2008; Garriga and Grana, 2004; Hargreaves et al., 2009; Kohoutek, 2009). Previous studies have found that P-TEFb plays a role in transcription from the HPV16 early promoter, and the viral transcriptional repressor E2 functions in part by preventing recruitment of P-TEFb to the early promoter (Yan et al., 2010). Our ChIP data show that the levels of CDK9 associated with elongating Pol II increase upon differentiation. Furthermore, pharmacological inhibition of CDK9 potently prevents late promoter upregulation. Our results indicate that CDK9 recruitment and activity are more likely to be points of regulation than Pol II recruitment and initiation, suggesting that the late promoter is regulated at the level of elongation.
We have observed that luciferase reporters containing the HPV16 late promoter can occasionally be induced by differentiation (Bodily et al., 2013), but this finding is not as consistent in the case of HPV16 as it is in HPV31 ((Bodily and Meyers, 2005) and unpublished observations). We do not understand why the two viruses differ in the differentiation-inducibility of their respective late promoter reporters. Luciferase reporters can effectively measure promoter activity, but would not capture other aspects of transcriptional regulation, such as elongation or transcript stability, that depend on elements found outside the promoter itself. Our results rule out a significant role for transcript stability in differentiation-dependent increase in transcripts from the late promoter. Data showing that elongation is likely the critical step may help explain inconsistency in the ability of late promoter reporters to be upregulated by differentiation.
Many factors are known to be able to recruit elongation machinery to promoters under various circumstances, but CDK8/Mediator (Donner et al., 2010; Donner et al., 2007b; Galbraith et al., 2013; Galbraith et al., 2010) and Brd4 (Hargreaves et al., 2009; Jang et al., 2005; Kohoutek, 2009; Lenasi and Barboric, 2010; Palermo et al., 2011; Patel et al., 2013; Yang et al., 2005) are the most important overall, which is why we chose to focus on these two factors. c-Myc can also bind and recruit P-TEFb to some promoters to promote transcriptional elongation (Rahl et al., 2010). We could detect c-Myc binding to the HPV16 genome, but binding did not change upon differentiation, and treatment with a c-Myc inhibitor did not interfere with upregulation of late promoter activity in MC (not shown). Our studies do not rule out a contribution by other factors to CDK9 recruitment to the viral genome upon differentiation, but they do demonstrate that CDK8 and Brd4 each have an important role.
Because of its role in integrating signals from transcription factors with control of Pol II recruitment and activation, Mediator may be considered as the endpoint of most signal transduction cascades (Allen and Taatjes, 2015). Mediator complexes containing CDK8 are structurally unable to bind to or activate Pol II, and so CDK8 can act as an inhibitor of Pol II recruitment and PIC formation (Allen and Taatjes, 2015). On the other hand, by disrupting the interaction between Mediator and the PIC, CDK8 facilitates the shift from an initiating complex to an elongation complex (Allen and Taatjes, 2015; Donner et al., 2010; Galbraith et al., 2013; Galbraith et al., 2010; Taatjes, 2010). In addition, CDK8 acts either alone or in cooperation with Brd4 to promote recruitment of P-TEFb/CDK9, facilitating transcriptional elongation (Donner et al., 2010; Donner et al., 2007b; Galbraith et al., 2013). For these reasons, CDK8 recruitment to promoters acts as a molecular switch controlling the transition from initiation to elongation (Allen and Taatjes, 2015). We observed Pol II and MED1 bound at the late promoter in undifferentiated cells. This suggests that the PIC may already be present at the late promoter under these conditions and that differentiation-dependent recruitment of the CDK8/kinase module and subsequent binding of CDK9 is the critical activating even. We observed that knockdown of CDK8 resulted in reduced late promoter activation whereas inhibition of CDK8 kinase activity did not. Other studies have also suggested that CDK8 plays a structural role in regulating genes that is distinct from its kinase activity (Gold et al., 1996), for example in the disparity between the effects of inhibiting the kinase activity of CDK8 as compared to knocking down the protein (Galbraith et al., 2013; Poss et al., 2016; Tsutsui et al., 2013).
Our data also show that one or more BET family proteins sensitive to JQ1 is critical for late promoter activation. Because of its ability to recruit P-TEFb to promoters (Hargreaves et al., 2009; Kohoutek, 2009) and its known association with HPV (Iftner et al., 2016), we chose to focus on Brd4. Brd4 regulates the HPV early promoter and viral DNA replication (Iftner et al., 2016; Schweiger et al., 2006; Wang et al., 2013; Yan et al., 2010), and the viral E2 repressor protein disrupts the interaction between Brd4 and P-TEFb (Yan et al., 2010). A mutation in E2 that prevents binding to Brd4 also prevents episomal maintenance of HPV16 (Gauson et al., 2015). The role of E2 in regulation of the late promoter is not known, nor is its impact on the association between CDK8 and CDK9. However, we were able to observe inhibition of early transcripts only at much higher doses of JQ1 than were needed to inhibit the late promoter, suggesting that early transcription is not as dependent on Brd4 as late transcription is. Although JQ1 clearly inhibited late promoter activation, our results are somewhat equivocal about the role of Brd4 binding to the viral genome. We observed increased binding to the early promoter upon differentiation, but we did not observe a corresponding increase in early promoter activity. Furthermore, JQ1 actually increased the association of Brd4 with the viral genome, but again without an increase in transcriptional activity. Thus the mechanistic role of Brd4 recruitment in viral transcription, or the impact of other BET family members, remains to be determined.
The relationship between the recruitment of Brd4, CDK8, and P-TEFb is not always clear. Brd4 can form a complex with CDK8 and P-TEFb (Wu and Chiang, 2007). CDK8 is needed to recruit Brd4 to HIF-1 target genes (Galbraith et al., 2013) and serum-stimulated genes (Donner et al., 2010). CDK8 knockdown also reduces occupancy by other components of the Mediator complex (Donner et al., 2010). On the other hand, at some promoters, binding to Brd4 is needed to mediate contact between P-TEFb and Mediator and promote transcription (Yang et al., 2005). Brd4 can also be found in both Mediator complexes and P-TEFb complexes (Donner et al., 2010; Galbraith et al., 2010; Jiang et al., 1998; Loven et al., 2013; Wu and Chiang, 2007), and is upstream of CDK8 function (Pelish et al., 2015). Inhibition of bromodomain binding with JQ1 has been shown to evict Mediator and CDK9 from binding to promoters (Bhagwat et al., 2016). Our studies show that JQ1 prevents recruitment of CDK8 to the viral genome in differentiated keratinocytes. They also show that unusual binding of Brd4 to the viral genome upon treatment with JQ1 is associated with increased CDK8 binding, and suggest that Brd4 may be directly involved in CDK8 recruitment. Finally, both CDK8 and Brd4 can bind to P-TEFb as part of the larger Mediator complex as well as separate complexes (Donner et al., 2010; Wu and Chiang, 2007). CDK8 and Brd4 can be found along the gene body, suggesting that they may be part of the elongation complex (Donner et al., 2007b). Additional work will be needed to more fully understand the distribution of these factors on the viral genome at sites other than the sites we chose to examine in this study, but we have found both CDK8 and Brd4 associating with the viral genome not only at the LCR and late promoter, but even at downstream sites such as the E5 region (Supplementary Figure 3b and not shown). What role these factors may play at later stages of the transcription cycle is a subject for further investigation.
Supplementary Material
Research Highlights.
The HPV16 late promoter is upregulated by transcriptional rather than posttranscriptional mechanisms.
Promoter upregulation upon differentiation is associated with increased occupancy of Pol II at the 3′ end of the early region.
Association of elongation factor CDK9 with the viral genome increases upon differentiation and CDK9 activity is required for promoter upregulation.
CDK8 and Brd4, two factors that facilitate CDK9 association with promoters, are recruited to the viral genome upon differentiation and contribute to promoter activity.
Acknowledgments
We thank Lucile Guion, Haley Ruther, and Cynthia Rodriguez for technical assistance, members of the Bodily laboratory for helpful discussion, and James Bradner (Dana Farber/Harvard Cancer Center) for the generous gift of JQ1.
Funding sources: This work was supported by grants from the National Institute of Allergy and Infectious Diseases (R01AI118904), the National Institute of General Medical Sciences (P30GM110703), and the Feist-Weiller Cancer Center. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or the Feist-Weiller Cancer Center.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adelman K, Lis JT. Promoter-proximal pausing of RNA polymerase II: emerging roles in metazoans. Nature Reviews Genetics. 2012;13:720–731. doi: 10.1038/nrg3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ai W, Narahari J, Roman A. Yin yang 1 negatively regulates the differentiation-specific E1 promoter of human papillomavirus type 6. Journal of Virology. 2000;74:5198–5205. doi: 10.1128/jvi.74.11.5198-5205.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ai W, Toussaint E, Roman A. CCAAT displacement protein binds to and negatively regulates human papillomavirus type 6 E6, E7, and E1 promoters. Journal of Virology. 1999;73:4220–4229. doi: 10.1128/jvi.73.5.4220-4229.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen BL, Taatjes DJ. The Mediator complex: a central integrator of transcription. Nature Reviews Molecular Cell Biology. 2015;16:155–166. doi: 10.1038/nrm3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bancerek J, Poss ZC, Steinparzer I, Sedlyarov V, Pfaffenwimmer T, Mikulic I, Dolken L, Strobl B, Muller M, Taatjes DJ, Kovarik P. CDK8 kinase phosphorylates transcription factor STAT1 to selectively regulate the interferon response. Immunity. 2013;38:250–262. doi: 10.1016/j.immuni.2012.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bataille AR, Jeronimo C, Jacques PE, Laramee L, Fortin ME, Forest A, Bergeron M, Hanes SD, Robert F. A universal RNA polymerase II CTD cycle is orchestrated by complex interplays between kinase, phosphatase, and isomerase enzymes along genes. Molecular Cell. 2012;45:158–170. doi: 10.1016/j.molcel.2011.11.024. [DOI] [PubMed] [Google Scholar]
- Bedell MA, Hudson JB, Golub TR, Turyk ME, Hosken M, Wilbanks GD, Laimins LA. Amplification of human papillomavirus genomes in vitro is dependent on epithelial differentiation. Journal of Virology. 1991;65:2254–2260. doi: 10.1128/jvi.65.5.2254-2260.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard HU. Regulatory elements in the viral genome. Virology. 2013;445:197–204. doi: 10.1016/j.virol.2013.04.035. [DOI] [PubMed] [Google Scholar]
- Bhagwat AS, Roe JS, Mok BY, Hohmann AF, Shi J, Vakoc CR. BET Bromodomain Inhibition Releases the Mediator Complex from Select cis-Regulatory Elements. Cell Reports. 2016;15:519–530. doi: 10.1016/j.celrep.2016.03.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodily J, Laimins LA. Persistence of human papillomavirus infection: keys to malignant progression. Trends in Microbiology. 2011;19:33–39. doi: 10.1016/j.tim.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodily JM, Hennigan C, Wrobel GA, Rodriguez CM. Regulation of the human papillomavirus type 16 late promoter by E7 and the cell cycle. Virology. 2013;443:11–19. doi: 10.1016/j.virol.2013.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodily JM, Mehta KP, Cruz L, Meyers C, Laimins LA. The E7 Open Reading Frame Acts in cis and in trans To Mediate Differentiation-Dependent Activities in the Human Papillomavirus Type 16 Life Cycle. Journal of Virology. 2011a;85:8852–8862. doi: 10.1128/JVI.00664-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodily JM, Mehta KP, Laimins LA. Human papillomavirus E7 enhances hypoxia-inducible factor 1-mediated transcription by inhibiting binding of histone deacetylases. Cancer Research. 2011b;71:1187–1195. doi: 10.1158/0008-5472.CAN-10-2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodily JM, Meyers C. Genetic analysis of the human papillomavirus type 31 differentiation-dependent late promoter. Journal of Virology. 2005;79:3309–3321. doi: 10.1128/JVI.79.6.3309-3321.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeing S, Rigault C, Heidemann M, Eick D, Meisterernst M. RNA polymerase II C-terminal heptarepeat domain Ser-7 phosphorylation is established in a mediator-dependent fashion. The Journal of Biological Chemistry. 2010;285:188–196. doi: 10.1074/jbc.M109.046565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carson A, Khan SA. Characterization of transcription factor binding to human papillomavirus type 16 DNA during cellular differentiation. Journal of Virology. 2006;80:4356–4362. doi: 10.1128/JVI.80.9.4356-4362.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Mar Pena LM, Laimins LA. Differentiation-dependent chromatin rearrangement coincides with activation of human papillomavirus type 31 late gene expression. Journal of Virology. 2001;75:10005–10013. doi: 10.1128/JVI.75.20.10005-10013.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Micco R, Fontanals-Cirera B, Low V, Ntziachristos P, Yuen SK, Lovell CD, Dolgalev I, Yonekubo Y, Zhang G, Rusinova E, Gerona-Navarro G, Canamero M, Ohlmeyer M, Aifantis I, Zhou MM, Tsirigos A, Hernando E. Control of embryonic stem cell identity by BRD4-dependent transcriptional elongation of super-enhancer-associated pluripotency genes. Cell reports. 2014;9:234–247. doi: 10.1016/j.celrep.2014.08.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donner AJ, Ebmeier CC, Taatjes DJ, Espinosa JM. CDK8 is a positive regulator of transcriptional elongation within the serum response network. Nature structural & molecular biology. 2010;17:194–201. doi: 10.1038/nsmb.1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donner AJ, Hoover JM, Szostek SA, Espinosa JM. Stimulus-specific transcriptional regulation within the p53 network. Cell cycle (Georgetown, Tex) 2007a;6:2594–2598. doi: 10.4161/cc.6.21.4893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donner AJ, Szostek S, Hoover JM, Espinosa JM. CDK8 is a stimulus-specific positive coregulator of p53 target genes. Molecular Cell. 2007b;27:121–133. doi: 10.1016/j.molcel.2007.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooley KE, Warburton A, McBride AA. Tandemly Integrated HPV16 Can Form a Brd4-Dependent Super-Enhancer-Like Element That Drives Transcription of Viral Oncogenes. mBio. 2016:7. doi: 10.1128/mBio.01446-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doorbar J, Quint W, Banks L, Bravo IG, Stoler M, Broker TR, Stanley MA. The biology and life-cycle of human papillomaviruses. Vaccine. 2012;30(Suppl 5):F55–70. doi: 10.1016/j.vaccine.2012.06.083. [DOI] [PubMed] [Google Scholar]
- Fehrmann F, Klumpp DJ, Laimins LA. Human papillomavirus type 31 E5 protein supports cell cycle progression and activates late viral functions upon epithelial differentiation. Journal of Virology. 2003;77:2819–2831. doi: 10.1128/JVI.77.5.2819-2831.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, Morse EM, Keates T, Hickman TT, Felletar I, Philpott M, Munro S, McKeown MR, Wang Y, Christie AL, West N, Cameron MJ, Schwartz B, Heightman TD, La Thangue N, French CA, Wiest O, Kung AL, Knapp S, Bradner JE. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forman D, de Martel C, Lacey CJ, Soerjomataram I, Lortet-Tieulent J, Bruni L, Vignat J, Ferlay J, Bray F, Plummer M, Franceschi S. Global burden of human papillomavirus and related diseases. Vaccine. 2012;30(Suppl 5):F12–23. doi: 10.1016/j.vaccine.2012.07.055. [DOI] [PubMed] [Google Scholar]
- Frattini MG, Lim HB, Laimins LA. In vitro synthesis of oncogenic human papillomaviruses requires episomal genomes for differentiation-dependent late expression. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:3062–3067. doi: 10.1073/pnas.93.7.3062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazer IH. Interaction of human papillomaviruses with the host immune system: a well evolved relationship. Virology. 2009;384:410–414. doi: 10.1016/j.virol.2008.10.004. [DOI] [PubMed] [Google Scholar]
- Fujita T, Ryser S, Piuz I, Schlegel W. Up-regulation of P-TEFb by the MEK1-extracellular signal-regulated kinase signaling pathway contributes to stimulated transcription elongation of immediate early genes in neuroendocrine cells. Molecular and Cellular Biology. 2008;28:1630–1643. doi: 10.1128/MCB.01767-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galbraith MD, Allen MA, Bensard CL, Wang X, Schwinn MK, Qin B, Long HW, Daniels DL, Hahn WC, Dowell RD, Espinosa JM. HIF1A employs CDK8-mediator to stimulate RNAPII elongation in response to hypoxia. Cell. 2013;153:1327–1339. doi: 10.1016/j.cell.2013.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galbraith MD, Donner AJ, Espinosa JM. CDK8: a positive regulator of transcription. Transcription. 2010;1:4–12. doi: 10.4161/trns.1.1.12373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garriga J, Grana X. Cellular control of gene expression by T-type cyclin/CDK9 complexes. Gene. 2004;337:15–23. doi: 10.1016/j.gene.2004.05.007. [DOI] [PubMed] [Google Scholar]
- Gauson EJ, Donaldson MM, Dornan ES, Wang X, Bristol M, Bodily JM, Morgan IM. Evidence supporting a role for TopBP1 and Brd4 in the initiation but not continuation of human papillomavirus 16 E1/E2-mediated DNA replication. Journal of Virology. 2015;89:4980–4991. doi: 10.1128/JVI.00335-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gielen V, Schmitt D, Thivolet J. HLA class I antigen (heavy and light chain) expression by Langerhans cells and keratinocytes of the normal human epidermis: ultrastructural quantitation using immunogold labelling procedure. Archives of Dermatological Research. 1988;280:131–136. doi: 10.1007/BF00456841. [DOI] [PubMed] [Google Scholar]
- Gold MO, Tassan JP, Nigg EA, Rice AP, Herrmann CH. Viral transactivators E1A and VP16 interact with a large complex that is associated with CTD kinase activity and contains CDK8. Nucleic Acids Research. 1996;24:3771–3777. doi: 10.1093/nar/24.19.3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grassmann K, Rapp B, Maschek H, Petry KU, Iftner T. Identification of a differentiation-inducible promoter in the E7 open reading frame of human papillomavirus type 16 (HPV-16) in raft cultures of a new cell line containing high copy numbers of episomal HPV-16 DNA. Journal of Virology. 1996;70:2339–2349. doi: 10.1128/jvi.70.4.2339-2349.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunasekharan V, Hache G, Laimins L. Differentiation-dependent changes in levels of C/EBPbeta repressors and activators regulate human papillomavirus type 31 late gene expression. Journal of Virology. 2012;86:5393–5398. doi: 10.1128/JVI.07239-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargreaves DC, Horng T, Medzhitov R. Control of inducible gene expression by signal-dependent transcriptional elongation. Cell. 2009;138:129–145. doi: 10.1016/j.cell.2009.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidemann M, Hintermair C, Voss K, Eick D. Dynamic phosphorylation patterns of RNA polymerase II CTD during transcription. Biochimica et Biophysica Acta. 2013;1829:55–62. doi: 10.1016/j.bbagrm.2012.08.013. [DOI] [PubMed] [Google Scholar]
- Hummel M, Hudson JB, Laimins LA. Differentiation-induced and constitutive transcription of human papillomavirus type 31b in cell lines containing viral episomes. Journal of Virology. 1992;66:6070–6080. doi: 10.1128/jvi.66.10.6070-6080.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iftner T, Haedicke-Jarboui J, Wu SY, Chiang CM. Involvement of Brd4 in different steps of the papillomavirus life cycle. Virus Research. 2016 doi: 10.1016/j.virusres.2016.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang MK, Mochizuki K, Zhou M, Jeong HS, Brady JN, Ozato K. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Molecular Cell. 2005;19:523–534. doi: 10.1016/j.molcel.2005.06.027. [DOI] [PubMed] [Google Scholar]
- Jeon S, Lambert PF. Integration of human papillomavirus type 16 DNA into the human genome leads to increased stability of E6 and E7 mRNAs: implications for cervical carcinogenesis. Proceedings of the National Academy of Sciences of the United States of America. 1995;92:1654–1658. doi: 10.1073/pnas.92.5.1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha S, Vande Pol S, Banerjee NS, Dutta AB, Chow LT, Dutta A. Destabilization of TIP60 by human papillomavirus E6 results in attenuation of TIP60-dependent transcriptional regulation and apoptotic pathway. Molecular Cell. 2010;38:700–711. doi: 10.1016/j.molcel.2010.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang YW, Veschambre P, Erdjument-Bromage H, Tempst P, Conaway JW, Conaway RC, Kornberg RD. Mammalian mediator of transcriptional regulation and its possible role as an end-point of signal transduction pathways. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:8538–8543. doi: 10.1073/pnas.95.15.8538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohoutek J. P-TEFb- the final frontier. Cell division. 2009;4:19. doi: 10.1186/1747-1028-4-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohoutek J, Blazek D. Cyclin K goes with Cdk12 and Cdk13. Cell Division. 2012;7:12. doi: 10.1186/1747-1028-7-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korb E, Herre M, Zucker-Scharff I, Darnell RB, Allis CD. BET protein Brd4 activates transcription in neurons and BET inhibitor Jq1 blocks memory in mice. Nature Neuroscience. 2015;18:1464–1473. doi: 10.1038/nn.4095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kukimoto I, Kanda T. Displacement of YY1 by differentiation-specific transcription factor hSkn-1a activates the P(670) promoter of human papillomavirus type 16. Journal of Virology. 2001;75:9302–9311. doi: 10.1128/JVI.75.19.9302-9311.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kukimoto I, Takeuchi T, Kanda T. CCAAT/enhancer binding protein beta binds to and activates the P670 promoter of human papillomavirus type 16. Virology. 2006;346:98–107. doi: 10.1016/j.virol.2005.10.025. [DOI] [PubMed] [Google Scholar]
- Lenasi T, Barboric M. P-TEFb stimulates transcription elongation and pre-mRNA splicing through multilateral mechanisms. RNA Bbiology. 2010;7:145–150. doi: 10.4161/rna.7.2.11057. [DOI] [PubMed] [Google Scholar]
- Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- Loven J, Hoke HA, Lin CY, Lau A, Orlando DA, Vakoc CR, Bradner JE, Lee TI, Young RA. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 2013;153:320–334. doi: 10.1016/j.cell.2013.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik S, Roeder RG. The metazoan Mediator co-activator complex as an integrative hub for transcriptional regulation. Nature Reviews Genetics. 2010;11:761–772. doi: 10.1038/nrg2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malumbres M, Barbacid M. Mammalian cyclin-dependent kinases. Trends in Biochemical Sciences. 2005;30:630–641. doi: 10.1016/j.tibs.2005.09.005. [DOI] [PubMed] [Google Scholar]
- Mayer A, Lidschreiber M, Siebert M, Leike K, Soding J, Cramer P. Uniform transitions of the general RNA polymerase II transcription complex. Nature Structural & Molecular Biology. 2010;17:1272–1278. doi: 10.1038/nsmb.1903. [DOI] [PubMed] [Google Scholar]
- Moscicki AB, Schiffman M, Burchell A, Albero G, Giuliano AR, Goodman MT, Kjaer SK, Palefsky J. Updating the natural history of human papillomavirus and anogenital cancers. Vaccine. 2012;30(Suppl 5):F24–33. doi: 10.1016/j.vaccine.2012.05.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagarajan S, Hossan T, Alawi M, Najafova Z, Indenbirken D, Bedi U, Taipaleenmaki H, Ben-Batalla I, Scheller M, Loges S, Knapp S, Hesse E, Chiang CM, Grundhoff A, Johnsen SA. Bromodomain protein BRD4 is required for estrogen receptor-dependent enhancer activation and gene transcription. Cell Reports. 2014;8:460–469. doi: 10.1016/j.celrep.2014.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nardelli B, Zaritskaya L, Semenuk M, Cho YH, LaFleur DW, Shah D, Ullrich S, Girolomoni G, Albanesi C, Moore PA. Regulatory effect of IFN-kappa, a novel type I IFN, on cytokine production by cells of the innate immune system. J Immunol. 2002;169:4822–4830. doi: 10.4049/jimmunol.169.9.4822. [DOI] [PubMed] [Google Scholar]
- Ozbun M, Meyers C. Human papillomavirus type 31b transcription during the differentiation-dependent viral life cycle. Current Topics in Virology. 1999;1:203–217. [Google Scholar]
- Ozbun MA, Meyers C. Temporal usage of multiple promoters during the life cycle of human papillomavirus type 31b. Journal of Virology. 1998;72:2715–2722. doi: 10.1128/jvi.72.4.2715-2722.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palermo RD, Webb HM, West MJ. RNA polymerase II stalling promotes nucleosome occlusion and pTEFb recruitment to drive immortalization by Epstein-Barr virus. PLoS Pathogens. 2011;7:e1002334. doi: 10.1371/journal.ppat.1002334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel MC, Debrosse M, Smith M, Dey A, Huynh W, Sarai N, Heightman TD, Tamura T, Ozato K. BRD4 coordinates recruitment of pause release factor P-TEFb and the pausing complex NELF/DSIF to regulate transcription elongation of interferon-stimulated genes. Molecular and Cellular Biology. 2013;33:2497–2507. doi: 10.1128/MCB.01180-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peh WL, Brandsma JL, Christensen ND, Cladel NM, Wu X, Doorbar J. The viral E4 protein is required for the completion of the cottontail rabbit papillomavirus productive cycle in vivo. Journal of Virology. 2004;78:2142–2151. doi: 10.1128/JVI.78.4.2142-2151.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelish HE, Liau BB, Nitulescu II, Tangpeerachaikul A, Poss ZC, Da Silva DH, Caruso BT, Arefolov A, Fadeyi O, Christie AL, Du K, Banka D, Schneider EV, Jestel A, Zou G, Si C, Ebmeier CC, Bronson RT, Krivtsov AV, Myers AG, Kohl NE, Kung AL, Armstrong SA, Lemieux ME, Taatjes DJ, Shair MD. Mediator kinase inhibition further activates super-enhancer-associated genes in AML. Nature. 2015;526:273–276. doi: 10.1038/nature14904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter DC, Farmaki E, Altilia S, Schools GP, West DK, Chen M, Chang BD, Puzyrev AT, Lim CU, Rokow-Kittell R, Friedhoff LT, Papavassiliou AG, Kalurupalle S, Hurteau G, Shi J, Baran PS, Gyorffy B, Wentland MP, Broude EV, Kiaris H, Roninson IB. Cyclin-dependent kinase 8 mediates chemotherapy-induced tumor-promoting paracrine activities. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:13799–13804. doi: 10.1073/pnas.1206906109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poss ZC, Ebmeier CC, Odell AT, Tangpeerachaikul A, Lee T, Pelish HE, Shair MD, Dowell RD, Old WM, Taatjes DJ. Identification of Mediator Kinase Substrates in Human Cells using Cortistatin A and Quantitative Phosphoproteomics. Cell reports. 2016;15:436–450. doi: 10.1016/j.celrep.2016.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, Sharp PA, Young RA. c-Myc regulates transcriptional pause release. Cell. 2010;141:432–445. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren K, Zhang W, Chen X, Ma Y, Dai Y, Fan Y, Hou Y, Tan RX, Li E. An Epigenetic Compound Library Screen Identifies BET Inhibitors That Promote HSV-1 and -2 Replication by Bridging P-TEFb to Viral Gene Promoters through BRD4. PLoS Pathogens. 2016;12:e1005950. doi: 10.1371/journal.ppat.1005950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruesch MN, Laimins LA. Human papillomavirus oncoproteins alter differentiation-dependent cell cycle exit on suspension in semisolid medium. Virology. 1998;250:19–29. doi: 10.1006/viro.1998.9359. [DOI] [PubMed] [Google Scholar]
- Ruesch MN, Stubenrauch F, Laimins LA. Activation of papillomavirus late gene transcription and genome amplification upon differentiation in semisolid medium is coincident with expression of involucrin and transglutaminase but not keratin-10. Journal of Virology. 1998;72:5016–5024. doi: 10.1128/jvi.72.6.5016-5024.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K, Takeuchi T, Kukimoto I, Mori S, Yasugi T, Yano T, Taketani Y, Kanda T. Human papillomavirus type 16 P670 promoter is negatively regulated by CCAAT displacement protein. Virus Genes. 2007;35:473–481. doi: 10.1007/s11262-006-0074-8. [DOI] [PubMed] [Google Scholar]
- Schroder S, Cho S, Zeng L, Zhang Q, Kaehlcke K, Mak L, Lau J, Bisgrove D, Schnolzer M, Verdin E, Zhou MM, Ott M. Two-pronged binding with bromodomain-containing protein 4 liberates positive transcription elongation factor b from inactive ribonucleoprotein complexes. The Journal of Biological Chemistry. 2012;287:1090–1099. doi: 10.1074/jbc.M111.282855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S. Papillomavirus transcripts and posttranscriptional regulation. Virology. 2013;445:187–196. doi: 10.1016/j.virol.2013.04.034. [DOI] [PubMed] [Google Scholar]
- Schweiger MR, You J, Howley PM. Bromodomain protein 4 mediates the papillomavirus e2 transcriptional activation function. Journal of Virology. 2006;80:4276–4285. doi: 10.1128/JVI.80.9.4276-4285.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senderowicz AM. Small-molecule cyclin-dependent kinase modulators. Oncogene. 2003;22:6609–6620. doi: 10.1038/sj.onc.1206954. [DOI] [PubMed] [Google Scholar]
- Spink KM, Laimins LA. Induction of the human papillomavirus type 31 late promoter requires differentiation but not DNA amplification. Journal of Virology. 2005;79:4918–4926. doi: 10.1128/JVI.79.8.4918-4926.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele BK, Meyers C, Ozbun MA. Variable expression of some “housekeeping” genes during human keratinocyte differentiation. Analytical Biochemistry. 2002;307:341–347. doi: 10.1016/s0003-2697(02)00045-3. [DOI] [PubMed] [Google Scholar]
- Taatjes DJ. The human Mediator complex: a versatile, genome-wide regulator of transcription. Trends in Biochemical Sciences. 2010;35:315–322. doi: 10.1016/j.tibs.2010.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terhune SS, Hubert WG, Thomas JT, Laimins LA. Early polyadenylation signals of human papillomavirus type 31 negatively regulate capsid gene expression. Journal of Virology. 2001;75:8147–8157. doi: 10.1128/JVI.75.17.8147-8157.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsutsui T, Fukasawa R, Shinmyouzu K, Nakagawa R, Tobe K, Tanaka A, Ohkuma Y. Mediator complex recruits epigenetic regulators via its two cyclin-dependent kinase subunits to repress transcription of immune response genes. The Journal of Biological Chemistry. 2013;288:20955–20965. doi: 10.1074/jbc.M113.486746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Doorslaer K, Tan Q, Xirasagar S, Bandaru S, Gopalan V, Mohamoud Y, Huyen Y, McBride AA. The Papillomavirus Episteme: a central resource for papillomavirus sequence data and analysis. Nucleic Acids Research. 2013;41:D571–578. doi: 10.1093/nar/gks984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Helfer CM, Pancholi N, Bradner JE, You J. Recruitment of Brd4 to the human papillomavirus type 16 DNA replication complex is essential for replication of viral DNA. Journal of Virology. 2013;87:3871–3884. doi: 10.1128/JVI.03068-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westrich JA, Warren CJ, Pyeon D. Evasion of host immune defenses by human papillomavirus. Virus Research. 2017;231:21–33. doi: 10.1016/j.virusres.2016.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson R, Fehrmann F, Laimins LA. Role of the E1^E4 Protein in the Differentiation-Dependent Life Cycle of Human Papillomavirus Type 31. Journal of Virology. 2005;79:6732–6740. doi: 10.1128/JVI.79.11.6732-6740.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson R, Laimins LA. Differentiation of HPV-containing cells using organotypic “raft” culture or methylcellulose. Methods in Molecular Medicine. 2005;119:157–169. doi: 10.1385/1-59259-982-6:157. [DOI] [PubMed] [Google Scholar]
- Woodman CB, Collins S, Winter H, Bailey A, Ellis J, Prior P, Yates M, Rollason TP, Young LS. Natural history of cervical human papillomavirus infection in young women: a longitudinal cohort study. Lancet. 2001;357:1831–1836. doi: 10.1016/S0140-6736(00)04956-4. [DOI] [PubMed] [Google Scholar]
- Wooldridge TR, Laimins LA. Regulation of human papillomavirus type 31 gene expression during the differentiation-dependent life cycle through histone modifications and transcription factor binding. Virology. 2008;374:371–380. doi: 10.1016/j.virol.2007.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SY, Chiang CM. The double bromodomain-containing chromatin adaptor Brd4 and transcriptional regulation. The Journal of Biological Chemistry. 2007;282:13141–13145. doi: 10.1074/jbc.R700001200. [DOI] [PubMed] [Google Scholar]
- Wu SY, Lee AY, Hou SY, Kemper JK, Erdjument-Bromage H, Tempst P, Chiang CM. Brd4 links chromatin targeting to HPV transcriptional silencing. Genes & Development. 2006;20:2383–2396. doi: 10.1101/gad.1448206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SY, Nin DS, Lee AY, Simanski S, Kodadek T, Chiang CM. BRD4 Phosphorylation Regulates HPV E2-Mediated Viral Transcription, Origin Replication, and Cellular MMP-9 Expression. Cell Reports. 2016;16:1733–1748. doi: 10.1016/j.celrep.2016.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Li Q, Lievens S, Tavernier J, You J. Abrogation of the Brd4-positive transcription elongation factor B complex by papillomavirus E2 protein contributes to viral oncogene repression. Journal of Virology. 2010;84:76–87. doi: 10.1128/JVI.01647-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Yik JH, Chen R, He N, Jang MK, Ozato K, Zhou Q. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Molecular Cell. 2005;19:535–545. doi: 10.1016/j.molcel.2005.06.029. [DOI] [PubMed] [Google Scholar]
- You J, Schweiger MR, Howley PM. Inhibition of E2 binding to Brd4 enhances viral genome loss and phenotypic reversion of bovine papillomavirus-transformed cells. Journal of Virology. 2005;79:14956–14961. doi: 10.1128/JVI.79.23.14956-14961.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.