Abstract
Non-small-cell lung cancer (NSCLC) is the leading cause of cancer-associated mortality in the United States. AXL, which is a member of the receptor tyrosine kinases, has been established as a strong candidate for the targeted therapy of cancer. Therefore, the present study aimed to investigate the role of AXL in NSCLC; in particular the molecular mechanisms underlying the involvement of AXL in the epithelial-to-mesenchymal transition (EMT). Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) and western blot analysis demonstrated that AXL, EMT-inducing Twist and the mesenchymal marker N-cadherin were upregulated, and the epithelial markers E-cadherin and β-cadherin were downregulated, in the PC9 NSCLC cell line. Furthermore, downregulation of AXL expression by RNA interference was shown to inhibit cell growth by inducing the apoptosis of PC9 cells, as demonstrated by MTT and flow cytometry analyses. Notably, inhibition of AXL attenuated the regulation of EMT-associated genes, specifically downregulating Twist and N-cadherin, and upregulating E-cadherin and β-cadherin. Conversely, downregulation of Twist did not affect the expression levels of AXL. These results suggested that AXL may inhibit the EMT by the regulation of EMT-associated genes in the PC9 cell line. The results of the present study indicated that AXL may have a role in the regulation of EMT and the cell cycle of the PC9 cells; thus suggesting that AXL may have clinical significance in the design of therapeutic strategies targeting NSCLC and EMT signaling pathways.
Keywords: AXL, epithelial-to-mesenchymal transition, non-small-cell lung cancer, RNA interference, quantitative polymerase chain reaction
Introduction
Lung cancer is one of the leading causes of cancer-associated mortalities worldwide, with ~1 million new cases annually in terms of incidence and mortality (1–4). Lung cancer accounted for 13% (1.6 million) of the total cases and 18% (1.8 million) of mortalities in 2008 (1). Non-small-cell lung cancer (NSCLC) accounts for ~80% of all lung cancer cases (5). The management of patients with NSCLC is based on systemic chemotherapy; the standard treatment for patients with advanced NSCLC includes platinum-based combination chemotherapy, which has demonstrated modest but significant improvements in survival rates over best supportive care (6). However, the survival rates remain low (7). In addition, although chemotherapy may prolong the survival of patients with advanced disease, clinically significant adverse side effects, including excessive toxicity, reduce its effectiveness (8). Therefore, novel agents are urgently required for the treatment of this disease.
The epithelial-to-mesenchymal transition (EMT) has an important role in numerous physiological and pathological processes in the human body, including regulating the transcription of genes involved in embryonic development (6), the inflammatory response (8), tissue regeneration (9), organ fibrosis (10,11), and tumor invasion and metastasis. During EMT, epithelial cells loosen and cell-cell adhesion is lost (12). Furthermore, EMT is characterized by the downregulation of E-cadherin and cytokeratins, and the upregulation of mesenchymal proteins, including vimentin, fibronectin and N-cadherin (13,14). A previous study demonstrated that activation of the epidermal growth factor (EGF) receptor (EGFR) by EGF promoted EMT in hepatocellular carcinoma cell lines by altering the expression levels and molecular structures of EMT-associated markers, including the E-cadherin/β-catenin complex (14).
AXL is a member of the receptor tyrosine kinase family (9). Previous studies demonstrated that downregulation of AXL expression in solid tumors by RNA interference (RNAi) was able to decrease cell invasion and proliferation, and increase the chemosensitivity of cells via the activation of AKT and mitogen-activated protein kinase (MAPK) pathways (10,11,13). In addition, AXL was identified as an oncogene in human leukemia cells (14–16), and the expression levels of AXL were shown to be increased in >50% of NSCLC cell lines (17). Similarly, a previous study reported that the expression levels of AXL were upregulated in 48.3% of lung adenocarcinoma tissues, in which they were correlated with lymph node metastasis and the stage of the disease (16). These reports highlight the clinical importance of AXL. Targeting of AXL with RNAi or specific monoclonal antibodies has previously been shown to inhibit the proliferation of NSCLC cells and tumor cells in a mouse xenograft model (18). A previous study on NSCLC reported that increased activation of AXL was associated with acquired resistance to EGFR-targeted therapy (19).
The present study aimed to investigate the role of AXL in NSCLC, in particular the molecular mechanism underlying its involvement in EMT. Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) and western blot analysis demonstrated that AXL was upregulated in NSCLC cells. Subsequently, inhibition of AXL by RNAi was conducted in order to investigate the effects of downregulation of AXL on cell viability and the expression levels of EMT-associated genes.
Materials and methods
Agents
Doxorubicin, paclitaxel, vincristine, cisplatin and 3-(4,5-dimethylthiazol-yl)-2,5-diphenyltetrazolium bromide (MTT) were from Sigma-Aldrich, Inc. (Shanghai, China). Dulbecco's modified Eagle's medium (DMEM) and RPMI-1640 medium were from Gibco (Thermo Fisher Scientific, Inc., Waltham, MA, USA).
Cell lines
The human H1299, A549 and PC9 NSCLC cell lines, and normal lung cells (as the control), were purchased from the Cancer Research Institute of China Medical University (Shenyang, China). The cells (1×105 cells/ml) were cultured in DMEM supplemented with 10% (m/v) fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc.), 100 U/ml penicillin and 100 µg/ml streptomycin (Sangong Biotech, Shanghai, China) at 37°C in a 5% CO2 atmosphere with stable humidity.
MTT assay
Cell viability was assessed using the MTT assay, as described in a previous study (20). Briefly, the cells were plated at a density of 3,000 cells/well into 96-well plates. At the end of treatment, the supernatant was removed using a pipette and 20 µl MTT and 270 ml fresh DMEM were added to the supernatant. Following incubation for 4 h at 37°C, 120 µl dimethyl sulfoxide was placed in each well to dissolve the tetrazolium crystals, after which the absorbance at 570 nm was recorded using a Multi-Well Plate Reader (Tecan Group, Ltd., Männedorf, Switzerland). Each experiment was performed four times. The results are presented as the percentage growth inhibition with respect to the untreated cells.
RNAi procedure
Double-strained small interfering RNA (siRNA) probes targeting the human AXL gene (GenBank accession no. NM-021913) and the Twist gene (GenBank accession no. NM_000474.3) were synthesized using the Silencer siRNA Construction kit (Ambion; Thermo Fisher Scientific, Inc.), as described in a previous study (21). Briefly, single-stranded gene-specific sense and antisense RNA oligomers were synthesized by in vitro transcription using T7 RNA polymerase (Sangong Biotech). In order to promote annealing of the siRNA, sense and antisense single-stranded RNA oligomers were mixed and incubated at 37°C overnight, after which the siRNA was ethanol-precipitated and resuspended in nuclease-free water. The integrity of the siRNA was assessed by gel electrophoresis (Sangong Biotech), and it was quantified of by measuring the absorbance at 260 nm. Subsequently, the cells were transfected with four pooled siRNA duplexes (20 nM; Invitrogen; Thermo Fisher Scientific, Inc.) using the TransIT-TKO® Transfection Reagent (Mirus Bio, LLC, Madison, WI, USA), in order to target the endogenous AXL and Twist genes. The cells were transfected twice at 3-day intervals, and were then collected at 72 h following the second transfection. Specific silencing of the target genes was confirmed using qPCR and western blot analysis. A mock transfection with the transfection reagent alone served as the control.
Flow cytometry (FCM) analysis
Log phase PC9 cells were collected at a final concentration of 2×105 cells/ml, washed with phosphate-buffered saline (PBS) and fixed with 70% ethanol. The cells were centrifuged at 8,000 × g for 5 min at 4°C to remove ethanol, washed with PBS, and stained with propidium iodide (PI; Sangong Biotech) in the dark for 30 min prior to FCM analysis. The FACSCalibur™ platform (BD Biosciences, Franklin Lakes, NJ, USA) was used to detect the cell cycle distribution. The cells were sampled using the CellQuest Pro software, version 3.0 (BD Biosciences), and the proportion of cells in the various stages of the cell cycle were quantified using ModFit LT 3.0 (Verity Software House, Inc., Topsham, ME, USA) (22). Each experiment was performed four times.
Evaluation of apoptosis
PC9 cells (1×106) were collected by centrifugation at 8,000 × g for 5 min at 4°C. The cell pellets were lysed using DNA lysis buffer (10 mM Tris, pH 7.5, 400 mM EDTA and 1% Triton X-100), followed by centrifugation at 6,000 × g for 8 min at 4°C. The supernatant was incubated overnight with proteinase K (0.1 mg/ml; Sangong Biotech), then with RNase (0.2 mg/ml; Sangong Biotech) for 2 h at 37°C. DNA was extracted using phenol:chloroform (1:1), separated by 2% agarose gel electrophoresis (Sangong Biotech), and visualized by ultraviolet illumination after staining with 10% ethidium bromide and washing twice with water. Quantitative assessment of apoptotic cells was conducted using the terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) method (BD ApoAlert DNA Fragmentation Assay kit; cat. no. 630107; BD Biosciences), which examines DNA-strand breaks during apoptosis.
RT-qPCR
RNA extraction from the cells was carried out using TRIzol reagent (Life Technologies; Thermo Fisher Scientific, Inc.) according to the manufacturer's recommendations. Genomic DNA was eliminated and RT conducted using the PrimeScript RT Reagent kit with gDNA Eraser, and qPCR was conducted using the SYBR Green PCR kit (both Takara Biotechnology Co., Ltd., Dalian, China) according to the manufacturer's recommendations. The process was conducted using a 7500 Real-Time PCR System (Applied Biosystems; Thermo Fisher Scientific, Inc. The primers used in the PCR were as follows: AXL forward, 5′-TCGCCAGTGGCATGGAGTATCTG-3′ and reverse, 5′-GGCGATACGTCCCTGGCGGTA-3′; Twist forward, 5′-CATCCACACCGTCCCCTCCC-3′ and reverse, 5′-GACTGGCGAGCTGGACACGTC-3′; and β-actin forward, 5′-TGAAGTACCCCATCGAGCAC-3′ and reverse, 5′-CTTGGGGTTCAGGGGGGCCT-3′ (GenBank accession no. BC004251.1). β-actin was used as an internal reference.
The reaction mix consisted of 4 µl cDNA, 0.5 µl forward and reverse primer mix (20 µM of each), 1 µl 50X ROX Reference Dye II and 25 µl 2X SYBR Green PCR mix in a final volume of 50 µl. All reactions were setup in triplicate and every sample was replicated in parallel three times to ensure statistical relevance. A negative control without cDNA template, and RT control without RT transcription were included. The following thermal conditions were used for all PCR reactions: 30 sec at 95°C, followed by 40 cycles of 30 sec at 95°C and 34 sec at 60°C. Primer specificity was confirmed by RT-PCR amplification prior to qPCR, which generated single amplicons of the expected size for each primer set. Furthermore, the amplicons were sequenced by a commercial sequencing service (BGI, Shenzhen, China) in order to validate their specific amplification, and the specificity of qPCR was confirmed by the presence of dissociation curves with single peaks and the sequencing of its products with unique bands of the expected size. Amplicon dissociation curves were obtained after cycle 40 using default settings suggested by the instrument. Data were analyzed using the SDS software (version 2.0.6; Applied Biosystems; Thermo Fisher Scientific, Inc.). All quantifications were normalized against the quantity of β-actin using the 2−ΔΔCq method (23).
Western blot analysis
Cell lysates were prepared in radioimmunoprecipitation assay buffer [50 mM Tris-HCl buffer, pH 7.4, 150 mM NaCl, 1% Triton X-100, 1% sodium deoxycholate, and 0.1% sodium dodecyl sulfate (SDS)], supplemented with 1X Halt protease inhibitor and 1X Halt phosphatase inhibitor cocktails (Pierce Biotechnology, Inc., Rockford, IL, USA). The Bio-Rad protein assay (cat. no. 500-0006; Bio-Rad Laboratories, Inc., Hercules, CA, USA) was used to determine protein concentrations. Proteins (100 ng) were separated by 10–12% SDS-polyacrylamide gel electrophoresis and transferred to polyvinylidene difluoride (PVDF) membranes (GE Healthcare Life Sciences, Chalfont, UK). The membranes were blocked for 1 h at room temperature using bovine serum albumin (BSA; Sangong Biotech). Subsequently, the membranes were incubated with specific primary antibodies (1:100 dilution) targeting Axl (cat. no. 4566), Twist (cat. no. 46702) and β-actin (cat. no. 4790; all from Cell Signaling Technology, Inc., Danvers, MA, USA) at 4°C overnight. After washing the membrane three times with TBST (5 min per wash), it was incubated with the recommended dilution (1:100) of horseradish peroxidase-conjugated secondary antibodies (cat. no. 7071; Cell Signaling Technology, Inc.) at room temperature for 1 h, and then washed as described above. The protein bands were visualized using the Immobilon Western Chemiluminescent HRP Substrate (EMD Millipore, Billerica, MA, USA). The chemiluminescence of proteins transferred to the PVDF membranes was detected using the ECL Plus Western Blot analysis Reagent (GE Healthcare Life Sciences). Relative protein expression levels were semi-quantified by densitometry using ImageJ software (version 2.1.4.7; National Institutes of Health, Bethesda, MA, USA).
Statistical analysis
Data were analyzed using SPSS statistical software, version 16.0 (SPSS, Inc., Chicago, IL, USA). The results are expressed as the mean ± standard deviation. The mean was compared using the Student's t-test or by analysis of variance. The statistical significance of the studies was determined using the parametric unpaired Student's t-test. P<0.05 was considered to indicate a statistically significant difference.
Results
AXL is upregulated in NSCLC cells
To evaluate the expression of AXL in the NSCLC cell lines, RT-qPCR and western blot analysis were conducted. AXL protein expression levels were markedly upregulated in the NSCLC cell lines compared with the control (Fig. 1A). Consistent with this, the mRNA expression levels of AXL were significantly upregulated in the NSCLC cells, as compared with the control (P<0.05; Fig. 1B). PC9 exhibited the highest level of AXL transcripts (~2-fold upregulation), as compared with the control; the AXL transcripts were ~1.7-fold upregulated in the A549 cells and 1.3-fold upregulated in the H1299 cells, as compared with the control (Fig. 1B). These results suggest that AXL is upregulated in NSCLC cells.
Figure 1.
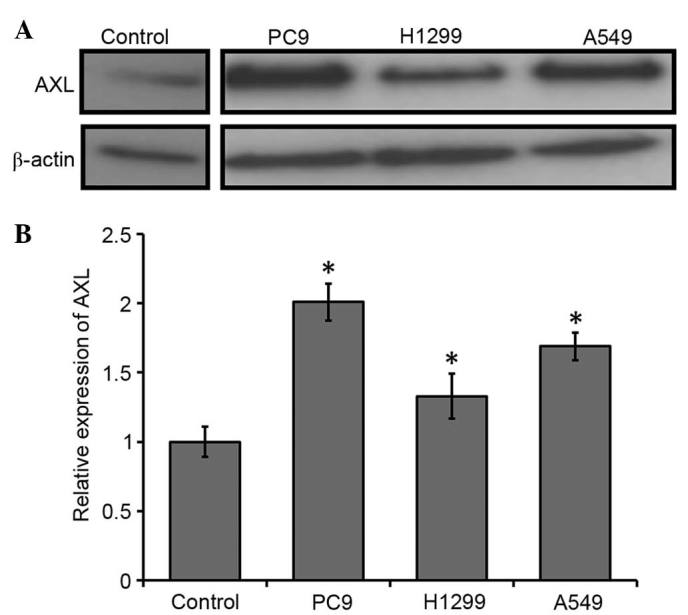
Determination of AXL expression levels in the NSCLC cell lines. (A) Western blot analysis and (B) reverse transcription-quantitative polymerase chain reaction demonstrated that AXL was upregulated in the three NSCLC cell lines, as compared with the control (normal lung cells). The PC9 cell line exhibited the highest expression levels of AXL. *P<0.05 vs. the control by analysis of variance. NSCLC, non-small-cell lung cancer.
AXL promotes NSCLC cell survival
To characterize the role of AXL in NSCLC cells, a gene-silencing experiment using double-stranded siRNA to silence AXL in the PC9 cells was conducted. PC9 cells were selected for further experimentation, as the greatest upregulation of AXL expression levels was observed in this cell line. PC9 cells were transfected with the siRNA and, after 3–7 days, RNA was extracted from control and transfected cell lines. A marked reduction was detected in the mRNA and protein expression levels of AXL in the siRNA-transfected cells, as compared with the control cells (Fig. 2A and B), and this was shown to be significant using RT-qPCR (P<0.01; Fig. 2C). In particular, ~1.83-fold downregulation of AXL was detected in the transfected cells, as compared with the control (Fig. 2). These results suggested that knockdown of AXL expression in the PC9 cells was successful.
Figure 2.
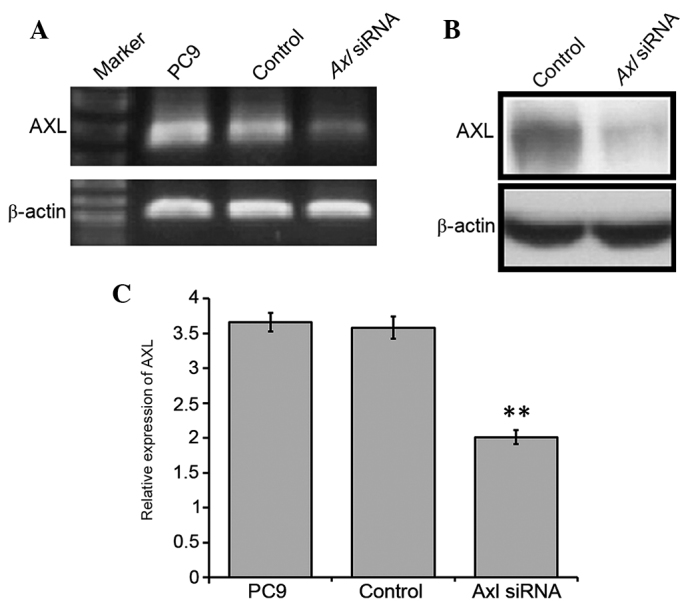
Silencing of AXL in the PC9 cell line was conducted by RNA interference, in which the PC9 NSCLC cell line was transfected with siRNA/AXl. (A) Total RNA was extracted and analyzed using RT-PCR using primers specific to AXL and β-actin. Cells were collected and lysed, and the expression levels of AXL were analyzed by (B) western blot analysis and (C) RT-qPCR. Mock transfection samples were used as the control. Normal PC9 cells were also used in the RT-PCR and RT-qPCR assays. **P<0.01 vs. the control by analysis of variance. NSCLC, non-small-cell lung cancer; siRNA, small interfering RNA; RT-PCR, reverse transcription-polymerase chain reaction; qPCR, quantitative-PCR.
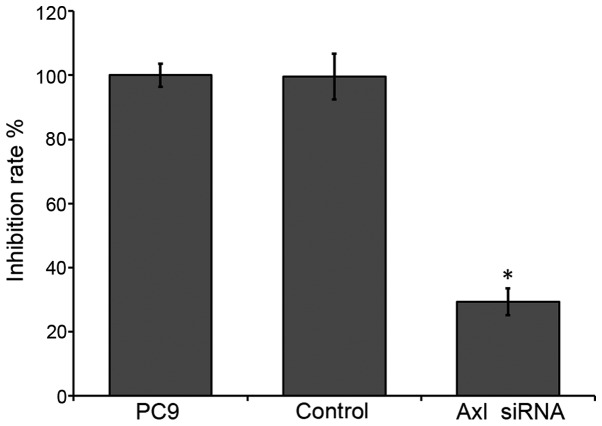
The viability of the PC9 cells was analyzed using the MTT assay. Downregulation of endogenous AXL in PC9 cells significantly reduced cell survival (~29.3%), as compared with the control (P<0.05; Fig. 3). These results suggest that inhibition of AXL by RNAi may reduce the survival of NSCLC cells.
Figure 3.
Inhibition of AXL in PC9 cells suppressed cell growth. Data are presented as mean ± standard deviation (n=4). Results are expressed as the percentage growth inhibition with respect to the control (mock transfection). *P<0.05 vs. control by analysis of variance. siRNA, small interfering RNA.
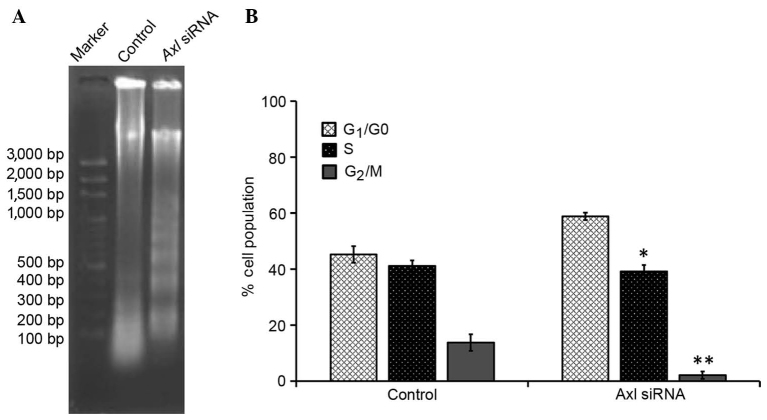
AXL is involved in NSCLC cell apoptosis. To further investigate the involvement of AXL in the inhibition of cell proliferation, cell-cycle distribution was evaluated using FCM analysis. As compared with the control group, the number of cells in the G1/G0 phase was significantly increased (P<0.05), and the number of cells in the S and G2/M phases was significantly decreased (P<0.05), in the PC9 cells treated with AXL siRNA; the majority of cells were in the G1 phase (Table I). Furthermore, the proportion of diploid cells slightly increased, with a reduction in the number of aneuploid cells, and the cell debris was markedly increased in the PC9 cells treated with AXL siRNA, as compared with the control group.
Table I.
Differences in cell cycle induced by downregulated AXL.
| Group | Cell debris (%) | Apoptosis (%) | Diploid (%) | Aneuploid (%) |
|---|---|---|---|---|
| Control | 1.88±0.2167 | 0.0267±0.0252 | 94.7867±4.5027 | 5.2133±4.5027 |
| Axl siRNA | 46.9533±5.0537 | 0.06934±0.0723 | 99.9101±0.1558 | 0.0901±0.1558 |
Data are presented as the mean ± standard deviation. Control, mock transfection samples. P<0.05 vs. the control by Student's t-test.
The effect of downregulating AXL expression on the induction of apoptosis in PC9 cells was investigated using a DNA fragmentation assay. Agarose gel electrophoresis demonstrated that knockdown of AXL resulted in the formation of DNA fragments in PC9 cells (Fig. 4A). In addition, a quantitative evaluation was conducted using TUNEL to detect DNA-strand breaks. As compared to with the control cells, 30.7% of PC9 G2 cells treated with AXL siRNA were apoptotic cells (Fig. 4B). These results suggest that interfering with AXL expression may inhibit cell-cycle progression in PC9 G2 cells, thereby resulting in a marked increase in the percentage of cells in the G1 phase.
Figure 4.
Inhibition of AXL accelerated cell-cycle arrest and apoptosis in PC9 cells. (A) DNA fragmentation was induced following AXL knockdown by siRNA in PC9 cells. (B) Quantitative evaluation of the terminal deoxynucleotidyl transferase dUTP nick end labeling assay by flow cytometry. Mock transfection samples were used as a control. Data are presented as the mean ± standard deviation of three determinations. *P<0.05 and **P<0.01 vs. the control by analysis of variance. siRNA, small interfering RNA.
Downregulation of AXL inhibits EMT-inducing Twist
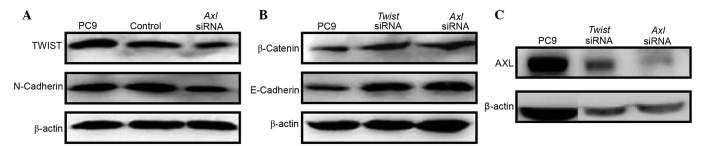
The acquisition of mesenchymal cell characteristics by epithelial cells, in particular the ability to migrate as single cells and invade the extracellular matrix, is the functional hallmark of EMT (24). EMT-inducing genes that have essential roles in EMT, including Twist and Snail, are termed master EMT genes (25). These genes function as transcriptional repressors of the cell-cell adhesion glycoprotein, E-cadherin whose functional loss is one of the hallmarks of EMT (26). Therefore, the status of Twist expression in PC9 cells was investigated in the present study. PC9 cells that were stably expressing Twist exhibited an EMT phenotype characterized by upregulation of the mesenchymal marker N-cadherin (Fig. 5A), and downregulation of the epithelial markers E-cadherin and β-cadherin (Fig. 5B). Notably, Axl was markedly upregulated in the Twist-expressing PC9 cells (Fig. 5C). Conversely, in the PC9 cells treated with AXL siRNA, downregulation of Twist and N-cadherin (Fig. 5A), and upregulation of E-cadherin and β-cadherin (Fig. 5B) were observed. However, knockdown of Twist did not affect the expression levels of AXL (Fig. 5C), despite the fact that E-cadherin and β-cadherin were upregulated (Fig. 5B). These results suggest that downregulation of AXL may abrogate EMT program induction by inhibiting the expression of Twist in NSCLC cells.
Figure 5.
Inhibition of AXL suppressed Twist expression. Upregulation of the master EMT gene Twist was accompanied by (A) upregulation of the mesenchymal marker N-cadherin, (B) downregulation of the epithelial markers E-cadherin and β-cadherin. and (C) upregulation of AXL. (A-C) These expression patterns were reversed by AXL knockdown using siRNA. Mock transfection samples were used as the control. siRNA, small interfering RNA.
Discussion
This study investigated the role of AXL in NSCLC cells, in particular the molecular mechanisms underlying the involvement of AXL in the EMT. AXL was shown to be upregulated in all NSCLC cell lines used in the present study, which is consistent with a previous study (27), suggesting that AXL may be used as a biomarker of NSCLC. Furthermore, in the present study, downregulation of AXL by RNAi was shown to inhibit PC9 cell survival, which may have been due to accelerated cell apoptosis or cell proliferation inhibition at the G1 stage. A previous study reported that human NSCLC with activating mutations in the EGFR did not respond to treatment with EGFR-targeted tyrosine kinase inhibitors (TKIs), due to the acquisition of resistance (27). Furthermore, activation of the AXL kinase was associated with resistance to EGFR-targeted therapy in patients with lung cancer (27), suggesting that increased AXL expression levels may contribute to drug resistance in lung cancer. Therefore, downregulation of AXL expression may be considered an effective tool for lung cancer therapy (28). Alterations in AXL-associated signaling pathways have been observed in ~20% of patients with acquired resistance to EGFR-TKI (29), although it remains to be determined whether these patients would benefit from AXL inhibition. In EGFR-TKI resistance, AXL may act as a bypass to activate downstream signals associated with cell survival and growth (30). Therefore, combined treatment with EGFR and AXL inhibitors may effectively abrogate the growth of tumor cells. A similar phenomenon was observed in hepatocyte growth factor receptor-mediated resistance (31).
EMT is an embryonic developmental process involving changes in cell morphology and expression of EMT-associated genes (25). EMT also occurs during the progression of several types of human cancer, in which it confers motility and invasiveness on cancer cells, leading them to acquire the ability to metastasize to distant sites (32). A previous study reported that lung cancer cell lines may be classified into mesenchymal and epithelial phenotypes based on the expression status of E-cadherin and Vimentin, which are markers of epithelial and mesenchymal phenotypes, respectively (33). A genetic study of early development reported the existence of a number of EMT-inducing genes encoding transcription factors that are capable of inducing EMT when ectopically expressed in epithelial cells (34). Therefore, the present study investigated the expression levels of several master EMT genes, as well as AXL, in order to elucidate the potential role of AXL in EMT. The master EMT gene Twist was upregulated in the PC9 NSCLC cells, which was accompanied by upregulation of AXL. Conversely, inhibition of AXL by RNAi resulted in the downregulation of Twist in PC9 cells; thus suggesting that downregulation of AXL inhibits EMT-inducing Twist. Therefore, AXL may regulate EMT via the EMT-associated genes. The results of the present study may have clinical significance in the design of therapeutic strategies targeting AXL and EMT signaling pathways.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 2.Pujol JL, Barlesi F, Daurès JP. Should chemotherapy combinations for advanced non-small cell lung cancer be platinum-based? A meta-analysis of phase III randomized trials. Lung Cancer. 2006;51:335–345. doi: 10.1016/j.lungcan.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 3.Le Chevalier T, Scagliotti G, Natale R, Danson S, Rosell R, Stahel R, Thomas P, Rudd RM, Vansteenkiste J, Thatcher N, et al. Efficacy of gemcitabine plus platinum chemotherapy compared with other platinum containing regimens in advanced non-small-cell lung cancer: A meta-analysis of survival outcomes. Lung Cancer. 2005;47:69–80. doi: 10.1016/j.lungcan.2004.10.014. [DOI] [PubMed] [Google Scholar]
- 4.Ettinger DS, Akerley W, Borghaei H, Chang AC, Cheney RT, Chirieac LR, D'Amico TA, Demmy TL, Ganti AK, Govindan R, et al. NCCN (National Comprehensive Cancer Network) Non-small cell lung cancer. J Natl Compr Canc Netw. 2012;10:1236–1271. doi: 10.6004/jnccn.2012.0130. [DOI] [PubMed] [Google Scholar]
- 5.Chevalier TL, Brisgand D, Douillard JY, Pujol JL, Alberola V, Monnier A, Riviere A, Lianes P, Chomy P, Cigolari S. Randomized study of vinorelbine and cisplatin versus vindesine and cisplatin versus vinorelbine alone in advanced non-small-cell lung cancer: Results of a European multicenter trial including 612 patients. J Clin Oncol. 1994;12:360–367. doi: 10.1200/JCO.1994.12.2.360. [DOI] [PubMed] [Google Scholar]
- 6.Non-small Cell Lung Cancer Collaborative Group, corp-author. Chemotherapy in non-small cell lung cancer: A meta-analysis using updated data on individual patients from 52 randomised clinical trials. BMJ. 1995;311:899–909. doi: 10.1136/bmj.311.7010.899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cappuzzo F, Hirsch FR, Rossi E, Bartolini S, Ceresoli GL, Bemis L, Haney J, Witta S, Danenberg K, Domenichini I, et al. Epidermal growth factor receptor gene and protein and gefitinib sensitivity in non-small-cell lung cancer. J Natl Cancer Inst. 2005;97:643–655. doi: 10.1093/jnci/dji112. [DOI] [PubMed] [Google Scholar]
- 8.Schiller JH, Harrington D, Belani CP, Langer C, Sandler A, Krook J, Zhu J, Johnson DH, Eastern Cooperative Oncology Group Comparison of four chemotherapy regimens for advanced non-small-cell lung cancer. N Engl J Med. 2002;346:92–98. doi: 10.1056/NEJMoa011954. [DOI] [PubMed] [Google Scholar]
- 9.Robinson DR, Wu YM, Lin SF. The protein tyrosine kinase family of the human genome. Oncogene. 2000;19:5548–5557. doi: 10.1038/sj.onc.1203957. [DOI] [PubMed] [Google Scholar]
- 10.Valverde P, Obin MS, Taylor A. Role of Gas6/Axl signaling in lens epithelial cell proliferation and survival. Exp Eye Res. 2004;78:27–37. doi: 10.1016/j.exer.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 11.Goruppi S, Ruaro E, Schneider C. Gas6, the ligand of Axl tyrosine kinase receptor, has mitogenic and survival activities for serum starved NIH3T3 fibroblasts. Oncogene. 1996;12:471–480. [PubMed] [Google Scholar]
- 12.Cano A, Pérez-Moreno MA, Rodrigo I, Locascio A, Blanco MJ, Del Barrio MG, Portillo F, Nieto MA. The transcription factor Snail controls epithelial-mesenchymal transitions by repressing E-cadherin expression. Nat Cell Biol. 2000;2:76–83. doi: 10.1038/35010506. [DOI] [PubMed] [Google Scholar]
- 13.Goruppi S, Ruaro E, Varnum B, Schneider C. Requirement of phosphatidylinositol 3-kinase-dependent pathway and Src for Gas6-Axl mitogenic and survival activities in NIH 3T3 fibroblasts. Mol Cell Biol. 1997;17:4442–4453. doi: 10.1128/MCB.17.8.4442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.O'Donnell K, Harkes IC, Dougherty L, Wicks IP. Expression of receptor tyrosine kinase Axl and its ligand Gas6 in rheumatoid arthritis: Evidence for a novel endothelial cell survival pathway. Am J Pathol. 1999;154:1171–1180. doi: 10.1016/S0002-9440(10)65369-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Janssen JW, Schulz AS, Steenvoorden AC, Schmidberger M, Strehl S, Ambros PF, Bartram CR. A novel putative tyrosine kinase receptor with oncogenic potential. Oncogene. 1991;6:2113–2120. [PubMed] [Google Scholar]
- 16.O'Bryan JP, Frye RA, Cogswell PC, Neubauer A, Kitch B, Prokop C, Espinosa R, Le Beau MM, Earp HS, Liu ET. axl, a transforming gene isolated from primary human myeloid leukemia cells, encodes a novel receptor tyrosine kinase. Mol Cell Biol. 1991;11:5016–5031. doi: 10.1128/MCB.11.10.5016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shieh YS, Lai CY, Kao YR, Shiah SG, Chu YW, Lee HS, Wu CW. Expression of axl in lung adenocarcinoma and correlation with tumor progression. Neoplasia. 2005;7:1058–1064. doi: 10.1593/neo.05640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ye X, Li Y, Stawicki S, Couto S, Eastham-Anderson J, Kallop D, Weimer R, Wu Y, Pei L. An anti-Axl monoclonal antibody attenuates xenograft tumor growth and enhances the effect of multiple anticancer therapies. Oncogene. 2010;29:5254–5264. doi: 10.1038/onc.2010.268. [DOI] [PubMed] [Google Scholar]
- 19.Meyer AS, Miller MA, Gertler FB, Lauffenburger DA. The receptor AXL diversifies EGFR signaling and limits the response to EGFR-targeted inhibitors in triple-negative breast cancer cells. Sci Signal. 2013;6:ra66. doi: 10.1126/scisignal.2004155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Janmaat ML, Kruyt FA, Rodriguez JA, Giaccone G. Response to epidermal growth factor receptor inhibitors in non-small cell lung cancer cells: Limited antiproliferative effects and absence of apoptosis associated with persistent activity of extracellular signal-regulated kinase or Akt kinase pathways. Clin Cancer Res. 2003;9:2316–2326. [PubMed] [Google Scholar]
- 21.Holland SJ, Pan A, Franci C, Hu Y, Chang B, Li W, Duan M, Torneros A, Yu J, Heckrodt TJ, et al. R428, a selective small molecule inhibitor of Axl kinase, blocks tumor spread and prolongs survival in models of metastatic breast cancer. Cancer Res. 2010;70:1544–1554. doi: 10.1158/0008-5472.CAN-09-2997. [DOI] [PubMed] [Google Scholar]
- 22.Danielsen T, Hvidsten M, Stokke T, Solberg K, Rofstad EK. Hypoxia induces p53 accumulation in the S-phase and accumulation of hypophosphorylated retinoblastoma protein in all cell cycle phases of human melanoma cells. Br J Cancer. 1998;78:1547–1558. doi: 10.1038/bjc.1998.722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-tie quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 24.Xu MM, Mao GX, Liu J, Li JC, Huang H, Liu YF, Liu JH. Low expression of the FoxO4 gene may contribute to the phenomenon of EMT in non-small cell lung cancer. Asian Pac J Cancer Prev. 2014;15:4013–4018. doi: 10.7314/APJCP.2014.15.9.4013. [DOI] [PubMed] [Google Scholar]
- 25.Christiansen JJ, Rajasekaran AK. Reassessing epithelial to mesenchymal transition as a prerequisite for carcinoma invasion and metastasis. Cancer Res. 2006;66:8319–8326. doi: 10.1158/0008-5472.CAN-06-0410. [DOI] [PubMed] [Google Scholar]
- 26.Vandewalle C, Van Roy F, Berx G. The role of the ZEB family of transcription factors in development and disease. Cell Mol Life Sci. 2009;66:773–787. doi: 10.1007/s00018-008-8465-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Z, Lee JC, Lin L, Olivas V, Au V, LaFramboise T, Abdel-Rahman M, Wang X, Levine AD, Rho JK, et al. Activation of the AXL kinase causes resistance to EGFR-targeted therapy in lung cancer. Nat Genet. 2012;44:852–860. doi: 10.1038/ng.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cerchia L, Esposito CL, Camorani S, Rienzo A, Stasio L, Insabato L, Affuso A, de Franciscis V. Targeting Axl with an high-affinity inhibitory aptamer. Mol Ther. 2012;20:2291–2303. doi: 10.1038/mt.2012.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hata A, Katakami N, Yoshioka H, Takeshita J, Tanaka K, Nanjo S, Fujita S, Kaji R, Imai Y, Monden K, et al. Rebiopsy of non-small cell lung cancer patients with acquired resistance to epidermal growth factor receptor-tyrosine kinase inhibitor: Comparison between T790M mutation-positive and mutation-negative populations. Cancer. 2013;119:4325–4332. doi: 10.1002/cncr.28364. [DOI] [PubMed] [Google Scholar]
- 30.Niederst MJ, Engelman JA. Bypass mechanisms of resistance to receptor tyrosine kinase inhibition in lung cancer. Sci Signal. 2013;6:10.1126/scisignal.2004652. doi: 10.1126/scisignal.2004652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qi J, McTigue MA, Rogers A, Lifshits E, Christensen JG, Jänne PA, Engelman JA. Multiple mutations and bypass mechanisms can contribute to development of acquired resistance to MET inhibitors. Cancer Res. 2011;71:1081–1091. doi: 10.1158/0008-5472.CAN-10-1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chaffer CL, Weinberg RA. A perspective on cancer cell metastasis. Science. 2011;331:1559–1564. doi: 10.1126/science.1203543. [DOI] [PubMed] [Google Scholar]
- 33.Takeyama Y, Sato M, Horio M, Hase T, Yoshida K, Yokoyama T, Nakashima H, Hashimoto N, Sekido Y, Gazdar AF, et al. Knockdown of ZEB1, a master epithelial-to-mesenchymal transition (EMT) gene, suppresses anchorage-independent cell growth of lung cancer cells. Cancer Lett. 2010;296:216–224. doi: 10.1016/j.canlet.2010.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mani SA, Guo W, Liao M-J, Eaton EN, Ayyanan A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cells. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]