Abstract
Purpose
The interaction of Porphyromonas gingivalis with commensal streptococci promotes P. gingivalis colonization of the oral cavity. We previously showed that a synthetic peptide (BAR) derived from Streptococcus gordonii potently inhibited the formation of P. gingivalis/S. gordonii biofilms (IC50 =1.3 µM) and reduced P. gingivalis virulence in a mouse model of periodontitis. Thus, BAR represents a novel therapeutic to control periodontitis by limiting P. gingivalis colonization of the oral cavity. Here, we sought to develop drug-delivery vehicles for potential use in the oral cavity that comprise BAR-modified poly(lactic-co-glycolic)acid (PLGA) nanoparticles (NPs).
Methods
PLGA-NPs were initially modified with palmitylated avidin and subsequently conjugated with biotinylated BAR. The extent of BAR modification was quantified using a fluorescent-labeled peptide. Inhibition of P. gingivalis adherence to S. gordonii by BAR-modified NPs was compared with free peptide using a two-species biofilm model.
Results
BAR-modified NPs exhibited an average size of 99±29 nm and a more positive surface charge than unmodified NPs (zeta potentials of −7 mV and −25 mV, respectively). Binding saturation occurred when 37 nmol BAR/mg of avidin-NPs was used, which resulted in a payload of 7.42 nmol BAR/mg NPs. BAR-modified NPs bound to P. gingivalis in a dose-dependent manner and more potently inhibited P. gingivalis/S. gordonii adherence and biofilm formation relative to an equimolar amount of free peptide (IC50 of 0.2 µM versus 1.3 µM). BAR-modified NPs also disrupted the preformed P. gingivalis/S. gordonii biofilms more effectively than free peptide. Finally, we demonstrate that BAR-modified NPs promoted multivalent association with P. gingivalis, providing an explanation for the increased effectiveness of NPs.
Conclusion
These results indicate that BAR-modified NPs deliver a higher local dose of peptide and may represent a more effective therapeutic approach to limit P. gingivalis colonization of the oral cavity compared to treatment with formulations of free peptide.
Keywords: nanoparticle, peptide delivery, multivalent, drug delivery, Porphyromonas gingivalis, periodontal disease
Introduction
The oral cavity is colonized by a complex microbial community that comprises bacteria, viruses, and fungi.1,2 Periodontal disease can arise when normal host–microbe homeostasis is disrupted.3 Porphyromonas gingivalis plays a pivotal role in this process and has been designated as a “keystone pathogen” that can induce dysbiosis through manipulation of the host innate immune response, leading to uncontrolled inflammation and tissue damage.4 P. gingivalis has also been associated with systemic illnesses such as heart disease5,6 and rheumatoid arthritis.7,8 Thus, limiting P. gingivalis colonization of the oral cavity may represent an effective approach to both alter the progression of periodontal inflammation and reduce the levels of systemic inflammation that constitute a risk factor for systemic conditions associated with P. gingivalis infection.
Our previous work suggested that adherence of P. gingivalis to oral streptococci facilitates P. gingivalis colonization of the oral cavity and showed that adherence requires a protein–protein interaction between the minor fimbrial antigen of P. gingivalis and antigen I/II of specific streptococcal species (eg, Streptococcus gordonii).9,10 From these studies, a synthetic peptide (designated BAR) was identified that potently inhibited P. gingivalis adherence to streptococci and the formation of P. gingivalis/S. gordonii biofilms (50% inhibitory concentration [IC50] =1.3 µM).11,12 BAR also significantly reduced P. gingivalis virulence in a mouse model of periodontitis.11–13 These results suggest that BAR may represent a novel therapeutic to limit P. gingivalis colonization of the oral cavity.
There has been growing interest in the use of engineered nanoparticles (NPs) as drug-delivery vehicles, nanomedicines, or dental material/devices.14,15 As delivery vehicles, polymer NPs are attractive options for the delivery of drugs, proteins, or genetic agents that target cells.16 Their small size enables more effective barrier penetration and drug accumulation at target sites. Furthermore, relative to hydroxyapatite and silver NPs that have been used to prevent dental caries and periodontal disease,17 polymeric NPs function as non-immunogenic, biologically stable carriers that can encapsulate and/or present a wide range of therapeutic agents on their surfaces. Considering this, the goal of this study was to increase the localized delivery of BAR by developing poly(lactic-co-glycolic)acid (PLGA) NPs that are surface-modified with BAR.
Material and methods
Peptide synthesis
BAR peptide (NH2-LEAAPKKVQDLLKKANITVK GAFQLFS-COOH) comprises residues 1,167 to 1,193 of the streptococcal surface protein B (SspB) sequence of S. gordonii.12 For conjugation with surface-modified avidin-NPs, BAR was synthesized with biotin covalently attached to the N-terminus. To quantify BAR density on the NPs surface, biotinylated-BAR was further modified to contain 6-carboxyfluorescein (Flc) covalently attached to the ε-amine of the lysine residue underlined in the aforementioned sequence. This peptide was designated BAR-Flc. For adherence inhibition experiments using free peptide, BAR peptide without biotin or fluorophore modifications was utilized. All peptides were synthesized by BioSynthesis, Inc. (Lewisville, TX, USA).
Growth of bacterial strains
P. gingivalis ATCC33277 was grown in Trypticase soy broth (Difco Laboratories Inc., Livonia, MI, USA) supplemented with 0.5% (w/v) yeast extract, 1 µg/mL menadione, and 5 µg/mL hemin. The medium was reduced for 24 h under anaerobic conditions (10% CO2, 10% H2, and 80% N2) and P. gingivalis was subsequently inoculated and grown anaerobically for 48 h at 37°C. S. gordonii DL-1 was cultured aerobically without shaking in brain–heart infusion broth (Difco Laboratories Inc.) supplemented with 1% yeast extract for 16 h at 37°C.
Synthesis of avidin-palmitate
NPs were initially conjugated with avidin-palmitate as previously described.18–20 Briefly, 10 mg of avidin was dissolved in 1.2 mL of 2% (w/v) sodium deoxycholate (NaDC) in PBS and warmed to 37°C. A 1 mg/mL palmitic acid-N-hydroxysuccinimide ester (PA-NHS; Sigma-Aldrich, St Louis, MO, USA) solution was prepared in 2% (w/v) NaDC and sonicated until well-mixed. Eight hundred microliters of the PA-NHS solution was added in drops to the reaction vial containing avidin and allowed to react overnight at 37°C. The reaction solution was then dialyzed in 1,200 mL of 0.15% (w/v) NaDC in PBS at 37°C using a 3,500 molecular weight cut-off dialysis tube to remove free PA-NHS. After overnight dialysis at 37°C, avidin-palmitate was transferred to a storage vial and stored at 4°C until use.
Nanoparticle synthesis
Unmodified and surface-modified NPs encapsulating the fluorescent dye Coumarin 6 (C6) were synthesized for binding studies. C6-containing NPs were synthesized using an oil-in-water (o/w) single-emulsion technique.19–21 Briefly, C6 was encapsulated in 50–200 mg PLGA carboxyl- terminated polymer (0.55–0.75 dL/g; LACTEL®; DURECT Corporation, Cupertino, CA, USA). C6 was dissolved in 200 µL dichloromethane (DCM) overnight at a concentration of 15 µg/mg PLGA. In parallel, 50 mg of PLGA crystals was dissolved in 2 mL of DCM overnight. The following day, the PLGA/DCM solution was vortexed while adding C6 DCM solution and was subsequently sonicated to attain a uniform suspension. Next, 2 mL of 5% (w/v) polyvinyl alcohol (PVA) in deionized water (diH2O) was mixed with 2 mL of 5 mg/mL avidin-palmitate to obtain a homogeneous suspension. Two milliliters of the PLGA/DCM/C6 solution was added in drops to the PVA/avidin-palmitate suspension with vortexing and subsequent sonication. Residual DCM was evaporated by adding the NPs solution to 50 mL of 0.3% PVA for 3 h while mixing. After solvent evaporation, the NPs solution was transferred to tubes and centrifuged at 13,000 rpm at 4°C and washed with PBS. The density of avidin on the NPs surface was determined using micro BCA assay (Thermo Fisher Scientific, Waltham, MA, USA). From the density of avidin incorporation and previous work suggesting that some avidin binding sites may be sterically blocked,18,21 we estimated that 18.5 nmol BAR/mg NPs represented a threefold molar excess of peptide. Thus, for BAR conjugation, NPs were suspended in 9 mL of diH2O and incubated with 18.5 nmol biotinylated-BAR/mg NPs in PBS for 30 min on a benchtop rotator. After conjugation, NPs were centrifuged at 13,000 rpm and washed three times with 20 mL diH2O to remove unbound peptides. NPs were then suspended in 9 mL of diH2O, frozen at −80°C for 3 h, lyophilized, and stored at −20°C. Unmodified C6 NPs were prepared similarly; however, 5% (w/v) PVA alone was added instead of PVA/avidin-palmitate solution.
Quantification of BAR payload on the NPs surface
To quantify and maximize the BAR payload of NPs, BAR-Flc was used to measure the density of peptide on the NPs surface. Avidin-NPs (5 mg) were mixed with increasing amounts (18.5–111.2 nmol/mg NPs) of BAR-Flc in 1 mL PBS for 45 min on a rocker platform in the dark. After conjugation, NPs were washed twice with diH2O and were frozen, lyophilized, and stored at −20°C. NPs from each sample were then suspended in 1× PBS to create a 1 mg/mL NPs solution, and the resulting samples were transferred to a microtiter plate in triplicate. Total NP-associated fluorescence was determined using a Victor3 multilabel spectrophotometer, and peptide quantity was determined from a standard curve of known BAR-Flc concentrations.
NPs characterization
Particle size and morphology were determined using scanning electron microscopy (SEM). Dry NPs were mounted on carbon tape and sputter coated with gold under vacuum. Average particle diameter and size distribution were determined from SEM images of at least 400 particles per batch using “ImageJ” image analysis software (version 1.5a, imageJ.nih.gov). Zeta potential and dynamic light scattering were measured with a Zetasizer Nano ZS (Malvern Instruments, Malvern, UK) in diH2O to determine particle charge and hydrated diameter.
Binding of BAR-modified NPs to P. gingivalis
Adherence to P. gingivalis cells was measured using BAR-modified NPs that encapsulated C6. P. gingivalis cells were washed with PBS and adjusted to a final optical density of 0.4 at 600 nm (OD600 nm). Subsequently, 1 mL aliquots of P. gingivalis cells were mixed with increasing amounts (1–10 µg) of BAR-modified C6 NPs for 60 min on a rocker platform in the dark. Negative controls consisted of P. gingivalis incubated with unmodified or avidin-modified C6 NPs and BAR-modified C6 NPs incubated in buffer without P. gingivalis to evaluate nonspecific binding. After incubation, samples were centrifuged at 5,600 rpm for 5 min, the cell pellets were suspended in 1× PBS, and cell-associated fluorescence was measured at 488 nm.
BAR-modified NPs inhibition of P. gingivalis/S. gordonii adherence
S. gordonii DL-1 cultures (10 mL) were harvested by centrifugation at 5,600 rpm and suspended in 1 mL PBS. Cells were labeled with 20 µL of 10 mM hexidium iodide (Thermo Fisher Scientific) for 15 min in the dark, centrifuged at 5,600 rpm for 5 min, and washed with PBS. For all experiments, the OD600 nm of S. gordonii cells was adjusted to 0.8, and 1 mL of the cell suspension was added to each well of a 12-well microtiter plate containing a glass coverslip. S. gordonii cells were incubated for 24 h under anaerobic conditions on a rocker platform in the dark to facilitate S. gordonii binding to the coverslip.
Similarly, P. gingivalis cultures (10 mL) were harvested as mentioned earlier, labeled with 20 µL of 4 mg/mL carboxyfluorescein–succinylester (Thermo Fisher Scientific) in the dark, and washed with PBS. For adherence inhibition assays, the OD600 nm was adjusted to 0.8 and the P. gingivalis cell suspension was subsequently diluted with an equal volume of an appropriate BAR-modified NPs suspension or free BAR peptide to generate a final OD600 nm of 0.4. BAR-modified NPs or free BAR was preincubated with labeled P. gingivalis cells at BAR peptide concentrations ranging from 0 to 1.7 µM at 25°C for 30 min before transferring to wells containing S. gordonii. The plates were then incubated for 24 h at 25°C under anaerobic conditions in the dark. Following incubation, the supernatant was removed and cells were washed with pre-reduced PBS to remove non-adherent and loosely bound bacteria. Adhered cells were fixed with 4% (w/v) paraformaldehyde and the cover glass was mounted on a glass slide. P. gingivalis-S. gordonii biofilms were visualized using an Olympus FluoView confocal laser scanning microscope (Olympus Corp., Center Valley, PA, USA) under 60× magnification. An argon or HeNe(G) laser was used to visualize fluorescein-labeled and hexidium iodide-labeled P. gingivalis and S. gordonii cells, respectively. Z-stack images of the biofilms were obtained from 30 randomly chosen frames using FluoView 500 Software (Olympus Corp., Center Valley, PA, USA) using a z-step size of 0.7 µm. Images were analyzed with Volocity image analysis software (version 6.3; Perkin Elmer, Waltham, MA, USA) to determine the ratio of green to red fluorescence (GR). Each peptide concentration was analyzed in triplicate and three independent frames were measured for each well. The mean and variation (SD) between samples were determined using analysis of variance (ANOVA) and differences were considered to be statistically significant when P<0.05. Percent inhibition of P. gingivalis adherence was calculated with the following formula: 1 – GR sample/GR control. For some experiments, P. gingivalis was allowed to adhere to immobilized streptococci in the absence of peptide inhibitor to demonstrate the ability of BAR-modified NPs to disrupt pre-established biofilms. The resulting P. gingivalis/S. gordonii biofilms were then treated for 1–3 h with BAR or BAR-modified NPs at various concentrations and processed and analyzed as described earlier.
Results
Nanoparticle characterization
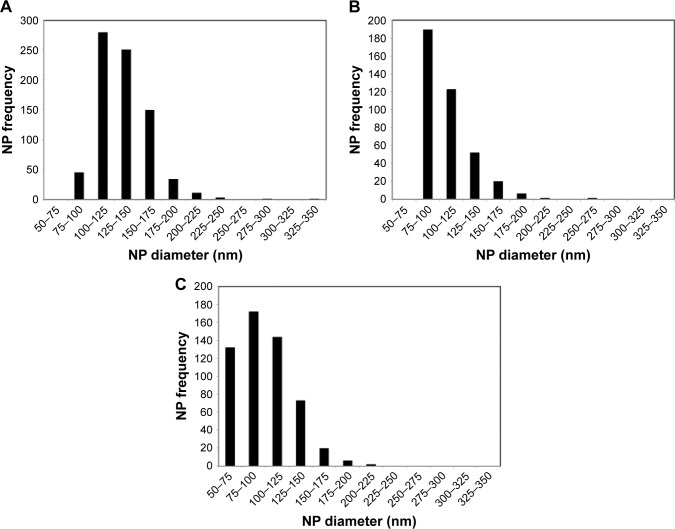
Previous studies demonstrated the utility of coupling peptides to NPs surfaces via avidin–biotin ligands.19,20 In this study, we synthesized unmodified, avidin-modified, and BAR-modified NPs and determined the overall NPs size distribution for each sample (Figure 1). The size distribution of unhydrated modified NPs trended smaller than unmodified NPs, but the average diameters of unmodified, avidin-modified, and BAR-modified NPs calculated from SEM images (134±28 nm, 107±25 nm, and 99±29 nm, respectively; Table 1) were not statistically significant (P>0.05). As expected, the diameters of the hydrated NPs were greater than the unhydrated NPs and did not differ significantly among the samples. The average hydrodynamic diameters determined for unmodified, avidin-modified, and BAR-modified NPs were 298±13 nm, 317±4 nm, and 329±10 nm, respectively (Table 1). In addition, no change was observed in the texture or morphology of NPs after conjugation with BAR (not shown). Finally, as shown in Table 1, unmodified NPs exhibited a negative surface charge of −25±1.6 mV, and the addition of positively charged avidin, or avidin and BAR to the NPs surface resulted in significantly greater positive zeta potentials (−9±0.5 mV and −7±1.3 mV, respectively; P<0.01), confirming avidin or avidin-BAR conjugation to the NPs surface.
Figure 1.
Size distribution of unhydrated, unmodified PLGA NPs (A), avidin-modified NPs (B), and avidin-NPs that were subsequently conjugated with BAR peptide (C).
Abbreviations: NPs, nanoparticles; PLGA, poly(lactic-co-glycolic)acid.
Table 1.
Physical characterization of NPs
| NPs type | Unhydrated diameter (nm) | Hydrated diameter (nm) | Zeta potential (mV) |
|---|---|---|---|
| Unmodified | 134±28 | 298±13 | −25±1.6 |
| Avidin-modified | 107±25 | 317±4 | −9±0.5 |
| BAR-modified | 99±29 | 329±10 | −7±1.3 |
Abbreviation: NPs, nanoparticles.
Quantification of the total BAR payload of BAR-modified NPs
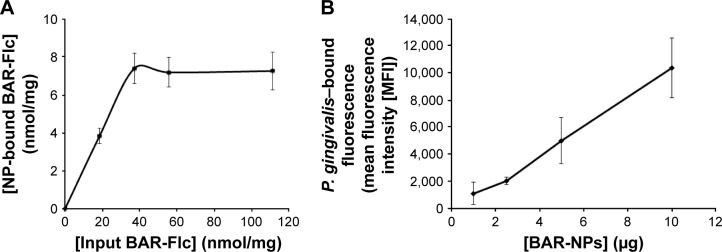
The avidin content of avidin-NPs was determined to be 3.1 nmol/mg NPs. This was similar to the total amount added to the NPs synthesis reaction (3.0 nmol/mg NPs), indicating that 100% of the input avidin was incorporated on the NPs surface. For avidin-NPs that were subsequently conjugated with biotinylated-BAR, the peptide payload was determined by incubating avidin-NPs with BAR-Flc and quantifying NP-bound fluorescence. As shown in Figure 2A, the incorporation of BAR-Flc on avidin-NPs directly correlated with the input concentration of BAR-Flc, and saturation occurred at an input concentration of 37.1 nmol/mg NPs. Mean fluorescence values did not significantly increase at higher input concentrations of BAR-Flc (Figure 2A). At saturation, 7.42 nmol of BAR-Flc was incorporated per milligram of avidin-NPs. Avidin has four binding sites, and with 3 nmol avidin incorporated per milligram NPs, a potential 12 nmol BAR-Flc could bind per milligram of NPs if all avidin sites were occupied. Our results suggest that only 62% of the avidin sites (7.42 of 12) were available for interaction with BAR-Flc.
Figure 2.
(A) The total payload of BAR peptide bound to the surface of avidin-NPs was determined by incubating avidin-NPs with increasing concentrations of BAR-Flc and measuring the NP-associated fluorescence. Binding saturation occurred at an input peptide concentration of 37.1 nmol/mg NPs. Under these conditions, BAR-modified NPs contained 7.42 nmol of BAR/mg NPs. (B) BAR-modified NPs interact with the P. gingivalis cell surface in a dose-dependent manner. Intact P. gingivalis cells were incubated with various amounts of BAR-modified NPs that encapsulated the fluorescent dye C6. Cell-associated fluorescence was then measured after washing to remove unbound NPs.
Abbreviations: NPs, nanoparticles; Flc, 6-carboxyfluorescein; P. gingivalis, Porphyromonas gingivalis; C6, Coumarin 6.
Binding of BAR-modified NPs to P. gingivalis and inhibition of P. gingivalis/S. gordonii biofilm development
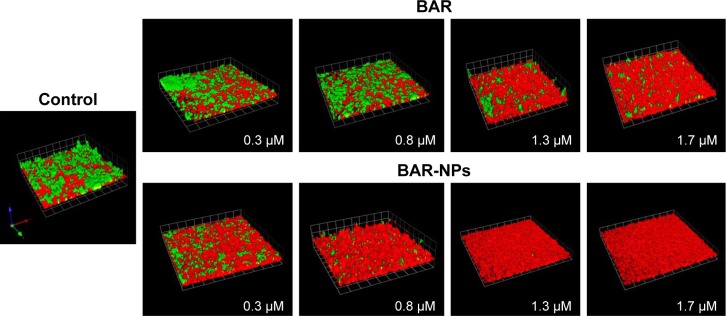
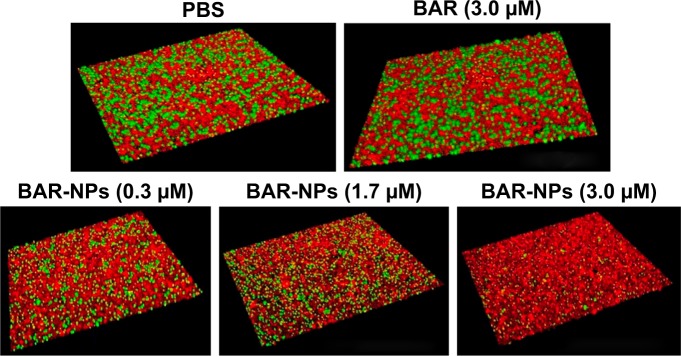
To determine whether the peptide presented by BAR-modified NPs was capable of interacting with P. gingivalis, bacterial cells were incubated with BAR-modified NPs encapsulating the fluorophore C6. As shown in Figure 2B, BAR-modified NPs bound to P. gingivalis cells in a specific and dose-dependent manner, indicating that immobilizing BAR on the NPs surface did not affect its interaction with the minor fimbrial antigen of P. gingivalis. Next, to determine whether BAR-modified NPs competitively inhibit P. gingivalis adherence to streptococci and prevent biofilm formation, P. gingivalis/S. gordonii biofilms were formed in the presence of increasing amounts of peptide (0.3–1.7 µM) delivered either by BAR-modified NPs or by supplying molar equivalents of free BAR peptide. Representative images of biofilms formed in the presence of BAR-modified NPs or soluble BAR are shown in Figure 3. As shown, BAR-modified NPs more potently inhibited P. gingivalis (green) biofilm formation with S. gordonii (red) than molar equivalents of free BAR peptide across all of the peptide concentrations that were tested. As shown in Figure 4, the IC50 for free BAR peptide was approximately 1.3 µM, consistent with the value originally reported by Daep et al.11 In contrast, the IC50 for inhibition of adherence by BAR-modified NPs was approximately 0.2 µM, which is 6.5-fold lower than the free BAR peptide. This result indicates that BAR-modified NPs are a significantly more potent inhibitor of P. gingivalis adherence to streptococci and subsequent biofilm formation than free peptide.
Figure 3.
BAR and BAR-modified NPs inhibit P. gingivalis adherence to S. gordonii and subsequent biofilm formation. Two-species biofilms were formed in the presence of 0.3–1.7 µM BAR associated with NPs or an equimolar amount of soluble BAR peptide. The resulting biofilms were visualized by confocal laser scanning microscopy and the ratio of green (P. gingivalis) to red (S. gordonii) fluorescence in Z-stack images was determined using Volocity image analysis software. The blocks shown in each image represent 21.3 µM.
Abbreviations: NPs, nanoparticles; P. gingivalis, Porphyromonas gingivalis; S. gordonii, Streptococcus gordonii.
Figure 4.
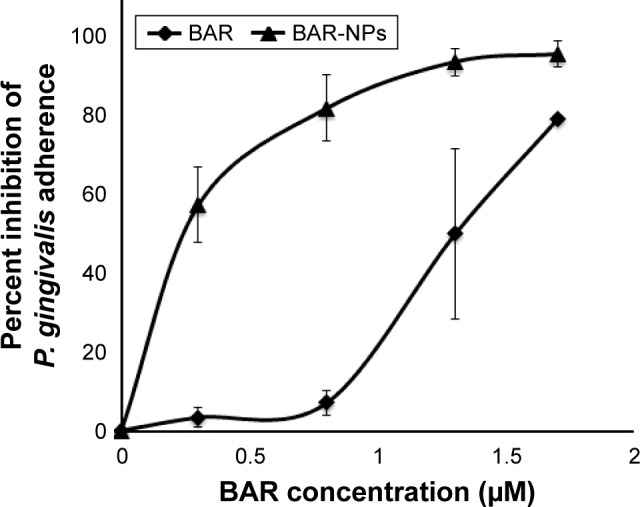
The inhibition profiles of BAR-modified NPs and free peptide indicate that BAR-modified NPs are 6.5-fold more potent in blocking the adherence of P. gingivalis to S. gordonii than soluble BAR.
Abbreviations: NPs, nanoparticles; P. gingivalis, Porphyromonas gingivalis; S. gordonii, Streptococcus gordonii.
BAR-modified NPs promote multivalent association with P. gingivalis
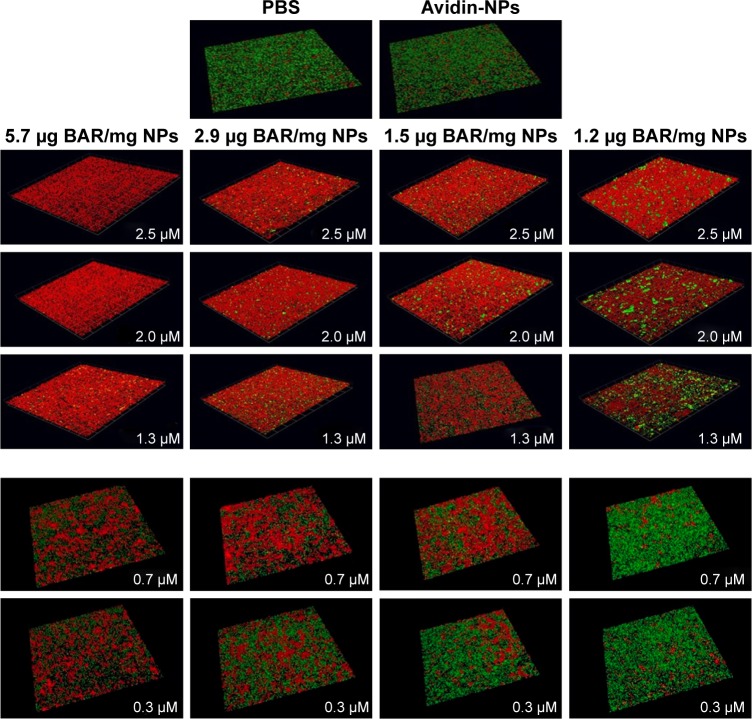
To determine whether the increase in potency exhibited by BAR-modified NPs could be explained by increased valency of the BAR/P. gingivalis interaction, four independent preparations of BAR-modified NPs were synthesized in parallel that contained BAR payloads of 5.7, 2.9, 1.5, or 1.2 µg peptide/mg NPs. Each preparation was tested for inhibition of P. gingivalis adherence to S. gordonii using a range of final BAR peptide concentrations of 0.3, 0.7, 1.3, 2.0, and 2.5 µM. As shown in Figure 5 and as summarized in Table 2, P. gingivalis adhered efficiently to S. gordonii in the PBS and avidin-NPs controls, indicating that NPs without BAR have no effect on P. gingivalis adherence to streptococci and subsequent biofilm formation. In contrast, P. gingivalis adherence was significantly reduced in the presence of BAR-modified NPs. As expected, inhibition of P. gingivalis adherence increased as the molar equivalent of BAR peptide delivered by each NPs preparation was increased (compare vertical columns for each NPs preparation in Figure 5 and Table 2). Moreover, as the density of BAR peptide on the NPs surface decreased from 5.7 to 1.2 µg peptide/mg NPs, inhibition of P. gingivalis adherence also decreased even though the final peptide concentration delivered by each NPs preparation was the same. This indicates that the potency of biofilm inhibition increases as a function of BAR peptide density contained on the NPs surface and suggests that BAR-modified NPs may promote multivalent association with P. gingivalis, increasing the avidity of the interaction and the effectiveness of the peptide.
Figure 5.
BAR-modified NPs promote multivalent interaction with P. gingivalis. Four independent preparations of BAR-modified NPs were synthesized that contained 5.7, 2.9, 1.5, and 1.2 µg BAR/mg NPs. Each preparation was tested for inhibition of P. gingivalis/S. gordonii biofilm formation by adding an appropriate amount of BAR-modified NPs to deliver a final peptide concentration of 0.3, 0.7, 1.3, 2.0, or 2.5 µM to the reaction mixture. For each preparation of NPs, inhibition of biofilm formation increased as a function of the final concentration of peptide delivered (see vertical columns). In addition, as shown in the horizontal rows, inhibition of biofilm formation increased as the total payload of BAR peptide present in NPs preparations increased, and this occurred for each of the final peptide concentrations that were tested.
Abbreviations: NPs, nanoparticles; PBS, phosphate-buffered saline; P. gingivalis, Porphyromonas gingivalis; S. gordonii, Streptococcus gordonii.
Table 2.
Percent inhibition of P. gingivalis adherence as a function of BAR payload
| BAR (µM) | BAR payload (µg/mg NPs)
|
|||
|---|---|---|---|---|
| 5.7 | 2.9 | 1.5 | 1.2 | |
| 2.5 | 95.3±2.6 | 81.7±1.5 | 68.4±1.8 | 59.0±3.3 |
| 2.0 | 92.9±7.3 | 78.1±4.7 | 65.9±5.5 | 52.7±3.1 |
| 1.3 | 89.6±3.5 | 75.1±5.0 | 63.7±3.6 | 43.8±7.2 |
| 0.7 | 62.4±2.3 | 40.0±7.1 | 40.7±8.4 | 17.3±8.7 |
| 0.3 | 32.1±6.8 | 29.5±7.4 | 25.3±9.8 | 14.1±11.3 |
Note: Data are presented as mean ± SD.
Abbreviation: NPs, nanoparticles.
BAR-modified NPs disrupt existing P. gingivalis/S. gordonii biofilms
As described in the “Materials and methods” section, our main approach to assess the activity of BAR involved using the peptide as a competitive inhibitor of P. gingivalis adherence to streptococci, an essential first step in biofilm formation. To determine whether BAR peptide is capable of disrupting a preexisting P. gingivalis/S. gordonii biofilm, dual species biofilms were formed in PBS in the absence of peptide inhibitor and were subsequently incubated for various time periods with peptide or BAR-modified NPs. As shown in Table 3, incubation of P. gingivalis/S. gordonii biofilms with free peptide at a concentration of 3.0 µM did not disrupt the biofilm until the exposure time was increased to 3 h. Since BAR-modified NPs more potently inhibited the adherence of P. gingivalis relative to free peptide, we next tested the activity of BAR-modified NPs against preexisting P. gingivalis/S. gordonii biofilms. As shown in Figure 6 and as summarized in Table 4, BAR-modified NPs disrupted established dual species biofilms in a dose-dependent manner under conditions where free BAR peptide exhibited little activity. Together with the earlier results, this indicates that BAR peptide delivered by surface-modified NPs more potently prevents P. gingivalis adherence to streptococci and also disrupts established biofilms, suggesting that surface-modified NPs may represent an attractive delivery vehicle to limit P. gingivalis colonization of the oral cavity.
Table 3.
Disruption of existing biofilms by BAR (3.0 µM)
| Exposure time (h) | Percent decrease |
|---|---|
| 1 | 8.7±7.9 |
| 2 | 6.1±3.9 |
| 3 | 54.6±3.3 |
Note: Data are presented as mean ± SD.
Figure 6.
Disruption of existing P. gingivalis/S. gordonii biofilms by BAR peptide and BAR-modified NPs. Two-species biofilms were formed and were subsequently incubated for 1 h with PBS or with PBS containing either free BAR peptide (3.0 µM) or BAR-modified NPs that delivered a final concentration of peptide of 0.3, 1.7, or 3.0 µM. Under these conditions, free peptide did not disrupt the dual-species biofilm. In contrast, a dose-dependent reduction of biofilm was observed with increasing peptide concentration delivered by BAR-modified NPs.
Abbreviations: NPs, nanoparticles; PBS, phosphate-buffered saline; P. gingivalis, Porphyromonas gingivalis; S. gordonii, Streptococcus gordonii.
Table 4.
Disruption of existing biofilms after 1 h of exposure to BAR-modified NPs
| Peptide | Concentration (µM) | Percent decrease |
|---|---|---|
| BAR | 3.0 | 5.1±5.0 |
| BAR-NPs | 0.3 | 4.0±5.2 |
| 1.7 | 27.1±5.8 | |
| 3.0 | 59.3±4.6 |
Abbreviation: NPs, nanoparticles.
Discussion
Previous studies have shown that P. gingivalis interacts with primary colonizing organisms such as S. gordonii22,23 and that this interaction may be an initial event that facilitates P. gingivalis colonization of the oral cavity. As such, it represents an ideal target for therapeutic intervention to limit P. gingivalis colonization. Indeed, our previous work showed that BAR peptide prevents P. gingivalis adherence to S. gordonii and reduced maxillary alveolar bone loss in mice when the peptide was administered simultaneously with P. gingivalis infection.12,13 However, our current results suggest that free BAR is less effective in disrupting established P. gingivalis/S. gordonii biofilms (50% inhibition at 3.0 µM) than preventing initial adherence of P. gingivalis to streptococci (50% inhibition at 1.3 µM). This represents a potential limitation in developing BAR as a therapeutic targeting P. gingivalis. One approach to address this problem is to increase the efficacy of the peptide by delivering BAR at higher localized concentration. In this study, this was accomplished using a reproducible and rapid preparation to synthesize BAR-modified NPs.
Previous studies demonstrated that avidin-biotin ligand conjugation provides one of the strongest non-covalent bonds, while offering a flexible, tunable, and efficient method to conjugate and alter ligand density on the NPs surface.18,21 Here, we produced spherical NPs with zeta potential values for unmodified (−25 mV) and BAR-modified NPs (−7 mV) that are consistent with other avidin-NPs studies.19,20 Since unmodified NPs have a negative surface charge, this difference can be attributed to the successful conjugation of positively charged avidin and BAR peptide to the NPs surface. In addition to confirming successful conjugation, the more positive surface charge exhibited by avidin-NPs and BAR-modified NPs may also facilitate electrostatic interactions of NPs with negatively charged moieties on the P. gingivalis cell surface.
To increase the local delivery of BAR, it is important to formulate NPs that contain maximal levels of the peptide. Although 100% of the input avidin was incorporated on the surface of NPs, only 62% of the avidin binding sites were available for interaction with BAR. A possible explanation for this is that the four biotin binding sites of avidin are in close proximity to each other and to the NPs surface, which may lead to steric hindrance. Nonetheless, BAR-modified NPs more potently inhibited P. gingivalis adherence to S. gordonii at all concentrations tested and exhibited a significantly lower IC50, relative to free BAR. This suggests that BAR-modified NPs may promote multivalent interaction with P. gingivalis, and our experimental evidence is consistent with this. Previous studies have demonstrated the utility of multivalency in a variety of biological systems to confer stronger and enhanced binding by enhancing the affinity (avidity) for a given target and decreasing the dissociation rate from that target.24,25 These concepts have been applied to the design of pharmaceutical agents and drug-delivery vehicles to significantly improve the therapeutic potential of active agents in a variety of applications.24–35 Similar to other studies reporting that NPs can increase drug efficacy by promoting a multivalent binding interface,25,30,36 it is likely that BAR-modified NPs exhibit a similar mechanism leading to increased efficacy. To further enhance multivalent interactions, future studies will focus on developing alternative approaches to enhance payload and efficacy, by increasing avidin modification in combination with increasing peptide concentrations. For example, while we achieved 100% avidin incorporation on the NPs surface, it is possible that the density of biotinylated-BAR may be enhanced by increasing the avidin content of NPs. In addition, other surface chemistries may increase the number of ligands incorporated on the NPs surface. From these formulations, we can determine if a “threshold” density of BAR exists that leads to maximal efficacy of BAR-modified NPs.
It is also important to identify the roles that ligand number and ligand type play in multivalent interactions between NP-ligands and a targeted receptor. Although the current approach did not provide comprehensive information about the binding kinetics of BAR-modified NPs, this study impacts real-world biopharmaceutical development by providing a theoretical framework for designing NPs that may be better suited for targeting other microorganisms in oral biofilms. Clearly, the etiology of periodontal disease is complex,37 and although recent evidence suggests that P. gingivalis may play an essential role in altering host–microbe homeostasis,3 other pathogens or pathogen interactions may have a significant impact on disease progression. Therefore, while these delivery approaches have been initially applied to target BAR to P. gingivalis, NPs that target multiple organisms could be achieved by comodifying the surface of NPs with several other antimicrobial agents targeting other bacteria in the oral cavity. It is possible that these NPs may facilitate the formulation of products such as an oral rinse or varnish to combat oral disease, but a limitation of this approach is that such products are only transiently applied in the oral cavity. Our work suggests that BAR-modified NPs represent one mechanism to deliver a high local dose of the inhibitory peptide to the oral cavity. In addition, we are currently developing NPs that encapsulate BAR and release peptide over a sustained period of time. We envision that sustained release NPs can be further comodified with cell surface adhesive proteins that adhere to streptococci in order to produce NPs that exhibit increased retention time and that deliver BAR to the same niche in the oral microbiome that P. gingivalis seeks to occupy. In summary, we have developed a reliable and well-defined method for modifying NPs with inhibitory peptides, and our results suggest that nanotechnology can be efficiently used to target specific oral organisms and combat oral diseases.
Conclusion
Our results indicate that BAR-modified NPs deliver a higher local dose of peptide and promote multivalent interaction with P. gingivalis. BAR-modified NPs may represent a more effective therapeutic approach to limit P. gingivalis colonization of the oral cavity than treatment with formulations of free peptide.
Acknowledgments
This work was supported by grants R01DE023206 and R21DE025345 from the National Institute for Dental and Cranialfacial Research. We will freely provide reagents and/or data generated in this study to academic investigators according to the policies of the University of Louisville.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Marsh PD. Dental plaque as a biofilm and a microbial community – implications for health and disease. BMC Oral Health. 2006;6(Suppl 1):S14. doi: 10.1186/1472-6831-6-S1-S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rosan B, Lamont RJ. Dental plaque formation. Microbes Infect. 2000;2:1599–1607. doi: 10.1016/s1286-4579(00)01316-2. [DOI] [PubMed] [Google Scholar]
- 3.Hajishengallis G, Lamont RJ. Beyond the red complex and into more complexity: the polymicrobial synergy and dysbiosis (PSD) model of periodontal disease etiology. Mol Oral Microbiol. 2012;27(6):409–419. doi: 10.1111/j.2041-1014.2012.00663.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hajishengallis G, Lamont RJ. Breaking bad: manipulation of the host response by Porphyromonas gingivalis. Eur J Immunol. 2014;44(2):328–338. doi: 10.1002/eji.201344202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Desvarieux M, Demmer RT, Rundek T, et al. Periodontal microbiota and carotid intima-media thickness: the Oral Infections and Vascular Disease Epidemiology Study (INVEST) Circulation. 2005;111(5):576–582. doi: 10.1161/01.CIR.0000154582.37101.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Genco RJ, Van Dyke TE. Prevention: reducing the risk of CVD in patients with periodontitis. Nat Rev Cardiol. 2010;7(9):479–480. doi: 10.1038/nrcardio.2010.120. [DOI] [PubMed] [Google Scholar]
- 7.Koziel J, Mydel P, Potempa J. The link between periodontal disease and rheumatoid arthritis: an updated review. Curr Rheumatol Rep. 2014;16(3):408. doi: 10.1007/s11926-014-0408-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lundberg K, Wegner N, Yucel-Lindberg T, Venables PJ. Periodontitis in RA – the citrullinated enolase connection. Nat Rev Rheumatol. 2010;6(12):727–730. doi: 10.1038/nrrheum.2010.139. [DOI] [PubMed] [Google Scholar]
- 9.Lamont RJ, El-Sabaeny A, Park Y, Cook GS, Costerton JW, Demuth DR. Role of the Streptococcus gordonii SspB protein in the development of Porphyromonas gingivalis biofilms on streptococcal substrates. Microbiology. 2002;148(Pt 6):1627–1636. doi: 10.1099/00221287-148-6-1627. [DOI] [PubMed] [Google Scholar]
- 10.Park Y, Simionato MR, Sekiya K, et al. Short fimbriae of Porphyromonas gingivalis and their role in coadhesion with Streptococcus gordonii. Infect Immun. 2005;73(7):3983–3989. doi: 10.1128/IAI.73.7.3983-3989.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Daep CA, James DM, Lamont RJ, Demuth DR. Structural characterization of peptide-mediated inhibition of Porphyromonas gingivalis biofilm formation. Infect Immun. 2006;74(10):5756–5762. doi: 10.1128/IAI.00813-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Daep CA, Lamont RJ, Demuth DR. Interaction of Porphyromonas gingivalis with oral streptococci requires a motif that resembles the eukaryotic nuclear receptor box protein-protein interaction domain. Infect Immun. 2008;76(7):3273–3280. doi: 10.1128/IAI.00366-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Daep CA, Novak EA, Lamont RJ, Demuth DR. Structural dissection and in vivo effectiveness of a peptide inhibitor of Porphyromonas gingivalis adherence to Streptococcus gordonii. Infect Immun. 2011;79(1):67–74. doi: 10.1128/IAI.00361-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Allaker RP, Ian Douglas CW. Non-conventional therapeutics for oral infections. Virulence. 2015;6(3):196–207. doi: 10.4161/21505594.2014.983783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Sousa FF, Ferraz C, Rodrigues LK, Nojosa Jde S, Yamauti M. Nanotechnology in dentistry: drug delivery systems for the control of biofilm-dependent oral diseases. Curr Drug Deliv. 2014;11(6):719–728. doi: 10.2174/156720181106141202115157. [DOI] [PubMed] [Google Scholar]
- 16.Masood F. Polymeric nanoparticles for targeted drug delivery system for cancer therapy. Mater Sci Eng C Mater Biol Appl. 2016;60:569–578. doi: 10.1016/j.msec.2015.11.067. [DOI] [PubMed] [Google Scholar]
- 17.Emmanuel R, Palaniswamy S, Chen SM, et al. Antimicrobial efficacy of green synthesized drug blended silver nanoparticles against dental caries and periodontal disease causing microorganisms. Mater Sci Eng C Mater Biol Appl. 2015;56:374–379. doi: 10.1016/j.msec.2015.06.033. [DOI] [PubMed] [Google Scholar]
- 18.Fahmy TM, Samstein RM, Harness CC, Mark Saltzman W. Surface modification of biodegradable polyesters with fatty acid conjugates for improved drug targeting. Biomaterials. 2005;26(28):5727–5736. doi: 10.1016/j.biomaterials.2005.02.025. [DOI] [PubMed] [Google Scholar]
- 19.Sims LB, Curtis LT, Frieboes HB, Stainbach-Rankins JM. Enhanced uptake and transport of PLGA-modified nanoparticles in cervical cancer. J Nanobiotechnology. 2016;14:33. doi: 10.1186/s12951-016-0185-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Steinbach JM, Seo YE, Saltzman WM. Cell penetrating peptide-modified poly(lactic-co-glycolic acid) nanoparticles with enhanced cell internalization. Acta Biomater. 2016;30:49–61. doi: 10.1016/j.actbio.2015.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martin DT, Steinbach JM, Liu J, et al. Surface-modified nanoparticles enhance transurothelial penetration and delivery of survivin siRNA in treating bladder cancer. Mol Cancer Ther. 2014;13(1):71–81. doi: 10.1158/1535-7163.MCT-13-0502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stinson MW, Haraszthy GG, Zhang XL, Levine MJ. Inhibition of Porphyromonas gingivalis adhesion to Streptococcus gordonii by human submandibular-sublingual saliva. Infect Immun. 1992;60(7):2598–2604. doi: 10.1128/iai.60.7.2598-2604.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wright CJ, Burns LH, Jack AA, et al. Microbial interactions in building of communities. Mol Oral Microbiol. 2013;28(2):83–101. doi: 10.1111/omi.12012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fasting C, Schalley CA, Weber M, et al. Multivalency as a chemical organization and action principle. Angew Chem Int Ed. 2012;51:10472–10498. doi: 10.1002/anie.201201114. [DOI] [PubMed] [Google Scholar]
- 25.Mammen M, Choi SK, Whitesides GM. Polyvalent interactions in biological systems: implications for design and use of multivalent ligands and inhibitors. Angew Chem Int Ed. 1998;37:2755–2794. doi: 10.1002/(SICI)1521-3773(19981102)37:20<2754::AID-ANIE2754>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 26.Carlson CB, Mowery P, Owen RM, Dykhuizen EC, Kiessling LL. Selective tumor cell targeting using low-affinity, multivalent interactions. ACS Chem Biol. 2007;2(2):119–127. doi: 10.1021/cb6003788. [DOI] [PubMed] [Google Scholar]
- 27.Davis ME, Chen Z, Shin DM. Nanoparticle therapeutics: an emerging treatment modality for cancer. Nat Rev Drug Discov. 2008;7(9):771–782. doi: 10.1038/nrd2614. [DOI] [PubMed] [Google Scholar]
- 28.Hall PR, Hjelle B, Brown DC, et al. Multivalent presentation of anti-hantavirus peptides on nanoparticles enhances infection blockade. Antimicrob Agents Chemother. 2008;52(6):2079–2088. doi: 10.1128/AAC.01415-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hennig R, Pollinger K, Veser A, Breunig M, Goepferich A. Nanoparticle multivalency counterbalances the ligand affinity loss upon PEGylation. J Control Release. 2014;194:20–27. doi: 10.1016/j.jconrel.2014.07.062. [DOI] [PubMed] [Google Scholar]
- 30.Hong S, Leroueil PR, Majaros IJ, Orr BG, Baker JR, Jr, Banaszak Holl MM. The binding avidity of a nanoparticle-based multivalent targeted drug delivery platform. Chem Biol. 2007;14(1):107–115. doi: 10.1016/j.chembiol.2006.11.015. [DOI] [PubMed] [Google Scholar]
- 31.Montet X, Funovics M, Montet-Abou K, Weissleder R, Josephson L. Multivalent effects of RGD peptides obtained by nanoparticle display. J Med Chem. 2006;49(20):6087–6093. doi: 10.1021/jm060515m. [DOI] [PubMed] [Google Scholar]
- 32.Rosca EV, Stukel JM, Gillies RJ, Vagner J, Caplan MR. Specificity and mobility of biomacromolecular, multivalent constructs for cellular targeting. Biomacromolecules. 2007;8(12):3830–3835. doi: 10.1021/bm700791a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stukel JM, Li RC, Maynard HD, Caplan MR. Two-step synthesis of multivalent cancer-targeting constructs. Biomacromolecules. 2010;11(1):160–167. doi: 10.1021/bm9010276. [DOI] [PubMed] [Google Scholar]
- 34.Wang J, Tian SM, Petros RA, Napier ME, DeSimone JM. The complex role of multivalency in nanoparticles targeting the transferrin receptor for cancer therapies. J Am Chem Soc. 2010;132(32):11306–11313. doi: 10.1021/ja1043177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weissleder R, Kelly K, Sun EY, Shtatland T, Josephson L. Cell-specific targeting of nanoparticles by multivalent attachment of small molecules. Nat Biotechnol. 2005;23(11):1418–1423. doi: 10.1038/nbt1159. [DOI] [PubMed] [Google Scholar]
- 36.Lane LA, Qian XM, Smith AM, Nie S. Physical chemistry of nano-medicine: understanding the complex behaviors of nanoparticles in vivo. Ann Rev Phys Chem. 2015;66:521–547. doi: 10.1146/annurev-physchem-040513-103718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Colombo AP, Boches SK, Cotton SL, et al. Comparisons of subgingival microbial profiles of refractory periodontitis, severe periodontitis, and periodontal health using the human oral microbe identification microarray. J Periodontol. 2009;80(9):1421–1432. doi: 10.1902/jop.2009.090185. [DOI] [PMC free article] [PubMed] [Google Scholar]