Abstract
Although microRNAs have emerged as key regulators in diverse cellular processes, the roles of microRNAs are poorly understood in human embryonic stem cells (hESCs) during differentiation into specialized cell types. In this study, we used a microRNA array with 799 human microRNA probes to examine the expression profiles of microRNAs in hESCs during differentiation into endodermal and mesodermal lineages in vitro. Among the microRNAs analyzed, 7 and 20 microRNAs were enriched in the developmental process of hESCs into mesodermal and endodermal lineages, respectively. In particular, the expression levels of miR-200 family, which is known to regulate the epithelial to mesenchymal transition (EMT), gradually increased in hESCs during differentiation into hepatocytes while they gradually decreased during differentiation into vascular endothelial cells. Downregulation of ZEB1, a direct target of miR-200 family, and E-CADHERIN, a target protein of ZEB1, was observed in hESCs during differentiation into endodermal and mesodermal lineages, respectively. These results indicate that miR-200 family has an important role in determining the cell fate between endodermal and mesodermal lineages from the pluripotent state.
Keywords: Human embryonic stem cells, microRNA, miR-200 family, EMT
Introduction
Human embryonic stem cells (hESCs) which have pluripotency and self-renewal capacity can be used as sources for numerous biomedical studies and cell therapies (1). Differentiation of hESCs into a specialized cell type might be regulated by complex networks of inter- and intracellular molecules (2). Additionally, it has been known that microRNAs function as key regulators in stem cells (3, 4). MicroRNAs that are single stranded, small (~22 nucleotides) and non-coding RNAs generated from endogenous transcripts regulate gene expressions (5) and have important roles in cellular functions, such as cell proliferation or survival (6), fat metabolism regulation (7), mesenchymal stem cell differentiation (8), epithelial to mesenchymal transition (9), and even in antiviral defense (10). Additionally, recent studies have revealed that microRNAs are critical regulators for the differentiation of hESCs and cell fate determination during development (11–14). For examples, miR-430/427/302 family is involved in mesoderm fate determination by regulating LEF1 and LEF2 (Lefty-1/2) (15), and miR-145 represses the expressions of stem cell marker genes OCT4, SOX2, and KLF4, thereby leading to the suppression of the stemness in hESCs (16). p53 protein promotes the differentiation of hESCs by targeting miR-34a and miR-145 (17). Recently, miR-34a was identified as a regulator of the cell fate to pluripotent state in mouse ESCs and induced pluripotent stem cells (18). We previously reported that different kinds of microRNAs may function in a developmental stage-specific manner during the differentiation of hESCs into hepatocytes (19). Although respective microRNAs are associated with the differentiation process of pluripotent stem cells, the roles of microRNAs have remained unclear in cell fate determination for lineage specification.
Here we found that different kinds of microRNAs were enriched between endodermal and mesodermal cells differentiated from hESCs. Interestingly, the expression levels of miR-200 family gradually increased in hESCs during differentiation into hepatocytes (endodermal lineage) while they gradually decreased in hESCs during differentiation into vascular endothelial cells (mesodermal lineage). The expression level of ZEB1, which is a target protein of miR-200 family and represses the transcriptional expression of E-CADHERIN (20), was decreased during the differentiation process of hESCs into the endodermal lineage. Our findings show the involvement of miR-200 family in endodermal specification in hESCs during in vi-tro differentiation.
Materials and Methods
Culture of hESCs
Human embryonic stem cells (CHA4-hES line) (21) were maintained on a layer of mitomycin-C treated mouse embryonic fibroblasts (MEFs) in ESC medium in a humidified atmosphere of 5% CO2 in air. The ESC medium consisted of Dulbecco’s DMEM/F12 supplemented with 20% knockout serum replacement (Invitrogen, CA), 0.1 mM β-mercaptoethanol (Sigma-Aldrich, MO, USA), 1% non-essential amino acids (Invitrogen), 1% penicillin-streptomycin (Invitrogen), and 4 ng/ml basic FGF (R&D Systems, MN). For the feeder-free culture, hESCs were cultured on Matrigel (BD Biosciences, CA) in conditioned medium which was obtained by culturing mitomycin-C treated MEFs.
Differentiation of hESCs into hepatocytes
hESCs were differentiated into hepatocytes as previously described (19). Briefly, for definitive endoderm (DE) induction, hESCs were cultured in RPMI1640 medium (Hyclone, UT) supplemented with 0.5 mg/ml albumin fraction V (Sigma-Aldrich) and 50 ng/ml Activin A (Peprotech, NJ, USA) for 1 d and 1% insulin-transferrin-selenium (ITS, Sigma-Aldrich) was added to this medium for an additional 4 d. For hepatocyte differentiation, hESC-derived DE cells were cultured in hepatocyte culture medium (HCM, Lonza, MD) containing 30 ng/ml FGF4 (Peprotech) and 20 ng/ml BMP2 (Peprotech) for 5 d, then incubated in HCM containing 20 ng/ml HGF (Peprotech) for 5 d, and further cultured in HCM supplemented with 10 ng/ml Oncostatin M (OSM, R&D systems) and 0.1 μM dexamethasone (DEX, Sigma-Aldrich) for 5 d. The medium was changed daily.
Differentiation of hESCs into vascular endothelial cells
Differentiation of hESCs into vascular endothelial cells was done as previously described (22). Briefly, hESCs were first treated with 50 μM PD98059 (Promega, Madison, WI) and 20 ng/ml BMP4 (Peprotech, Rocky Hill, NJ) to induce the mesodermal lineage for 3 d. The mesodermal cells were committed to CD34+ vascular progenitor cells by treatment with 100 ng/ml VEGF-A and 50 ng/ml bFGF (R&D Systems, Minneapolis, MN) for 6 d. hESC-derived CD34+ cells were isolated by with CD34+ microbeads (Miltenyi Biotech, Bergisch Gladbach, Germany). To obtain vascular endothelial cells, the hESC-derived CD34+ cells were cultured in EGM-2MV medium (CAMBREX, Rutherford, NJ) supplemented with 100 ng/ml VEGF-A and 50 ng/ml bFGF for 10 d.
Microarray analysis
Total RNAs were extracted from cells lysed in Trizol (Invitrogen) according to the manufacturer’s instruction. For the microRNA microarray, microRNA Complete Labeling and Hyb kit (Agilent Technology, GE5190-0456) was used, following the manufacturer’s instruction. Briefly, 100 ng of total RNA were incubated with CIP at 37°C for 30 min. Dephosphorylated RNA sample was denatured with DMSO at 100°C for 5 min. Ligation master mix was prepared following the manufacturer’s instructions (10×T4 RNA ligase buffer, Cyanine3-pCp, T4 RNA ligase) and added to each sample. Samples were resuspended with blocking agent and Hi-RPM hybridization buffer, and incubated at 100°C for 5 min, and then loaded onto Agilent’s Human microRNA microarray (Agilent Technology, G4112F). Human miRNA Microarray Kit (Aglent Technology, G2545A) was used for hybridization. Samples were hybridized at 55°C and 20×rpm for 20 h on Agilent Hybridization oven (Agilent Technology, G2545A). The hybridized microarrays were washed following the manufacturer’s washing instructions (Agilent Technology).
Acquisition of the microarray data
The hybridized images were scanned with Agilent’s DNA microarray scanner (Agilent Technology, G2505B) and quantified with Feature Extraction Software (Agilent Technology). All data normalization and selection of fold-changed genes were performed with GeneSpringGX 7.3 (Agilent Technology). The averages of normalized ratios were calculated by dividing the average of the normalized signal channel intensity by the average of the normalized control channel intensity. Cluster and the TreeView program (Eisen Lab, http://rana.lbl.gov/eisen/) were used for clustering and visualizing the expression level of the microRNAs. Hierarchical clustering was performed with average linkage and correlation similarity with the software listed above.
Quantitative RT-PCR
For extracting total RNAs, hESCs were homogenized in TRIzol (Invitrogen) according to the manufacturer’s instructions. Approximately 1 μg of total RNAs was used to generate first-strand cDNA using Superscript II reverse transcriptase (Invitrogen), and cDNA was amplified by PCR with the PCR PreMix (GENETBIO, Daejeon, Korea). The specific primers used in this study are listed in Supplementary Table 1. The expression levels of the respective genes were measured by quantitative RT-PCR using Prime Q-Master mix with SYBR Green I (GENETBIO). The relative expression level was analyzed with an iCycler iQ5 Real-Time detection system (Bio-Rad Laboratories, Hercules, CA). The reaction parameters for the quantitative RT-PCR analysis were 95°C for 10 min followed by 40 cycles of 95°C for 30 sec, 60°C for 30 sec, and 72°C for 30 sec, and a final elongation step at 72°C for 5 min. All reactions were performed in duplicate. For the comparative quantification, the transcriptional levels of respective genes were normalized to that of GAPDH, and expressed as fold-change relative to the expression level in undifferentiated hESCs. The sample ΔCt (SΔCt) value was calculated as the difference between the Ct values of GAPDH and the target. The ΔCt value of the undifferentiated hESCs was used as the control ΔCt (CΔCt) value. The relative gene expression levels between the sample and control were determined with the formula 2−(SΔCt−CΔCt).
For quantitative RT-PCR of the microRNAs, TaqMan microRNA assays (Applied Biosystems) were used to quantify the level of mature microRNAs according to the manufacturer’s instructions. Reactions were done by BioRad iQ5 real-time PCR system. The reaction parameters for the quantitative RT-PCR analysis were 95°C for 10 min followed by 40 cycles of 95°C for 10 sec, 60°C for 10 sec, and 72°C for 10 sec. The sample ΔCt (SΔCt) value was calculated as the difference between the Ct value of RNU6B and the target. The ΔCt value of the undifferentiated hESCs was used as the control ΔCt (CΔCt) value. The relative gene expression levels between the sample and control were determined with the formula 2−(SΔCt−CΔCt). Quantitative RT-PCR of the microRNAs was performed in duplicate.
Western blot
For protein purification, cells were mechanically harvested in Western blot PRO-PREP lysis buffer (Intron Biotechnology) and homogenized with Vibra cellTM VCX-750 (SONICS & MATERIALS Inc., Newton, CT) if required. The proteins were separated on 10% SDS-PAGE gels and transferred to nitrocellulose membranes. The protein-bound membranes were blocked with 4% BSA in TBST for 1 h at RT followed by overnight incubation with a primary antibody at 4°C. The primary antibodies used in this study were as follows: ZEB1 (1:200, Santa Cruz Biotechnology), E-CADHERIN (1:1000, BD Biosciences), and HRP-conjugated actin (1:1000, Santa Cruz Biotechnology). After washing several times with TBST, the membranes were incubated with horseradish peroxidase (HRP)-conjugated secondary antibody (1:5,000, Santa Cruz Biotechnology). The membranes were developed with ECL (Elpis biotech) and exposed on a Fujifilm LAS-4000 CCD camera system (Fujifilm, Tokyo, Japan) for quantitative imaging. Band intensities were measured with the ImageJ program. Quantification of the protein bands was normalized by that of actin.
Statistical analysis
The statistical significance of the real-time RT-PCR data rate was evaluated with one-way ANOVA. Values of p<0.05 were considered significant.
Results and Discussion
Differentiation of hESCs into specific lineages
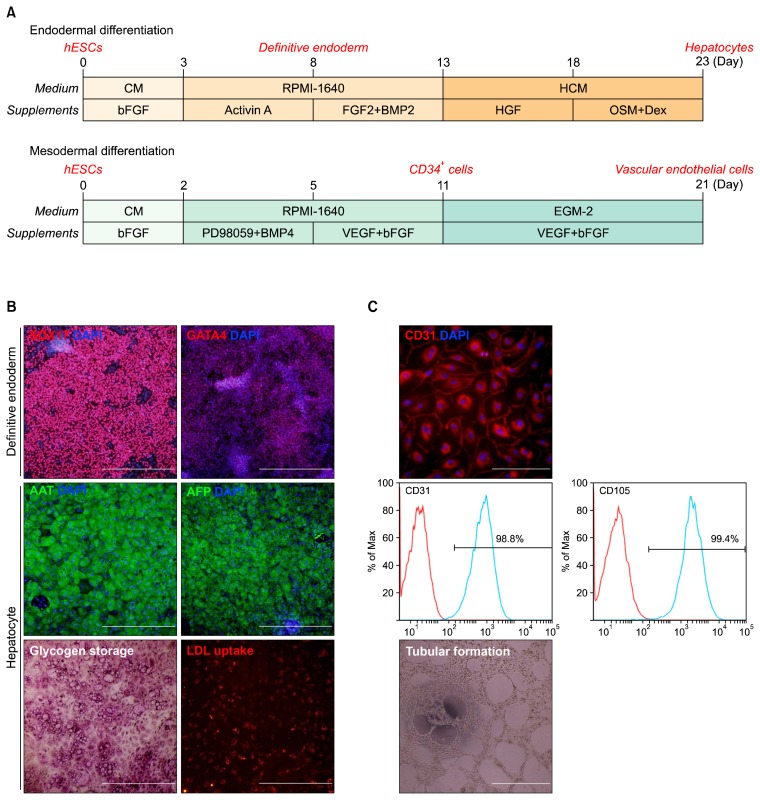
To compare expression profiles of microRNAs between two lineages, hESCs were differentiated into endodermal and mesodermal lineages, respectively (Fig. 1A). For endodermal differentiation, hESCs were induced to definitive endoderm (DE) cells expressing DE markers SOX17 and GATA4 (Fig. 1B, upper) and then further differentiated into hepatocytes. hESC-derived hepatocytes expressed hepatic markers AAT and AFP (Fig. 1B, middle) and showed the functionality of glycogen storage and LDL uptake (Fig. 1B, lower). For mesodermal differentiation, hESCs were developed to CD34-positive cells (CD34+ cells) and then the sorted CD34+ cells were differentiated into vascular endothelial cells with high efficiency (Fig. 1C, upper and middle). These hESC-derived vascular endothelial cells had tubular formation competence (Fig. 1C, lower). By microarray analysis using 799 human microRNA probes and quantitative RT-PCR of specific microRNAs, the expression profiles of the microRNAs were analyzed in hESCs, intermediate cells (DE and CD34+ cells), and terminally differentiated cells (hepatocytes and vascular endothelial cells), respectively.
Fig. 1.
Differentiation of hESCs. (A) Overall scheme for the differentiation of hESCs into hepatocytes or vascular endothelial cells. (B) Characterization of endodermal lineage cells derived from hESCs. Immunocytochemistry of SOX17 and GATA4 (up), AAT and AFP (middle), and PAS staining and LDL uptake assay (lower). AAT, anti-α trypsin; AFP, α-fetoprotein; LDL, low-density lipoprotein. Scale bar, 500 μm. (C) Characterization of mesodermal lineage cells derived from hESCs; Immunocytochemistry of CD31 (upper), FACS analysis of vascular endothelial cells (middle), and tubular formation of vascular endothelial cells (lower). Scale bar, 500 μm.
Expression patterns of the microRNAs in hESCs during differentiation into hepatocytes and vascular endothelial cells
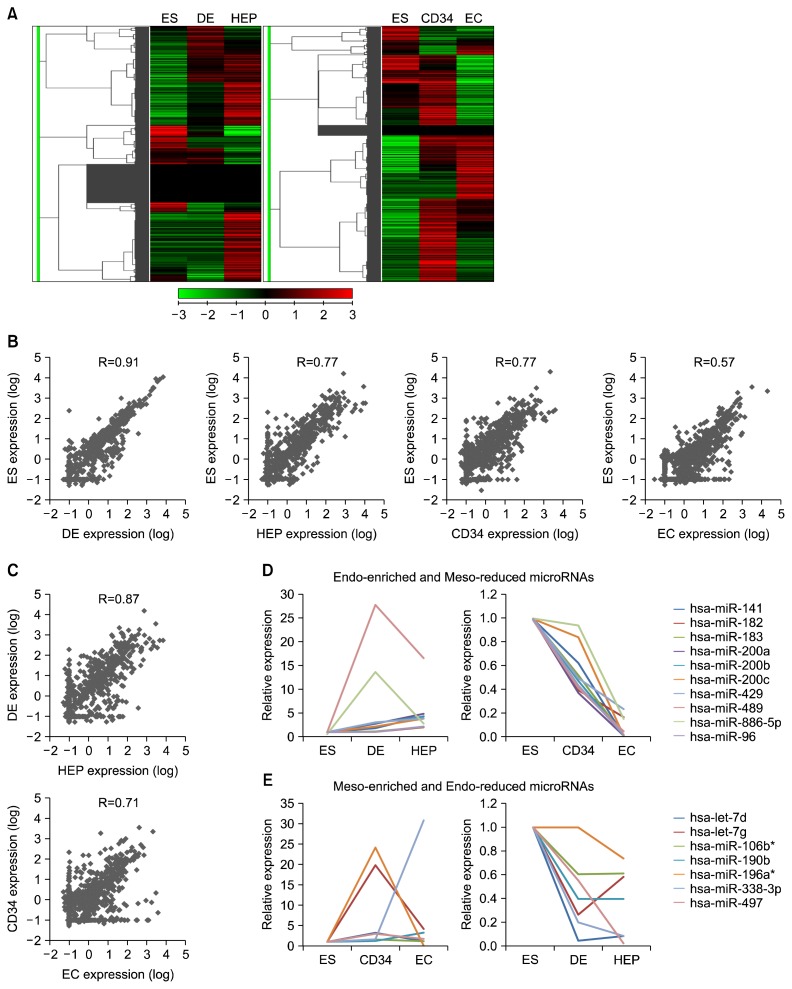
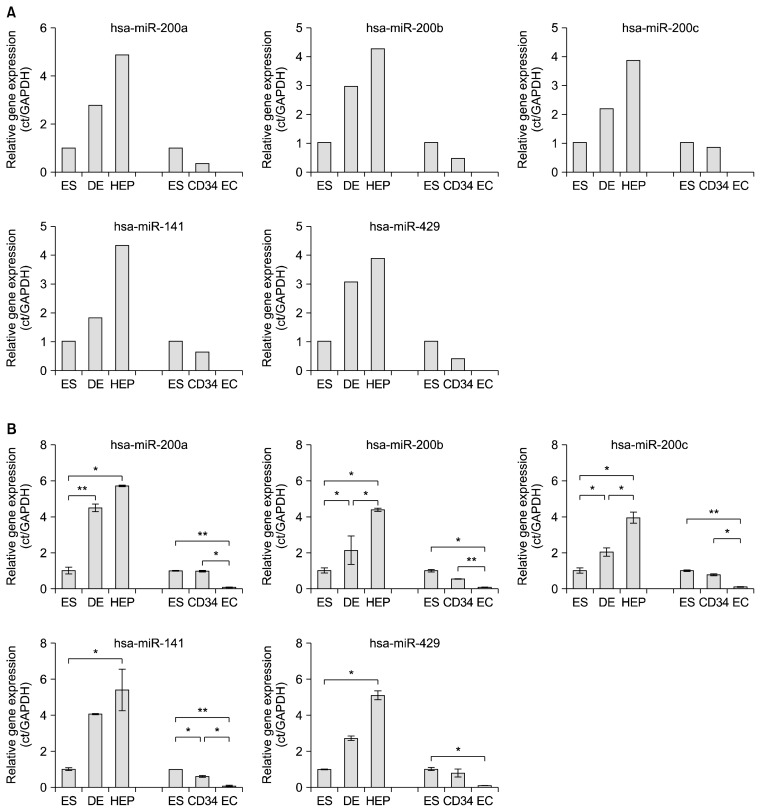
The heatmaps for the total microRNAs showed differential expression patterns in both endodermal and mesodermal lineages during in vitro differentiation of hESCs (Fig. 2A). Additionally, the overall microRNA expression levels of the different cell types derived from hESCs showed differential profiles in each lineage compared to those of hESCs (Fig. 2A). As the developmental stage of hESCs advanced, the correlation of the microRNA expression patterns decreased in both endodermal (R square value; 0.91 vs 0.77) and mesodermal lineages (0.77 vs 0.57) (Fig. 2B). In addition, the correlation of the microRNA expression patterns between the intermediate cells and terminally differentiated cells was relatively high in both lineages compared to the microRNA expression profiles between hESCs and intermediate cells (Fig. 2C). These results imply that the expression patterns of the microRNA are continuously changed in hESCs during the differentiation process. Among the microRNAs showing various expression patterns during in vitro differentiation of hESCs, 20 and 7 microRNAs were enriched in the endodermal and mesodermal lineages, respectively (Table 1). Out of 20 endodermal-enriched microRNAs, interestingly, the expressions of 10 microRNAs (miR-141, 182, 183, 201a, 200b, 200c, 429, 489, 886-5p, and 96) were increased in hESCs during the endodermal development whereas they were decreased during the mesodermal development (Fig. 2D). The expression of mir-182 and miR-886-5p were enormously enhanced in DE cells and then decreased in hepatocytes, and the expressions of the others were gradually increased during endodermal differentiation of hESCs in vitro (Fig. 2D, left diagram). Intriguingly, the transcriptional activities of all 10 endodermal-enriched microRNAs were gradually reduced in hESCs during the mesodermal development (Fig. 2D, right diagram). Moreover, 7 mesodermal-enriched microRNAs showed inverse expression patterns in the developmental process of hESCs between the endodermal and mesodermal lineages (Fig. 2E). let-7g, miR-196a*, and miR-497 had the highest expressions in CD34+ cells, and the others (let-7d, miR-106b*, miR-190b, and miR-338-3p) gradually increased during mesodermal differentiation (Fig. 2E, left diagram). In endodermal differentiation, 6 microRNAs except miR-196a* were drastically decreased in DE cells (Fig. 2E, right diagram). These results could be considered as silencing of those microRNAs at the early mesodermal differentiation stage. The microRNAs particularly enriched in the endodermal lineage were categorized into three groups: 1) miR-200 family (miR-141, miR-200a, miR-200b, miR-200c, and miR-429), 2) miR-183 family (miR-182, miR-183, and miR-96), and 3) others (miR-489 and miR-886-5p). Among the microRNAs enriched in the endodermal lineage, the expression profiles of miR-200 family were obviously changed in the differentiation process of hESCs between the mesodermal and endodermal lineages (Fig. 3A). These differential expression patterns of miR-200 family were clarified again by quantitative RT-PCR (Fig. 3B). Our findings show that the expression of miR-200 family is associated with the endodermal development of hESCs in vitro.
Fig. 2.
Expression profiles of microRNAs in hESCs during differentiation into endodermal and mesodermal lineages. (A) A heatmap showing hierarchical clustering of microRNA expression level. ES, embryonic stem cells; DE, definitive endoderm; HEP, hepatocytes; CD34, CD34+ cells; EC, vascular endothelial cells. (B) Correlation of microRNA expression levels (log) between hESCs and differentiated cells in each stage, respectively. R value was calculated by Pearson’s correlation coefficient. HEP, hepatocytes; EC, vascular endothelial cells. (C) Correlation of microRNA expression levels (log) between the cells at an intermediate status and the finally differentiated cells. R value was calculated by Pearson’s correlation coefficient. (D) Relative expression levels of microRNAs enriched in the endodermal lineage cells. (E) Relative expression levels of microRNAs enriched in the mesodermal lineage cells.
Table 1.
List of microRNAs enriched in a specific lineage
| Lineage specificity | Enriched microRNAs | ||
|---|---|---|---|
| Mesodermal differentiation | hsa-let-7d | hsa-miR-130a* | hsa-miR-338-3p |
| hsa-let-7g | hsa-miR-190b | hsa-miR-497 | |
| hsa-miR-106b* | hsa-miR-196a* | ||
| Endodermal differentiation | hsa-let-7f-1* | hsa-miR-33a | hsa-miR-599 |
| hsa-miR-141 | hsa-miR-340* | hsa-miR-605 | |
| hsa-miR-182 | hsa-miR-421 | hsa-miR-628-5p | |
| hsa-miR-183 | hsa-miR-429 | hsa-miR-886-5p | |
| hsa-miR-200a | hsa-miR-432 | hsa-miR-942 | |
| hsa-miR-200b | hsa-miR-489 | hsa-miR-96 | |
| hsa-miR-200c | hsa-miR-518d-3p |
Fig. 3.
Quantification of microRNAs using qRT-PCR. (A) Relative expression levels of miR-200 family from the microarray results. ES, embryonic stem cells; DE, definitive endoderm; HEP, hepatocytes; CD34, CD34+ cells; EC, vascular endothelial cells. (B) Comparative quantification of miR-200 family in hESCs during differentiation into hepatocytes and vascular endothelial cells. *p<0.05, **p<0.01, n=3.
MiR-200 family and EMT
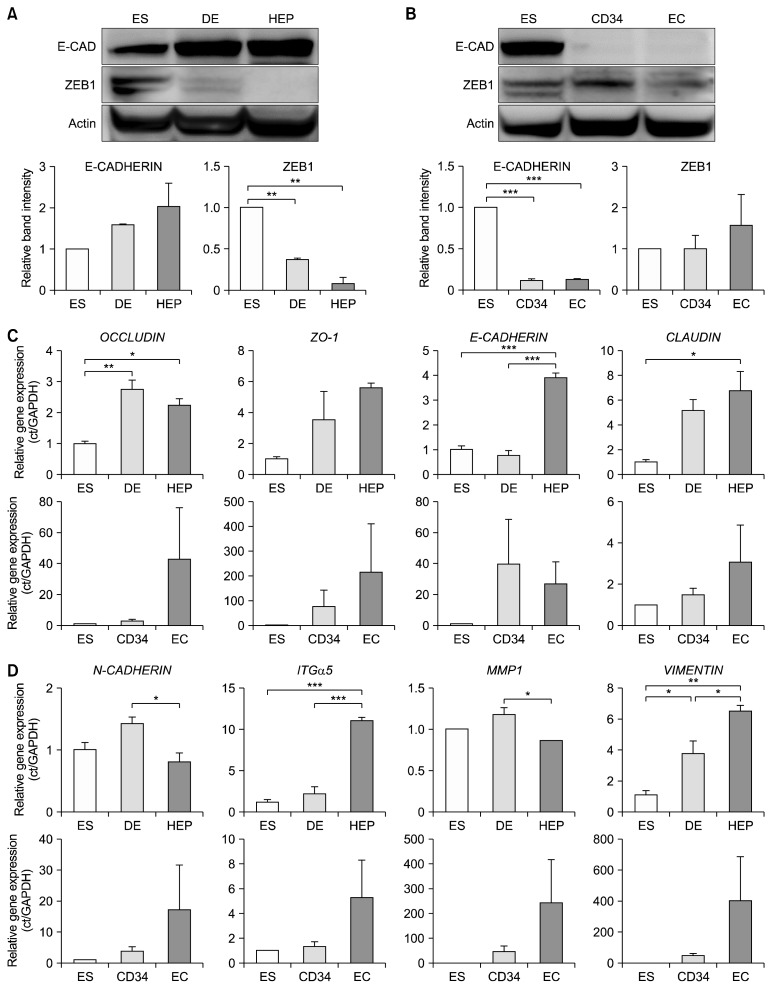
The next question was how the expression of miR-200 family is involved in cell fate determination in hESCs during in vitro differentiation. It is well known that miR-200 family suppresses the epithelial to mesenchymal transition (EMT) process which has an important role in mammalian development (9). To determine whether miR-200 family actually functions in hESCs during differentiation into specialized lineages, the expression levels of ZEB1 and E-CADHERIN were examined. The expression level of ZEB1, a direct target of miR-200 family in the EMT process, was gradually decreased during the differentiation of hESCs into hepatocytes, and a gradual increment of E-CADHERIN, a target protein of ZEB1, was detected (Fig. 4A). In contrast, the expression of ZEB1 and the repression of E-CADHERIN were observed in the differentiation of hESCs into the mesodermal lineage (Fig. 4B). These results show that expression of miR-200 family is critical for determining the endodermal specification through the EMT process during in vitro differentiation of hESCs. The transcriptional activities of epithelial marker genes (OCCLUDIN, ZO-1, E-CADHERIN, and CLAUDIN1) were increased in hESCs during endodermal differentiation while their activities were not significant during mesodermal differentiation (Fig. 4C). The mesenchymal marker genes (INTEGRIN-α5, MMP-1, N-CA-DHERIN, and VIMENTIN) did not exhibit any consistent patterns in the transcriptional expression level during differentiation of hESCs into both lineages (Fig. 4D). It has been reported that the knockdown of miR-200c inhibits NANOG and upregulates GATA4, thereby regulating self-renewal and differentiation in hESCs (23). Transfection with a group of microRNAs including miR-200c, miR-302s, and miR-376s family successfully induced somatic cells into induced pluripotent stem cells (24). Thus, miR-200c appears to be associated with the self-renewal of hESCs. In contrast to previous studies, our results provide new insight showing that miR-200 family is involved in the endodermal development during the differentiation of hESCs in vitro.
Fig. 4.
Expression of miR-200 family-downstream target genes and proteins in hESCs during in vitro differentiation. (A) Expression levels of miR-200 family target proteins in the endodermal lineage cells. Band intensities were measured using ImageJ program. Quantification of protein bands was normalized by that of actin. **p<0.01, n=3. ES, embryonic stem cells; DE, definitive endoderm; HEP, hepatocytes; CD34, CD34+ cells; EC, vascular endothelial cells; E-CAD, E-CADHERIN. (B) Expression levels of miR-200 family target proteins in mesodermal lineage cells. ***p<0.005, n=3. (C) Transcriptional expression levels of the epithelial marker genes in hESCs during in vitro differentiation. *p<0.05; **p<0.01; ***p<0.005, n=3. (D) Transcriptional expression levels of the mesenchymal marker genes in hESCs during in vitro differentiation. *p<0.05; **p<0.01; ***p<0.005, n=3.
This study reports for the first time that specific microRNAs or a microRNA family has an important role in the lineage determination of hESCs during in vitro differentiation. In particular, we found that miR-200 family was considered the critical microRNAs for endodermal determination in hESCs during early development in vitro. Our results indicate that endodermal lineage in hESCs may be determined through the suppression of the EMT process by expression of miR-200 family which down-regulate the target protein (ZEB1). Studies on the dynamics of microRNAs in hESCs will be helpful in understanding cell fate and lineage specification in the developmental process.
Supplementary Information
Acknowledgments
This work was supported by the NRF Stem Cell Program (2011-0019509) funded by MSIP, Republic of Korea.
Footnotes
Potential conflict of interest
The authors declare that there is no conflict of interest.
Supplementary data including one table can be found with this article online at http://pdf.medrang.co.kr/paper/pdf/IJSC/IJSC-10-s17013.pdf.
References
- 1.Pera MF, Trounson AO. Human embryonic stem cells: prospects for development. Development. 2004;131:5515–5525. doi: 10.1242/dev.01451. [DOI] [PubMed] [Google Scholar]
- 2.Odorico JS, Kaufman DS, Thomson JA. Multilineage differentiation from human embryonic stem cell lines. Stem Cells. 2001;19:193–204. doi: 10.1634/stemcells.19-3-193. [DOI] [PubMed] [Google Scholar]
- 3.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 4.Gangaraju VK, Lin H. MicroRNAs: key regulators of stem cells. Nat Rev Mol Cell Biol. 2009;10:116–125. doi: 10.1038/nrm2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014;15:509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 6.Brennecke J, Cohen SM. Towards a complete description of the microRNA complement of animal genomes. Genome Biol. 2003;4:228. doi: 10.1186/gb-2003-4-9-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu P, Vernooy SY, Guo M, Hay BA. The Drosophila microRNA Mir-14 suppresses cell death and is required for normal fat metabolism. Curr Biol. 2003;13:790–795. doi: 10.1016/S0960-9822(03)00250-1. [DOI] [PubMed] [Google Scholar]
- 8.Martin EC, Qureshi AT, Dasa V, Freitas MA, Gimble JM, Davis TA. MicroRNA regulation of stem cell differentiation and diseases of the bone and adipose tissue: Perspectives on miRNA biogenesis and cellular transcriptome. Biochimie. 2016;124:98–111. doi: 10.1016/j.biochi.2015.02.012. [DOI] [PubMed] [Google Scholar]
- 9.Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y, Goodall GJ. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 10.Lecellier CH, Dunoyer P, Arar K, Lehmann-Che J, Eyquem S, Himber C, Saïb A, Voinnet O. A cellular micro-RNA mediates antiviral defense in human cells. Science. 2005;308:557–560. doi: 10.1126/science.1108784. [DOI] [PubMed] [Google Scholar]
- 11.Alvarez-Garcia I, Miska EA. MicroRNA functions in animal development and human disease. Development. 2005;132:4653–4662. doi: 10.1242/dev.02073. [DOI] [PubMed] [Google Scholar]
- 12.Chen K, Rajewsky N. The evolution of gene regulation by transcription factors and microRNAs. Nat Rev Genet. 2007;8:93–103. doi: 10.1038/nrg1990. [DOI] [PubMed] [Google Scholar]
- 13.Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol. 2005;6:376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- 14.Heinrich EM, Dimmeler S. MicroRNAs and stem cells: control of pluripotency, reprogramming, and lineage commitment. Circ Res. 2012;110:1014–1022. doi: 10.1161/CIRCRESAHA.111.243394. [DOI] [PubMed] [Google Scholar]
- 15.Rosa A, Spagnoli FM, Brivanlou AH. The miR-430/427/302 family controls mesendodermal fate specification via species-specific target selection. Dev Cell. 2009;16:517–527. doi: 10.1016/j.devcel.2009.02.007. [DOI] [PubMed] [Google Scholar]
- 16.Xu N, Papagiannakopoulos T, Pan G, Thomson JA, Kosik KS. MicroRNA-145 regulates OCT4, SOX2, and KLF4 and represses pluripotency in human embryonic stem cells. Cell. 2009;137:647–658. doi: 10.1016/j.cell.2009.02.038. [DOI] [PubMed] [Google Scholar]
- 17.Jain AK, Allton K, Iacovino M, Mahen E, Milczarek RJ, Zwaka TP, Kyba M, Barton MC. p53 regulates cell cycle and microRNAs to promote differentiation of human embryonic stem cells. PLoS Biol. 2012;10:e1001268. doi: 10.1371/journal.pbio.1001268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Choi YJ, Lin CP, Risso D, Chen S, Kim TA, Tan MH, Li JB, Wu Y, Chen C, Xuan Z, Macfarlan T, Peng W, Lloyd KC, Kim SY, Speed TP, He L. Deficiency of microRNA miR-34a expands cell fate potential in pluripotent stem cells. Science. 2017;355 doi: 10.1126/science.aag1927. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim N, Kim H, Jung I, Kim Y, Kim D, Han YM. Expression profiles of miRNAs in human embryonic stem cells during hepatocyte differentiation. Hepatol Res. 2011;41:170–183. doi: 10.1111/j.1872-034X.2010.00752.x. [DOI] [PubMed] [Google Scholar]
- 20.Sánchez-Tilló E, Lázaro A, Torrent R, Cuatrecasas M, Vaquero EC, Castells A, Engel P, Postigo A. ZEB1 represses E-cadherin and induces an EMT by recruiting the SWI/SNF chromatin-remodeling protein BRG1. Oncogene. 2010;29:3490–3500. doi: 10.1038/onc.2010.102. [DOI] [PubMed] [Google Scholar]
- 21.Yeo S, Jeong S, Kim J, Han JS, Han YM, Kang YK. Characterization of DNA methylation change in stem cell marker genes during differentiation of human embryonic stem cells. Biochem Biophys Res Commun. 2007;359:536–542. doi: 10.1016/j.bbrc.2007.05.120. [DOI] [PubMed] [Google Scholar]
- 22.Park SW, Jun Koh Y, Jeon J, Cho YH, Jang MJ, Kang Y, Kim MJ, Choi C, Sook Cho Y, Chung HM, Koh GY, Han YM. Efficient differentiation of human pluripotent stem cells into functional CD34+ progenitor cells by combined modulation of the MEK/ERK and BMP4 signaling pathways. Blood. 2010;116:5762–5772. doi: 10.1182/blood-2010-04-280719. [DOI] [PubMed] [Google Scholar]
- 23.Huang HN, Chen SY, Hwang SM, Yu CC, Su MW, Mai W, Wang HW, Cheng WC, Schuyler SC, Ma N, Lu FL, Lu J. miR-200c and GATA binding protein 4 regulate human embryonic stem cell renewal and differentiation. Stem Cell Res. 2014;12:338–353. doi: 10.1016/j.scr.2013.11.009. [DOI] [PubMed] [Google Scholar]
- 24.Miyoshi N, Ishii H, Nagano H, Haraguchi N, Dewi DL, Kano Y, Nishikawa S, Tanemura M, Mimori K, Tanaka F, Saito T, Nishimura J, Takemasa I, Mizushima T, Ikeda M, Yamamoto H, Sekimoto M, Doki Y, Mori M. Reprogramming of mouse and human cells to pluripotency using mature microRNAs. Cell Stem Cell. 2011;8:633–638. doi: 10.1016/j.stem.2011.05.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.