Abstract
Background
Application of competent cells such as mesenchymal stem cells (MSCs) for treatment of musculoskeletal disorders in equine athletes is increasingly needed. Moreover, similarities of horse and human in size, load and types of joint injuries, make horse as a good model for MSCs therapy studies. This study was designed to isolate and characterize stemness signature of equine bone marrow-derived mesenchymal stem cells (BM-MSCs).
Methods
BM of three mares was aspirated and the mononuclear cells (MNCs) were isolated using density gradient. The primary MNCs were cultured and analyzed after tree passages (P3) for growth characteristics, differentiation potentials, and the expression of genes including CD29, CD34, CD44, CD90, CD105, MHC-I, MHC-II and pluripotency related genes (Nanog, Oct-4, Sox-2, SSEA-1, -3, -4) using RT-PCR or immunocytochemistry techniques.
Results
The isolated cells in P3 were adherent and fibroblast-like in shape with doubling times of 78.15 h. Their clonogenic capacity was 8.67±4% and they were able to differentiate to osteogenic, adipogenic and chondrogenic lineages. Cells showed expression of CD29, CD44, CD90, MHC-I and Sox-2 while no expression for CD34, MHC-II, CD105, and pluripotency stemness markers was detected.
Conclusions
In conclusion, data showed that isolated cells have the basic and minimal criteria for MSCs, however, expressing only one pluripotency gene (sox-2).
Keywords: Mesenchymal stem cells, Bone marrow, Equine, Pluripotency, Stemness markers
Introduction
Most of degenerative diseases including spinal cord, musculoskeletal, and tendon lesions are not repairable with the routine methods, due to loss of tissue cellular content and inability to replace them (1). Musculoskeletal disorders in equine athletes have a major influence on equine performance, and are the main reason for financial loss in racing industry (2). Fast and full recovery of chronic musculoskeletal disorders and returning to race is of primary importance. New hopes have been raised by improvement in stem cell research field to treat these kinds of incurable equine diseases. To date, there are several studies that show the application of different types of stem cells in the treatment of equine musculoskeletal disorders (3, 4).
Furthermore, suitability of horse as an animal model for human studies in musculoskeletal disorders, especially in cartilage and tendon injuries has been previously emphasized (5, 6). The characteristics of horse joint and cartilage, including size, anatomy, and morphology of the joints, the thickness and composition of cartilage, and low intrinsic capability for repair are similar to those in human (5–7). In addition, the hierarchical structure, matrix composition, the function and the nature of the injuries of weight-bearing tendons (e.g. Achilles tendon) in the human athlete and horse have many similarities (8). On the other hand, the homology of equine immune system proteins to human ones is about 60~98% (9). Moreover, regeneration techniques applicable to human may need validation in a large animal model. A report from U.S. Food and Drug Administration (FDA) has also concluded that the horse is the most appropriate animal model for testing and analyzing the clinical effects of mesenchymal stem cell (MSC)-based therapies for joint injuries in human due to similarities in size, load and types of joint injuries (10).
Complete identification and stemness signature of adult stem cells, in particular MSCs, is the first critical step for their clinical application. MSCs can be obtained from different tissues such as bone marrow (BM) stroma, periosteum, umbilical cord blood, fat, and skin (11). Despite of isolation of equine MSCs from many tissues, bone marrow is the most common source and it is the most investigated source. It has been proven that these multipotent cells are able to differentiate to cartilage, muscle, tendon, ligament, fat and also some other type of cells such as neurons and hepatocytes which show their pluripotent capacity (12). In addition, it has been reported that MSCs are hypoimmunogenic and immunomodulator and escape from allogeneic rejection (13) and can be used for allogeneic cell therapy. Evidence show that transplantations of allogeneic MSCs have therapeutic effects similar to autologous transplantation (14).
Although some studies have identified the characteristics of equine MSCs, some of their features, in particular the expression of embryonic stemness markers and their multipotency or pluripotency abilities are not completely shown (3, 15). De Schauwer et al. (3) has determined minimal criteria for identification of equine MSCs: 1) adherence to the plastic culture dish in normal condition, 2) expression of surface antigens CD29, CD44 and CD90 and no expression of CD14, CD79α, and MHC-II, and 3) in vitro differentiation to three kinds of cells (osteoblasts, chondrocytes and adipocytes) under standard differentiation conditions. Expression of Oct4, SSEA-3, SSEA-4, Nanog and Tra-1-60 (as embryonic stem cell markers) has been shown in some studies (16–20), while some others have reported negative results (15). There are some reports that MSCs are able to differentiate into various cell types of non-mesodermal lineages such as hepatocytes (21) in rat, neuron in mice, and epithelial and endothelial cells in human (22). Thus, it has been suggested that MSCs are in mid-stage between pluripotent embryonic stem cells and lineage-restricted adult stem cells and have a great capacity for differentiation.
Therefore, this study aimed to isolate and characterize equine BM-MSCs and to identify their growth features, differentiation potentials and gene expression profile including embryonic stemness markers (SSEA-1, SSEA-3, SSEA-4, Nanog and Sox-2).
Materials and Methods
The experiment protocol was approved by Animal Welfare Committee of Veterinary faculty, Ferdowsi University of Mashhad, Iran. All the reagents were provided by Sigma-Aldrich (Germany) company, except for those which are mentioned later on. All the culture dishes and polypropylene tubes were purchased from SPL (Korea).
Sample Collection and Cell Culture
Bone Marrow Aspiration
Bone marrow (BM) was aspirated from 3 healthy crossbreed mares, aged 6, 9 and 10 years old. To strain horse, xylazine (0.5 mg/kg) was injected intravenously and aspiration location in sternum was determined by ultrasonography. Then, 10 ml of lidocaine with adrenalin was used in the aspiration area for local anesthesia. Using a surgery knife, a surface cut made and by Jamshidi needle (gauge 13 and 100 mm length) 10~15 ml of bone marrow was aspirated into a 20 ml syringe which contained 1000 IU/ml heparin. The samples, in less than 4 hours, were carried on ice to the laboratory.
Cell Isolation and Culture
In laboratory and in sterilized condition, the BM samples were diluted by basal medium (DMEM-HG, 10% FBS, 1% Penicillin-Streptomycin and 0.1% Amphotericin B) twice the volume of the samples. Then, it was gently poured onto density gradient media (Histopaque® 1077) to the ratio of 2:1. The tubes containing samples were centrifuged at 400 g in 4°C for 30 min (in brake off mode). Afterwards, monolayer nucleated cells (MNCs) were carefully gathered and rinsed twice by Dulbecco’s Phosphate Buffer Saline (DPBS-) by centrifugation in 600 g in 4°C for 5 min. The final cell pellet was diluted in 4 ml basal medium and cell population was counted using Neubauer counting chamber. Then, cells were cultured at the density of 8×105 cells/cm2 as the primary culture or passage 0 (P0) in 37°C humidified atmosphere containing 5% CO2 condition. Culture medium was replaced with fresh medium every 3 days. At confluency of 80~90%, cells came into passage 1 (P1) using trypsinization where the cells were seeded at the density of 5×103. Similar process was conducted until passages 3 (P3). P3 cells were collected for the next experiments.
Growth Characteristics
Growth Curve
In order to assess cell growth rate, the cells were seeded at 3×104 cells/well in 2 twelve-well plates. Every 24 hours, the cell numbers of 3 wells were counted and the mean was calculated until 8th day and the growth curve was drawn based on the gathered data.
Colony Forming Unit-Fibroblast (CFU-F) and Plating Efficiency (PE)
For colony formation evaluation, 5×102 cells were seeded in each well of a six-well plate. Every 3 days the mediums were changed and after 12 days, culture medium was discarded and the cells were rinsed with PBS. The colonies were then stained by 0.5% Crystal Violet for 10 min. Afterwards, the wells were rinsed by tap water and the colonies containing over 15 cells were counted under invert microscope. The following formula was utilized to determine the plating efficiency:
Cell Population Doubling Time
In each of the passages, the number of cells was recorded in the beginning and the end of passage. In order to calculate the proliferation of cells per day, the following formula was used, where Ni=indicates initial seeded cells, Nf is the harvested cell number, CT is the culture time, PD is cell population doubling per day and DT shows the proper time for doubling cells (hours).
Characterization of Mesenchymal Stem Cells
Trilineage differentiation potential
P3 cells of all 3 mares were used for trilineage differentiation.
Osteogenic differentiation
3×105 cells were seeded in 3 wells of a six-well plate. At 50% confluency, basal medium was discarded and two of them were replaced with osteogenic medium (basal medium supplemented with 0.1 μM dexamethasone, 10 mM β-Glycerophosphate disodium salt hydrate and 50 μM 2-Phospho-L-ascorcbic acid trisodium salt) and the remaining one was left unchanged as control. The media were changed every 3 days. After 21 days, the cells were fixed with 10% neutral buffered formalin and then stained using 2% Alizarian Red S in order to analyze differentiation.
Adipogenic differentiation
3×105 cells were cultured in three wells of a six-well plate. After 80% confluency, the medium was discarded and two of the wells were replaced with adipogenic medium (basal medium supplemented with 0.1 μM dexamethasone, 100 μM indomethacin, 500 μM 3-isobutyl-1-methyl xanthine, 1% Insulin-Transferrin-selenium-X) and the other one was replaced with basal medium. Every 3 days the mediums were changed until the 21st day, on which the wells were rinsed by PBS, fixed and stained by fresh Oil Red O to determine adipogenic differentiation.
Chondrogenic differentiation
5×105 cells were transferred into each of the 15-ml polypropylene conical Falcon tubes, and then centrifuged at 260 g for 5 min (at room temperature). Supernatant was discarded and 2 ml basal medium added to the falcons and let them to adapt for 24 hours. The following day, basal medium was substituted with chondrogenic differentiation medium (basal medium supplemented with 0.1 μM dexamethasone, 50 μ M 2-Phospho-L-ascorcbic acid trisodium salt, 10 mM β-Glycerophosphate disodium salt hydrate, 1 mg/ml Bovine Serum Albumin (BSA), 10 ng/ml human Transforming Growth Factor-β3 (TGF-β3) and 10 ng/ml Bone Morphogenetic Protein-6 (BMP6)) for the treated group and basal medium was added to the control group wells. The mediums of each group were replaced with fresh ones every four days until the end of culture (21 days). Afterwards, the pellets were stained by Alician Blue for histological examination.
Gene expression profiling
Reverse Transcription - Polymerase Chain Reaction (RT-PCR)
RNA Extraction and cDNA synthesis
Total RNA of cells was extracted by DenaZist kit (Iran) under the manufacture protocol. Briefly, each sample was lysed and homogenized in 1 ml G1 buffer. The homogenate was incubated at room temperature (20°C) for 10 minutes. Then, 200 μl chloroform was added and sample was centrifuged at 12,000 g for 15 min at 4°C and upper phase containing RNA was precipitated with equivalent of half the volume aqueous phase of the isopropyl alcohol and the same volume from G2 buffer. Afterwards washing was performed by 75% ethanol and sample was dried in contact with air, and resuspended in diethyl pyrocarbonate (DEPC)-treated water. In order to remove any possible genomic DNA, five unit RNase free DNAse I (Roche, Germany) was added per each 20 μg of RNA and incubated at 34°C for 20 min followed by adding 0.8 μl 0.5 M EDTA and heat inactivation of the enzyme at 75 °C for 10 min. RNA concentration, purity and quality were appraised using NanoDrop 2000 (Thermo Scientific, USA) and gel electrophoresis. Then cDNA was synthesized by AccuPower® RT Premix kit (Bioneer, USA). 1 μg of RNA was mixed with 0.5 μg Oligo(dT)18 Primer (Fermentas, USA) and it was added to the kit, then reached 20 μl using diethyl pyrocarbonate (DEPC)-treated water. The kit was incubated at 42°C for 60 min and finally at 70°C for 10 min to deactivate reverse transcriptase enzyme.
Polymerase Chain Reaction (PCR)
Specific primers of GAPDH, CD29, CD34, CD44, CD90, CD105, Sox-2, Oct-4 and Nanog genes were designed based on the available sequences in GeneBank (NCBI) using Primer Premier software (PREMIER Biosoft International, USA) (Table 1). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) used as internal control.
Table 1.
Characteristics of primer pairs which were used in the experiment
| Gene | Accession Number | Primer Sequence | Annealing Temperature | Product Size (bp) |
|---|---|---|---|---|
| Equine GAPDH | NM_001163856 | Fa: TGTCATCAACGGAAAGGC | 56 | cDNAc=183 |
| NC_009149 | Rb: GCATCAGCAGAAGGAGCA | gDNAd=429 | ||
| Equine CD29 | XM_001492665 | F: AATCGGGACAAGTTACCTCA | 56 | 234 |
| R: CTTCCAAATCAGCAGCAAT | ||||
| Equine CD34 | XM_001491596 | F: TGATGAATCGCCGTAGT | 56 | cDNA=204 |
| R: CGGGTTGTCTCGCTGA | gDNA=907 | |||
| Equine CD44 | NM_001085435 | F: AACCTCGGGTCCCATAC | 56 | 193 |
| R: TCCATTGAGCCCACTTGC | ||||
| Equine CD90 | XM_001503225 | F: AGAATACCACCGCCACA | 51 | 155 |
| R:GGATAAGTAGAGGACCTTGATG | ||||
| Equine CD105 | XM_003364144 | F: GACGCCAATCACAACATACA | 60 | 158 |
| R: TCCACATAGGACGCTACGAC | ||||
| Equine MHC-I | NM_001123381 | F: CTGGGTCTCCCTGTCGTTG | 56 | 110 |
| R: CCTTGGGCACTGTCACTG | ||||
| Equine MHC-II | NM_001142816 | F: GGAACGGGCAGCAGGACAT | 56 | 184 |
| R: AAGCCATTCACAGAGCAGACCA | ||||
| Equine Sox-2 | XM_003363345 | F: TGGACCAACGGAGGCTATG | 56 | 198 |
| R: CCCTTGCTGGGAGTACGAC | ||||
| Oct-4 | F: GTTGTCCGGGTCTGGTTCT | 57 | 189 | |
| R: GTGGAAAGGTGGCATGTAGAC | ||||
| Nanog | F: CAGCAGACCTCTCCTTGACC | 55 | 187 | |
| R: TTCCTTGTCCCACTCTCACC |
F: Forward primer;
R: Reverse primer;
cDNA: complementary DNA;
gDNA: genomic DNA.
PCR was performed in 25 μl final volume with tag polymerase enzyme (Pars Tous, Iran) at the following condition: initial denaturation at 95°C for 5 min, 30 cycles at 95°C for 30 s (denaturation), 51~61°C for 45 s (annealing for different primers), 72°C for 1 min (elongation) and final extension at 72°C for 10 min, and then cooling to room temperature. PCR products were visualized with ethidium bromide (Cinnagen, Iran) on a 1.5% agarose gel (Cinnagen, Iran). A 100 bp DNA ladder (Fermentas, USA) used as marker to determine the size of amplified products.
Immunocytochemistry
To detect expression of some specific markers in isolated MSCs, immunocytochemistry method was performed. 6×104 undifferentiated cells at P3 were grown overnight on coverslips and then fixed with 4% paraformaldehyde for 15 min at room temperature. Afterward cells were permeabilized with Triton X-100 diluted in PBS for 10 min. After washing 3 times with PBS, endogenous peroxidases were blocked with 3% H2O2 (Merck, Germany) in PBS for 30 min, then cells were incubated in 4% BSA in PBS for 1 h at room temperature to prevent the nonspecific binding of the antibodies. Coverslips were incubated overnight with different primary antibodies: Nanog (1:100, Santa Cruz Biotechnology, USA), SSEA-1 (1:100, Santa Cruz Biotechnology, USA), SSEA-3 (1:100, Abcam, USA) and SSEA-4 (1:100, Santa Cruz Biotechnology, USA) diluted in 1% BSA in PBS at 4°C in humid chamber. After washing 3 times, the cells were incubated 2 h with secondary IgM (Jackson Immunoresearch, USA) or IgG (Dianova, Germany) antibodies conjugated to horseradish peroxidase (HRP) diluted 1:500 in 1% BSA in PBS. Reaction between antigen and antibody was visualized by 3,3′-Diaminobenzidine tetrahydrochloride hydrate (DAB) substrate. The NTERA2 (human testicular embryonic carcinoma cell line) was used as positive control for Nanog, SSEA-3, and SSEA-4 and P19 cells (cell line derived from a teratocarcinoma in mice) was used as positive control for SSEA-1.
Results
Morphology of cultured cells
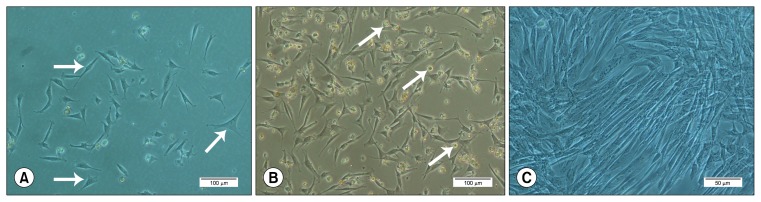
MNCs containing MSCs were cultured as P0 at day 0. Dead or non-attachable cells were removed by the first medium change after 3 days in culture. The first colonies of MSCs were observed 4 days after MNCs seeding. The morphology of MSCs in colonies was heterogeneous and had spindle-shaped fibroblast-like, asteroid and triangular shape (Fig. 1A). In addition, considerable number of round-shaped cells were observed at P1 (Fig. 1B). At the end of P3, an almost homogeneous fibroblast-like population of MSCs was obtained (Fig. 1C) which was used for further analysis.
Fig. 1.
The morphology of equine BM-MSCs. (A) Heterogeneous, spindle-shaped fibroblast-like, asteroid and, triangular shape of equine MSCs at P1. (B) Round-shape cells among fibroblast-like cells at early passages, and (C) almost homogeneous fibroblast-like population of equine MSCs at the end of P3.
Growth Characteristics
Growth Curve
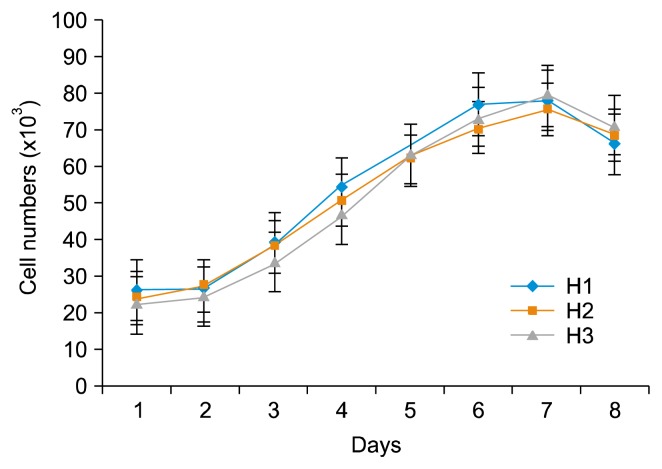
Growth curve of BM-MSCs showed an initial lag phase of 3 days, followed by an exponential phase for 3 days and then the cells were entered to plateau phase at the day 6 or 7. On day 8, the cell population immediately starts to decrease (Fig. 2).
Fig. 2.
The growth curve of the equine BM-MSCs at P3. Cells enter the 3 days of log phase after 3 days of lag phase, and reach the plateau phase at the day 6 or 7.
Colony Forming Unit-Fibroblast (CFU-F) and Plating Efficiency (PE)
Cultured MSCs in wells with density of 5×102 produced averagely 43.3±20.1 colonies which show their colonogenic capability (Table 2). The mean percentage of PE BM-MSCs at the P3 was 8.67±4% and the details of different horses are also presented in Table 2.
Table 2.
Clonogenic capacity and mean of plating efficiency percentage (PE %) of equine mesenchymal stem cells at passage 3
| Number of colonies (mean±SD) | PE (%) | |
|---|---|---|
| Cells from animal 1 | 49±24 | 9.8±4.8 |
| Cells from animal 2 | 42.3±15.3 | 8.46±3 |
| Cells from animal 3 | 38.6±21.2 | 7.73±4.2 |
| Total average | 43.3±20.1 | 8.67±4 |
Plating efficiency was calculated as the percentage of colonies formed from the total number of seeded cells.
Cell Population Doubling Time
PD (PD=CD/CT) of cells from 0.5 in passage 1 was decreased to 0.32 in passage 3. Consequently, the needed time for doubling of population (DT) were increased from 48.62 hours in passage 1 to 78.15 hours in passage 3 (Table 3).
Table 3.
Population doubling per day (PD) and doubling time (DT) for equine mesenchymal stem cells at passage 1 to 3
| Passage 1 | Passage 2 | Passage 3 | ||||
|---|---|---|---|---|---|---|
|
|
|
|
||||
| PD | DTb | PD | DT | PD | DT | |
| H1a | 0.52 | 47.10 | 0.42 | 59.11 | 0.32 | 76.51 |
| H2 | 0.47 | 51.22 | 0.38 | 63.62 | 0.38 | 64.16 |
| H3 | 0.51 | 47.54 | 0.43 | 55.65 | 0.26 | 93.79 |
| Mean | 0.50 | 48.62 | 0.41 | 59.46 | 0.32 | 78.15 |
H1, 2, 3 indicate different horses.
DT was calculated as hours at each passage.
Conformation of MSC characteristics
To identify the isolated cells as MSCs, their trilineage differentiation potentials and gene expression profiling were analyzed. In addition, the expression of embryonic stemness markers was investigated to clarify their potency.
Trilineage differentiation potential
Osteogenic differentiation
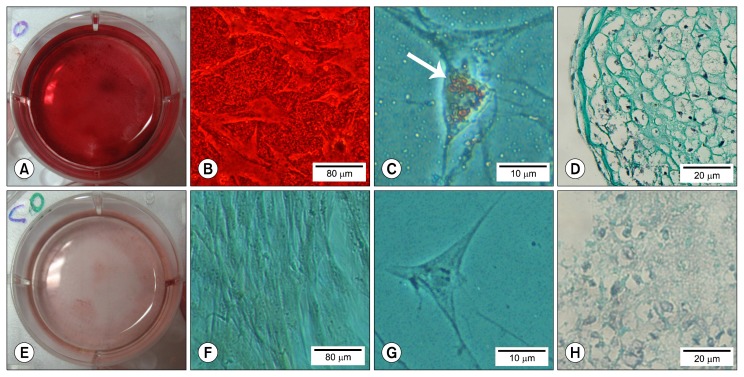
The cultured cells after exposure to osteogenic inductive medium for 21 days, secreted and deposited extracellular calcium crystals which were identified by Alizarian Red S staining (Fig. 3A and 3B).
Fig. 3.
Tri-lineage differentiation of equine BM-MSCs. Macroscopic (A) and microscopic (B, ×40) view of Alizarin Red S stained differentiated BM-MSCs. Oil Red O stained fat droplet in adipogenic treatment (C, ×400). Proteoglycans are stained with Alcian Blue in a pellet of chondrogenic differentiation group (D, ×200). E–H are the control for A–D in which cultured with basal growth medium.
Adipogenic differentiation
Adipogenic differentiating inductive medium resulted in appearance of cytosolic lipid droplets after 21 days, which were identified and stained with Oil Red O staining (Fig. 3C).
Chondrogenic differentiation
Histological staining and analysis of extracellular-matrix proteoglycans with Alcian Blue showed chondrogenic differentiation in micropellets which were exposed to chondrogenic inductive medium for 21 days. The morphology of cells had changed from fibroblast-like cells to round cells within lacunae in treated groups (Fig. 3D).
In all trilineage differentiation assays, control groups were treated with basal growth medium without any additive inductive components (Fig. 3E~H).
Gene expression profiling
Expression of MSC marker genes at mRNA level
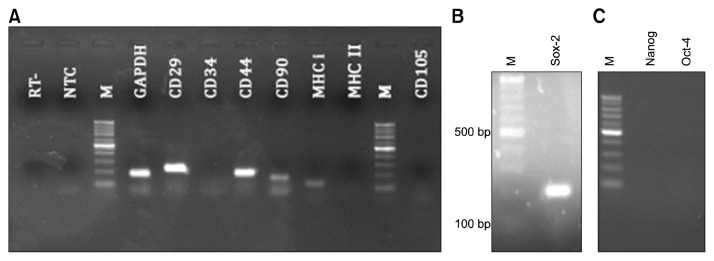
For RT-PCR analysis, GAPDH was used as internal control. GAPDH primers sets would amplify two fragments (183 bp and 429 bp) from genomic DNA, and only one fragment (183 bp) form cDNA. Hence, observation of only 183 bp amplicon in PCR products confirmed the absence of any DNA contamination in all samples. In addition, primer sets for CD34 were also designed as intron-spanning primers giving rise to different PCR fragments from DNA and cDNA.
Observation of PCR products of undifferentiated P3 MSCs from each horse revealed that they expressed CD29, CD44, CD90, and MHC I. However, they did not showed expression of CD34, CD105 (hematopoietic marker) and MHC-II (Fig. 4A).
Fig. 4.
Expression of surface and pluripotency markers of MSCs at mRNA level. RT-PCR showed the expression of GAPDH, CD29, CD44, CD90, MHC-I and lack of expression of CD34, CD105 and MHC-II. In NTC, dH2O was used for template as a control negative, and in RT sample, RNA was used instead of template to control genomic DNA contamination (A). The expression of SOX-2 gene was observed (B), and no expression of Oct-4 and Nanog was detected (C). M: 100 bp DNA ladder.
Expression of embryonic stemness genes at mRNA and/or protein levels
mRNA expression analysis of Nanog, Oct-4 and Sox-2 in P3 cells showed that the cells express only Sox-2 as a pluripotentcy marker (Fig. 4B and 4C).
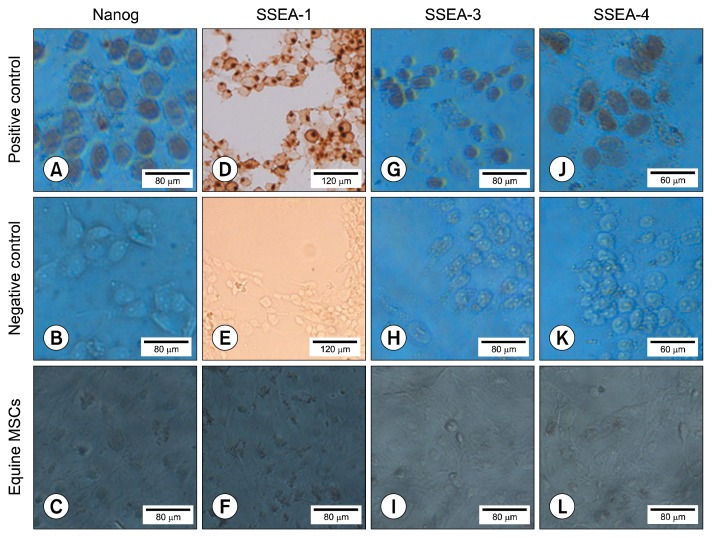
Immunocytochemical analysis of all samples was done to investigate the expression of SSEA-1, -3, -4 and Nanog at protein level. No expression of these mentioned markers was found in the cells. Groups were stained with DAB substrate in the presence of HRP-conjugated secondary antibody. Positive and negative controls were included in the immunostaining process of all markers (Fig. 5).
Fig. 5.
Immunocytochemical analysis of pluripotency markers (Nanog, SSEA-1, -3 and -4) in equine MSCs at passage 3. The first row belongs to the positive control samples (A, G and J: NTERA2 cell line, and D: P19 cell line); the cells at the second row (B, E, H and K) are the same cells of first row without using secondary antibody, and the third row cells (C, F, I and L) are equine MSCs at P 3. No expression of pluripotency markers was found in equine MSCs.
Discussion
In this study we succeeded to isolate and characterize equine BM-MSCs. The isolated cells were fibroblast-like in shape, were attached to the culture dish and they expressed putative equine MSCs markers including CD29, CD44, CD90 and MHC-I and had no expression of CD34 and MHC-II (3). These cells properly differentiated to three osteogenic, adipogenic and chondrogenic lineages under specific differentiating mediums. Embryonic and pluripotency markers such as SOX-2 were detected at RNA level, but no expression was found for SSEA-1, -3, -4 and Nanog at protein level. The average of population doubling time (PDT) of cells during three passages was 62.07 hours and the PE% was 8.67%.
The plastic-adherence ability of MSCs is a general feature (3). MSCs population was heterogeneous in morphology at first passage. The heterogeneity degree of cells was decreased in later passages and fibroblast-like cells with two narrow ends were dominant in agreement with previous study (23).
Regarding growth characteristics, the lag phase (2 days) probably is due to stress of centrifugation and trypsinization of cells before the culture in 12-well plates. This lag time is similar to the results reported by Baghban Eslaminejad et al. (24). However, Lange–Consiglio et al. (25) reported 6 days for lag phase of BM-MSCs and less than 24 hours for adipose-derived MSCs (AMSCs). Finally, they found that the growth rate of AMSCs is higher than BM-MSCs. Moreover, Alipour ae al. (26) observed 3 days for lag time and 8 days for log phase of AMSCs. It seems that the growth rate of AMSCs is more than BM-MSCs. Moreover, different log phase of BM-MSCs in different studies might show that cells with different characteristics, at least regarding growth rate capabilities, have been isolated in these studies.
Assessment of colony forming unit (PE%), as one of the MSCs ability (27) which indicates their self-renewal capacity and quality of cell preparations (28), was 8.67% in 12 days. Lovati et al. (29) and Bourzac et al. (30) calculated 42% and 32% for equine BM-MSCs in 28 days, respectively. The difference is probably due to the time for colony formation or different characteristics of isolated cells. In addition, it has been shown that the number of colonies is affected by cell isolation method (30). Based on colony formation assay, it seems that BM-MSCs have more self-renewal capacity than AMSCs, as Alipour et al. (26) reported 5.5% PE for AMSCs. Population doubling time (PDT) which assay cell expansion in vitro (31), was decreased with increasing the passage numbers. Vidal et al. (23) and Burk et al. (32) observed 33.6 and 98 hours for PDT of BM-MSCs, respectively. Ranera et al. (33) reported 59 hours of PDT for equine BM-MSCs and 52 hours for AMSCs. In another study, PDT for equine AMSCs was between 40 to 46 hours (26). In addition to difference between species, the culture conditions, growth factors and cell seeding number are affecting PDT.
Differentiation of human and equine MSCs toward osteogenic, adipogenic and chondrogenic lineages under the influence of defined medium, is one of the minimal criteria for their confirmation (3, 34). In our study, BM-MSCs at P3 were differentiated to these three lineages similar to other studies (29, 35). This finding approves the multi-potency of our isolated BM-MSCs.
The expression of specific CD markers for equine MSCs was examined by RT-PCR at the mRNA level. A comparative study showed that the mRNA level of equine CD markers, follows the same patterns of relevant protein expression, then in the lack of proper antibody, RT-PCR is a valuable alternative (36). Gene expression analysis of isolated cells at P3 confirmed a specific expression profile for BM-MSCs. Despite of CD29 and CD44 is not included in ISCT minimal criteria for BM-MSCs (34), but De Schauwer et al. (3) stated that these markers are also important for characterization of the equine MSCs (3). Sox-2 gene expression was also analyzed by RT-PCR as a pluripotency marker. Sox-2 is an essential transcription factor in undifferentiated embryonic stem cells for maintaining pluripotency (37). Expression of sox-2 in our isolated cells is agreement with other studies which reported sox-2 expression at mRNA level for equine BM-, FAT- and Cord Blood-MSCs (38, 39). Marfe et al. (40) only reported 15% expression of Sox-2 in equine blood-derived stem cells at protein level.
Expression of pluripotency markers in equine MSCs is some studies confirmed the expression of Oct-4 (as another critical transcription factor for self-renewal in undifferentiated embryonic stem cells) in equine MSCs at protein level (17–19), whereas other studies observed no expression in equine MSCs (15). There are also positive results for SSEA-1, -3, -4, TRA-1-60 and TRA-1-81 expression (as other embryonic markers) in equine MSCs (17, 18), but Guest et al. (15) reported no expression of these markers in equine MSCs. In this study, no expression of Nanog, SSEA-1, -3 and -4 was observed in equine MSCs at protein level. The used mABs and control cell lines in our study were belonged to human and mice. The lack of specific mAB and appropriate control group, probably play an important role in these negative results (3). Moreover, different results in various study might show that the isolated cell population in these studies are different each other with specific characteristics. So, identification and purification of specific cells using more techniques and markers are strongly suggested. The isolated cells in this study also did not expressed Nanog and Oct-4 mRNA. Nevertheless, Ranera et al. (38) and Violini et al. (39) showed the expression of Nanog using qRT-PCR, and Marfe et al. (40) showed that 15% of cells were able to express Nanog.
In conclusion, this study succeed to isolate and characterize equine BM-MSCs with detailed data regarding their growth characteristics, differentiation capacities, expression of specific genes and pluripotency stemness markers. The results show that isolated BM-MSCs have the minimal defined criteria of MSCs.
Acknowledgments
This work was financially supported by Ferdowsi University of Mashhad.
Footnotes
Potential Conflict of Interest
The authors have no conflicting financial interest.
References
- 1.Metsäranta M, Kujala UM, Pelliniemi L, Österman H, Aho H, Vuorio E. Evidence for insufficient chondrocytic differentiation during repair of full-thickness defects of articular cartilage. Matrix Biol. 1996;15:39–47. doi: 10.1016/S0945-053X(96)90125-0. [DOI] [PubMed] [Google Scholar]
- 2.Paris DB, Stout TA. Equine embryos and embryonic stem cells: defining reliable markers of pluripotency. Theriogenology. 2010;74:516–524. doi: 10.1016/j.theriogenology.2009.11.020. [DOI] [PubMed] [Google Scholar]
- 3.De Schauwer C, Meyer E, Van de Walle GR, Van Soom A. Markers of stemness in equine mesenchymal stem cells: a plea for uniformity. Theriogenology. 2011;75:1431–1443. doi: 10.1016/j.theriogenology.2010.11.008. [DOI] [PubMed] [Google Scholar]
- 4.Taylor SE, Smith RK, Clegg PD. Mesenchymal stem cell therapy in equine musculoskeletal disease: scientific fact or clinical fiction? Equine Vet J. 2007;39:172–180. doi: 10.2746/042516407X180868. [DOI] [PubMed] [Google Scholar]
- 5.Frisbie DD, Kawcak CE, Werpy NM, McIlwraith CW. Evaluation of bone marrow derived stem cells and adipose derived stromal vascular fraction for treatment of osteoarthitis using an equine experimental model. USA, Texas. AAEP Proceedings; 2006. pp. 420–421. [Google Scholar]
- 6.Koch TG, Betts DH. Stem cell therapy for joint problems using the horse as a clinically relevant animal model. Expert Opin Biol Ther. 2007;7:1621–1626. doi: 10.1517/14712598.7.11.1621. [DOI] [PubMed] [Google Scholar]
- 7.Ahern BJ, Parvizi J, Boston R, Schaer TP. Preclinical animal models in single site cartilage defect testing: a systematic review. Osteoarthritis Cartilage. 2009;17:705–713. doi: 10.1016/j.joca.2008.11.008. [DOI] [PubMed] [Google Scholar]
- 8.Smith RK, Webbon PM. Harnessing the stem cell for the treatment of tendon injuries: heralding a new dawn? Br J Sports Med. 2005;39:582–584. doi: 10.1136/bjsm.2005.015834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ibrahim S, Saunders K, Kydd JH, Lunn DP, Steinbach F. Screening of anti-human leukocyte monoclonal antibodies for reactivity with equine leukocytes. Vet Immunol Immunopathol. 2007;119:63–80. doi: 10.1016/j.vetimm.2007.06.034. [DOI] [PubMed] [Google Scholar]
- 10.CTGTA Committee. Cellular products for joint surface repair-Briefing document. 2005. [Google Scholar]
- 11.Ribitsch I, Burk J, Delling U, Geißler C, Gittel C, Jülke H, Brehm W. Basic science and clinical application of stem cells in veterinary medicine. Adv Biochem Eng Biotechnol. 2010;123:219–263. doi: 10.1007/10_2010_66. [DOI] [PubMed] [Google Scholar]
- 12.Lakshmipathy U, Verfaillie C. Stem cell plasticity. Blood Rev. 2005;19:29–38. doi: 10.1016/j.blre.2004.03.001. [DOI] [PubMed] [Google Scholar]
- 13.Ryan JM, Barry FP, Murphy JM, Mahon BP. Mesenchymal stem cells avoid allogeneic rejection. J Inflamm (Lond) 2005;2:8. doi: 10.1186/1476-9255-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guest DJ, Smith MR, Allen WR. Monitoring the fate of autologous and allogeneic mesenchymal progenitor cells injected into the superficial digital flexor tendon of horses: preliminary study. Equine Vet J. 2008;40:178–181. doi: 10.2746/042516408X276942. [DOI] [PubMed] [Google Scholar]
- 15.Guest DJ, Ousey JC, Smith MR. Defining the expression of marker genes in equine mesenchymal stromal cells. Stem Cells Cloning. 2008;1:1–9. doi: 10.2147/sccaa.s3824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Horwitz EM, Le Blanc K, Dominici M, Mueller I, Slaper-Cortenbach I, Marini FC, Deans RJ, Krause DS, Keating A International Society for Cellular Therapy. Clarification of the nomenclature for MSC: the international society for cellular therapy position statement. Cytotherapy. 2005;7:393–395. doi: 10.1080/14653240500319234. [DOI] [PubMed] [Google Scholar]
- 17.Hoynowski SM, Fry MM, Gardner BM, Leming MT, Tucker JR, Black L, Sand T, Mitchell KE. Characterization and differentiation of equine umbilical cord-derived matrix cells. Biochem Biophys Res Commun. 2007;362:347–353. doi: 10.1016/j.bbrc.2007.07.182. [DOI] [PubMed] [Google Scholar]
- 18.Li X, Zhou SG, Imreh MP, Ahrlund-Richter L, Allen WR. Horse embryonic stem cell lines from the proliferation of inner cell mass cells. Stem Cells Dev. 2006;15:523–531. doi: 10.1089/scd.2006.15.523. [DOI] [PubMed] [Google Scholar]
- 19.Reed SA, Johnson SE. Equine umbilical cord blood contains a population of stem cells that express Oct4 and differentiate into mesodermal and endodermal cell types. J Cell Physiol. 2008;215:329–336. doi: 10.1002/jcp.21312. [DOI] [PubMed] [Google Scholar]
- 20.Reed SA, Johnson SE. Refinement of culture conditions for maintenance of undifferentiated equine umbilical cord blood stem cells. J Equine Vet Sci. 2012;32:360–366. doi: 10.1016/j.jevs.2011.12.004. [DOI] [Google Scholar]
- 21.Lange C, Bassler P, Lioznov MV, Bruns H, Kluth D, Zander AR, Fiegel HC. Liver-specific gene expression in mesenchymal stem cells is induced by liver cells. World J Gastroenterol. 2005;11:4497–4504. doi: 10.3748/wjg.v11.i29.4497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoon YS, Wecker A, Heyd L, Park JS, Tkebuchava T, Kusano K, Hanley A, Scadova H, Qin G, Cha DH, Johnson KL, Aikawa R, Asahara T, Losordo DW. Clonally expanded novel multipotent stem cells from human bone marrow regenerate myocardium after myocardial infarction. J Clin Invest. 2005;115:326–338. doi: 10.1172/JCI200522326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vidal MA, Kilroy GE, Johnson JR, Lopez MJ, Moore RM, Gimble JM. Cell growth characteristics and differentiation frequency of adherent equine bone marrow-derived mesenchymal stromal cells: adipogenic and osteogenic capacity. Vet Surg. 2006;35:601–610. doi: 10.1111/j.1532-950X.2006.00197.x. [DOI] [PubMed] [Google Scholar]
- 24.Baghaban Eslaminejad MR, Taghiyar L, Dehghan MM, Falahi F, Kazemi Mehrjerdi H. Equine marrow-derived mesenchymal stem cells: isolation, differentiation and culture optimization. Iran J Vet Res. 2009;10:1–11. [Google Scholar]
- 25.Lange–Consiglio A, Corradetti B, Meucci A, Perego R, Bizzaro D, Cremonesi F. Characteristics of equine mesenchymal stem cells derived from amnion and bone marrow: in vitro proliferative and multilineage potential assessment. Equine Vet J. 2013;45:737–744. doi: 10.1111/evj.12052. [DOI] [PubMed] [Google Scholar]
- 26.Alipour F, Parham A, Kazemi Mehrjerdi H, Dehghani H. Equine adipose-derived mesenchymal stem cells: phenotype and growth characteristics, gene expression profile and differentiation potentials. Cell J. 2015;16:456–465. doi: 10.22074/cellj.2015.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Prockop DJ. Marrow stromal cells as stem cells for non-hematopoietic tissues. Science. 1997;276:71–74. doi: 10.1126/science.276.5309.71. [DOI] [PubMed] [Google Scholar]
- 28.Mensing N, Gasse H, Hambruch N, Haeger JD, Pfarrer C, Staszyk C. Isolation and characterization of multipotent mesenchymal stromal cells from the gingiva and the periodontal ligament of the horse. BMC Vet Res. 2011;7:42. doi: 10.1186/1746-6148-7-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lovati AB, Corradetti B, Lange Consiglio A, Recordati C, Bonacina E, Bizzaro D, Cremonesi F. Comparison of equine bone marrow-, umbilical cord matrix and amniotic fluid-derived progenitor cells. Vet Res Commun. 2011;35:103–121. doi: 10.1007/s11259-010-9457-3. [DOI] [PubMed] [Google Scholar]
- 30.Bourzac C, Smith LC, Vincent P, Beauchamp G, Lavoie JP, Laverty S. Isolation of equine bone marrow-derived mesenchymal stem cells: a comparison between three protocols. Equine Vet J. 2010;42:519–527. doi: 10.1111/j.2042-3306.2010.00098.x. [DOI] [PubMed] [Google Scholar]
- 31.Baghaban Eslaminejad M, Mardpour S, Ebrahimi M. Growth kinetics and in vitro aging of mesenchymal stem cells isolated from rat adipose versus bone marrow tissues. Iran J Vet Surg. 2008;3:9–20. [Google Scholar]
- 32.Burk J, Ribitsch I, Gittel C, Juelke H, Kasper C, Staszyk C, Brehm W. Growth and differentiation characteristics of equine mesenchymal stromal cells derived from different sources. Vet J. 2013;195:98–106. doi: 10.1016/j.tvjl.2012.06.004. [DOI] [PubMed] [Google Scholar]
- 33.Ranera B, Ordovás L, Lyahyai J, Bernal ML, Fernandes F, Remacha AR, Romero A, Vázquez FJ, Osta R, Cons C, Varona L, Zaragoza P, Martín-Burriel I, Rodellar C. Comparative study of equine bone marrow and adipose tissue-derived mesenchymal stromal cells. Equine Vet J. 2012;44:33–42. doi: 10.1111/j.2042-3306.2010.00353.x. [DOI] [PubMed] [Google Scholar]
- 34.Dominici M, Le Blanc K, Mueller I, Slaper-Cortenbach I, Marini F, Krause D, Deans R, Keating A, Prockop Dj, Horwitz E. Minimal criteria for defining multipotent mesenchymal stromal cells. The international society for cellular therapy position statement. Cytotherapy. 2006;8:315–317. doi: 10.1080/14653240600855905. [DOI] [PubMed] [Google Scholar]
- 35.Koch TG, Heerkens T, Thomsen PD, Betts DH. Isolation of mesenchymal stem cells from equine umbilical cord blood. BMC Biotechnol. 2007;7:26. doi: 10.1186/1472-6750-7-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Radcliffe CH, Flaminio MJ, Fortier LA. Temporal analysis of equine bone marrow aspirate during establishment of putative mesenchymal progenitor cell populations. Stem Cells Dev. 2010;19:269–282. doi: 10.1089/scd.2009.0091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Masui S, Nakatake Y, Toyooka Y, Shimosato D, Yagi R, Takahashi K, Okochi H, Okuda A, Matoba R, Sharov AA, Ko MS, Niwa H. Pluripotency governed by Sox2 via regulation of Oct3/4 expression in mouse embryonic stem cells. Nat Cell Biol. 2007;9:625–635. doi: 10.1038/ncb1589. [DOI] [PubMed] [Google Scholar]
- 38.Ranera B, Remacha AR, Álvarez-Arguedas S, Romero A, Vázquez FJ, Zaragoza P, Martín-Burriel I, Rodellar C. Effect of hypoxia on equine mesenchymal stem cells derived from bone marrow and adipose tissue. BMC Vet Res. 2012;8:142. doi: 10.1186/1746-6148-8-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Violini S, Ramelli P, Pisani LF, Gorni C, Mariani P. Horse bone marrow mesenchymal stem cells express embryo stem cell markers and show the ability for tenogenic differentiation by in vitro exposure to BMP-12. BMC Cell Biol. 2009;10:29. doi: 10.1186/1471-2121-10-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marfe G, Massaro-Giordano M, Ranalli M, Cozzoli E, Di Stefano C, Malafoglia V, Polettini M, Gambacurta A. Blood derived stem cells: an ameliorative therapy in veterinary ophthalmology. J Cell Physiol. 2012;227:1250–1256. doi: 10.1002/jcp.22953. [DOI] [PubMed] [Google Scholar]