Abstract
Purpose
A retrospective study was performed to analyze the relationship between uridine diphosphate glucuronosyltransferase 1A1 (UGT1A1) *6/*28 gene polymorphisms and adverse reactions associated with irinotecan (CPT-11)-based chemotherapy. The correlation between UGT1A1 polymorphisms and the clinical efficacy of CPT-11 was also analyzed, along with the influence of age and tumor type.
Patients and methods
Patients administered a CPT-11-based regimen in the Beijing Cancer Hospital from April 2015 to September 2016 were included in our study (n=81). Blood samples for detecting UGT1A1 were collected from each patient after various administration regimens.
Results
Colorectal cancer patients with the UGT1A1*6 mutant genotype had a significantly higher risk of severe delayed diarrhea than that of wild-type individuals when administered a CPT-11 dose ≥130 mg/m2 (P=0.042); the same phenomenon was observed when the UGT1A1*6 and UGT1A1*28 mutant genotypes were considered together (P=0.028). However, in lung cancer patients administered a low dose of CPT-11, UGT1A1*6/*28 variants were not significantly associated with severe neutropenia or delayed diarrhea. Furthermore, adult patients with the UGT1A1*6 mutation were more likely to develop severe delayed diarrhea than did wild-type adults (P=0.013); however, the difference was not significant in elderly patients. No significant differences in tumor response were found among the different genotypes (P>0.05).
Conclusion
Thus, age and tumor type influence our ability to predict adverse reactions based on UGT1A1 gene polymorphisms in cancer patients. Further, UGT1A1 gene polymorphisms are not correlated with the efficacy of CPT-11-based regimens.
Keywords: CPT-11, uridine diphosphate glucuronosyltransferase 1A1, SN-38, digital fluorescence molecular hybridization
Plain language summary
Pharmacogenetic testing of uridine diphosphate glucuronosyltransferase 1A1 (UGT1A1) *6/*28 is recommended in clinical practice prior to the administration of irinotecan (CPT-11)-based regimens; however, the research results are not uniform. We need to conduct more research and perform more rigorous experiments to verify the results. In this study, we analyzed the relationship between UGT1A1*6/*28 gene polymorphisms and adverse reactions, as well as the clinical efficacy of CPT-11 and the influence of age and tumor type. We found that colorectal cancer patients with the UGT1A1*6 mutant genotype had a significantly higher risk of severe delayed diarrhea than that of wild-type individuals when administered a CPT-11 dose ≥130 mg/m2 (P=0.042). Further, the same phenomenon was observed when the UGT1A1*6 and UGT1A1*28 mutant genotypes were considered together (P=0.028); however, no significant difference in lung cancer patients was observed (all P>0.05). Furthermore, adult patients with the UGT1A1*6 mutation were more likely to develop severe delayed diarrhea than did wild-type adults (P=0.013). No significant differences in tumor response were found among the different genotypes (P>0.05). Moreover, UGT1A1 gene polymorphisms were not correlated with the efficacy of CPT-11-based regimens.
Introduction
Irinotecan (CPT-11), a semisynthetic camptothecin (CPT) derivative, was initially isolated from Camptotheca acuminata in the early 1960s.1 It exerts high antineoplastic activity via inhibition of topoisomerase I (topo I) and is widely used to treat solid tumors in lung, colorectal, gastric, esophageal, cervical, ovarian, and other types of cancers.2–4 However, CPT-11 can cause many adverse reactions, including delayed diarrhea and neutropenia, which have limited its clinical application.
CPT-11 is hydrolyzed to 7-ethyl-10-hydroxy-camptothecin (SN-38) by carboxylesterases. SN-38 is an active form of CPT-11 and a thousand times more toxic than CPT-11.5–7 SN-38 is converted into an inactive SN-38 glucuronide (SN-38G) by uridine diphosphate-glucuronosyl transferases (UGTs) in the liver. Therefore, UGTs play a major role in SN-38 glucuronidation and may be related to CPT-11-induced adverse reactions.
Recently, single nucleotide polymorphisms (SNPs) have been found to be essential genomic resources that can significantly influence responses to pharmacotherapy, and they can be used to predict whether a drug will produce adverse reactions. As UGTs play an important role in the metabolism of CPT-11, mutations in the UGT genes may decrease UGT activity, which subsequently affects the pharmacokinetics and toxicity of CPT-11.8,9
UGTs are divided into 2 families (UGT1 and UGT2), which in turn are further divided into 3 subfamilies (UGT1A, UGT2A, and UGT2B).1,10 The UGT1A enzyme is present as 3 isozymes, namely, UGT1A1, UGT1A7, and UGT1A9.11 UGT1A1*6 and UGT1A1*28 are considered to be the most important alleles for preventing severe SN-38-induced adverse reactions such as neutropenia and delayed diarrhea. Asians with mutant genotypes are more likely to develop neutropenia and delayed diarrhea; however, Caucasians with mutant genotypes are more likely to develop neutropenia.12 Further, the prevalence of the UGT1A1*6 polymorphism is higher in Asian populations than that in Caucasian populations. Therefore, we considered the UGT1A1*6 polymorphism in Asian populations. Previously, studies have shown that UGT1A1*6/*28 gene polymorphisms can be used to assess the risk of neutropenia; Chinese people are more likely to suffer delayed diarrhea than Japanese and Thai people.12–16
In this study, we used digital fluorescence molecular hybridization (DFMH) to determine the UGT1A1 genotypes of cancer patients treated with CPT-11 in our hospital. We determined the relationship between UGT1A1*6 and UGT1A1*28 gene polymorphisms and the clinical efficacy and toxicity of CPT-11. In addition, we categorized the patients according to age and tumor type to examine the influence of these factors on the efficacy and toxicity of CPT-11.
Patients and methods
Patients
All 81 patients administered a CPT-11-based regimen in the Beijing Cancer Hospital from April 2015 to September 2016 were recruited. For inclusion in our study, patients were required to have at least 2 cycles of CPT-11-based chemotherapy. The study was reviewed and approved by the research and medical ethics committee of Beijing Cancer Hospital. Written informed consent was obtained from each patient after a brief description of the purpose and protocols of the study.
Patient treatments
Lung cancer
The following treatments were used for lung cancer patients: 1) irinotecan mono-therapy, which included intravenous infusion of 60 mg/m2 CPT-11 on days 1, 8, and 15 (repeated every 4 weeks); 2) irinotecan plus cisplatin (IP) regimen, which included intravenous infusion of 60 mg/m2 CPT-11 on days 1, 8, and 15 and 60 mg/m2 cisplatin on days 1 and 2 (repeated every 4 weeks); and 3) IP regimen plus bevacizumab, which included the same dosage regimen as IP and 5 mg/kg bevacizumab on day 1 (repeated every 3 weeks) (Table 1).
Table 1.
Treatment regimens
| Tumor | Regimen | No. of patients (n=81) | Dose of CPT-11 (mg/m2) | Days on which CPT-11 was administered | Cycle (weeks) |
|---|---|---|---|---|---|
| Lung cancer | IP | 36 | 60 | 1, 8, 15 | 4 |
| CPT-11 monotherapy | 8 | 60 | 1, 8, 15 | 4 | |
| IP plus bevacizumab | 1 | 60 | 1, 8, 15 | 3 | |
| Colorectal cancer | IP | 1 | 130 | 1 | 3 |
| FOLFIRI | 22 | 180 | 1 | 2 | |
| FOLFIRI plus bevacizumab/cetuximab | 2 | 180 | 1 | 2 | |
| CPT-11 plus bevacizumab | 1 | 180 | 1 | 3 | |
| CPT-11, capecitabine plus bevacizumab | 1 | 180 | 1 | 2 | |
| Esophageal cancer | IP | 2 | 130 | 1 | 3 |
| FOLFIRI | 1 | 180 | 1 | 2 | |
| CPT-11 plus apatinib mesylate | 6 | 150 | 1 | 2 |
Abbreviations: CPT-11, irinotecan; IP, irinotecan plus cisplatin; FOLFIRI, folinic acid, 5-fluorouracil plus irinotecan.
Colorectal cancer
The following treatments were used for colorectal cancer patients: 1) FOLFIRI regimen, which included intravenous infusion of 180 mg/m2 CPT-11, 300 mg folinic acid, and 400 mg/m2 5-fluorouracil (FU) on day 1, then continuous infusion of 2,500 mg/m2 5-FU by infusion pump for 46 h (repeated every 2 weeks); 2) FOLFIRI regimen plus bevacizumab/cetuximab, which included the same dosage regimen as FOLFIRI, plus intravenous infusion of 250 mg/m2 cetuximab on day 1, repeated each week, or 5 mg/kg bevacizumab on day 1, repeated every 2 weeks; 3) IP regimen, which included intravenous infusion of 130 mg/m2 CPT-11 on day 1 and 70 mg/m2 cisplatin on days 1 and 2 (repeated every 3 weeks); 4) CPT-11 plus bevacizumab, which included intravenous infusion of 180 mg/m2 CPT-11 on day 1 and 5 mg/kg bevacizumab on day 1 (repeated every 3 weeks); and 5) CPT-11, capecitabine plus bevacizumab, which included intravenous infusion of 180 mg/m2 CPT-11 on day 1, 1,000 mg/m2 capecitabine on days 1–7, and 5 mg/kg bevacizumab on day 1 (repeated every 2 weeks) (Table 1).
Esophageal cancer
The following treatments were used for esophageal cancer patients: 1) IP regimen, which included intravenous infusion of 130 mg/m2 CPT-11 on day 1 and 60 mg/m2 cisplatin on days 2 and 3 (repeated every 3 weeks); 2) CPT-11 plus apatinib mesylate, which included intravenous infusion of 150 mg/m2 CPT-11 on day 1 and 250 mg apatinib mesylate once daily (repeated every 2 weeks); and 3) FOLFIRI regimen, which included the same dosage regimen as previously mentioned (Table 1).
Evaluation criteria for drug toxicity
Toxicity was evaluated based on the National Cancer Institute Common Toxicity Criteria for Adverse Events (NCI-CTCAE) Version 3.0. Adverse reactions attributable to CPT-11 include neutropenia, leukopenia, delayed diarrhea, nausea, and vomiting. These reactions were divided into 4 stages in accordance with the NCI-CTCAE, with stages III and IV representing severe reactions.
Evaluation criteria for tumor response
The Response Evaluation Criteria in Solid Tumors (RECIST) is a standardized set of criteria for measuring tumor responses. Tumor response was assessed by computed tomography (CT) after every 2 cycles of treatment. The cases were defined as a complete response (CR), partial response (PR), or progressive disease (PD).
UGT1A1 gene detection
Blood samples were collected from each patient and placed in blood collection tubes containing ethylenediaminetetraacetic acid (EDTA) to prevent coagulation. The blood samples were then transferred to an Eppendorf (EP) tube and lysed using ammonium chloride (Sino-Era Jiyin Tech Co., Ltd., Beijing, China) for 5 min. The samples were then centrifuged at 700× g for 5 min to obtain plasma samples. Leukocyte genomic DNA was extracted directly from the blood samples using a nucleic acid purification kit (Sino-Era Jiyin Tech Co., Ltd.) and vortexed for 1 min. After 15 min of stasis, universal sequencing kits (Sino-Era Jiyin Tech Co., Ltd.) for individual gene loci were used to determine the UGT1A1 genotype. UGT1A1*6 and UGAT1A1*28 gene polymorphisms were then identified by DFMH using fluorescent probes (Tianlong Science and Technology Co., Ltd., Xi’an, China).
DFMH is a new technique that is based on the principle of fluorescence in situ hybridization (FISH). Specific DNA strains containing fluorophore-labeled nucleotides can be used as probes to identify complementary sequences.
The conditions for sequencing by fluorescence molecular hybridization were as follows: 1) for the UGT1A1*6 system, initial pre-degeneration at 95°C was performed for 10 min, followed by 50 cycles of degeneration at 95°C for 30 s and revival at 62°C for 75 s and 2) for the UGT1A1*28 system, initial pre-degeneration at 95°C was performed for 10 min, followed by 50 cycles of degeneration at 95°C for 30 s and revival at 64°C for 75 s.
UGT1A1 genotype analysis
UGT1A1*6 (G/G, G/A, and A/A) and UGT1A1*28 (TA6/6, TA6/7, and TA7/7) genotypes were used as quality control samples (Sino-Era Jiyin Tech Co., Ltd.) to set the range of parameters, which were determined by measuring the time to reach the specific fluorescent signal intensity (ST). ST1 and ST2 values reflect the interaction between the 2 probes and template and the different times to reach the specific fluorescent intensity, respectively; therefore, we set the parameter range by calculating the difference using the following formula:
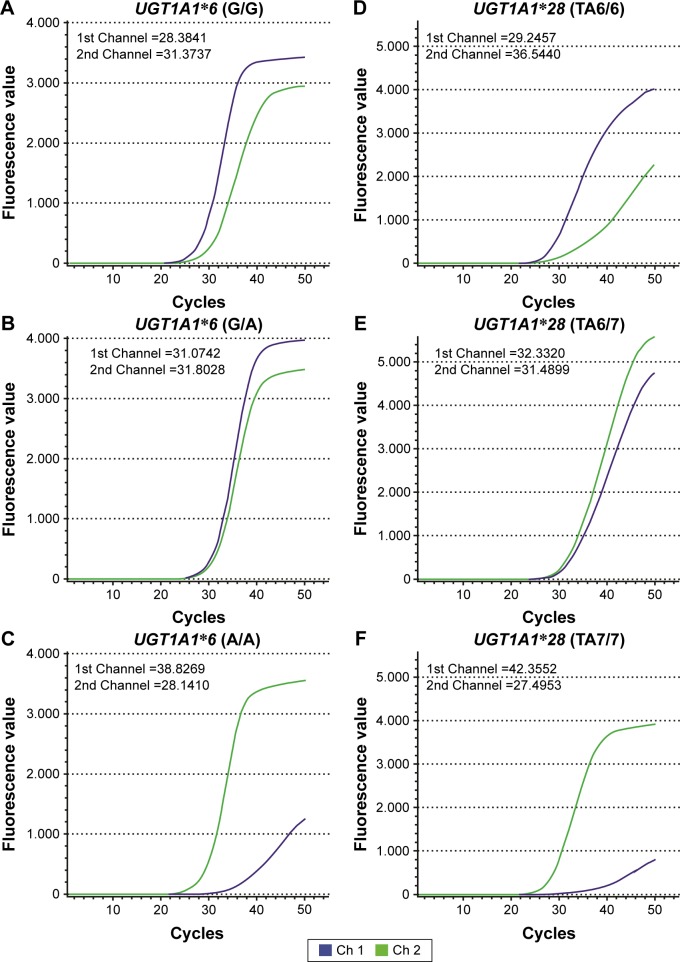
Depending on the ΔST value, a threshold for the fluorescent signal can be set to facilitate genotype determination. The ΔST value ranges for the UGT1A1*6/*28 genotypes were calculated using quality control samples. The specific genotype corresponds to a specific range of ΔST. Therefore, we determined the genotype of each patient using the ΔST value ranges of the UGT1A1*6 and UGT1A1*28 genotypes. Results of UGT1A1*6/*28 quality control gene polymorphism tests are shown in Figure 1A–F.
Figure 1.
Detection of gene polymorphisms in UGT1A1*6 and UGT1A1*28 quality control genes by DFMH.
Notes: UGT1A1 gene polymorphisms were detected using different fluorescent probes specific for UGT1A1*6 or UGT1A1*28 wild-type or mutant genotypes. The time to reach the specific fluorescence ST was determined for each channel using the fluorescence detector software, Microseq (Tianlong Science and Technology Co., Ltd., Xi’an, China). ΔST was calculated to set the range of ΔST values indicative of each UGT1A1 polymorphism, which could then be used to determine the genotype of each patient. The results were as follows: (A) UGT1A1*6 (G/G), ΔST =−2.9896; (B) UGT1A1*6 (G/A), ΔST =−0.7286; (C) UGT1A1*6 (A/A), ΔST =10.6859; (D) UGT1A1*28 (TA6/6), ΔST =−7.2983; (E) UGT1A1*28 (TA6/7), ΔST =0.8421; and (F) UGT1A1*28 (TA7/7), ΔST =14.8599. The maximum value of ΔST is 5. Therefore, the ΔST values of UGT1A1*6 (G/A), UGT1A1*6 (G/G), and UGT1A1*6 (A/A) were between −2.0657 and 5.000, <−2.0657, and >5.000, respectively, and the ΔST values of UGT1A1*28 (TA6/7), UGT1A1*28 (TA6/6), and UGT1A1*6 (TA7/7) were between −3.5868 and 5.000, <−3.5868, and >5.000, respectively.
Abbreviations: DFMH, digital fluorescence molecular hybridization; ST, signal intensity.
Statistics
Data were analyzed using SPSS Version 22.0 software (IBM Corporation, Armonk, NY, USA). Genotyping data were analyzed for deviation from the Hardy–Weinberg equilibrium using the chi-square test. Differences in the incidence of adverse reactions and the clinical efficacy of CPT-11 were also analyzed using the chi-square test and Fisher’s exact test. All statistical analyses were 2-sided tests, and a P-value ≤0.05 was defined as statistically significant.
Results
Distribution of UGT1A1 gene polymorphisms
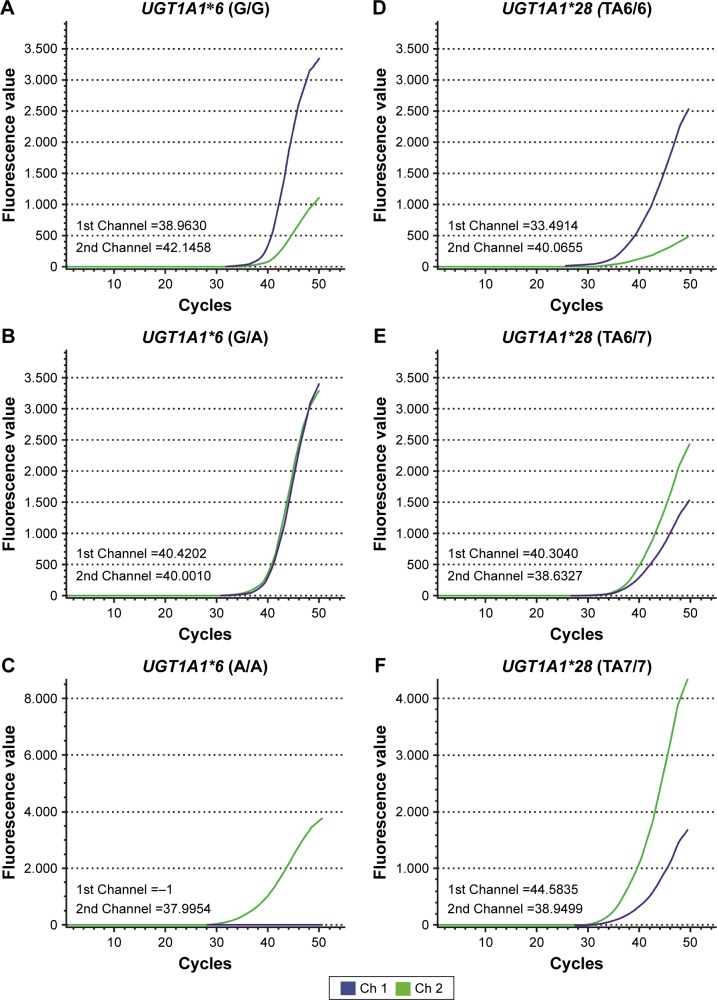
Complete genotyping was performed for all 81 patients, of whom 67 were men (82.72%) and 14 were women (17.28%). Thirty three patients (40.74%) were ≥60 years old, and 48 patients (59.26%) were <60 years old; the median patient age was 58 years (range: 28–79 years). The clinical characteristics of the patients are shown in Table 2, and results of the UGT1A1*6 and UGT1A1*28 genotyping are shown in Figure 2A–F. The UGT1A1*28 genotype was divided into 3 groups: wild-type (TA6/6), heterozygous mutant (TA6/7), and homozygous mutant (TA7/7), with 61 (75.31%), 16 (19.75%), and 4 (4.49%) of the cases, respectively. The UGT1A1*6 genotype was similarly divided into wild-type (G/G), heterozygous mutant (G/A), and homozygous mutant (A/A) groups, with 62 (76.54%), 17 (20.99%), and 2 (2.47%) of the cases, respectively. In total, there were 9 possible genotype combinations when both UGT1A1*6 and UGT1A1*28 were taken into consideration. These genotypes were divided into 3 categories depending on the number of mutations: double wild-type (G/G and TA6/6, n=44, 54.32%), single-site mutation (G/G and TA6/7 or G/A and TA6/6, n=29, 35.80%), and 2-site mutations (G/G and TA7/7, G/A and TA6/7, or A/A and TA6/6, n=8, 9.88%). However, we were unable to find the remaining 3 combinations (G/A and TA7/7, A/A and TA6/7, and A/A and TA7/7) in this study. The distribution of UGT1A1 gene polymorphisms is shown in Table 3. The genotype distributions of UGT1A1*6/*28 were in accordance with the Hardy–Weinberg equilibrium (P>0.05).
Table 2.
Clinical characteristics of patients with irinotecan-based treatment in this study
| Characteristics | No. of patients (n=81) |
Frequency (%) |
|---|---|---|
| Mean age, years (range) | 58 (28–79) | |
| Gender | ||
| Male | 67 | 82.72 |
| Female | 14 | 17.28 |
| ECOG performance status | ||
| 0 | 47 | 58.02 |
| 1 | 31 | 38.27 |
| 2 | 2 | 2.47 |
| 3 | 1 | 1.23 |
| Type of tumor | ||
| Lung cancer | 45 | 55.56 |
| Colorectal cancer | 27 | 33.33 |
| Esophageal cancer | 9 | 11.11 |
| Habit of smoking and drinking | ||
| Smoking | 20 | 24.69 |
| Drinking | 5 | 6.17 |
| Smoking and drinking | 31 | 38.27 |
| None | 25 | 34.57 |
| Line of treatment | ||
| First line | 30 | 37.04 |
| Second line | 47 | 58.02 |
| Third line | 4 | 4.94 |
| Cycle of treatment | ||
| Second cycle | 34 | 41.98 |
| Third cycle | 24 | 29.63 |
| ≥ fourth cycle | 23 | 28.40 |
Abbreviation: ECOG, Eastern Cooperative Oncology Group.
Figure 2.
UGT1A1 gene polymorphisms in patients.
Notes: Blood samples from the patients were analyzed by DFMH, and the results are as follows: (A) ΔST =−3.1828, representing UGT1A1*6 (G/G); (B) ΔST =0.4192, representing UGT1A1*6 (G/A); (C) ΔST =12.0046, representing UGT1A1*6 (A/A); (D) ΔST =−6.5741, representing UGT1A1*28 (TA6/6); (E) ΔST =1.6713, representing UGT1A1*28 (TA6/7); and (F) ΔST =5.6336, representing UGT1A1*28 (TA7/7).
Abbreviations: DFMH, digital fluorescence molecular hybridization; ST, signal intensity.
Table 3.
Distribution of UGT1A1 gene polymorphisms
| Genotype | No. of patients (n=81) | Ratio (%) |
|---|---|---|
| UGT1A1*6 | ||
| G/G | 62 | 76.54 |
| G/A | 17 | 20.99 |
| A/A | 2 | 2.47 |
| UGT1A1*28 | ||
| TA6/6 | 61 | 75.31 |
| TA6/7 | 16 | 19.75 |
| TA7/7 | 4 | 4.94 |
| UGT1A1 combinations | ||
| G/G TA6/6 | 44 | 54.32 |
| G/G TA6/7 | 14 | 17.28 |
| G/A TA6/6 | 15 | 18.52 |
| G/G TA7/7 | 4 | 4.94 |
| G/A TA6/7 | 2 | 2.47 |
| A/A TA6/6 | 2 | 2.47 |
Correlation between UGT1A1 genotype and adverse reactions
The different adverse reactions attributable to CPT-11 in patients, such as leukopenia, neutropenia, delayed diarrhea, and nausea and vomiting, were recorded in this study. In total, 32 patients (39.51%) suffered from leukopenia, 28 (34.57%) from neutropenia, 39 (48.15%) from nausea and vomiting, and 26 (32.10%) from delayed diarrhea. The results show that mutant UGT1A1*6 and UGT1A1*28 genotypes, as well as mutant UGT1A1*6/*28 genotypes considered together, increase the risk of severe diarrhea over that of wild-type individuals; however, the differences were not significant (P=0.305, P=0.707, and P=0.354, respectively). In addition, there were no significant differences in the incidence of severe leukopenia, neutropenia, or nausea and vomiting in patients with UGT1A1 gene polymorphisms (Table 4).
Table 4.
Relationship between UGT1A1*6/*28 genotype and the incidence of adverse reactions associated with CPT-11
| Genotyping | Leukopenia, n (%)
|
Neutropenia, n (%)
|
Nausea and vomiting, n (%)
|
Diarrhea, n (%)
|
||||
|---|---|---|---|---|---|---|---|---|
| Grades I–II | Grades III–IV | Grades I–II | Grades III–IV | Grades I–II | Grades III–IV | Grades I–II | Grades III–IV | |
| G/G (n=62) | 19 (30.65) | 7 (11.29) | 14 (22.58) | 11 (17.74) | 26 (41.94) | 4 (6.45) | 17 (27.41) | 3 (4.84) |
| G/A (n=17) | 5 (29.41) | 1 (5.88) | 0 (0) | 3 (17.65) | 8 (47.06) | 0 (0) | 3 (17.65) | 3 (17.65) |
| A/A (n=2) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (50.00) | 0 (0) | 0 (0) | 0 (0) |
| P-value | 1.000 | 1.000 | 0.067 | 1.000 | 0.908 | 0.614 | 0.739 | 0.305 |
| TA6/6 (n=61) | 18 (29.51) | 7 (11.48) | 10 (16.39) | 11 (18.03) | 27 (44.26) | 3 (4.92) | 15 (24.59) | 4 (6.56) |
| TA6/7 (n=16) | 5 (31.25) | 1 (6.25) | 3 (18.75) | 2 (12.50) | 7 (43.75) | 1 (6.35) | 4 (25.00) | 2 (12.50) |
| TA7/7 (n=4) | 1 (25.00) | 0 (0) | 1 (25.00) | 1 (25.00) | 1 (25.00) | 0 (0) | 1 (25.00) | 0 (0) |
| P-value | 1.000 | 1.000 | 0.757 | 0.646 | 0.844 | 1.000 | 1.000 | 0.707 |
| Wild-type (n=44) | 13 (29.55) | 6 (13.64) | 10 (22.73) | 8 (18.18) | 19 (43.18) | 3 (6.38) | 12 (27.27) | 2 (4.55) |
| Single-site mutant (n=29) | 10 (34.48) | 2 (9.60) | 3 (10.34) | 5 (17.24) | 13 (44.83) | 1 (3.45) | 7 (24.13) | 3 (10.34) |
| Two-site mutant (n=8) | 1 (12.50) | 0 (0) | 1 (12.50) | 1 (12.50) | 3 (37.50) | 0 (0) | 1 (12.50) | 1 (12.50) |
| P-value | 0.483 | 0.357 | 0.365 | 0.926 | 1.000 | 1.000 | 0.670 | 0.354 |
Correlation between UGT1A1 genotype and adverse reactions with different tumors
The patients were divided into 2 groups according to tumor type. One group included 45 lung cancer patients treated with a low dose of CPT-11 (<130 mg/m2), and the other group comprised 27 colorectal cancer patients treated with a high dose of CPT-11 (≥130 mg/m2). The relationship between UGT1A1 genotype and adverse reactions with different cancers is shown in Table 5. In lung cancer patients, the risk of grade I/II nausea and vomiting was higher in those with the UGT1A1*6 mutant genotypes (40.00%; 4/10) than those with the wild-type UGT1A1 (17.14%; 6/35). UGT1A1*28 mutant genotypes (18.18%; 2/11) resulted in a higher risk of grade I/II neutropenia than the wild-type genotype (5.88; 2/34); however, the difference was not statistically significant (P=0.247). Thus, neither the wild-type nor mutant genotypes were significantly associated with severe adverse reactions in lung cancer patients. However, the outcome was different in the colorectal cancer group. The risk of severe delayed diarrhea was significantly higher in patients with the UGT1A1*6 mutant genotypes (42.86%; 3/7) than those with the wild-type genotype (5.00%; 1/20) (P=0.042). Similarly, severe delayed diarrhea was higher in patients with the mutant UGT1A1*28 genotypes (28.57%; 2/7) than those with the wild-type genotype (10.00%; 2/20); however, the difference was not significant (P=0.269). When the UGT1A1*6 and UGT1A1*28 mutant genotypes were considered together, the incidence of severe delayed diarrhea was significantly higher than that of the wild-type genotype (P=0.028).
Table 5.
Relationship between UGT1A1*6/*28 genotype and the incidence of adverse reactions associated with different tumors
| Tumor | Genotyping | Leukopenia, n (%)
|
Neutropenia, n (%)
|
Nausea and vomiting, n (%)
|
Diarrhea, n (%)
|
||||
|---|---|---|---|---|---|---|---|---|---|
| Grades I–II | Grades III–IV | Grades I–II | Grades III–IV | Grades I–II | Grades III–IV | Grades I–II | Grades III–IV | ||
| Lung cancer | G/G (n=35) | 7 (20.00) | 6 (17.14) | 4 (11.43) | 4 (11.43) | 6 (17.14) | 4 (11.43) | 10 (28.57) | 2 (5.71) |
| G/A A/A (n=10) | 3 (30.00) | 0 (0) | 0 (0) | 0 (0) | 4 (40.00) | 0 (0) | 2 (20.00) | 0 (0) | |
| P-value | 0.811 | 0.379 | 0.561 | 0.561 | 0.270 | 0.561 | 0.893 | 1.000 | |
| Colorectal cancer | G/G (n=20) | 7 (35.00) | 1 (5.00) | 5 (25.00) | 6 (30.00) | 15 (75.00) | 0 (0) | 5 (25.00) | 1 (5.00) |
| G/A A/A (n=7) | 1 (14.29) | 0 (0) | 0 (0) | 1 (14.29) | 4 (57.14) | 0 (0) | 1 (14.29) | 3 (42.86) | |
| P-value | 0.633 | 1.000 | 0.283 | 0.633 | 0.633 | NG | 1.000 | 0.042 | |
| Lung cancer | TA6/6 (n=34) | 7 (20.59) | 5 (14.71) | 2 (5.88) | 3 (8.82) | 8 (23.53) | 3 (8.82) | 8 (23.53) | 2 (5.88) |
| TA6/7 TA7/7 (n=11) | 3 (27.27) | 1 (9.09) | 2 (18.18) | 1 (9.09) | 2 (18.18) | 1 (9.09) | 4 (36.36) | 0 (0) | |
| P-value | 0.963 | 1.000 | 0.247 | 1.000 | 1.000 | 1.000 | 0.657 | 1.000 | |
| Colorectal cancer | TA6/6 (n=20) | 6 (30.00) | 1 (5.00) | 4 (20.00) | 5 (25.00) | 14 (70.00) | 0 (0) | 5 (25.00) | 2 (10.00) |
| TA6/7 TA7/7 (n=7) | 2 (28.57) | 0 (0) | 1 (14.28) | 2 (28.57) | 5 (71.43) | 0 (0) | 1 (14.29) | 2 (28.57) | |
| P-value | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | NG | 1.000 | 0.269 | |
| Lung cancer | Wild-type (n=24) | 4 (16.67) | 5 (20.83) | 2 (8.33) | 3 (12.50) | 3 (12.50) | 3 (12.50) | 6 (25.00) | 2 (8.33) |
| Mutant type (n=21) | 6 (28.57) | 1 (4.76) | 2 (9.52) | 1 (4.76) | 6 (28.57) | 1 (4.76) | 6 (28.57) | 0 (0) | |
| P-value | 0.549 | 0.253 | 1.000 | 0.700 | 0.331 | 0.700 | 1.000 | 0.491 | |
| Colorectal cancer | Wild-type (n=15) | 5 (33.33) | 1 (6.67) | 4 (26.67) | 4 (26.67) | 11 (73.33) | 0 (0) | 4 (26.67) | 0 (0) |
| Mutant type (n=12) | 3 (25.00) | 0 (0) | 1 (8.33) | 3 (25.00) | 8 (66.67) | 0 (0) | 2 (16.67) | 4 (33.33) | |
| P-value | 0.696 | 1.000 | 0.342 | 1.000 | 1.000 | NG | 0.662 | 0.028 | |
Abbreviation: NG, not given.
Correlation of UGT1A1 genotype and adverse reactions in aging patients
The patients were divided into 2 groups depending on their age. One group included 33 elderly patients (≥60 years old), and the other comprised 48 adult patients (<60 years old). The relationship between age and adverse reactions associated with different UGT1A1 genotypes is shown in Table 6. In adult patients, the incidence of severe delayed diarrhea was higher in patients with the UGT1A1*6 mutant genotypes than those with the wild-type genotype (P=0.013), while the UGT1A1*28 genotype had no significant effect on delayed diarrhea. When the UGT1A1*6 and UGT1A1*28 mutant genotypes were considered together (12.50%; 3/24), the risk of delayed diarrhea was higher than that of the wild-type genotype (0%; 0/24); however, the difference was not statistically significant (P=0.233). In elderly patients, the incidence of grade I/II nausea and vomiting was higher in patients with the UGT1A1*6 mutant genotypes than those with the wild-type genotype (P=0.039). Patients with UGT1A1*28 polymorphisms were more likely to suffer from grade I/II neutropenia; however, the difference was not significant (P=0.093). The incidence of severe delayed diarrhea in elderly patients carrying a UGT1A1*28 mutation (16.67%; 1/6) was higher than in those with wild-type UGT1A1 (7.41%; 2/27; P=0.464). No significant differences in adverse reactions associated with CPT-11 in elderly patients were observed when UGT1A1*6 and UGT1A1*28 were considered together.
Table 6.
Relationship between UGT1A1 genotype and the incidence of adverse reactions in patients of different ages
| Age | Genotyping | Leukopenia, n (%)
|
Neutropenia, n (%)
|
Nausea and vomiting, n (%)
|
Diarrhea, n (%)
|
||||
|---|---|---|---|---|---|---|---|---|---|
| Grades I–II | Grades III–IV | Grades I–II | Grades III–IV | Grades I–II | Grades III–IV | Grades I–II | Grades III–IV | ||
| <60 | G/G (n=36) | 13 (36.11) | 5 (13.89) | 6 (16.67) | 7 (19.44) | 16 (44.44) | 0 (0) | 10 (27.78) | 0 (0) |
| G/A A/A (n=12) | 3 (27.27) | 1 (9.09) | 0 (0) | 2 (18.18) | 3 (27.27) | 0 (0) | 0 (0) | 3 (25.00) | |
| P-value | 0.724 | 1.000 | 0.313 | 1.000 | 0.394 | NG | 0.101 | 0.013 | |
| ≥60 | G/G (n=26) | 6 (23.08) | 2 (7.69) | 8 (30.77) | 4 (15.38) | 10 (38.46) | 4 (15.38) | 7 (26.92) | 3 (11.54) |
| G/A A/A (n=7) | 2 (33.33) | 0 (0) | 0 (0) | 1 (16.67) | 6 (85.71) | 0 (0) | 1 (14.28) | 0 (0) | |
| P-value | 1.000 | 1.000 | 0.154 | 1.000 | 0.039 | 0.555 | 0.652 | 1.000 | |
| <60 | TA6/6 (n=34) | 11 (32.35) | 5 (14.71) | 6 (17.65) | 6 (17.65) | 11 (32.35) | 0 (0) | 7 (20.59) | 2 (5.88) |
| TA6/7 TA7/7 (n=14) | 3 (27.27) | 1 (9.09) | 1 (9.09) | 3 (21.43) | 6 (42.86) | 0 (0) | 4 (28.57) | 1 (7.14) | |
| P-value | 0.684 | 0.810 | 0.626 | 1.000 | 0.719 | NG | 0.826 | 1.000 | |
| ≥60 | TA6/6 (n=27) | 7 (25.93) | 2 (7.41) | 4 (14.81) | 5 (18.52) | 16 (59.26) | 3 (11.11) | 8 (29.63) | 2 (7.41) |
| TA6/7 TA7/7 (n=6) | 3 (50.00) | 0 (0) | 3 (50.00) | 0 (0) | 2 (40.00) | 1 (20.00) | 1 (16.67) | 1 (16.67) | |
| P-value | 0.336 | 1.000 | 0.093 | 0.556 | 0.375 | 1.000 | 1.000 | 0.464 | |
| <60 | Wild-type (n=24) | 7 (29.17) | 4 (16.67) | 5 (20.83) | 4 (16.67) | 9 (37.50) | 0 (0) | 6 (25.00) | 0 (0) |
| Mutant type (n=24) | 6 (33.33) | 2 (11.11) | 1 (5.56) | 5 (20.83) | 9 (37.50) | 0 (0) | 5 (20.83) | 3 (12.50) | |
| P-value | 0.754 | 0.663 | 0.190 | 1.000 | 1.000 | NG | 1.000 | 0.233 | |
| ≥60 | Wild-type (n=20) | 6 (30.00) | 2 (10.00) | 5 (25.00) | 4 (20.00) | 10 (50.00) | 3 (15.00) | 7 (35.00) | 2 (10.00) |
| Mutant type (n=13) | 5 (38.46) | 0 (0) | 3 (23.08) | 1 (9.09) | 7 (53.85) | 1 (9.09) | 3 (23.07) | 1 (7.69) | |
| P-value | 0.714 | 0.508 | 1.000 | 0.625 | 1.000 | 1.000 | 0.701 | 1.000 | |
Abbreviation: NG, not given.
Correlation of UGT1A1 genotype and tumor response
Tumor response was assessed in 30 (66.67%; 30/45) patients with lung cancer and 19 (70.37%; 19/27) patients with colorectal cancer, respectively (Table 7). PR, stable disease (SD), and PD were observed in 6, 17, and 7 lung cancer cases and 4, 12, and 3 colorectal cancer cases, respectively. Colorectal patients with UGT1A1*28 mutant genotypes had a good tumor response in the PR group; however, the difference was not significant (P=0.178).
Table 7.
Relationship between UGT1A1 genotypes and tumor response
| Tumor | Genotyping | PR, n (%) | P-value | SD, n (%) | P-value | PD, n (%) | P-value |
|---|---|---|---|---|---|---|---|
| Lung cancer | G/G (n=21) | 3 (14.28) | 0.320 | 13 (61.90) | 0.443 | 5 (23.81) | 1.000 |
| G/A A/A (n=9) | 3 (33.33) | 4 (44.44) | 2 (22.22) | ||||
| TA6/6 (n=23) | 5 (21.74) | 1.000 | 13 (56.52) | 1.000 | 5 (21.74) | 1.000 | |
| TA6/7 TA7/7 (n=7) | 1 (14.29) | 4 (57.14) | 2 (28.57) | ||||
| Wild-type (n=14) | 2 (14.29) | 0.657 | 9 (64.29) | 0.484 | 3 (21.43) | 1.000 | |
| Mutant type (n=16) | 4 (25.00) | 8 (50.00) | 4 (25.00) | ||||
| Colorectal cancer | G/G (n=14) | 4 (28.57) | 0.530 | 8 (57.14) | 0.603 | 2 (14.28) | 1.000 |
| G/A A/A (n=5) | 0 (0) | 4 (80.00) | 1 (20.00) | ||||
| TA6/6 (n=15) | 2 (13.33) | 0.178 | 10 (66.67) | 0.603 | 3 (20.00) | 1.000 | |
| TA6/7 TA7/7 (n=4) | 2 (50.00) | 2 (50.00) | 0 (0) | ||||
| Wild-type (n=11) | 2 (18.18) | 1.000 | 7 (63.64) | 1.000 | 2 (18.18) | 1.000 | |
| Mutant type (n=8) | 2 (25.00) | 5 (62.50) | 1 (12.50) |
Abbreviations: PR, partial response; SD, stable disease; PD, progressive disease.
Discussion
CPT-11 causes obvious adverse reactions, including myelosuppression and delayed diarrhea, which limit its clinical application. The rapid growth of pharmacogenetics has shown that differences in drug metabolism between individuals are associated with genetic polymorphisms. Pharmacogenetic research has shown that genetic disparities play a major role in pharmacokinetics and can explain the clinical profile of many drugs, especially antineoplastic agents.17–19 For example, UGT1A1 plays an important role in SN-38 metabolism. The different UGT1A1*6/*28 genotypes result in different metabolic rates, and elevated SN-38 blood concentrations induce adverse reactions. Therefore, it may be possible to predict the likelihood of SN-38-induced adverse reactions in patients using UGT1A1*6/*28 genotyping.
Recently, the association between UGT1A1 gene polymorphisms and adverse reactions, particularly delayed diarrhea and neutropenia, has been of interest worldwide. Many studies have shown that UGT1A1*28 is significantly associated with CPT-11-induced toxicity, especially neutropenia.20–24 Caucasians with mutant genotypes are more likely to have neutropenia.12 However, UGT1A1*6 is more prevalent in Asian countries than in Caucasian populations, suggesting that UGT1A1*6 polymorphisms should be considered in addition to UGT1A1*28 polymorphisms for more individualized CPT-11-based chemotherapy. In Japan, Hoskins et al25 discovered that the risk of severe hematologic toxicity is higher in patients with UGT1A1*28 homozygous mutations than in patients with wild-type or heterozygous genotypes at a dose >150 mg/m2, but not <125 mg/m2. However, Hirasawa et al26 found that UGT1A1*6/*28 mutations are associated with an increased risk of neutropenia and delayed diarrhea at a low dose of CPT-11. In China, Xu et al3 discovered that mutant UGT1A1*6/*28 genotypes significantly increase the incidence of delayed diarrhea, and the incidence of neutropenia is significantly increased when the UGT1A1*6 and UGT1A1*28 genotypes are considered together. Further, Chinese patients with UGT1A1*6/*28 mutations are more likely to suffer delayed diarrhea than Japanese patients. Our results demonstrate that the UGT1A1*6 mutant genotypes are associated with delayed diarrhea in colorectal cancer patients (P=0.042). In addition, when both UGT1A1*6 and UGT1A1*28 genotypes are taken into consideration, we discovered that the risk of severe delayed diarrhea is significantly higher in colorectal patients with mutant genotypes (P=0.028). However, in our study, there was no association between UGT1A1 mutations and severe hematologic toxicity. Besides, we did not find DPYD mutations (rs55886062, rs67376798, or rs3918290) in our study; therefore, we could eliminate the effects of fluorouracil drug metabolism on adverse reactions.
The occurrence of cancer is increasing because of population growth and aging.27 Hence, more attention should be paid to elderly patients, and individualized medication programs should be implemented to reduce the incidence of adverse reactions. In our study, UGT1A1*6 mutations (G/A and A/A) increased the risk of grade I/II nausea and vomiting specifically in elderly patients (P=0.039). In contrast, adult patients with UGT1A1*6 mutations had a significantly higher risk of severe delayed diarrhea (P=0.013); however, there were no significant differences in elderly patients. Differences in drug metabolism may not be obvious in elderly patients owing to reduced body function and metabolic enzyme activity.
There are currently no uniform conclusions regarding the correlation between UGT1A1 gene polymorphisms and the clinical efficacy of CPT-11. Some studies have demonstrated that CPT-11 is more efficacious in patients with mutant genotypes.28,29 However, most studies have found no correlation between different UGT1A1 genotypes and clinical efficacy.30,31 Further, a meta-analysis of 6,087 patients found that both homozygous and heterozygous UGT1A1*28 mutant types had a higher response than wild-type patients, particularly Caucasians.12 In our study, we observed that the tumor response in patients with UGT1A1*28 mutant genotypes with colorectal cancer was better than in those with the wild-type genotype; however, the difference was not statistically significant (P=0.178).
A limitation of this study is its retrospective design; therefore, we did not conduct patient follow-ups once they discontinued treatment in our hospital. Consequently, we did not conduct a survival analysis in addition to the tumor response analysis to evaluate efficacy. Further, the small sample size of this study may impact the results. Owing to the low UGT1A1*6 and UGT1A1*28 mutation rates, we need to increase sample size and perform more rigorous experiments to verify the results of this study.
Conclusion
UGT1A1 is a potential pharmacogenetic marker for detecting the occurrence of severe delayed diarrhea, especially in colorectal cancer patients receiving a high dose of CPT-11. However, neither UGT1A1*6 nor UGT1A1*28 mutations were associated with severe neutropenia. UGT1A1 gene polymorphisms cannot predict the clinical outcome of CPT-11-based regimens.
Acknowledgments
We would like to thank Editage (www.editage.com) for English language editing and publication support.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Mathijssen R, van Alphen RJ, Verweij J, et al. Clinical pharmacokinetics and metabolism of irinotecan (CPT-11) Clin Cancer Res. 2001;7(8):2182–2194. [PubMed] [Google Scholar]
- 2.Xiao XG, Xia S, Zou M, et al. The relationship between UGT1A1 gene polymorphism and irinotecan effect on extensive-stage small-cell lung cancer. Onco Targets Ther. 2015;8:3575–3583. doi: 10.2147/OTT.S95149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xu C, Tang X, Qu Y, Keyoumu S, Zhou N, Tang Y. UGT1A1 gene polymorphism is associated with toxicity and clinical efficacy of irinotecan-based chemotherapy in patients with advanced colorectal cancer. Cancer Chemother Pharmacol. 2016;78(1):119–130. doi: 10.1007/s00280-016-3057-z. [DOI] [PubMed] [Google Scholar]
- 4.Kim M, Keam B, Kim TM, et al. Phase II study of irinotecan and cisplatin combination chemotherapy in metastatic, unresectable esophageal cancer. Cancer Res Treat. 2016;49(2):416–422. doi: 10.4143/crt.2016.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lopes F, Smith R, Nash S, Mitchell RT, Spears N. Irinotecan metabolite SN38 results in germ cell loss in the testis but not in the ovary of pre-pubertal mice. Mol Hum Reprod. 2016;22(11):745–755. doi: 10.1093/molehr/gaw051. Epub 2016 Jul 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fujita K, Sparreboom A. Pharmacogenetics of irinotecan disposition and toxicity: a review. Curr Clin Pharmacol. 2010;5(3):209–217. doi: 10.2174/157488410791498806. [DOI] [PubMed] [Google Scholar]
- 7.Kawato Y, Aonuma M, Hirota Y, Kuga H, Sato K. Intracellular roles of SN-38, a metabolite of the camptothecin derivative CPT-11, in the antitumor effect of CPT-11. Cancer Res. 1991;51(16):4187–4191. [PubMed] [Google Scholar]
- 8.Miyata Y, Touyama T, Kusumi T, et al. UDP-glucuronosyltransferase 1A1*6 and *28 polymorphisms as indicators of initial dose level of irinotecan to reduce risk of neutropenia in patients receiving FOLFIRI for colorectal cancer. Int J Clin Oncol. 2016;21(4):696–703. doi: 10.1007/s10147-015-0937-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yeh YS, Tsai HL, Huang CW, et al. Prospective analysis of UGT1A1 promoter polymorphism for irinotecan dose escalation in metastatic colorectal cancer patients treated with bevacizumab plus FOLFIRI as the first-line setting: study protocol for a randomized controlled trial. Trials. 2016;17:46. doi: 10.1186/s13063-016-1153-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shimoyama S. Pharmacogenetics of irinotecan: an ethnicity-based prediction of irinotecan adverse events. World J Gastrointest Surg. 2010;2(1):14–21. doi: 10.4240/wjgs.v2.i1.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Inoue K, Sonobe M, Kawamura Y, et al. Polymorphisms of the UDP-glucuronosyl transferase 1A genes are associated with adverse events in cancer patients receiving irinotecan-based chemotherapy. Tohoku J Exp Med. 2013;229(2):107–114. doi: 10.1620/tjem.229.107. [DOI] [PubMed] [Google Scholar]
- 12.Liu XH, Lu J, Duan W, et al. Predictive value of UGT1A1*28 polymorphism in irinotecan-based chemotherapy. J Cancer. 2017;8(4):691–703. doi: 10.7150/jca.17210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li M, Wang Z, Guo J, et al. Clinical significance of UGT1A1 gene polymorphisms on irinotecan-based regimens as the treatment in metastatic colorectal cancer. Onco Targets Ther. 2014;23(7):1653–1661. doi: 10.2147/OTT.S67867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao J, Zhou J, Li Y, Lu M, Jia R, Shen L. UGT1A1 6/28 polymorphisms could predict irinotecan-induced severe neutropenia not diarrhea in Chinese colorectal cancer patients. Med Oncol. 2013;30(3):604. doi: 10.1007/s12032-013-0604-x. [DOI] [PubMed] [Google Scholar]
- 15.Takano M, Yamamoto K, Tabata T, et al. Impact of UGT1A1 genotype upon toxicities of combination with low-dose irinotecan plus platinum. Asia Pac J Clin Oncol. 2016;12(2):115–124. doi: 10.1111/ajco.12453. [DOI] [PubMed] [Google Scholar]
- 16.Atasilp C, Chansriwong P, Sirachainan E, et al. Correlation of UGT1A1(*)28 and (*)6 polymorphisms with irinotecan-induced neutropenia in Thai colorectal cancer patients. Drug Metab Pharmacokinet. 2016;31(1):90–94. doi: 10.1016/j.dmpk.2015.12.004. [DOI] [PubMed] [Google Scholar]
- 17.Deeken JF, Figg WD, Bates SE, Sparreboom A. Toward individualized treatment: prediction of anticancer drug disposition and toxicity with pharmacogenetics. Anticancer Drugs. 2007;18(2):111–126. doi: 10.1097/CAD.0b013e3280109411. [DOI] [PubMed] [Google Scholar]
- 18.Lee W, Lockhart AC, Kim RB, Rothenberg ML. Cancer pharmacogenomics: powerful tools in cancer chemotherapy and drug development. Oncologist. 2005;10(2):104–111. doi: 10.1634/theoncologist.10-2-104. [DOI] [PubMed] [Google Scholar]
- 19.Deeken JF, Slack R, Marshall JL. Irinotecan and uridine diphosphate glucuronosyltransferase 1A1 pharmacogenetics: to test or not to test, that is the question. Cancer. 2008;113(7):1502–1510. doi: 10.1002/cncr.23777. [DOI] [PubMed] [Google Scholar]
- 20.Strassburg CP, Kalthoff S, Ehmer U. Variability and function of family 1 uridine-5′-diphosphate glucuronosyltransferases (UGT1A) Crit Rev Clin Lab Sci. 2008;45(6):485–530. doi: 10.1080/10408360802374624. [DOI] [PubMed] [Google Scholar]
- 21.Shulman K, Cohen I, Barnett-Griness O, et al. Clinical implications of UGT1A1*28 genotype testing in colorectal cancer patients. Cancer. 2011;117(14):3156–3162. doi: 10.1002/cncr.25735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gagne JF, Montminy V, Belanger P, Journault K, Gaucher G, Guillemette C. Common human UGT1A polymorphisms and the altered metabolism of irinotecan active metabolite 7-ethyl-10-hydroxycamptothecin (SN-38) Mol Pharmacol. 2002;62(3):608–617. doi: 10.1124/mol.62.3.608. [DOI] [PubMed] [Google Scholar]
- 23.Wang Y, Shen L, Xu N, et al. UGT1A1 predicts outcome in colorectal cancer treated with irinotecan and fluorouracil. World J Gastroenterol. 2012;18(45):6635–6644. doi: 10.3748/wjg.v18.i45.6635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim TW, Innocenti F. Insights, challenges, and future directions in irinogenetics. Ther Drug Monit. 2007;29(3):265–270. doi: 10.1097/FTD.0b013e318068623b. [DOI] [PubMed] [Google Scholar]
- 25.Hoskins JM, Goldberg RM, Qu P, Ibrahim JG, McLeod HL. UGT1A1*28 genotype and irinotecan-induced neutropenia: dose matters. J Natl Cancer Inst. 2007;99(17):1290–1295. doi: 10.1093/jnci/djm115. [DOI] [PubMed] [Google Scholar]
- 26.Hirasawa A, Zama T, Akahane T, et al. Polymorphisms in the UGT1A1 gene predict adverse effects of irinotecan in the treatment of gynecologic cancer in Japanese patients. J Hum Genet. 2013;58(12):794–798. doi: 10.1038/jhg.2013.105. [DOI] [PubMed] [Google Scholar]
- 27.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 28.Douillard JY, Cunningham D, Roth AD, et al. Irinotecan combined with fluorouracil compared with fluorouracil alone as first-line treatment for metastatic colorectal cancer: a multicentre randomised trial. Lancet. 2000;355(9212):1041–1047. doi: 10.1016/s0140-6736(00)02034-1. [DOI] [PubMed] [Google Scholar]
- 29.Saltz LB, Cox JV, Blanke C, et al. Irinotecan plus fluorouracil and leucovorin for metastatic colorectal cancer. Irinotecan Study Group. N Engl J Med. 2000;343(13):905–914. doi: 10.1056/NEJM200009283431302. [DOI] [PubMed] [Google Scholar]
- 30.McLeod HL, Sargent DJ, Marsh S, et al. Pharmacogenetic predictors of adverse events and response to chemotherapy in metastatic colorectal cancer: results from North American Gastrointestinal Intergroup Trial N9741. J Clin Oncol. 2010;28(20):3227–3233. doi: 10.1200/JCO.2009.21.7943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Glimelius B, Garmo H, Berglund A, et al. Prediction of irinotecan and 5-fluorouracil toxicity and response in patients with advanced colorectal cancer. Pharmacogenomics J. 2011;11(1):61–71. doi: 10.1038/tpj.2010.10. [DOI] [PMC free article] [PubMed] [Google Scholar]