Abstract
The recent identification of distinct genetic and epigenetic features in each glioma entity is leading to a multilayered, integrated diagnostic approach combining histologic features with molecular genetic information. Somatic mutations in isocitrate dehydrogenase (IDH) and receptor tyrosine kinase (RTK) pathways are key oncogenic events in diffuse gliomas, including lower grade (grade II and III) gliomas (LGG) and the highly lethal brain tumor glioblastoma (GBM) respectively, where they reprogram the epigenome, transcriptome and metabolome to drive tumor growth. However, the mechanisms by which these genetic aberrations are translated into the aggressive nature of gliomas through metabolic reprogramming have just begun to be unraveled. The intricate interactions between the oncogenic signaling and cancer metabolism have also been recently demonstrated. Here we describe a set of recent discoveries on cancer metabolism driven by IDH mutation and mutations in RTK pathways, highlighting the integration of genetic mutations, metabolic reprogramming and epigenetic shifts, potentially providing new therapeutic opportunities.
Keywords: genetic-metabolism interaction, IDH, glioma, metabolic reprogramming, RTK, molecular genetics
Introduction – genetic aberrations drive cancer metabolism in gliomas
The identification of distinct genetic and epigenetic profiles in different types of gliomas has revealed novel diagnostic, prognostic, and predictive molecular biomarkers for refining glioma classification [29, 42]. However, the elucidation of how these genetic abnormalities drive glioma pathogenesis is still a work in progress. Metabolic reprogramming is re-emerging as a central hallmark of cancer [16, 39]. Nearly 100 years ago, Otto Warburg demonstrated that cancer cells convert the majority of glucose into lactate even in the presence of sufficient oxygen, and this biochemical adaptation, termed ‘the Warburg effect,’ has once again assumed a central role in framing cancer as a metabolic disease [41, 44]. However, the Warburg effect alone cannot account for the full spectrum of metabolic changes required for tumor growth [22, 46]. Glutaminolysis, the catabolism of glutamine to support tumor cell proliferation, is also a central feature of cancer metabolic reprogramming [18]. Additionally, tumor cells require large amounts of lipid and nucleotides for membrane biogenesis, signal transduction, cell proliferation and potentially as an energy source [1]. The metabolic adaptations that reprogram how cancer cells take up and utilize nutrients to drive tumor growth are activated by profound changes in signaling and epigenetic/transcriptional networks induced by activated oncogenes (e.g. EGFR, RAS, MYC) and deactivated tumor suppressor proteins (e.g. TP53) [13, 46]. In addition, mutations in enzymes that regulate metabolite flux are also implicated in cancer development, as highlighted by the discovery of isocitrate dehydrogenase 1 (IDH1), or less commonly IDH2 gene mutations in more than 70% of diffusely infiltrating World Health Organization (WHO) grade II and grade III astrocytic and oligodendroglial gliomas, as well as in a small fraction of glioblastomas (GBMs), particularly those that develop from lower grade gliomas (LGGs) [3, 38, 51]. Therefore, unraveling the molecular mechanisms by which mutations in the growth factor receptor signaling system and IDH reprogram glioma cell metabolism will shed new light on the contribution of genetic aberrations to the glioma pathogenesis.
Here we review a set of recent discoveries on cancer metabolism involving IDH-mutated LGGs and IDH wild-type GBMs primarily driven by mutations in receptor tyrosine kinase (RTK) pathways. These highlight the integration of genetic aberrations with altered signaling, metabolic reprogramming, and epigenetic changes downstream of common cancer mutations, potentially providing new therapeutic opportunities for these deadly types of brain tumors.
Metabolic reprogramming as a basis for glioma pathogenesis
IDH at the crossroad of genetics, metabolism and epigenetics in gliomas
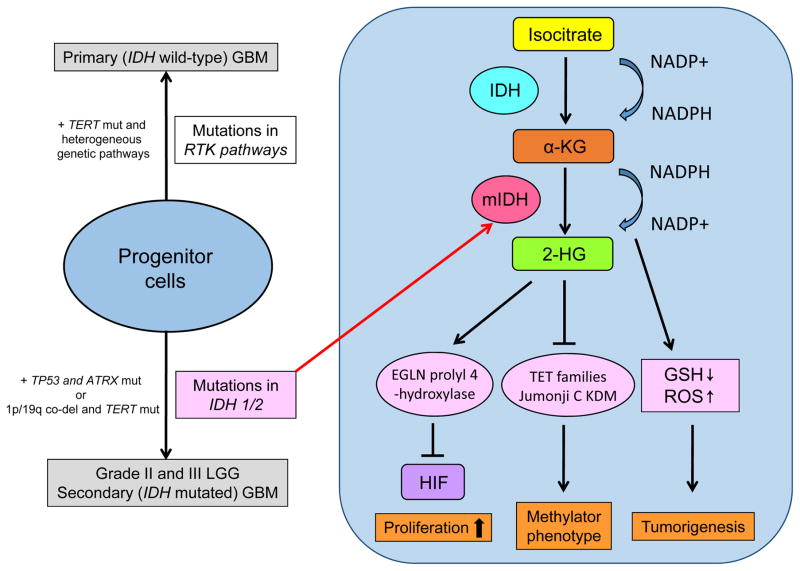
The IDH enzymes normally catalyze the oxidative carboxylation of isocitrate to α-ketoglutarate (α-KG), resulting in the reduction of nicotinamide adenine dinucleotide phosphate (NADP+) to NADPH. The tumorigenic potential of mutant IDH is primarily associated with a metabolic shift in glioma cells (Fig. 1). It has been shown that mutant IDH acquires a neomorphic activity that converts α-KG to D(R)-2-hydroxyglutarate (D-2-HG) in an NADPH-consuming reduction, leading to the intriguing idea that D-2-HG acts as “oncometabolites.” 2-HG in turn inhibits α-KG-dependent dioxygenases [50], eventually altering the genome-wide histone and DNA methylome in gliomas as will be further described. Others reported that increased production of 2-HG stimulates the activity of egl-9 family hypoxia-inducible factor (EGLN) prolyl 4-hydroxylases, which leads to reduced levels of hypoxia-inducible factor (HIF) and enhanced proliferation of human astrocytes [21]. Additionally, IDH mutation decreases intracellular NADPH levels required for the reduction of glutathione disulfide (GSSG) to GSH, thereby causing increased oxidative stress that promotes tumorigenesis but also increases therapy sensitivity [33]. In line with this hypothesis, oxidative stress may promote further genetic changes, such as TP53 mutation or t(1;19) translocation, leading to development of either astrocytoma or oligodendroglioma [53], and IDH mutation is associated with better response to cytotoxic therapy and longer survival in malignant glioma patients [8, 43, 48]. The specific nature of IDH mutation in gliomas may be further exploited for 2-HG-targeting diagnostics [9] and mutant IDH-targeting therapeutics [40].
Figure 1. Metabolic reprogramming in IDH-mutated gliomas.
Mutations in IDH, identified as an early genetic event in grade II/III LGG and secondary GBM, play an important role in gliomas through its neomorphic activity that converts α-KG to an oncometabolite 2-HG. 2-HG stimulates activity of EGLN prolyl 4-hydroxylases enhancing cellular proliferation through the degradation of HIF, and also inhibits α-KG-dependent dioxygenases including Jumonji C histone lysine demethylases (KDMs) and the ten-eleven translocation (TET) family of 5′-methlycytosine hydroxylases leading to methylator phenotypes including G-CIMP and aberrant histone methylation. Additionally, conversion of α-KG to 2-HG is a NADPH-consuming reduction, decreasing intracellular NADPH levels required for the reduction of oxidative stress that promotes tumorigenesis.
mut, mutation; mIDH, mutant form of IDH enzymes; GSH, reduced glutathione; ROS, reactive oxygen species; TERT, telomerase reverse transcriptase; ATRX, alpha thalassemia/mental retardation syndrome X-linked; 1p/19q co-del, chromosomes 1p and 19q co-deletion.
In comparison with IDH-mutated LGGs, primary GBM (IDH wild-type GBM) and IDH wild-type LGGs are characterized by a clinically aggressive behavior with a dismal prognosis [2, 6]. Understanding how IDH wild-type diffuse gliomas promote metabolic reprogramming may yield crucial insights into glioma pathogenesis, and hypoxia may be a key factor to drive cancer metabolism in this type of tumor. Recent studies have revealed that the D-2-HG enantiomer L(S)-2-HG is generated by hypoxia in IDH wild-type tumors and both 2-HG enantiomers have similar structures as α-KG and can competitively inhibit α-KG-dependent enzymes [45]. Further, while glucose provides the acetyl coenzyme A (acetyl-CoA) to support citrate production under normal oxygen tension, tricarboxylic acid (TCA) cycle anaplerosis is maintained primarily by glutamine, and hypoxic cells are able to maintain cell proliferation through wild-type IDH-dependent reductive carboxylation of glutamine-derived α-KG, despite a profound reduction in glucose-dependent citrate production [32, 34]. Interestingly, the increased glutamine-derived α-KG in hypoxia is also associated with a concomitant increased synthesis of 2-HG by wild-type IDH, and the reductive carboxylation of glutamine is part of the metabolic reprogramming associated with HIF [49].
RTK pathway aberration in GBM metabolic reprogramming: therapeutic implication
The emergence of next-generation sequencing technologies provide exquisite sensitivity and resolution. Recent progress in multi-disciplinary molecular analyses of cancers based on novel large-scale DNA methylation profiling and next-generation sequencing approaches have enabled the molecular stratification of GBM by the combination of molecular genetic signatures, as opposed to assessing the status of individual markers [29, 42]. The Cancer Genome Atlas (TCGA) Research Network has been established to generate the comprehensive catalog of genomic abnormalities driving tumorigenesis, and has revealed biologically relevant alterations in three core pathways: RTK/RAS/PI3K signaling, p53 and Rb pathways in GBM [7, 35]. Among these, the genomic characterization of IDH wild-type GBM reveals frequent genetic alterations of key components of the growth factor receptor-PI3K-Akt signaling pathway that activate mechanistic target of rapamycin (mTOR) signaling [7, 10].
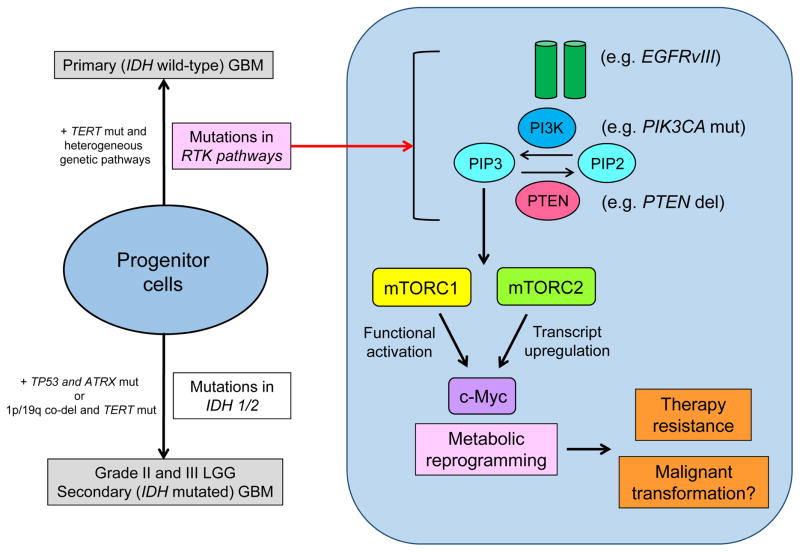
One of the master regulators of cancer metabolism is the oncogenic transcription factor, c-Myc [12]. c-Myc is controlled in a multi-layered way that includes gene rearrangement and amplification [11]. Only recently has it been determined how the mutations in growth factor receptor signaling pathways, such as epidermal growth factor receptor (EGFR) mutations which are most commonly detected in IDH wild-type GBM, cooperate with c-Myc to promote tumorigenesis. This is a critical question since c-Myc is rarely amplified or mutated in GBM [7], despite its potential importance in GBM pathogenesis. Recent studies identify a set of interlacing molecular mechanisms by which EGFRvIII, a constitutively activating mutant form of EGFR, co-opts c-Myc to reprogram cellular metabolism and drive tumor proliferation. This involves the serine/threonine kinase mTOR complexes 1 and 2 (mTORC1 and mTORC2) [4, 30] (Fig. 2). Importantly, failure to inhibit mTOR signaling can render GBM cells resistant to PI3K or Akt targeted therapies by maintaining elevated levels of c-Myc [30]. On the contrary, the convergence of multiple upstream signaling pathways on c-Myc raises the possibility that bromodomain and extraterminal (BET) bromodomain inhibitors that interfere with BET family protein binding to lysine-acetylated histone tails to suppress c-Myc-dependent target gene expression [14], may have a role in solid tumors with PI3K and mTOR activation including GBM. Interestingly, a recent genomic characterization demonstrated that activation of Myc and the RTK-RAS-PI3K pathway are also involved in IDH1-mutant glioma malignant progression [5, 20], and RTK- and Myc-dependent metabolic reprogramming might be involved in this process. Targeted therapies against Myc-dependent metabolism might then be effective for all types of high grade gliomas, regardless of their mutational status of IDH.
Figure 2. Metabolic reprogramming in GBM with mutations in RTK pathways.
Genetic alterations of key components of the growth factor receptor-PI3K-Akt signaling pathway are frequently observed in primary (IDH wild-type) GBM, which eventually activate mTOR signaling. c-Myc, a master regulator of cancer metabolism is transcriptionally and functionally regulated by two distinct mTOR complexes, mTORC1 and mTORC2. This circuit of metabolic shifting causes GBM cell resistance to molecularly targeted therapies by maintaining elevated levels of c-Myc. Interestingly, RTK- and Myc-dependent metabolic reprogramming might be also involved in malignant progression of IDH-mutant gliomas.
del, deletion; EGFRvIII, epidermal growth factor receptor variant III; mut, mutation; PI3K, phosphoinositide 3-kinase; PIK3CA, phosphatidylinositol (4,5)-bisphosphate 3-kinase catalytic subunit alpha; PIP2, phosphatidylinositol (4,5)-bisphosphate; PIP3, phosphatidylinositol (3,4,5)-trisphosphate; PTEN, phosphatase and tensin homolog deleted on chromosome 10.
When genetics and metabolism intersect – the emerging concept
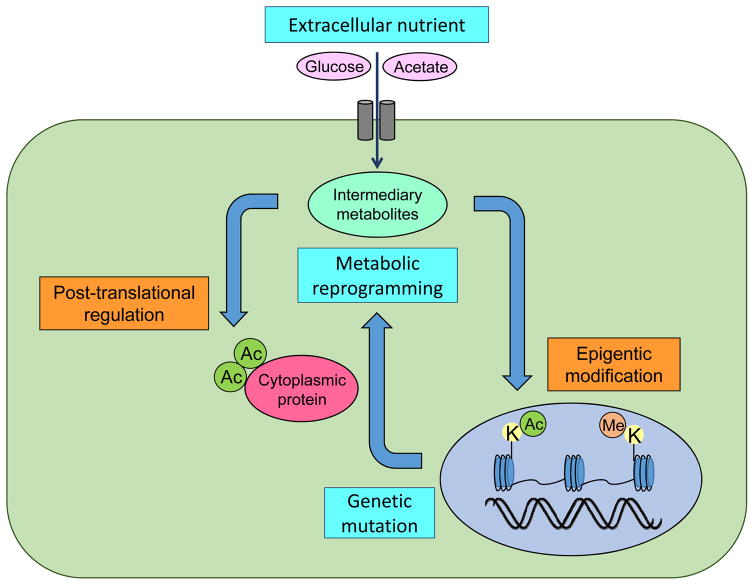
Cancer metabolic reprogramming has been shown to be a consequence of upstream mutations in IDH in LGGs and the RTK signaling network in IDH wild-type GBM. However, do glioma cells also adapt their genetic signaling in response to a shift in metabolism; that is, do genetics and metabolism interplay reciprocally in gliomas? Metabolic reprogramming results in changes in intracellular nutrient levels which can affect oncogenic signaling via control of epigenetics and consequently by globally altering gene transcription [19, 25, 28] (Fig. 3). Mutations in IDH, which were identified as early genetic events in gliomagenesis, play an important role in gliomas through neomorphic activity that converts α-KG to 2-HG as aforementioned, which inhibits α-KG-dependent dioxygenases including histone demethylases and the TET family of 5′-methlycytosine hydroxylases [50]. The presence of IDH mutations thus leads to a distinct subgroup of glioma with a CpG island methylator phenotype (G-CIMP) and aberrant histone methylation [36], which possibly locks differentiation-related genes in an inactive state [26], disrupts chromosomal topology [15] and changes the landscape of enhancers [8], eventually contributing to tumorigenesis. Subsequent discovery of germline or somatic mutations in genes coding for metabolic enzymes (succinate dehydrogenase and fumarate hydratase) that are associated with tumor susceptibility by altering the epigenome further supports the idea of oncometabolites to “tailor the genetics” [37]. On the other hand, in EGFR-mutant GBMs which do not usually possess the mutations in IDH or H3 histone family 3A (H3F3A) to potentially change the epigenetics, constitutive PI3K activation could engage the epigenetic machinery through several complementary routes. First, EGFR activation causes the glycolytic enzyme pyruvate kinase isozymes M2 (PKM2) to translocate to the nucleus where it phosphorylates histone 3 at Thr11, causing dissociation of histone deacetylase 3 (HDAC3) and promote histone acetylation to regulate transcription of the cancer-promoting genes including c-Myc and cyclin D1 [52]. Interestingly, epigenetic regulation of c-Myc may be a central mechanism for its overexpression in GBM [23]. Second, through integrated epigenome and transcriptome analyses, we showed that EGFRvIII remodels an epigenome and transcription factor network that regulates c-Myc, including via mTORC2 which controls GBM metabolism [24].
Figure 3. Reciprocal interaction of genetics, metabolism and environment in gliomas.
Mutations in the cardinal genes for gliomagenesis including IDH and RTK pathway components are the central force to drive metabolic reprogramming in gliomas. By promoting cancer metabolisms, glioma cells avidly take up extracellular nutrients such as glucose and acetate and metabolize them into intermediary metabolites which in turn tailor the genetic signaling by shifting epigenetics as well as post-translationally modifying oncogenic proteins in the cytosol.
Ac, acetyl-group; Me, methyl-group; K, lysine residues.
In addition to the change in intracellular nutrient, we recently made the surprising discovery that exogenous glucose or acetate, two “fuel sources” that are widely available in the brain and readily taken up by tumor cells [27], are required for mutant EGFR signaling. Under EGFR signaling, glucose and acetate activate mTORC2 and promote GBM resistance to molecularly targeted therapies through acetyl-CoA-dependent acetylation of Rictor, a core component of the mTORC2 signaling complex [31]. These results have a number of potentially important and unanticipated implications. First, extracellular nutrients can maintain oncogenic signaling in GBM cells through the post-translational modification of nuclear as well as non-nuclear proteins (Fig. 3), and the environment of GBM patients such as hyperglycemia would affect the therapeutic efficacy including chemotherapeutics [47] and molecularly targeted treatments [31]. Second, these works raise the question of how lifestyle changes, including diet, can potentially shift tumor cell metabolism and promote cancerous growth by altering the genomic and epigenomic landscapes, and there may be more interplay between oncogenic signaling and the environment than previously thought.
Conclusion and future perspective
Cancer is a disease of endogenous somatic mutations. Glioma is no exception, and the traditional phenotypic classification of diffuse gliomas has changed its direction to add genetics based on recent identification of distinct genetic and epigenetic profiles in different types of gliomas. One of the key mechanisms to link the genetic aberrations with the biology of gliomas including epigenetic changes and therapeutics resistance is through cancer metabolic reprogramming. An accumulation of recent evidence suggests that cancer metabolism in IDH-mutated LGGs is potentially linked to the global change in the epigenetic landscape including the shift in the genome-wide methylome through the production of oncometabolites. Metabolism in IDH wild-type GBM is mainly reprogrammed by hypoxia and RTK-dependent c-Myc upregulation to modulate the cellular metabolome and cause resistance to cancer therapeutics. This notion has been further supported by a recent large-scale genomic, epigenomic, transcriptomic and proteomic analysis of diffuse gliomas, demonstrating that methylome, transcriptome and functional copy number variations connect to the status of IDH mutations (i.e. IDH-mutated LGGs) whereas the expression of protein, or metabolome, is strongly related to WHO grading or aggressiveness rather than IDH mutations (i.e. IDH wild-type GBMs) [8]. Tumor development, progression and response to therapy are profoundly influenced by tumor cells’ intracellular metabolism and the exogenous tumor environment. The biochemical environment can shape the behavior of tumor cells in a genotype-specific fashion, potentially by altering the relative fitness of cells bearing a mutation to grow within that metabolic niche and also by directly regulating downstream signaling. Further, the heterogeneity in tumor metabolism and the strong influence of the microenvironment which potentially contribute to the tumor’s utilization of alternative fuels should be taken into consideration [17]. Future studies are needed to determine precisely how chief genetic mutations specific in each glioma entity facilitate cancer metabolic reprogramming and how at the same time extracellular nutrients modulate oncogenic signaling, in order to translate these insights into more effective treatments for glioma patients.
Acknowledgments
This work is supported by National Institute for Neurological Diseases and Stroke Grant NS73831; the Defeat GBM Research Collaborative, a subsidiary of National Brain Tumor Society, National Cancer Institute Grant CA119347; The Ben and Catherine Ivy Foundation; generous donations from the Ziering Family Foundation in memory of Sigi Ziering; and a grant provided by The Novartis Foundation (Japan) for the Promotion of Science, The Cell Science Research Foundation, Grant-in-Aid from the Tokyo Biochemical Research Foundation and JSPS KAKENHI Grant Number 15K19067. The authors declare no conflicts of interest.
References
- 1.Agnihotri S, Zadeh G. Metabolic reprogramming in glioblastoma: the influence of cancer metabolism on epigenetics and unanswered questions. Neuro Oncol. 2016;18:160–172. doi: 10.1093/neuonc/nov125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aldape K, Zadeh G, Mansouri S, Reifenberger G, von Deimling A. Glioblastoma: pathology, molecular mechanisms and markers. Acta Neuropathol. 2015;129:829–848. doi: 10.1007/s00401-015-1432-1. [DOI] [PubMed] [Google Scholar]
- 3.Arita H, Narita Y, Yoshida A, Hashimoto N, Yoshimine T, Ichimura K. IDH1/2 mutation detection in gliomas. Brain Tumor Pathol. 2015;32:79–89. doi: 10.1007/s10014-014-0197-x. [DOI] [PubMed] [Google Scholar]
- 4.Babic I, Anderson ES, Tanaka K, Guo D, Masui K, Li B, Zhu S, Gu Y, Villa GR, Akhavan D, Nathanson D, Gini B, Mareninov S, Li R, Camacho CE, Kurdistani SK, Eskin A, Nelson SF, Yong WH, Cavenee WK, Cloughesy TF, Christofk HR, Black DL, Mischel PS. EGFR mutation-induced alternative splicing of Max contributes to growth of glycolytic tumors in brain cancer. Cell Metab. 2013;17:1000–1008. doi: 10.1016/j.cmet.2013.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bai H, Harmancı AS, Erson-Omay EZ, Li J, Coşkun S, Simon M, Krischek B, Özduman K, Omay SB, Sorensen EA, Turcan Ş, Bakırcığlu M, Carrión-Grant G, Murray PB, Clark VE, Ercan-Sencicek AG, Knight J, Sencar L, Altınok S, Kaulen LD, Gülez B, Timmer M, Schramm J, Mishra-Gorur K, Henegariu O, Moliterno J, Louvi A, Chan TA, Tannheimer SL, Pamir MN, Vortmeyer AO, Bilguvar K, Yasuno K, Günel M. Integrated genomic characterization of IDH1-mutant glioma malignant progression. Nat Genet. 2016;48:59–66. doi: 10.1038/ng.3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brat DJ, Verhaak RG, Aldape KD, Yung WK, Salama SR, Cooper LA, Rheinbay E, Miller CR, Vitucci M, Morozova O, Robertson AG, Noushmehr H, Laird PW, Cherniack AD, Akbani R, Huse JT, Ciriello G, Poisson LM, Barnholtz-Sloan JS, Berger MS, Brennan C, Colen RR, Colman H, Flanders AE, Giannini C, Grifford M, Iavarone A, Jain R, Joseph I, Kim J, Kasaian K, Mikkelsen T, Murray BA, O’Neill BP, Pachter L, Parsons DW, Sougnez C, Sulman EP, Vandenberg SR, Van Meir EG, von Deimling A, Zhang H, Crain D, Lau K, Mallery D, Morris S, Paulauskis J, Penny R, Shelton T, Sherman M, Yena P, Black A, Bowen J, Dicostanzo K, Gastier-Foster J, Leraas KM, Lichtenberg TM, Pierson CR, Ramirez NC, Taylor C, Weaver S, Wise L, Zmuda E, Davidsen T, Demchok JA, Eley G, Ferguson ML, Hutter CM, Mills Shaw KR, Ozenberger BA, Sheth M, Sofia HJ, Tarnuzzer R, Wang Z, Yang L, Zenklusen JC, Ayala B, Baboud J, Chudamani S, Jensen MA, Liu J, Pihl T, Raman R, Wan Y, Wu Y, Ally A, Auman JT, Balasundaram M, Balu S, Baylin SB, Beroukhim R, Bootwalla MS, Bowlby R, Bristow CA, Brooks D, Butterfield Y, Carlsen R, Carter S, Chin L, Chu A, Chuah E, Cibulskis K, Clarke A, Coetzee SG, Dhalla N, Fennell T, Fisher S, Gabriel S, Getz G, Gibbs R, Guin R, Hadjipanayis A, Hayes DN, Hinoue T, Hoadley K, Holt RA, Hoyle AP, Jefferys SR, Jones S, Jones CD, Kucherlapati R, Lai PH, Lander E, Lee S, Lichtenstein L, Ma Y, Maglinte DT, Mahadeshwar HS, Marra MA, Mayo M, Meng S, Meyerson ML, Mieczkowski PA, Moore RA, Mose LE, Mungall AJ, Pantazi A, Parfenov M, Park PJ, Parker JS, Perou CM, Protopopov A, Ren X, Roach J, Sabedot TS, Schein J, Schumacher SE, Seidman JG, Seth S, Shen H, Simons JV, Sipahimalani P, Soloway MG, Song X, Sun H, Tabak B, Tam A, Tan D, Tang J, Thiessen N, Triche T, Van Den Berg DJ, Veluvolu U, Waring S, Weisenberger DJ, Wilkerson MD, Wong T, Wu J, Xi L, Xu AW, Zack TI, Zhang J, Aksoy BA, Arachchi H, Benz C, Bernard B, Carlin D, Cho J, DiCara D, Frazer S, Fuller GN, Gao J, Gehlenborg N, Haussler D, Heiman DI, Iype L, Jacobsen A, Ju Z, Katzman S, Kim H, Knijnenburg T, Kreisberg RB, Lawrence MS, Lee W, Leinonen K, Lin P, Ling S, Liu W, Liu Y, Lu Y, Mills G, Ng S, Noble MS, Paull E, Rao A, Reynolds S, Saksena G, Sanborn Z, Sander C, Schultz N, Senbabaoglu Y, Shen R, Shmulevich I, Sinha R, Stuart J, Sumer SO, Sun Y, Tasman N, Taylor BS, Voet D, Weinhold N, Weinstein JN, Yang D, Yoshihara K, Zheng S, Zhang W, Zou L, Abel T, Sadeghi S, Cohen ML, Eschbacher J, Hattab EM, Raghunathan A, Schniederjan MJ, Aziz D, Barnett G, Barrett W, Bigner DD, Boice L, Brewer C, Calatozzolo C, Campos B, Carlotti CG, Chan TA, Cuppini L, Curley E, Cuzzubbo S, Devine K, DiMeco F, Duell R, Elder JB, Fehrenbach A, Finocchiaro G, Friedman W, Fulop J, Gardner J, Hermes B, Herold-Mende C, Jungk C, Kendler A, Lehman NL, Lipp E, Liu O, Mandt R, McGraw M, Mclendon R, McPherson C, Neder L, Nguyen P, Noss A, Nunziata R, Ostrom QT, Palmer C, Perin A, Pollo B, Potapov A, Potapova O, Rathmell WK, Rotin D, Scarpace L, Schilero C, Senecal K, Shimmel K, Shurkhay V, Sifri S, Singh R, Sloan AE, Smolenski K, Staugaitis SM, Steele R, Thorne L, Tirapelli DP, Unterberg A, Vallurupalli M, Wang Y, Warnick R, Williams F, Wolinsky Y, Bell S, Rosenberg M, Stewart C, Huang F, Grimsby JL, Radenbaugh AJ, Network CGAR. Comprehensive, Integrative Genomic Analysis of Diffuse Lower-Grade Gliomas. N Engl J Med. 2015;372:2481–2498. doi: 10.1056/NEJMoa1402121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brennan CW, Verhaak RG, McKenna A, Campos B, Noushmehr H, Salama SR, Zheng S, Chakravarty D, Sanborn JZ, Berman SH, Beroukhim R, Bernard B, Wu CJ, Genovese G, Shmulevich I, Barnholtz-Sloan J, Zou L, Vegesna R, Shukla SA, Ciriello G, Yung WK, Zhang W, Sougnez C, Mikkelsen T, Aldape K, Bigner DD, Van Meir EG, Prados M, Sloan A, Black KL, Eschbacher J, Finocchiaro G, Friedman W, Andrews DW, Guha A, Iacocca M, O’Neill BP, Foltz G, Myers J, Weisenberger DJ, Penny R, Kucherlapati R, Perou CM, Hayes DN, Gibbs R, Marra M, Mills GB, Lander E, Spellman P, Wilson R, Sander C, Weinstein J, Meyerson M, Gabriel S, Laird PW, Haussler D, Getz G, Chin L, Network TR. The somatic genomic landscape of glioblastoma. Cell. 2013;155:462–477. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ceccarelli M, Barthel FP, Malta TM, Sabedot TS, Salama SR, Murray BA, Morozova O, Newton Y, Radenbaugh A, Pagnotta SM, Anjum S, Wang J, Manyam G, Zoppoli P, Ling S, Rao AA, Grifford M, Cherniack AD, Zhang H, Poisson L, Carlotti CG, Tirapelli DP, Rao A, Mikkelsen T, Lau CC, Yung WK, Rabadan R, Huse J, Brat DJ, Lehman NL, Barnholtz-Sloan JS, Zheng S, Hess K, Rao G, Meyerson M, Beroukhim R, Cooper L, Akbani R, Wrensch M, Haussler D, Aldape KD, Laird PW, Gutmann DH, Noushmehr H, Iavarone A, Verhaak RG, Network TR. Molecular Profiling Reveals Biologically Discrete Subsets and Pathways of Progression in Diffuse Glioma. Cell. 2016;164:550–563. doi: 10.1016/j.cell.2015.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choi C, Ganji SK, DeBerardinis RJ, Hatanpaa KJ, Rakheja D, Kovacs Z, Yang XL, Mashimo T, Raisanen JM, Marin-Valencia I, Pascual JM, Madden CJ, Mickey BE, Malloy CR, Bachoo RM, Maher EA. 2-hydroxyglutarate detection by magnetic resonance spectroscopy in IDH-mutated patients with gliomas. Nat Med. 2012;18:624–629. doi: 10.1038/nm.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ciriello G, Miller ML, Aksoy BA, Senbabaoglu Y, Schultz N, Sander C. Emerging landscape of oncogenic signatures across human cancers. Nat Genet. 2013;45:1127–1133. doi: 10.1038/ng.2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dang CV. MYC on the path to cancer. Cell. 2012;149:22–35. doi: 10.1016/j.cell.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dang CV. MYC, metabolism, cell growth, and tumorigenesis. Cold Spring Harb Perspect Med. 2013:3. doi: 10.1101/cshperspect.a014217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008;7:11–20. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 14.Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, Kastritis E, Gilpatrick T, Paranal RM, Qi J, Chesi M, Schinzel AC, McKeown MR, Heffernan TP, Vakoc CR, Bergsagel PL, Ghobrial IM, Richardson PG, Young RA, Hahn WC, Anderson KC, Kung AL, Bradner JE, Mitsiades CS. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146:904–917. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Flavahan WA, Drier Y, Liau BB, Gillespie SM, Venteicher AS, Stemmer-Rachamimov AO, Suvà ML, Bernstein BE. Insulator dysfunction and oncogene activation in IDH mutant gliomas. Nature. 2016;529:110–114. doi: 10.1038/nature16490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 17.Hensley CT, Faubert B, Yuan Q, Lev-Cohain N, Jin E, Kim J, Jiang L, Ko B, Skelton R, Loudat L, Wodzak M, Klimko C, McMillan E, Butt Y, Ni M, Oliver D, Torrealba J, Malloy CR, Kernstine K, Lenkinski RE, DeBerardinis RJ. Metabolic Heterogeneity in Human Lung Tumors. Cell. 2016;164:681–694. doi: 10.1016/j.cell.2015.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hensley CT, Wasti AT, DeBerardinis RJ. Glutamine and cancer: cell biology, physiology, and clinical opportunities. J Clin Invest. 2013;123:3678–3684. doi: 10.1172/JCI69600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaelin WG, McKnight SL. Influence of metabolism on epigenetics and disease. Cell. 2013;153:56–69. doi: 10.1016/j.cell.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kamoun A, Idbaih A, Dehais C, Elarouci N, Carpentier C, Letouzé E, Colin C, Mokhtari K, Jouvet A, Uro-Coste E, Martin-Duverneuil N, Sanson M, Delattre JY, Figarella-Branger D, de Reyniès A, Ducray F network P. Integrated multi-omics analysis of oligodendroglial tumours identifies three subgroups of 1p/19q co-deleted gliomas. Nat Commun. 2016;7:11263. doi: 10.1038/ncomms11263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koivunen P, Lee S, Duncan CG, Lopez G, Lu G, Ramkissoon S, Losman JA, Joensuu P, Bergmann U, Gross S, Travins J, Weiss S, Looper R, Ligon KL, Verhaak RG, Yan H, Kaelin WG. Transformation by the (R)-enantiomer of 2-hydroxyglutarate linked to EGLN activation. Nature. 2012;483:484–488. doi: 10.1038/nature10898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koppenol WH, Bounds PL, Dang CV. Otto Warburg’s contributions to current concepts of cancer metabolism. Nat Rev Cancer. 2011;11:325–337. doi: 10.1038/nrc3038. [DOI] [PubMed] [Google Scholar]
- 23.Kozono D, Li J, Nitta M, Sampetrean O, Gonda D, Kushwaha DS, Merzon D, Ramakrishnan V, Zhu S, Zhu K, Matsui H, Harismendy O, Hua W, Mao Y, Kwon CH, Saya H, Nakano I, Pizzo DP, VandenBerg SR, Chen CC. Dynamic epigenetic regulation of glioblastoma tumorigenicity through LSD1 modulation of MYC expression. Proc Natl Acad Sci U S A. 2015;112:E4055–4064. doi: 10.1073/pnas.1501967112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu F, Hon GC, Villa GR, Turner KM, Ikegami S, Yang H, Ye Z, Li B, Kuan S, Lee AY, Zanca C, Wei B, Lucey G, Jenkins D, Zhang W, Barr CL, Furnari FB, Cloughesy TF, Yong WH, Gahman TC, Shiau AK, Cavenee WK, Ren B, Mischel PS. EGFR Mutation Promotes Glioblastoma through Epigenome and Transcription Factor Network Remodeling. Mol Cell. 2015;60:307–318. doi: 10.1016/j.molcel.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu C, Thompson CB. Metabolic regulation of epigenetics. Cell Metab. 2012;16:9–17. doi: 10.1016/j.cmet.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu C, Ward PS, Kapoor GS, Rohle D, Turcan S, Abdel-Wahab O, Edwards CR, Khanin R, Figueroa ME, Melnick A, Wellen KE, O’Rourke DM, Berger SL, Chan TA, Levine RL, Mellinghoff IK, Thompson CB. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature. 2012;483:474–478. doi: 10.1038/nature10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mashimo T, Pichumani K, Vemireddy V, Hatanpaa KJ, Singh DK, Sirasanagandla S, Nannepaga S, Piccirillo SG, Kovacs Z, Foong C, Huang Z, Barnett S, Mickey BE, DeBerardinis RJ, Tu BP, Maher EA, Bachoo RM. Acetate is a bioenergetic substrate for human glioblastoma and brain metastases. Cell. 2014;159:1603–1614. doi: 10.1016/j.cell.2014.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Masui K, Cavenee WK, Mischel PS. mTORC2 in the center of cancer metabolic reprogramming. Trends Endocrinol Metab. 2014;25:364–373. doi: 10.1016/j.tem.2014.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Masui K, Mischel PS, Reifenberger G. Molecular classification of gliomas. Handb Clin Neurol. 2016;134:97–120. doi: 10.1016/B978-0-12-802997-8.00006-2. [DOI] [PubMed] [Google Scholar]
- 30.Masui K, Tanaka K, Akhavan D, Babic I, Gini B, Matsutani T, Iwanami A, Liu F, Villa GR, Gu Y, Campos C, Zhu S, Yang H, Yong WH, Cloughesy TF, Mellinghoff IK, Cavenee WK, Shaw RJ, Mischel PS. mTOR complex 2 controls glycolytic metabolism in glioblastoma through FoxO acetylation and upregulation of c-Myc. Cell Metab. 2013;18:726–739. doi: 10.1016/j.cmet.2013.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Masui K, Tanaka K, Ikegami S, Villa GR, Yang H, Yong WH, Cloughesy TF, Yamagata K, Arai N, Cavenee WK, Mischel PS. Glucose-dependent acetylation of Rictor promotes targeted cancer therapy resistance. Proc Natl Acad Sci U S A. 2015;112:9406–9411. doi: 10.1073/pnas.1511759112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Metallo CM, Gameiro PA, Bell EL, Mattaini KR, Yang J, Hiller K, Jewell CM, Johnson ZR, Irvine DJ, Guarente L, Kelleher JK, Vander Heiden MG, Iliopoulos O, Stephanopoulos G. Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature. 2012;481:380–384. doi: 10.1038/nature10602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Molenaar RJ, Radivoyevitch T, Maciejewski JP, van Noorden CJ, Bleeker FE. The driver and passenger effects of isocitrate dehydrogenase 1 and 2 mutations in oncogenesis and survival prolongation. Biochim Biophys Acta. 2014;1846:326–341. doi: 10.1016/j.bbcan.2014.05.004. [DOI] [PubMed] [Google Scholar]
- 34.Mullen AR, Wheaton WW, Jin ES, Chen PH, Sullivan LB, Cheng T, Yang Y, Linehan WM, Chandel NS, DeBerardinis RJ. Reductive carboxylation supports growth in tumour cells with defective mitochondria. Nature. 2012;481:385–388. doi: 10.1038/nature10642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Network CGAR. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Noushmehr H, Weisenberger DJ, Diefes K, Phillips HS, Pujara K, Berman BP, Pan F, Pelloski CE, Sulman EP, Bhat KP, Verhaak RG, Hoadley KA, Hayes DN, Perou CM, Schmidt HK, Ding L, Wilson RK, Van Den Berg D, Shen H, Bengtsson H, Neuvial P, Cope LM, Buckley J, Herman JG, Baylin SB, Laird PW, Aldape K, Network CGAR. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17:510–522. doi: 10.1016/j.ccr.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nowicki S, Gottlieb E. Oncometabolites: tailoring our genes. FEBS J. 2015;282:2796–2805. doi: 10.1111/febs.13295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Siu IM, Gallia GL, Olivi A, McLendon R, Rasheed BA, Keir S, Nikolskaya T, Nikolsky Y, Busam DA, Tekleab H, Diaz LA, Hartigan J, Smith DR, Strausberg RL, Marie SK, Shinjo SM, Yan H, Riggins GJ, Bigner DD, Karchin R, Papadopoulos N, Parmigiani G, Vogelstein B, Velculescu VE, Kinzler KW. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pavlova NN, Thompson CB. The Emerging Hallmarks of Cancer Metabolism. Cell Metab. 2016;23:27–47. doi: 10.1016/j.cmet.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rohle D, Popovici-Muller J, Palaskas N, Turcan S, Grommes C, Campos C, Tsoi J, Clark O, Oldrini B, Komisopoulou E, Kunii K, Pedraza A, Schalm S, Silverman L, Miller A, Wang F, Yang H, Chen Y, Kernytsky A, Rosenblum MK, Liu W, Biller SA, Su SM, Brennan CW, Chan TA, Graeber TG, Yen KE, Mellinghoff IK. An inhibitor of mutant IDH1 delays growth and promotes differentiation of glioma cells. Science. 2013;340:626–630. doi: 10.1126/science.1236062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Seyfried TN, Flores RE, Poff AM, D’Agostino DP. Cancer as a metabolic disease: implications for novel therapeutics. Carcinogenesis. 2014;35:515–527. doi: 10.1093/carcin/bgt480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sturm D, Witt H, Hovestadt V, Khuong-Quang DA, Jones DT, Konermann C, Pfaff E, Tönjes M, Sill M, Bender S, Kool M, Zapatka M, Becker N, Zucknick M, Hielscher T, Liu XY, Fontebasso AM, Ryzhova M, Albrecht S, Jacob K, Wolter M, Ebinger M, Schuhmann MU, van Meter T, Frühwald MC, Hauch H, Pekrun A, Radlwimmer B, Niehues T, von Komorowski G, Dürken M, Kulozik AE, Madden J, Donson A, Foreman NK, Drissi R, Fouladi M, Scheurlen W, von Deimling A, Monoranu C, Roggendorf W, Herold-Mende C, Unterberg A, Kramm CM, Felsberg J, Hartmann C, Wiestler B, Wick W, Milde T, Witt O, Lindroth AM, Schwartzentruber J, Faury D, Fleming A, Zakrzewska M, Liberski PP, Zakrzewski K, Hauser P, Garami M, Klekner A, Bognar L, Morrissy S, Cavalli F, Taylor MD, van Sluis P, Koster J, Versteeg R, Volckmann R, Mikkelsen T, Aldape K, Reifenberger G, Collins VP, Majewski J, Korshunov A, Lichter P, Plass C, Jabado N, Pfister SM. Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer Cell. 2012;22:425–437. doi: 10.1016/j.ccr.2012.08.024. [DOI] [PubMed] [Google Scholar]
- 43.Takano S, Ishikawa E, Sakamoto N, Matsuda M, Akutsu H, Noguchi M, Kato Y, Yamamoto T, Matsumura A. Immunohistochemistry on IDH 1/2, ATRX, p53 and Ki-67 substitute molecular genetic testing and predict patient prognosis in grade III adult diffuse gliomas. Brain Tumor Pathol. 2016;33:107–116. doi: 10.1007/s10014-016-0260-x. [DOI] [PubMed] [Google Scholar]
- 44.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wahl DR, Venneti S. 2-Hydoxyglutarate: D/Riving Pathology in gLiomaS. Brain Pathol. 2015;25:760–768. doi: 10.1111/bpa.12309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ward PS, Thompson CB. Metabolic reprogramming: a cancer hallmark even warburg did not anticipate. Cancer Cell. 2012;21:297–308. doi: 10.1016/j.ccr.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weiler M, Blaes J, Pusch S, Sahm F, Czabanka M, Luger S, Bunse L, Solecki G, Eichwald V, Jugold M, Hodecker S, Osswald M, Meisner C, Hielscher T, Rübmann P, Pfenning PN, Ronellenfitsch M, Kempf T, Schnölzer M, Abdollahi A, Lang F, Bendszus M, von Deimling A, Winkler F, Weller M, Vajkoczy P, Platten M, Wick W. mTOR target NDRG1 confers MGMT-dependent resistance to alkylating chemotherapy. Proc Natl Acad Sci U S A. 2014;111:409–414. doi: 10.1073/pnas.1314469111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weller M, Weber RG, Willscher E, Riehmer V, Hentschel B, Kreuz M, Felsberg J, Beyer U, Löffler-Wirth H, Kaulich K, Steinbach JP, Hartmann C, Gramatzki D, Schramm J, Westphal M, Schackert G, Simon M, Martens T, Boström J, Hagel C, Sabel M, Krex D, Tonn JC, Wick W, Noell S, Schlegel U, Radlwimmer B, Pietsch T, Loeffler M, von Deimling A, Binder H, Reifenberger G. Molecular classification of diffuse cerebral WHO grade II/III gliomas using genome- and transcriptome-wide profiling improves stratification of prognostically distinct patient groups. Acta Neuropathol. 2015;129:679–693. doi: 10.1007/s00401-015-1409-0. [DOI] [PubMed] [Google Scholar]
- 49.Wise DR, Ward PS, Shay JE, Cross JR, Gruber JJ, Sachdeva UM, Platt JM, DeMatteo RG, Simon MC, Thompson CB. Hypoxia promotes isocitrate dehydrogenase-dependent carboxylation of α-ketoglutarate to citrate to support cell growth and viability. Proc Natl Acad Sci U S A. 2011;108:19611–19616. doi: 10.1073/pnas.1117773108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xu W, Yang H, Liu Y, Yang Y, Wang P, Kim SH, Ito S, Yang C, Xiao MT, Liu LX, Jiang WQ, Liu J, Zhang JY, Wang B, Frye S, Zhang Y, Xu YH, Lei QY, Guan KL, Zhao SM, Xiong Y. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of α-ketoglutarate-dependent dioxygenases. Cancer Cell. 2011;19:17–30. doi: 10.1016/j.ccr.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, Kos I, Batinic-Haberle I, Jones S, Riggins GJ, Friedman H, Friedman A, Reardon D, Herndon J, Kinzler KW, Velculescu VE, Vogelstein B, Bigner DD. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360:765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang W, Xia Y, Hawke D, Li X, Liang J, Xing D, Aldape K, Hunter T, Alfred Yung WK, Lu Z. PKM2 phosphorylates histone H3 and promotes gene transcription and tumorigenesis. Cell. 2012;150:685–696. doi: 10.1016/j.cell.2012.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang C, Moore LM, Li X, Yung WK, Zhang W. IDH1/2 mutations target a key hallmark of cancer by deregulating cellular metabolism in glioma. Neuro Oncol. 2013;15:1114–1126. doi: 10.1093/neuonc/not087. [DOI] [PMC free article] [PubMed] [Google Scholar]