Abstract
To maintain homeostasis in the face of intrinsic and extrinsic insults, cells have evolved elaborate quality control networks to resolve damage at multiple levels. Interorganellar communication is a key requirement for this maintenance, however the underlying mechanisms of this communication have remained an enigma. Here we integrate the outcome of transcriptomic, proteomic, and metabolomics analyses of genotypes including ceh1, a mutant with constitutively elevated levels of both the stress-specific plastidial retrograde signaling metabolite methylerythritol cyclodiphosphate (MEcPP) and the defense hormone salicylic acid (SA), as well as the high MEcPP but SA deficient genotype ceh1/eds16, along with corresponding controls. Integration of multi-omic analyses enabled us to delineate the function of MEcPP from SA, and expose the compartmentalized role of this retrograde signaling metabolite in induction of distinct but interdependent signaling cascades instrumental in adaptive responses. Specifically, here we identify strata of MEcPP-sensitive stress response cascades, among which we focus on selected pathways including organelle-specific regulation of jasmonate biosynthesis; simultaneous induction of synthesis and breakdown of SA; and MEcPP-mediated alteration of cellular redox status in particular glutathione redox balance.
Collectively, these integrated multi-omic analyses provided a vehicle to gain an in-depth knowledge of genome-metabolism interactions, and to further probe the extent of these interactions and delineate their functional contributions. Through this approach we were able to pinpoint stress-mediated transcriptional and metabolic signatures and identify the downstream processes modulated by the independent or overlapping functions of MEcPP and SA in adaptive responses.
Keywords: multi-omics, methylerythritol cyclodiphosphate (MEcPP), plastidial retrograde signaling, salicylic acid (SA), glutathione redox status, jasmonate biosynthesis, metabolic signature of stress, Arabidopsis thaliana
Introduction
To maintain cellular homeostasis in the face of frequent changes in their internal and external environment, plants must delicately balance and fine-tune adaptive responses at the tissue, cellular, and organellar levels. To achieve this homeostasis, tight regulation of a variety of cellular processes, ranging from changes in nuclear gene expression to alterations in mRNA or protein stability is required. Of relevance to these adaptive processes, inter-organellar communication via signals such as the general-stress-induced plastidial retrograde metabolite 2-C-methyl-D-erythritol-2,4-cyclodiphosphate (MEcPP) have a key function in coordinating stress responses (Xiao et al., 2012).
MEcPP is a bifunctional chemical entity that serves both as a metabolic intermediate of the methyl erythritol phosphate (MEP) pathway responsible for production of isoprenoid precursors, and as a signaling metabolite. Constitutively expressing HYDROPEROXIDE LYASE 1 (ceh1) mutant plants were found to accumulate high levels of MEcPP, which in turn results in a constitutive production of the otherwise defense-associated hormone salicylic acid (SA) (Xiao et al., 2012). Generation of a double mutant containing high MEcPP and negligible levels of SA via introducing the SA defective mutant eds16 (mutant in ISOCHORISMATE SYNTHASE1, also known as sid2) into the ceh1 background unraveled distinct signaling roles of MEcPP (Xiao et al., 2012). These specific roles include induction of constitutive expression of one branch of jasmonic acid (JA) responsive genes (Lemos et al., 2016), a constitutive general stress response (Benn et al., 2016) (GSR), and induction of gene expression of a branch of the unfolded protein response (UPR) in the endoplasmic reticulum (Walley et al., 2015). Collectively, these findings identify MEcPP as an interorganellar communication signal, with an as-yet-unknown mechanism of action.
As an initial step to untangle the role and mechanism of MEcPP action, we characterized the ceh1 mutant using microarrays (Walley et al., 2015), followed by expanding these data through RNA-seq on ceh1, ceh1/eds16, eds16, and parent lines. Subsequently, we performed proteomic analyses on these genotypes to address the well-established regulatory function of translation in stress responses (Mustroph et al., 2009; Zanetti et al., 2005; Juntawong et al., 2014; Walley et al., 2015). We finally extended the analyses to metabolomics to gain insight into the metabolic status of these lines. The integration of transcriptomics, proteomics, and metabolomics data resulted in the most comprehensive analyses of the function of plastidial retrograde signaling, while circumventing reported deficiencies of the individual approaches (Wanichthanarak et al., 2015; Rajasundaram and Selbig, 2016; Larsen et al., 2015; Rajasundaram et al., 2014; Hyun et al., 2014; Zeng et al., 2013; Hirai et al., 2004; Wilson et al., 2015; Vijayakumar et al., 2016).
Collectively, the integration of three omics outputs delineates the role of MEcPP from SA in regulation of adaptive responses, and reveals a MEcPP-mediated function in controlling SA production and decay, organelle-specific regulation of JA biosynthesis, as well as enhanced oxidation in the pool of glutathione.
Results and Discussion
Gene Set Enrichment Analysis of Transcriptome and Proteome Data
To determine the nature of metabolic disruptions in the ceh1 mutant, we undertook gene set enrichment analysis (GSEA) to identify metabolic pathways/processes with altered transcription/translation. To evaluate the differences in both transcription and translation, and to differentiate which of these are due directly to MEcPP vs. the elevated SA in ceh1, we conducted GSEA analyses on both proteomics and transcriptomics data, comparing each of ceh1, ceh1/eds16, and eds16 to parental controls. Transcriptomics data detected more genes (7884, 551 of which were differentially regulated in at least one genotype) than proteomics (4192 proteins of which 35 were differentially regulated in at least one genotype) with a greater number differentially regulated, indicative of the greater sensitivity of this technique. These analyses revealed a number of robustly altered pathways (Table S1, gene lists of relevant pathways in Table S2). Few pathways were significantly modified in eds16 relative to parent, so for simplicity induced and suppressed pathways at the RNA and protein levels in only ceh1 and ceh1/eds16 are shown in Figure 1.
Figure 1. Differentially regulated pathways in ceh1 and ceh1/eds16 relative to parent line.
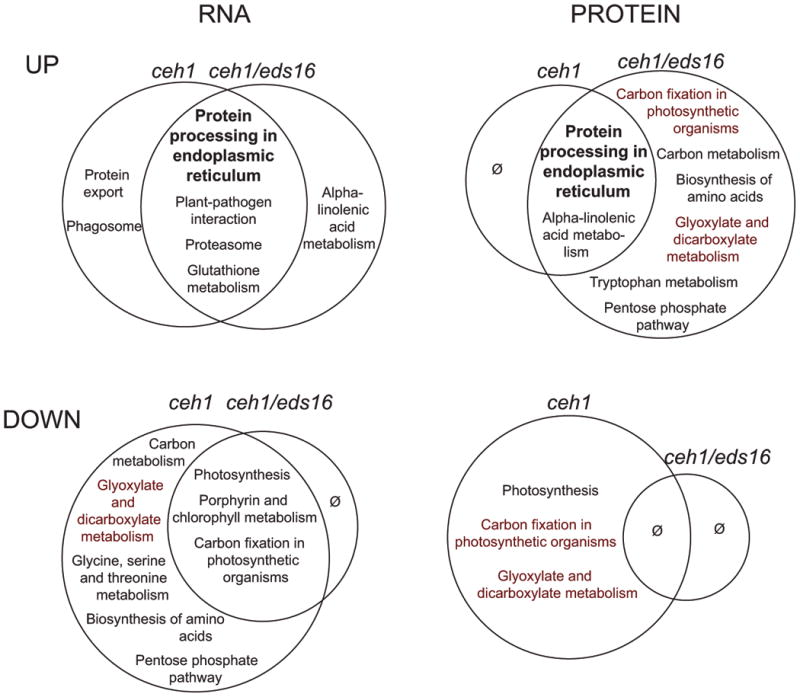
Depicted modulated pathways that are up (upper row) or down (lower row) regulated in ceh1 (left side of Venn diagrams) and ceh1/eds16 (right side of Venn diagrams) were identified. For RNAseq data (left column) pathways with an FDR-corrected p value of less than 0.05 are shown. For proteomics data, due to lower dynamic range, uncorrected p values were used as a less reliable indicator of significance, particularly when in concordance with transcriptomic data. Pathways in bold are those similarly regulated at RNA and protein level in both displayed genotypes, while pathways in red are inversely regulated between ceh1 and ceh1/eds16. Ø indicates no significantly differentially regulated pathways.
Among the upregulated pathways are several which are similarly altered in ceh1 and ceh1/eds16, indicating that they are changed primarily by MEcPP, with little or no input of SA. Of particular note is “Protein processing in the endoplasmic reticulum,” which is upregulated in both ceh1 and ceh1/eds16 at both the RNA and protein level. In addition to this previously reported role of MEcPP in induction of the unfolded protein response in the ER of ceh1 (Walley et al., 2015), using GSEA analyses on transcriptomics data enabled us to identify “Protein export” and “Phagosome” as differentially modified pathways in ceh1 and “Proteasome” in both ceh1 and ceh1/eds16, thereby extending the portfolio of disrupted protein folding and stability in response to MEcPP and associated SA accumulation. Other upregulated pathways identified only in transcriptomics data include “Plant-pathogen interaction” and “Glutathione metabolismThis may suggest translational or post-translational suppression of these pathways, or merely be the consequence of an intrinsically different detection limit between the two ‘omics approaches – indeed more than half of the proteins associated with these pathways were not detected in any genotype (Walley et al., 2015).
Among suppressed pathways are “Photosynthesis” and “Carbon fixation in photosynthetic organisms,” downregulated in both ceh1 and ceh1/eds16 at the RNA level, indicating a role of MEcPP in repressing these pathways independently of SA. In general, these disruptions in the ceh1 mutant may be considered more likely to be due to specific signaling by MEcPP than perturbation of the MEP pathway, which theoretically is believed to be redundant with the cytosolic mevalonate pathway. In plants, a large proportion of the produced isoprenoids participate in photosynthetic processes and only a minor of the isoprenoid flux contribute to the synthesis of hormones (Vranová et al., 2012; Vranová et al., 2013). The light activation of the MEP-pathways genes as opposed to light-inhibition of the MVA pathway genes, concomitant with increased synthesis of isoprenoid-derived metabolites such as phytyl chains and carotenoids after illumination is a prime example of distinct function of the two isoprenoid biosynthesis pathways (Vranová et al., 2013; Ghassemian et al., 2006; Rodríguez-Concepción, 2006). As such, the disruption of photosynthesis-related pathways in ceh1 may in part be due to perturbation of the MEP pathway and by extension chlorophyll biosynthesis, as represented by the downregulation of “Porphyrin and chlorophyll metabolism” in both ceh1 and ceh1/eds16 (Figure 1). In contrast to RNA levels, protein levels of “Photosynthesis” and “Carbon fixation in photosynthetic organisms” in ceh1/eds16 as compared to ceh1 are unaffected and enhanced, respectively. This is in line with published results that SA signaling affects protein translation or stability of these pathways (Rivas-San Vicente and Plasencia, 2011).
Alpha-linolenic acid metabolism, which comprises mostly JA biosynthesis, is among those pathways notably affected by both MEcPP and SA. This pathway is upregulated in both ceh1 and ceh1/eds16 at the protein level, but only ceh1/eds16 at the RNA level. These data support previous observations that while SA exerts an inhibitory function, MEcPP induces jasmonate biosynthesis and responses (Lemos et al., 2016). Finally, a number of pathways upregulated in ceh1/eds16 specifically at the protein level are, conversely, downregulated in ceh1 at the protein and RNA level, highlighted in red in Figure 1. These pathways are largely associated with central metabolism, and are likely modulated by SA levels, known to inhibit both photosynthesis and respiration via both transcriptional and post-translational mechanisms (Rivas-San Vicente and Plasencia, 2011).
Independent analyses of transcriptomic and proteomic data allowed identification of lowly-expressed pathways utilizing RNA-seq while leveraging proteomic data to identify specific protein level modulation and increase confidence in transcriptomic data (Table S1 and S2). In summary we identified several classes of differentially regulated pathways: those affected a) solely by MEcPP, such as protein stability; b) primarily by MEcPP with partial SA contribution, including stress signaling pathways such as jasmonate biosynthesis, pathogen response and glutathione metabolism; and c) predominantly by SA with some contribution of MEcPP, primarily related to central metabolism.
Selected metabolic disruptions caused by HDS mutation
The substantial alteration of metabolic pathways identified by transcriptomic and proteomic analyses prompted us to extend our studies to metabolomics, by first assaying a panel of 90 small molecules primarily associated with central carbon metabolism in all the genotypes, of which 82 accumulated differentially among the four tested genotypes (Table S3). A subset of these results is shown in Figure 2, with the remaining metabolites' information in Figure S1. As a confirmatory analysis, we initially examined intermediates of the MEP pathway, finding the expected accumulation of MEcPP upstream of HDS in both ceh1 and ceh1/eds16. In contrast with previously published results (Gonzalez-Cabanelas et al., 2015), we found similar over-accumulation of earlier pathway intermediates, up to and including MEP (Figure 2A). Of note, dimethylallyl pyrophosphate (DMAPP) is not reduced in lines with mutated HDS, reflective of function of the redundant mevalonate pathway in producing this metabolite. We did not detect significant changes in mevalonate pathway genes/proteins, but this does not rule out post-translational regulation of this pathway.
Figure 2. Selected metabolite sets accumulating differentially among ceh1, ceh1/eds16, and eds16.
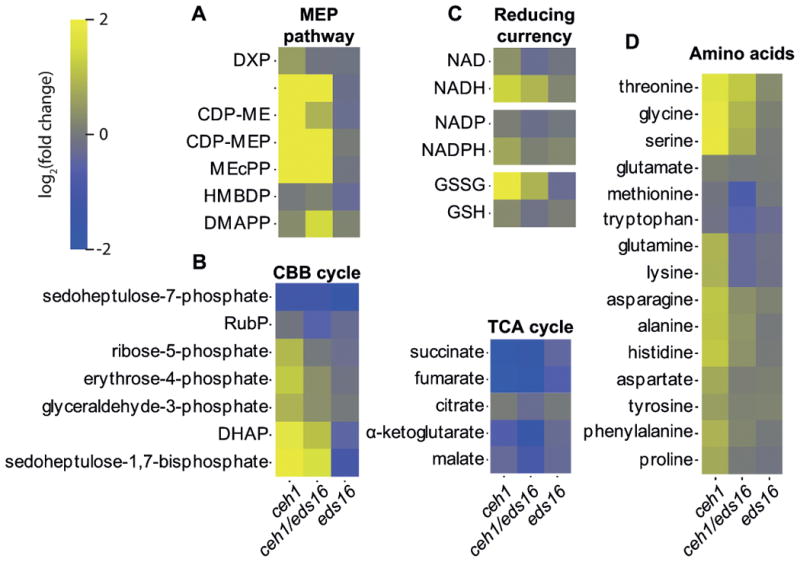
(A) MEP pathway intermediates, (B) central metabolism related Calvin Benson Bassham (CBB) and tricarboxylic acid (TCA) cycle metabolites, (C) Reducing currencies and (D) amino acids differentially accumulate in the displayed genotypes relative to parent lines. Shading represents log2-fold change for each metabolite (rows) relative to wild type for each genotype (columns), from -2 (darkest) to 2 (lightest). Abbreviations: DXP: Deoxy-xylulose phosphate, MEP: methyl-erythritol phosphate, CDP-ME: diphosphocytidyl-methylerythritol, CDP-MEP: diphosphocytidyl-methylerythritol phosphate, MEcPP: methyl-erythritol cyclodiphosphate, HMBDP: hydroxymethylbutenyl pyrophosphate, DMAPP: dimethylallyl pyrophosphate, RubP: ribulose 1,5 bisphosphate, DHAP: dihydroxyacetone phosphate, NAD: nicotinamide adenine dinucleotide, NADP: nicotinamide adenine dinucleotide phosphate.
Additional notable disruptions to central metabolism were in the Calvin-Benson-Bassham (CBB) cycle and the tricarboxylic acid (TCA) cycle, with an overall buildup of CBB intermediates and depletion of TCA cycle intermediates in ceh1 and ceh1/eds16 (Figure 2B), consistent with a potential block in the CBB cycle, and reduced sugar availability (Rojas-González et al., 2015). More information about this block may be inferred by the fact that while sedoheptulose 1,7-biosphosphate accumulates in ceh1 and ceh1/eds16, sedoheptulose 7-phosphate is depleted relative to WT levels. The gene encoding sedoheptulose 1,7 bisphosphatase, which mediates this conversion, is not only downregulated in ceh1 and ceh1/eds16 at the transcript and protein level (Table S4, Walley et al., 2015), it is also post-translationally regulated by chloroplast conditions including pH, Mg2+, feedback inhibition, and redox status (Raines et al., 1999). Thus the evident lack of sedoheptulose 1,7 bisphosphatase activity may suggest differences in the chloroplastic environment of MEcPP-overaccumulating lines. Intriguingly, the stronger modulation of a number of CBB intermediates in ceh1, as compared to ceh1/eds16, clearly differentiate input of SA from MEcPP in the process (Figure 2B), corroborating the observation that pathway “Carbon fixation in photosynthetic organisms” is induced in ceh1/eds16 and conversely suppressed in ceh1 (Figure 1). However, in the TCA cycle, most metabolites are similarly disrupted in ceh1/eds16 as in ceh1 alone, suggesting some effect on these pathways either of the MEcPP signal or the disruption of the MEP pathway, but independently of SA accumulation.
Another noted trend is the redox status of the various reducing currencies of the cell (Figure 2C). While NAD and NADP are present at lower levels, their reduced forms NADH and NADPH are present at greater levels in ceh1 and ceh1/eds16. In contrast, the existing glutathione pool is more oxidized, with more GSSG and less GSH in ceh1 and ceh1/eds16. Collectively, these data suggest altered redox potential in response to accumulation of MEcPP.
Finally, a number of assayed amino acids are differentially accumulated in ceh1 and ceh1/eds16 (Figure 2D). Specifically, the levels of over half of the amino acids show a strong SA-dependent trend (Figure 2D), present at greater levels in ceh1 than in ceh1/eds16. This corroborates the previous finding that SA increases free amino acid concentrations in algae (Kovacik et al., 2010), perhaps via effects on protein synthesis and/or breakdown. MEcPP also influences amino acid levels independently of SA; a subset of amino acids, including tryptophan and methionine, are reduced in both ceh1 and ceh1/eds16. Of these tryptophan is of note since the tryptophan breakdown pathway “Tryptophan metabolism” is upregulated in ceh1/eds16 at the protein level, reflecting dampening effects of SA (Figure 1). Additionally, constitutively high SA in ceh1 may further contribute to sequestration/reduction ofthe pool of chorismate, a precursor shared by SA and tryptophan biosynthetic pathways. These processes combined present an example of how transcriptomic/proteomic data may inform metabolomics data analyses. Finally a subset of assayed amino acids, including glycine and serine, is present at greater levels in both ceh1 and ceh1/eds16, but with stronger enrichment in ceh1 than ceh1/eds16. Notably, these amino acids play an important role in the rescue of Rubisco oxygenase activity, photorespiration. Photorespiration is not represented directly by a unique KEGG pathway, but is a part of the pathways “Glycine, serine, and threonine metabolism,” and “Glyoxylate and decarboxylate metabolism,” both of which are downregulated in ceh1 by the combined input from SA and MEcPP (Figure 1).
Collectively, the integration of transcriptomics, proteomics and metabolomics analyses enabled us to confirm expected results, as in MEcPP accumulation, to suggest underlying mechanisms, as in tryptophan depletion, and to identify areas for further investigation, as in glutathione redox balance and potential impact on photorespiration.
MEcPP regulation of salicylic acid biosynthesis and metabolism
High SA levels in the ceh1 mutant (Xiao et al., 2012), prompted us to construct a “SA biosynthesis and metabolism” pathway using Pathvisio (van Iersel et al., 2008; Kutmon et al., 2015). This was used to visualize ceh1 and ceh1/eds16 transcript, protein, and metabolite levels relative to control (Figure 3), and the data from eds16 are shown as a separate Pathvisio output (Figure S2).
Figure 3. Salicylic acid biosynthesis and degradation is regulated by MEcPP.
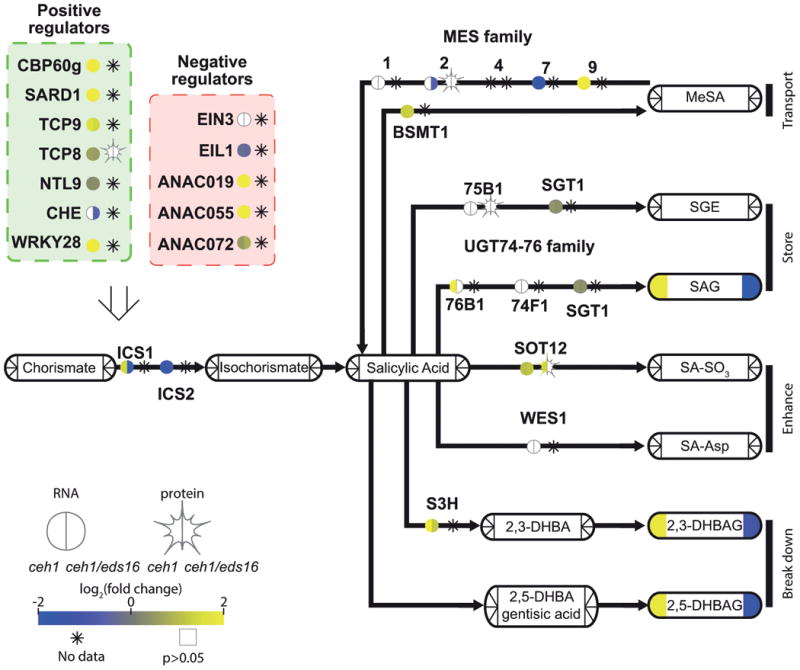
A pathway schematic for SA biosynthesis and breakdown, with transcription factors/enzymes shown by bare text with a circle, representing RNA, and star, representing protein levels. Metabolites are shown by rounded rectangles. For metabolites, RNA, and protein, the log2-fold change for ceh1 vs parent (left) and ceh1/eds16 vs. parent (right) is represented by shading, from -2 (darkest) to 2 (lightest). Non-detectable products in each section are shown by an asterisk, while empty symbols with grey borders represent non-significant fold changes relative to parent. Arrows represent chemical conversions, catalyzed by the enzymes shown on the line. The open arrow from transcription factors to ICS1 represents direct transcriptional regulation of this enzyme by these transcription factors. Abbreviations: CBP60g: calmodulin-binding protein 60g, SARD1: systemic acquired resistance deficient 1, TCP8/9: teosinte branched 1, cycloidea, PCF domain containing 8/9, NTL9: NTM1-like 9, CHE: CCA1 hiking expedition, EIN3: ethylene-insensitive 3, EIL1: EIN3-like1, ANAC019,055,072: Arabidopsis Nam/ATAF1,CUC2-domain containing TF 19/55/72, ICS1/2: isochorismate synthase ½, MES: methyl esterase, BSMT: SA methyl transferase, MeSA: methyl-salicylic acid, UGT: UDP-glucose transferase, SGT: SA glucose transferase, SGE: salicylate glucose ester, SAG: salicylic acid glucoside, SOT12: sulphotransferase 12, SA-SO3: sulphosalicylic acid, WES1: =GH3.5 WESO1, S3H: salicylate-3-hydroxylase, DHBA: dihydroxybenzoic acid, DHBAG: DHBA glucoside
The data confirmed the previously reported enhanced levels of ICS1 expression in ceh1 (Xiao et al., 2012). To better understand mechanism(s) involved in this regulation, we assayed transcript and protein levels of known regulators of ICS1. Direct transcriptional regulation of this gene has only recently begun to be elucidated (Chen et al., 2009; Zhang et al., 2010; Wang et al., 2015; Zheng et al., 2012; Van Verk et al., 2011; Zheng et al., 2015), and the interactions or hierarchy of ICS1-binding transcription factors remains to be discovered. In ceh1, the majority of published positive regulators of ICS1, namely CBP60g, SARD1, TCP8 and 9, NTL9, CHE and WRKY28 are themselves upregulated, suggesting some upstream mechanisms of MEcPP-mediated induction of ICS1. Intriguingly, the 3 NAC domain transcription factors (ANAC019, 055 and 072) known to suppress accumulation of SA and mediate coronatine (JA-Ile mimic)-induced re-opening of stomata are also upregulated (Zheng et al., 2012). This suggests priority of one or more of the ICS1 inducers over these TFs, with implications for robustness of SA production.
Upregulation of ICS1 in ceh1 has been shown to result in increased levels of total SA, as measured via gas chromatography/mass spectrometry (Xiao et al., 2012). Fine-tuned analyses using liquid chromatography/mass spectrometry determined that the majority of the total SA previously measured in ceh1 is in fact conjugated SA, namely, SA glucoside (SAG, Figure 3 and Figure S2). This is consistent with previous reports showing that total SA is largely present as SAG (Vlot et al., 2009), a more soluble derivative of SA facilitating vacuolar storage (Hennig et al., 2002; Klessig and Malamy, 1994; Seo et al., 1995). This led us to examine other known SA derivatives and their respective modifying enzymes in MEcPP-overaccumulating backgrounds, including transport form methyl SA (MeSA)(Park et al., 2007), SAG and salicylate glucose ester (SGE) as SA storage forms (Song et al., 2008; Dean and Delaney, 2008), the less well understood potential SA enhancers SA-SO3 (Baek et al., 2010) and SA-Asp (Zhang et al., 2007), as well as the SA breakdown products dihydroxy benzoic acids (DHBA)(Bartsch et al., 2010; Zhang et al., 2013; Li et al., 2014). Most of these metabolites were below detection level in our analyses, though undetectable levels of MeSA are most likely due to the volatile nature of this compound resulting in its loss during sample preparation.
The most abundant SA derivatives were the abovementioned SAG and DHBA glucosides (DHBAG, Table S3). Like SA, and consistent with previous reports (Bartsch et al., 2010), DHBA was detected solely as a sugar ester. In Arabidopsis, DHBA conjugates with both glucose and xylose have been detected, with the xylose activity dominant; as such DHBAG is detected primarily in the absence of the relevant xylosyltransferase, UGT89A2 (Li et al., 2014). UGT89A2 is downregulated in the ceh1 mutant (Table S5), and interestingly, Col-0, the parent genotype for ceh1, typically accumulates DHBA xylosides, rather than DHBAG. The mechanism and functional consequences of this shift are not yet clear, but suggest some differential function of these two glycosides in SA breakdown or signaling. The glucosyltransferase mediating DHBAG formation is not yet known, but several candidate UDP-glucosyltransferases with activity on DHBA in vitro (Lim et al., 2002) are upregulated in ceh1 (Table S5).
In spite of only negligible total levels of SA in ceh1/eds16, the large majority of the SA conjugation and breakdown enzymes differentially regulated in ceh1 are similarly regulated in the double mutant. This suggests that the differential regulation of these enzymes in ceh1 is not simply the established consequence of accumulation of SA (Seo et al., 1995), but rather an MEcPP-mediated specific response. The rationale and mechanism behind MEcPP inducing both SA production and breakdown/conjugation remains to be elucidated, but may be related to perturbation of other stress signals in the ceh1 mutant, such as jasmonate signaling and redox balance.
Misregulation of chloroplast-localized steps of jasmonate biosynthesis
The KEGG pathway “Alpha-linolenic acid metabolism,” largely composed of jasmonate biosynthesis, is among upregulated pathways in both ceh1 and ceh1/eds16. Biosynthesis of jasmonic acid (JA), an oxylipin stress hormone, begins in the chloroplast with deesterification of fatty acids, followed by their oxidation and subsequent cyclization. The cyclic intermediate 12-oxophytodienoic acid (OPDA) is then transported from the chloroplast to the peroxisome to generate JA. JA itself is then transported from the peroxisome to the cytosol, where it may be conjugated to isoleucine to generate the active signaling molecule JA-Ile, or to a methyl-group to generate methyl JA (MeJA) a long-distance transportable derivative (Figure 4). OPDA, the jasmonate intermediate, has been shown to act as an independent signaling molecule, with partially overlapping responses with JA (Savchenko et al., 2014). To understand regulation of jasmonate biosynthesis in ceh1 with and without SA accumulation, we overlaid transcript, protein, and metabolite levels of jasmonate synthesis to gain insight into the network connecting MEcPP to jasmonate biosynthesis and signaling pathways, in the presence and absence of high SA levels (Figure 4). Most genes of this pathway were not differentially expressed in eds16 (Figure S3), and thus for simplicity only differences of ceh1 and ceh1/eds16 relative to parent are presented in Figure 4.
Figure 4. MEcPP and SA alter jasmonate biosynthesis and signaling.
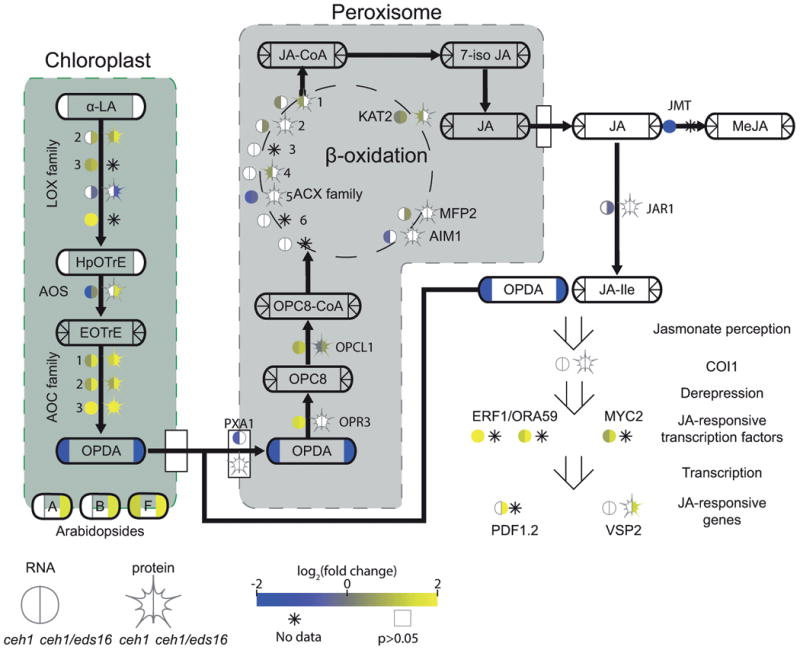
A pathway schematic for JA biosynthesis and breakdown, with transcription factors/enzymes shown by bare text with a circle, representing RNA, and star, representing protein levels. Metabolites are shown by rounded rectangles. For metabolites, RNA, and protein, the log2-fold change for ceh1 vs parent (left) and ceh1/eds16 vs. parent (right) is represented by shading, from -2 (darkest) to 2 (lightest). Non-detectable products in each section are shown by an asterisk, while empty symbols with grey borders represent non-significant fold changes relative to parent. Arrows represent chemical conversions, catalyzed by the enzymes shown on the line, or metabolite transport. Peroxisome-localized steps in conversion of 12-OPDA to JA are shown in the grey box, while simplified chloroplast-localized steps leading to 12-OPDA synthesis are in the green. Open arrows represent metabolite/protein, protein/protein, and protein/DNA (transcription regulation) interactions. Abbreviations: α-LA: α-linolenic acid, LOX: lipoxygenase, HpOTrE: hydroperoxyoctadecatrienoic acid, AOS: allene oxide synthase, EOTrE: epoxyoctadecatrienoic acid, AOC: allene oxide cyclase, OPDA: oxophytodienoic acid, PXA1: peroxisomal ABC transporter 1, OPR3: OPDA reductase 1, OPC8: oxopentenylcyclopentenyloctanoate, OPCL1: OPC8 coA ligase1, ACX: acyl-coA oxidase, KAT2: ketoacyl-coA thiolase 2, MFP2: multifunctional protein 2, AIM1: abnormal inflorescence meristem 1, MeJA: methyl-JA, COI1: coronatine insensitive 1, ERF1: ethylene response factor 1, ORA59: octadecanoid-responsive Arabidopsis AP2/ERF 59, MYC2: (=JIN1) MYC-domain containing transcription factor, PDF1.2: plant defensing 1.2, VSP2: vegetative storage protein 2.
We previously reported that OPDA, but not JA itself, is present at greater levels in ceh1 and ceh1/eds16 than parent plants (Lemos et al., 2016). Here, LC-MS based metabolomics analyses presented no increase in free OPDA in ceh1 and ceh1/eds16, but rather an increase in arabidopsides, in which OPDA and dinor-OPDA are esterified to glycerol backbones (Stelmach et al., 2001; Hisamatsu et al., 2003; Hisamatsu et al., 2005). Consistent with previous results, these arabidopsides accounted for the majority (∼90%) of detected OPDA (Table S3)(Nilsson et al., 2012). Arabidopsides have been found thus far only in the genus Arabidopsis (Mosblech et al., 2009) and Cirsium arvense (Hartley et al., 2015), where they have been shown to increase in response to wounding (Kourtchenko et al., 2007) and in the hypersensitive response (Andersson et al., 2006). They are suggested to act as a pool of OPDA to facilitate prolonged or rapid jasmonate response, as well as direct signaling and chemical roles (Mosblech et al., 2009; Andersson et al., 2006; Stelmach et al., 2001). Arabidopsides have faced scrutiny for potentially being artifacts of hormone extraction techniques; although conditions known to produce these artifacts were not part of extraction (Nilsson et al., 2012), the previously detected OPDA increase in ceh1 (by GC-MS) may be partially due to contamination from arabidopside-localized OPDA, de-esterified from the glycerol backbone during sample preparation for gas chromatography.
Strong upregulation of jasmonate biosynthesis at both protein and RNA levels in ceh1 and ceh1/eds1 is observed in the majority of enzymes catalyzing reactions up to OPDA biosynthesis in the chloroplast (e.g., LOX and AOC family members). Inversely, of enzymes catalyzing the conversion of OPDA to JA, only the first step, OPR3, is strongly upregulated; all other steps are only minutely altered. This differential regulation, while consistent with production of arabidopsides, also presents a bottleneck that may contribute to the previously published elevated levels of OPDA. Of note, the proteins of JA biosynthesis that are differentially regulated are predominantly chloroplast localized, as opposed to the peroxisome-located steps of JA biosynthesis that are slightly, if at all altered, thus suggestive of a compartment-specific role of MEcPP in altering jasmonate biosynthesis, and thereby determining the relative levels of jasmonates species to tailor plant adaptive responses.
Finally, in addition to differential regulation of jasmonate biosynthesis, OPDA localization may also be altered. In order to proceed to JA biosynthesis, OPDA must exit the chloroplast (through a mechanism(s) not yet fully understood) and be imported into the peroxisome byperoxisomal ATP-binding cassette (ABC) transporter 1 (PXA1, (Theodoulou et al., 2005), whose transcription is downregulated in the ceh1 mutant, thus potentially contributing to OPDA accumulation. This mechanism is not inconsistent with OPDA sequestration into membranes as arabidopsides, but the effect of MEcPP must be interpreted with caution; not only is CGI58 (Park et al., 2013) a PXA1-interacting protein also involved in OPDA transport upregulated in MEcPP-overaccumulating lines (Table S4), but also the downregulation of PXA1 is significant only in ceh1, in the presence of elevated SA. This suggests one underlying regulatory mechanism of antagonistic cross talk between SA and JA.
Higher levels of jasmonate in ceh1/eds16as compared to that of the ceh1 is a clear display of SA's functional footprint in each of the observed pathways including reduced PXA1 and enhanced OPDA biosynthesis enzyme and arabidopside accumulation as ceh1/eds16, allowing greater jasmonate accumulation. Finally, in accordance with previous results (Lemos et al., 2016), jasmonate-responsive genes are more highly upregulated in ceh1/eds16 than ceh1 (Figure 4). Antagonism between SA and JA is well established, but nuances are still being revealed. Although initial reports suggested that SA inhibits JA biosynthesis (Pena-Cortés et al., 1993; Harms et al., 1998), recent studies have suggested instead that SA does not affect JA biosynthesis but rather JA signaling (Van der Does et al., 2013; Spoel et al., 2003; Ndamukong et al., 2007), and that SA/JA interactions may not be solely antagonistic, but even synergistic depending on timing and relative concentrations (Mur et al., 2006). Surprisingly, our results suggest that in the context of MEcPP-induced constitutive SA and OPDA production, SA does have a dampening effect on jasmonate biosynthesis, refining our view of, and offering mechanistic insight into interplay between two key defense pathways.
Altered Glutathione redox coupling and impact in ceh1
Interplay between SA and JA is partially mediated by redox homeostasis, key in SA signaling and an important component of SA responses. Several other lines of evidence suggest altered redox homeostasis in ceh1: the pool of glutathione in ceh1 and ceh1/eds16 is largely oxidized in comparison to the parent, while other reducing equivalents show the opposite trend (Figure 5A, Supplemental Table S6). Moreover, “Glutathione metabolism” was identified as an upregulated pathway in ceh1 and ceh1/eds16 at the RNA level. Accordingly, we investigated glutathione metabolism under elevated MEcPP.
Figure 5. MEcPP influences glutathione redox balance and function.
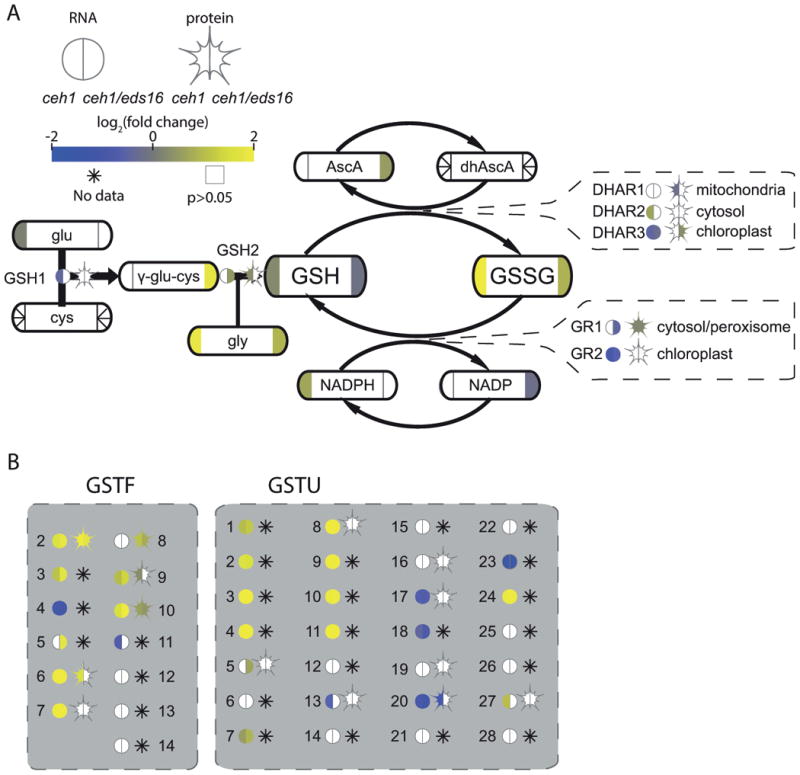
(A) A partial simplified pathway schematic for glutathione biosynthesis and function, with enzymes shown by bare text with a circle, representing RNA, and star, representing protein levels. Metabolites are shown by rounded rectangles. For metabolites, RNA, and protein, the log2-fold change for ceh1 vs parent (left) and ceh1/eds16 vs. parent (right) is represented by shading, from -2 (darkest) to 2 (lightest). Non-detectable products in each section are shown by an asterisk, while empty symbols with grey borders represent non-significant fold changes relative to parent. Arrows represent chemical conversions, catalyzed by the enzymes shown on the line. Callouts for the DHAR and GR enzyme families also show subcellular localization of these enzymes. (B) Glutathione S-transferases of the phi (GSTF) and tau (GSTU) families are shown in grey boxes. Abbreviations: glu: glutamate, cys: cysteine, GSH1: (=γ-ECS1, RML1, PAD2) glutathione synthetase 1, gly: glycine, GSH1: glutathione synthetase 2, GSH: reduced glutathione, GSSG: oxidized glutathione, AscA: ascorbic acid, dhAscA: dehydroxyascorbic acid, DHAR: dehydroxyascorbic acid reductase, GR: glutathione reductase, GSTF: glutathiose S-transferase phi family, GSTU: glutathione S-transferase tau family.
We first utilized the KEGG “Glutathione metabolism” pathway, identifying sub-pathways for glutathione biosynthesis, oxidation/reduction, and conjugation as schematically displayed (Figure 5A), with complete glutathione-associated genes available for all assayed lines (Figure S4). The altered reduction status of the glutathione pool in MEcPP-overaccumulating genotypes is presented by comparison of oxidized glutathione (GSSG, increased levels in ceh1/eds16 and more strongly increased in ceh1 relative to parent) to reduced glutathione (GSH, slightly increased in ceh1 but decreased in ceh1/eds16 relative to parent). The GSH/GSSG ratio, although not a perfect proxy for the reduction potential of this redox couple or its compartment-specific effect on cellular redox potential (Schafer and Buettner, 2001; Diaz-Vivancos et al., 2015), can suggest important changes in glutathione-mediated redox status, and the GSH/GSSG ratio in MEcPP-overaccumulating lines is only approximately 1/4 to 1/3 that of parent lines (Supplemental Table S6). These results were repeated using independent tissue and LC-MS conditions with a modified extraction, yielding similar fold changes in glutathione redox status (Method S1, Figure S5, Table S6). To investigate underlying mechanisms we first questioned whether diminished GSH biosynthesis may contribute to greater glutathione oxidation. The major regulators of glutathione synthesis are considered to be post-translationally regulated GSH1 enzyme activity and cysteine availability (Noctor et al., 2012; Queval et al., 2009). While GSH1 enzyme activity was not examined here and cysteine levels are not readily detected by the utilized LC-MS conditions, neither ceh1 nor ceh1/eds16 display altered levels of GSH1 protein, and in fact ceh1/eds16 even has enhanced levels of the γ-glutamylcysteine intermediate (Figure 5A), supporting the notion that glutathione biosynthesis is not a bottle neck.
A significant difference however is observed in glutathione redox regulation. Glutathione serves many key signaling and detoxifying roles in plant cells, one of which is regenerating reduced ascorbic acid (AscA) from dehydroascorbate (dhAscA), followed by subsequent regeneration of GSH from NADPH (Figure 5A). These reactions, catalyzed by the dehydroascorbate reductase (DHAR) and glutathione reductase (GR) gene families, provide a major reactive oxygen species (ROS) detoxification mechanism for the cell and as such these reducing currencies are generally tightly balanced (Mittler et al., 2004). A possible explanation for the imbalance in ceh1 is transcriptional regulation of GR1 and 2, the two differentially compartmentalized plant glutathione reductases which are both downregulated in ceh1/eds16, while only GR2, the chloroplastic isoform is downregulated in ceh1 (Yu et al., 2013; Tzafrir et al., 2004; Marty et al., 2009). Interestingly, a recent report has shown that the stress-induced upregulation of GR1 and 2 is dependent on signals originating in functional chloroplasts (Garnik et al., 2016), which may be disrupted ceh1 and ceh1/eds16.The root cause of the observed altered GSH/GSSG ratio in MEcPP-accumulating lines is not yet clear. However, one could assume ROS as a causative agent, simply because of the evolutionarily conserved role of this signaling molecule in response to unfavorable cues (Mittler et al., 2011).
Aside from direct roles as a reducing agent, glutathione serves in glutathionylation of various substrates, controlling protein redox status and function, and metabolite solubility and degradation. These functions also appear to be disrupted in ceh1. Among the families of glutathione-S-transferases (GSTs) and glutaredoxins (GRXs) responsible for these processes, the plant-specific tau (GSTU) and phi (GSTF) families possessed a large number of upregulated genes (Figure 5B, Figure S4). The mechanism and physiological implications of upregulation of GSTF and GSTU in ceh1 is not immediately obvious; indeed the regulation and function of the majority of GSTs and GRXs, and specifically that of the GSTU and GSTF families, is elusive, partly due to their large and partially redundant families (Sappl et al., 2009). A subset of GSTs has been demonstrated to be upregulated by SA, bacterial attack, and hydrogen peroxide (Sappl et al., 2009), but there is no clear overlap between these stress-induced GSTs and GSTs induced in ceh1. Efforts directed at identification of GSTU and GSTF substrates have indicated roles for these family members in glutathionylating small molecules, thereby rendering them more soluble, e.g. for xenobiotic detoxification (Noctor et al., 2012; Noctor et al., 2011; Krajewski et al., 2013). However, the individual substrate specificity in planta, and the relevance thereof, is not clear. Thus, potential effects of GSTU and GSTF upregulation in ceh1 remain to be discovered. One effect may include depletion of the reduced glutathione pool, contributing to general glutathione oxidation. Glutathionylation of small molecules often leads to degradation, of both the molecule and the conjugated amino acids. Thus, upregulation of the GSTU and GSTF families may both be a response to diminished availability of reduced glutathione, and contribute to this problem by diverting reduced glutathione to eventual degradation.
Exogenous reduced glutathione does not rescue ceh1 phenotypes
The ceh1 mutant is dwarfed relative to parent plants and this phenotype is not consistently rescued in ceh1/eds16, suggesting that although constitutive SA signaling can result in dwarfism, this is not the sole reason for compromised growth in ceh1. In contrast, double mutant lines of ceh1 and the key general stress response (GSR) regulator calmodulin-binding transcriptional activator CAMTA3 (Benn et al., 2014) (ceh1/camta3), with greatly diminished GSR, do grow to a larger size than ceh1, though still smaller than parent plants (Benn et al., 2016). The difference between phenotypes of ceh1/eds16 vs. ceh1/camta3 may lie in the specific vs. general nature of the signaling pathways – both UPR and SA responses are partially reduced in ceh1/camta3. Like the GSR, redox balance, and particularly glutathione, has numerous connections with stress transduction pathways disrupted in ceh1: redox signaling in fact induces the GSR (Bjornson et al., 2014), the ER stress response both induces and is induced by redox signals (Fedoroff, 2006; Ozgur et al., 2014), SA has oxidizing and reducing interactions with the redox status of the cell and with glutathione specifically, depending on timing and concentration (Herrera-Vasquez et al., 2015), and redox balance of glutathione influences SA/JA cross-inhibition (Ndamukong et al., 2007). Thus, redox balance and particularly glutathione redox status may represent a common pathway influencing the multitude of stress signaling pathways disrupted in ceh1.
To address this possibility and based on the report that application of exogenous GSH can lead to a larger and more reduced glutathione pool (Cheng et al., 2015; Tausz et al., 2004); we grew ceh1 on exogenous GSH to assay for rescue of plant size (Figure 6). Because of the effects of redox on the GSR, we also assayed for the GSR in these conditions using the 4xRapid Stress Response Element:LUCIFERASE (4xRSRE:LUC) reporter line (Walley et al., 2007). The previously reported constitutive expression of 4xRSRE:LUC in ceh1 and ceh1/eds16 (Benn et al., 2016), was not altered in plants grown on GSH-supplemented media as measured by LUC activity driven by RSRE (Figure 6A). In fact, glutathione supplementation only led to significant changes in eds16 and ceh1/eds16, where it actually increased luminescence. This surprising result may not be the effect of increased 4xRSRE:LUC transcription, but rather LUC enzyme activity; indeed, LUC is more stable and active in more reduced environments (Czupryna and Tsourkas, 2011). The change in LUC activity specifically in SA deficient lines is well aligned with the interplay between this hormone and redox homeostasis, although the underlying mechanism is still unknown (Herrera-Vasquez et al., 2015).Additionally, in the presence of GSH, only ceh1/eds16 and eds16 experience a marginal but significant increase in plant size (Figure 6B). This would also be consistent with exogenously supplied glutathione compensating for a redox imbalance in the absence of SA signaling.
Figure 6. Exogenous glutathione does not rescue GSR or size phenotypes of ceh1.
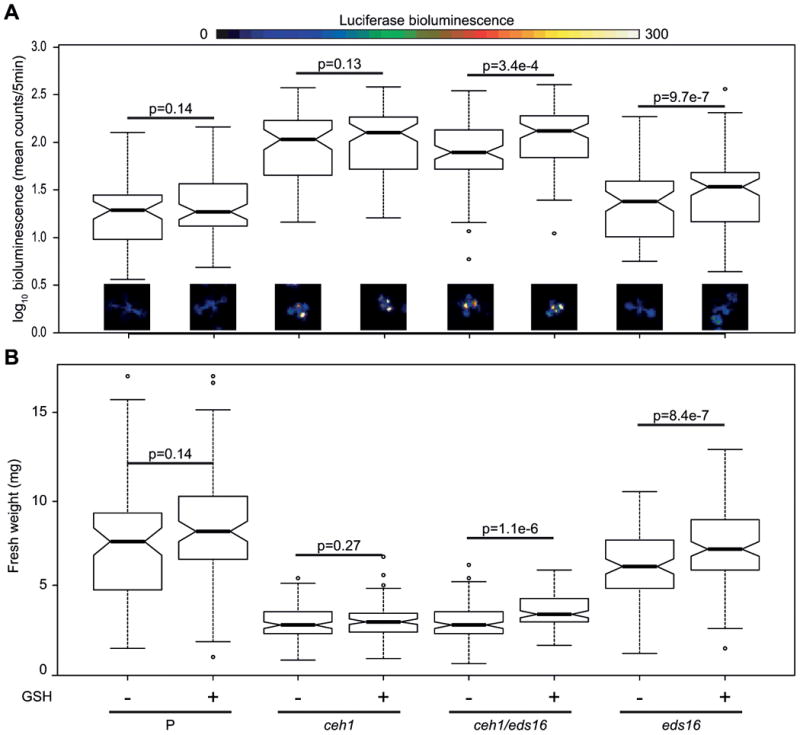
Parent (P), ceh1, ceh1/eds16, and eds16 seedlings were grown in media with or without reduced glutathione supplementation. The GSR, as measured via 4xRSRE:LUC activity (A) was examined in each line, represented by notched boxplots of quantified luminescence (log10 transformed to achieve normality) and by false-color images, with a scale bar above showing conversion from luminescence to color. Notched box plots show possible outliers (circles), the interquartile range (box), 95% confidence interval (notch), and median. Non-overlapping notches is considered strong evidence for statistically different populations. Following imaging, seedlings were harvested and weighed (B). For each genotype, p values for comparisons between (+) and (-) glutathione were calculated using a one-way ANOVA, blocking by experimental replicate in the statistical program R, n>75 seedlings per genotype/treatment combination over three experiments.
The lack of an increase in ceh1 size under glutathione supplementation suggests that one or more stress signaling pathway(s) is still active in reduced glutathione-supplemented ceh1, with the extension that MEcPP activates SA, JA, and GSR signaling independently of its effect on redox balance of glutathione. We cannot rule out the possibility that disruption of the MEP pathway leads to some size effect in ceh1 independently of MEcPP signaling, but the partial rescue of plant size in ceh1/camta3 mutants argues against this being the sole cause of the reduced size of ceh1. Rather, accumulation of MEcPP may result in activation of each pathway individually, or via some as-yet-undiscovered coordinating signaling node. Indeed, it has been shown that Ca2+ signaling is required for exogenously applied MEcPP GSR induction (Benn et al., 2016), suggesting the existence of at least one intermediary for MEcPP modulation of cellular status.
Conclusions
Over-accumulation of MEcPP in the ceh1 mutant leads to numerous biochemical and physiological stress response phenotypes. The interrelation and interface among stress transduction pathways prevents the drawing of irrefutable conclusions by analyses of each phenotype in isolation. Through integration of multi-omics outputs and pathway-oriented analyses, we have identified mechanisms leading to misregulation of ICS1 and reduced conversion of OPDA to JA - a clear indication of a compartmentalized stress response cascade - as well as other perturbations, including regulation of SA breakdown and imbalances in central metabolism, amino acid levels, and redox status. Integrated analysis of these complementary omics datasets has allowed us to generate a more nuanced picture of retrograde mediated stress signaling strata, interrelation, and compartmentalization, and to identify targets of investigation in the uncharted territories of interorganellar stress signaling.
Experimental Procedures
Plant growth conditions
For all “-omics” experiments, Arabidopsis thaliana plants of the genotypes pHPL:LUC (parent), ceh1, eds16, and ceh1/eds16 were grown under 16h light / 8h dark, and at 23-25°C. Three weeks old seedlings were harvested and flash-frozen in liquid nitrogen.
For 4xRSRE:LUC visualization, seedlings were grown on ½ strength MS media, without or with 500μM glutathione. Seedlings were imaged for 4xRSRE:LUC activity as described previously (Walley et al., 2007; Bjornson et al., 2014; Benn et al., 2014).
RNA-seq
Libraries for RNA-seq were prepared according to the HTR method (Kumar et al., 2012), and sequenced on an Illumina HiSeq200 in 50bp single end read mode. Reads were mapped to the Arabidopsis genome using TopHat2 (Kim et al., 2013) on the public usegalaxy.org instance of Galaxy (Afgan et al., 2016) (counts by gene and statistical analyses available in Table S4). RNAseq data is available through the Gene Expression Omnibus GSE96666.
Proteomics
Proteomics sample and data preparation has been described (Walley et al., 2015).
Metabolomics
Metabolites extraction and profiling from five biological replicates per genotype was as previously reported (Balcke et al., 2011; Treutler et al., 2016), with additional alterations described in Supporting Information Method S1. MS conditions and MRM transitions for the targeted approach are presented in Supporting Information Table S7. MS1 and MS2 raw spectra were deconvoluted using MS-DIAL (Tsugawa et al., 2015). Families of altered metabolites were identified using the web application MetFamily (Treutler et al., 2016), and levels of selected metabolites were quantified using the software Multiquant 3.0 (Sciex).
Gene Set Enrichment Analysis
Gene set enrichment analysis (GSEA) was carried out using the R (Team, 2016) package Generally Applicable Gene set Enrichment analysis (GAGE), available through Bioconductor (Luo et al., 2009). For gene sets we used the KEGG database of Arabidopsis thaliana metabolic pathways (Ogata et al., 1999; Kanehisa et al., 2016).
Analysis was carried out for RNA and protein. Briefly, for RNA counts were normalized using DESeq2 (Love et al., 2014), and the normalized counts were used as input for GAGE.
For proteins, a similar procedure was followed, using iTRAQ intensities (Walley et al., 2015) and paired biological replicates. Due to the smaller dynamic range of protein measurements, uncorrected p values were occasionally considered when providing support to RNA-seq results, as noted in Table S1.
Heatmap
Assayed metabolites were first sorted according to KEGG pathway/molecular category, then heat maps were created using the ggplot2 (Wickham, 2009) package in R. For consistency, limits of log2 fold change -2 (blue) to 2 (yellow) were used throughout the heatmap and pathway visualizations, although numerical values are available in Tables S3, S4, and S6.
Pathway visualization
With the exception of SA biosynthesis and breakdown, pathway visualizations are based on KEGG (Ogata et al., 1999; Kanehisa et al., 2016) pathway templates. Relevant steps and Arabidopsis family members were highlighted and expanded in custom-made Pathvisio (Kutmon et al., 2015; van Iersel et al., 2008) pathways, with metabolite data converted to fold change and corrected p values calculated in R.
Supplementary Material
Supporting Information Figure S1. Metabolite accumulation in ceh1, ceh1/eds16, and eds16.
Supporting Information Figure S2. SA biosynthesis and metabolism is largely downregulated in eds16.
Supporting Information Figure S3. JA biosynthesis is largely unchanged in eds16.
Supporting Information Figure S4. Only particular aspects of glutathione biosynthesis and use are altered in MEcPP-overaccumulating genotypes.
Supporting Information Figure S5. Independent analysis confirms MEcPP effect on glutathione reduction status.
Supporting Information Table S1. Statistical identification of differentially regulated pathways in ceh1, ceh1/eds16, and eds16.
Supporting Information Table S2. Gene lists of identified KEGG pathways
Supporting Information Table S3. Metabolite peak values and comparison among genotypes.
Supporting Information Table S4. Normalized read counts and comparisons among genotypes for RNAseq data
Supporting Information Table S5. Expression of UGT glycosyltransferases with in vitro activity against benzoates.
Supporting Information Table S6. Calculated ratio, percent reduction and redox potentials for reducing equivalents
Supporting Information Table S7. MRM parameters for targeted analysis of charged hydrophilic metabolites.
Supporting Information Method S1. Metabolite extraction, profiling and analyses.
Acknowledgments
The authors wish to thank Miguel de Lucas, Geoffrey Benn, and Mark Lemos for critical input on this work. This work was supported by National Science Foundation Grants IOS-1036491 and IOS-1352478; and NIH R01GM107311 to KD.
Footnotes
Conflict of interest: The authors have no conflict of interest to declare
References
- Afgan E, Baker D, Beek M, van den, et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Res. 2016;44 doi: 10.1093/nar/gkw343. Available at: http://nar.oxfordjournals.org/lookup/doi/10.1093/nar/gkw343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson MX, Hamberg M, Kourtchenko O, Brunnström Å, McPhail KL, Gerwick WH, Göbel C, Feussner I, Ellerström M. Oxylipin profiling of the hypersensitive response in Arabidopsis thaliana: Formation of a novel oxo-phytodienoic acid-containing galactolipid, arabidopside E. J Biol Chem. 2006;281:31528–31537. doi: 10.1074/jbc.M604820200. [DOI] [PubMed] [Google Scholar]
- Baek D, Pathange P, Chung JS, et al. A stress-inducible sulphotransferase sulphonates salicylic acid and confers pathogen resistance in Arabidopsis. Plant, Cell Environ. 2010;33:1383–1392. doi: 10.1111/j.1365-3040.2010.02156.x. [DOI] [PubMed] [Google Scholar]
- Balcke GU, Kolle SN, Kamp H, Bethan B, Looser R, Wagner S, Landsiedel R, Ravenzwaay B van. Linking energy metabolism to dysfunctions in mitochondrial respiration - A metabolomics in vitro approach. Toxicol Lett. 2011;203:200–209. doi: 10.1016/j.toxlet.2011.03.013. Available at: http://dx.doi.org/10.1016/j.toxlet.2011.03.013. [DOI] [PubMed] [Google Scholar]
- Bartsch M, Bednarek P, Vivancos PD, Schneider B, Roepenack-Lahaye E Von, Foyer CH, Kombrink E, Scheel D, Parker JE. Accumulation of isochorismate-derived 2,3-dihydroxybenzoic 3-O-β-D-xyloside in Arabidopsis resistance to pathogens and ageing of leaves. J Biol Chem. 2010;285:25654–25665. doi: 10.1074/jbc.M109.092569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benn G, Bjornson M, Ke H, Souza A De, Balmond EI, Shaw JT, Dehesh K. Plastidial metabolite MEcPP induces a transcriptionally centered stress-response hub via the transcription factor CAMTA3. Proc Natl Acad Sci. 2016 doi: 10.1073/pnas.1602582113. 201602582. Available at: http://www.pnas.org/lookup/doi/10.1073/pnas.1602582113. [DOI] [PMC free article] [PubMed]
- Benn G, Wang CQ, Hicks D, Stein J, Guthrie C, Dehesh K. A key general stress response motif is regulated non- uniformly by CAMTA transcription factors. Plant J. 2014;80:82–92. doi: 10.1111/tpj.12620. Available at: http://dx.doi.org/10.1111/tpj.12620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjornson M, Benn G, Song X, Comai L, Franz AK, Dandekar A, Drakakaki G, Dehesh K. Distinct roles for MAPK signaling and CAMTA3 in regulating the peak time and amplitude of the plant general stress response. [Accessed August 27, 2014];Plant Physiol. 2014 166:988–996. doi: 10.1104/pp.114.245944. Available at: http://www.ncbi.nlm.nih.gov/pubmed/25157030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Xue L, Chintamanani S, et al. ETHYLENE INSENSITIVE3 and ETHYLENE INSENSITIVE3-LIKE1 Repress SALICYLIC ACID INDUCTION DEFICIENT2 Expression to Negatively Regulate Plant Innate Immunity in Arabidopsis. Plant Cell. 2009;21:2527–2540. doi: 10.1105/tpc.108.065193. Available at: http://www.plantcell.org/cgi/doi/10.1105/tpc.108.065193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng MC, Ko K, Chang WL, Kuo WC, Chen GH, Lin TP. Increased glutathione contributes to stress tolerance and global translational changes in Arabidopsis. Plant J. 2015;83:926–939. doi: 10.1111/tpj.12940. [DOI] [PubMed] [Google Scholar]
- Czupryna J, Tsourkas A. Firefly luciferase and rluc8 exhibit differential sensitivity to oxidative stress in apoptotic cells. PLoS One. 2011;6 doi: 10.1371/journal.pone.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean JV, Delaney SP. Metabolism of salicylic acid in wild-type, ugt74f1 and ugt74f2 glucosyltransferase mutants of Arabidopsis thaliana. Physiol Plant. 2008;132:417–425. doi: 10.1111/j.1399-3054.2007.01041.x. [DOI] [PubMed] [Google Scholar]
- Diaz-Vivancos P, Simone A De, Kiddle G, Foyer CH. Glutathione - Linking cell proliferation to oxidative stress. Free Radic Biol Med. 2015;89:1154–1164. doi: 10.1016/j.freeradbiomed.2015.09.023. Available at: http://dx.doi.org/10.1016/j.freeradbiomed.2015.09.023. [DOI] [PubMed] [Google Scholar]
- Does D Van der, Leon-Reyes A, Koornneef A, et al. Salicylic acid suppresses jasmonic acid signaling downstream of SCFCOI1-JAZ by targeting GCC promoter motifs via transcription factor ORA59. [Accessed October 6, 2014];Plant Cell. 2013 25:744–61. doi: 10.1105/tpc.112.108548. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3608790&tool=pm686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedoroff N. Redox regulatory mechanisms in cellular stress responses. Ann Bot. 2006;98:289–300. doi: 10.1093/aob/mcl128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnik EY, Belkov VI, Tarasenko VI, Korzun MA, Konstantinov YM. Glutathione Reductase Gene Expression Depends on Chloroplast Signals in Arabidopsis thaliana. 2016;81:364–372. doi: 10.1134/S0006297916040064. [DOI] [PubMed] [Google Scholar]
- Ghassemian M, Lutes J, Tepperman JM, Chang HS, Zhu T, Wang X, Quail PH, Markus Lange B. Integrative analysis of transcript and metabolite profiling data sets to evaluate the regulation of biochemical pathways during photomorphogenesis. Arch Biochem Biophys. 2006;448:45–59. doi: 10.1016/j.abb.2005.11.020. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Cabanelas D, Wright LP, Paetz C, Onkokesung N, Gershenzon J, Rodriguez-Concepcion M, Phillips MA. The diversion of 2-C-methyl-d-erythritol-2,4-cyclodiphosphate from the 2-C-methyl-d-erythritol 4-phosphate pathway to hemiterpene glycosides mediates stress responses in Arabidopsis thaliana. Plant J. 2015;82:122–137. doi: 10.1111/tpj.12798. [DOI] [PubMed] [Google Scholar]
- Harms K, Ramirez I, Pena-Cortes H. Inhibition of wound-induced accumulation of allene oxide synthase transcripts in flax leaves by aspirin and salicylic acid. Plant Physiol. 1998;118:1057–65. doi: 10.1104/pp.118.3.1057. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=34779&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley SE, Eschen R, Horwood JM, Gange AC, Hill EM. Infection by a foliar endophyte elicits novel arabidopside-based plant defence reactions in its host, Cirsium arvense. New Phytol. 2015;205:816–827. doi: 10.1111/nph.13067. [DOI] [PubMed] [Google Scholar]
- Hennig J, Malamy J, Grynkiewicz G. Interconversion of the salicylic acid signal and its glucoside in tobacco. Plant J. 2002;4:593–600. doi: 10.1046/j.1365-313x.1993.04040593.x. Available at: http://onlinelibrary.wiley.com/doi/10.1046/j.1365-313X.1993.04040593.x/abstract. [DOI] [PubMed] [Google Scholar]
- Herrera-Vasquez A, Salinas P, Holuigue L. Salicylic acid and reactive oxygen species interplay in the transcriptional control of defense genes expression. Front Plant Sci. 2015;6:171. doi: 10.3389/fpls.2015.00171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai MY, Yano M, Goodenowe DB, Kanaya S, Kimura T, Awazuhara M, Arita M, Fujiwara T, Saito K. Integration of transcriptomics and metabolomics for understanding of global responses to nutritional stresses in Arabidopsis thaliana. Proc Natl Acad Sci U S A. 2004;101:10205–10. doi: 10.1073/pnas.0403218101. Available at: /pmc/articles/PMC454188/?report=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hisamatsu Y, Goto N, Hasegawa K, Shigemori H. Arabidopsides A and B, two new oxylipins from Arabidopsis thaliana. Tetrahedron Lett. 2003;44:5553–5556. [Google Scholar]
- Hisamatsu Y, Goto N, Sekiguchi M, Hasegawa K, Shigemori H. Oxylipins arabidopsides C and D from Arabidopsis thaliana. J Nat Prod. 2005;68:600–603. doi: 10.1021/np0495938. [DOI] [PubMed] [Google Scholar]
- Hyun TK, Lee S, Rim Y, Kumar R, Han X, Lee SY, Lee CH, Kim JY. De-novo RNA sequencing and metabolite profiling to identify genes involved in anthocyanin biosynthesis in Korean black raspberry (Rubus coreanus Miquel) PLoS One. 2014;9:1–13. doi: 10.1371/journal.pone.0088292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iersel MP van, Kelder T, Pico AR, Hanspers K, Coort S, Conklin BR, Evelo C. Presenting and exploring biological pathways with PathVisio. BMC Bioinformatics. 2008;9:399. doi: 10.1186/1471-2105-9-399. Available at: http://www.ncbi.nlm.nih.gov/pubmed/18817533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juntawong P, Girke T, Bazin J, Bailey-Serres J. Translational dynamics revealed by genome-wide profiling of ribosome footprints in Arabidopsis. [Accessed January 21, 2014];Proc Natl Acad Sci U S A. 2014 111:E203–12. doi: 10.1073/pnas.1317811111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016;44:D457–D462. doi: 10.1093/nar/gkv1070. Available at: http://nar.oxfordjournals.org/lookup/doi/10.1093/nar/gkv1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=4053844&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klessig DF, Malamy J. The salicylic acid signal in plants. Plant Mol Biol. 1994;26:1439–1458. doi: 10.1007/BF00016484. [DOI] [PubMed] [Google Scholar]
- Kourtchenko O, Andersson MX, Hamberg M, Brunnstrom A, Gobel C, McPhail KL, Gerwick WH, Feussner I, Ellerstrom M. Oxo-Phytodienoic Acid-Containing Galactolipids in Arabidopsis: Jasmonate Signaling Dependence. Plant Physiol. 2007;145:1658–1669. doi: 10.1104/pp.107.104752. Available at: http://www.plantphysiol.org/cgi/doi/10.1104/pp.107.104752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacik J, Klejdus B, Hedbavny J, Backor M. Effect of copper and salicylic acid on phenolic metabolites and free amino acids in Scenedesmus quadricauda (Chlorophyceae) Plant Sci. 2010;178:307–311. [Google Scholar]
- Krajewski MP, Kanawati B, Fekete A, Kowalski N, Schmitt-Kopplin P, Grill E. Analysis of Arabidopsis glutathione-transferases in yeast. Phytochemistry. 2013;91:198–207. doi: 10.1016/j.phytochem.2012.04.016. Available at: http://dx.doi.org/10.1016/j.phytochem.2012.04.016. [DOI] [PubMed] [Google Scholar]
- Kumar R, Ichihashi Y, Kimura S, Chitwood DH, Headland LR, Peng J, Maloof JN, Sinha NR. A High-Throughput Method for Illumina RNA-Seq Library Preparation. Front Plant Sci. 2012;3:202. doi: 10.3389/fpls.2012.00202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutmon M, Iersel MP van, Bohler A, Kelder T, Nunes N, Pico AR, Evelo CT. PathVisio 3: An Extendable Pathway Analysis Toolbox. PLoS Comput Biol. 2015;11:1–13. doi: 10.1371/journal.pcbi.1004085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen PE, Sreedasyam A, Trivedi G, Desai S, Dai Y, Cseke LJ, Collart FR. Multi-Omics Approach Identifies Molecular Mechanisms of Plant-Fungus Mycorrhizal Interaction. Front Plant Sci. 2015;6:1061. doi: 10.3389/fpls.2015.01061. Available at: http://journal.frontiersin.org/article/10.3389/fpls.2015.01061/abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemos M, Xiao Y, Bjornson M, Wang J, Hicks D, Souza A De. The plastidial retrograde signal methyl erythritol cyclopyrophosphate is a regulator of salicylic acid and jasmonic acid crosstalk. J Exp Bot. 2016 doi: 10.1093/jxb/erv550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Svedin E, Mo H, Atwell S, Dilkes BP, Chapple C. Exploiting natural variation of secondary metabolism identifies a gene controlling the glycosylation diversity of dihydroxybenzoic acids in Arabidopsis thaliana. Genetics. 2014;198:1267–1276. doi: 10.1534/genetics.114.168690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim EK, Doucet CJ, Li Y, Elias L, Worrall D, Spencer SP, Ross J, Bowles DJ. The activity of Arabidopsis glycosyltransferases toward salicylic acid, 4-hydroxybenzoic acid, and other benzoates. J Biol Chem. 2002;277:586–592. doi: 10.1074/jbc.M109287200. [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. Available at: http://genomebiology.biomedcentral.com/articles/10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W, Friedman MS, Shedden K, Hankenson KD, Woolf PJ. GAGE: generally applicable gene set enrichment for pathway analysis. BMC Bioinformatics. 2009;10:161. doi: 10.1186/1471-2105-10-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marty L, Siala W, Schwarzländer M, et al. The NADPH-dependent thioredoxin system constitutes a functional backup for cytosolic glutathione reductase in Arabidopsis. Proc Natl Acad Sci U S A. 2009;106:9109–9114. doi: 10.1073/pnas.0900206106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler R, Vanderauwera S, Gollery M, Breusegem F Van. Reactive oxygen gene network of plants. Trends Plant Sci. 2004;9:490–498. doi: 10.1016/j.tplants.2004.08.009. [DOI] [PubMed] [Google Scholar]
- Mittler R, Vanderauwera S, Suzuki N, Miller G, Tognetti VB, Vandepoele K, Gollery M, Shulaev V, Breusegem F Van. ROS signaling: the new wave? [Accessed July 20, 2012];Trends Plant Sci. 2011 16:300–9. doi: 10.1016/j.tplants.2011.03.007. Available at: http://www.ncbi.nlm.nih.gov/pubmed/21482172. [DOI] [PubMed] [Google Scholar]
- Mosblech A, Feussner I, Heilmann I. Oxylipins: Structurally diverse metabolites from fatty acid oxidation. Plant Physiol Biochem. 2009;47:511–517. doi: 10.1016/j.plaphy.2008.12.011. Available at: http://dx.doi.org/10.1016/j.plaphy.2008.12.011. [DOI] [PubMed] [Google Scholar]
- Mur LaJ, Kenton P, Atzorn R, Miersch O, Wasternack C. The outcomes of concentration-specific interactions between salicylate and jasmonate signaling include synergy, antagonism, and oxidative stress leading to cell death. Plant Physiol. 2006;140:249–62. doi: 10.1104/pp.105.072348. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=1326048&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustroph A, Zanetti ME, Jang CJH, Holtan HE, Repetti PP, Galbraith DW, Girke T, Bailey-Serres J. Profiling translatomes of discrete cell populations resolves altered cellular priorities during hypoxia in Arabidopsis. Proc Natl Acad Sci U S A. 2009;106:18843–8. doi: 10.1073/pnas.0906131106. Available at: http://www.pnas.org/content/106/44/18843.abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ndamukong I, Abdallat AAl, Thurow C, Fode B, Zander M, Weigel R, Gatz C. SA-inducible Arabidopsis glutaredoxin interacts with TGA factors and suppresses JA-responsive PDF1.2 transcription. Plant J. 2007;50:128–139. doi: 10.1111/j.1365-313X.2007.03039.x. [DOI] [PubMed] [Google Scholar]
- Nilsson AK, Fahlberg P, Ellerstroem M, Andersson MX. Oxo-phytodienoic acid (OPDA) is formed on fatty acids esterified to galactolipids after tissue disruption in Arabidopsis thaliana. FEBS Lett. 2012;586:2483–2487. doi: 10.1016/j.febslet.2012.06.010. Available at: http://dx.doi.org/10.1016/j.febslet.2012.06.010. [DOI] [PubMed] [Google Scholar]
- Noctor G, Mhamdi A, Chaouch S, Han Y, Neukermans J, Marquez-Garcia B, Queval G, Foyer CH. Glutathione in plants: An integrated overview. Plant, Cell Environ. 2012;35:454–484. doi: 10.1111/j.1365-3040.2011.02400.x. [DOI] [PubMed] [Google Scholar]
- Noctor G, Queval G, Mhamdi A, Chaouch S, Foyer CH. Glutathione. Arab B. 2011;9:1–32. doi: 10.1199/tab.0142. Available at: http://www.bioone.org/doi/abs/10.1199/tab.0142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata H, Goto S, Sato K, Fujibuchi W, Bono H, Kanehisa M. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 1999;27:29–34. doi: 10.1093/nar/27.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozgur R, Turkan I, Uzilday B, Sekmen AH. Endoplasmic reticulum stress triggers ROS signalling, changes the redox state, and regulates the antioxidant defence of Arabidopsis thaliana. J Exp Bot. 2014;65:1377–1390. doi: 10.1093/jxb/eru034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S, Gidda SK, James CN, et al. The α/β hydrolase CGI-58 and peroxisomal transport protein PXA1 coregulate lipid homeostasis and signaling in Arabidopsis. Plant Cell. 2013;25:1726–39. doi: 10.1105/tpc.113.111898. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3694702&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S, Kaimoyo E, Kumar D, Mosher S. Methyl salicylate is a critical mobile signal for plant systemic acquired resistance. Science (80- ) 2007;318:113–6. doi: 10.1126/science.1147113. Available at: http://science.sciencemag.org/content/318/5847/113.short. [DOI] [PubMed] [Google Scholar]
- Pena-Cortés H, Albrecht T, Prat S. Aspirin prevents wound-induced gene expression in tomato leaves by blocking jasmonic acid biosynthesis. [Accessed September 9, 2014];Planta. 1993 2 Available at: http://link.springer.com/article/10.1007/BF00240903. [Google Scholar]
- Queval G, Thominet D, Vanacker H, Miginiac-Maslow M, Gakire B, Noctor G. H2O2-activated up-regulation of glutathione in arabidopsis involves induction of genes encoding enzymes involved in cysteine synthesis in the chloroplast. Mol Plant. 2009;2:344–356. doi: 10.1093/mp/ssp002. [DOI] [PubMed] [Google Scholar]
- Raines CA, Lloyd JC, Dyer TA. New insights into the structure and function of but neglected Calvin cycle enzyme. 1999;50:1–8. [Google Scholar]
- Rajasundaram D, Runavot JL, Guo X, Willats WGT, Meulewaeter F, Selbig J. Understanding the Relationship between Cotton Fiber Properties and Non-Cellulosic Cell Wall Polysaccharides. PLoS One. 2014;9:e112168. doi: 10.1371/journal.pone.0112168. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=4226482&tool=pmcentrez&rendertype=abstract%5Cnhttp://dx.plos.org/10.1371/journal.pone.0112168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajasundaram D, Selbig J. More effort - more results: Recent advances in integrative “omics” data analysis. Curr Opin Plant Biol. 2016;30:57–61. doi: 10.1016/j.pbi.2015.12.010. Available at: http://dx.doi.org/10.1016/j.pbi.2015.12.010. [DOI] [PubMed] [Google Scholar]
- Rivas-San Vicente M, Plasencia J. Salicylic acid beyond defence: Its role in plant growth and development. J Exp Bot. 2011;62:3321–3338. doi: 10.1093/jxb/err031. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Concepción M. Early steps in isoprenoid biosynthesis: Multilevel regulation of the supply of common precursors in plant cells. Phytochem Rev. 2006;5:1–15. [Google Scholar]
- Rojas-González JA, Soto-Súarez M, García-Díaz Á, et al. Disruption of both chloroplastic and cytosolic FBPase genes results in a dwarf phenotype and important starch and metabolite changes in Arabidopsis thaliana. J Exp Bot. 2015;66:2673–2689. doi: 10.1093/jxb/erv062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sappl PG, Carroll AJ, Clifton R, Lister R, Whelan J, Harvey Millar A, Singh KB. The Arabidopsis glutathione transferase gene family displays complex stress regulation and co-silencing multiple genes results in altered metabolic sensitivity to oxidative stress. Plant J. 2009;58:53–68. doi: 10.1111/j.1365-313X.2008.03761.x. [DOI] [PubMed] [Google Scholar]
- Savchenko T, Kolla Va, Wang CCQ, et al. Functional Convergence of Oxylipin and Abscisic Acid Pathways Controls Stomatal Closure in Response to Drought. Plant Physiol. 2014;164:1151–1160. doi: 10.1104/pp.113.234310. Available at: http://www.plantphysiol.org/cgi/doi/10.1104/pp.113.234310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer FQ, Buettner GR. Redox environment of the cell as viewed through the redox state of the glutathione disulfide/glutathione couple. Free Radic Biol Med. 2001;30:1191–1212. doi: 10.1016/s0891-5849(01)00480-4. [DOI] [PubMed] [Google Scholar]
- Seo S, Ishizuka K, Ohashi Y. Induction of salicylic acid .beta.-glucosidase in tobacco leaves by exogenous salicylic acid. Plant Cell Physiol. 1995;36:447–453. [Google Scholar]
- Song JT, Koo YJ, Seo HS, Kim MC, Choi Y Do, Kim JH. Overexpression of AtSGT1, an Arabidopsis salicylic acid glucosyltransferase, leads to increased susceptibility to Pseudomonas syringae. Phytochemistry. 2008;69:1128–1134. doi: 10.1016/j.phytochem.2007.12.010. [DOI] [PubMed] [Google Scholar]
- Spoel SH, Koornneef A, Claessens SMC, et al. NPR1 modulates cross-talk between salicylate- and jasmonate-dependent defense pathways through a novel function in the cytosol. Plant Cell. 2003;15:760–70. doi: 10.1105/tpc.009159. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=150028&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelmach BA, Mueller A, Hennig P, Gebhardt S, Schubert-Zsilavecz M, Weiler EW. A Novel Class of Oxylipins, sn1-O-(12-Oxophytodienoyl)-sn2-O-(hexadecatrienoyl)-monogalactosyl Diglyceride, from Arabidopsis thaliana. J Biol Chem. 2001;276:12832–12838. doi: 10.1074/jbc.M010743200. [DOI] [PubMed] [Google Scholar]
- Tausz M, Pilch B, Rennenberg H, Grill D, Herschbach C. Root uptake, transport, and metabolism of externally applied glutathione in Phaseolus vulgaris seedlings. J Plant Physiol. 2004;161:347–349. doi: 10.1078/0176-1617-00658. [DOI] [PubMed] [Google Scholar]
- Team RC. R: A language and environment for statistical computing. R Found Stat Comput. 2016 Available at: https://www.r-project.org/
- Theodoulou FL, Job K, Slocombe SP, Footitt S, Holdsworth M, Baker A, Larson TR, Graham IA. Jasmonic acid levels are reduced in COMATOSE ATP-binding cassette transporter mutants. Implications for transport of jasmonate precursors into peroxisomes. Plant Physiol. 2005;137:835–40. doi: 10.1104/pp.105.059352. Available at: http://www.plantphysiol.org.gate1.inist.fr/content/137/3/835.long. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treutler H, Tsugawa H, Porzel A, Gorzolka K, Tissier A, Neumann S, Balcke GU. Discovering regulated metabolite families in untargeted metabolomics studies. Anal Chem. 2016;88:8082–8090. doi: 10.1021/acs.analchem.6b01569. Available at: http://pubs.acs.org/doi/abs/10.1021/acs.analchem.6b01569. [DOI] [PubMed] [Google Scholar]
- Tsugawa H, Cajka T, Kind T, et al. MS-DIAL: data-independent MS/MS deconvolution for comprehensive metabolome analysis. Nat Methods. 2015;12:523–526. doi: 10.1038/nmeth.3393. Available at: http://dx.doi.org/10.1038/nmeth.3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzafrir I, Pena-muralla R, Dickerman A, et al. Identification of Genes Required for Embryo Development in Arabidopsis 1 [ w ] Plant Physiol. 2004;135:1206–1220. doi: 10.1104/pp.104.045179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verk M Van, Bol J, Linthorst H. WRKY transcription factors involved in activation of SA biosynthesis genes. BMC Plant Biol. 2011;11:89. doi: 10.1186/1471-2229-11-89. Available at: http://www.biomedcentral.com/1471-2229/11/89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijayakumar V, Liebisch G, Buer B, Xue L, Gerlach N, Blau S, Schmitz J, Bucher M. Integrated multi-omics analysis supports role of lysophosphatidylcholine and related glycerophospholipids in the Lotus japonicus-Glomus intraradices mycorrhizal symbiosis. Plant, Cell Environ. 2016;39:393–415. doi: 10.1111/pce.12624. [DOI] [PubMed] [Google Scholar]
- Vlot AC, Dempsey DA, Klessig DF. Salicylic Acid, a Multifaceted Hormone to Combat Disease. Annu Rev Phytopathol. 2009;47:177–206. doi: 10.1146/annurev.phyto.050908.135202. [DOI] [PubMed] [Google Scholar]
- Vranová E, Coman D, Gruissem W. Network analysis of the MVA and MEP pathways for isoprenoid synthesis. Annu Rev Plant Biol. 2013;64:665–700. doi: 10.1146/annurev-arplant-050312-120116. Available at: http://www.ncbi.nlm.nih.gov/pubmed/23451776. [DOI] [PubMed] [Google Scholar]
- Vranová E, Coman D, Gruissem W. Structure and dynamics of the isoprenoid pathway network. Mol Plant. 2012;5:318–333. doi: 10.1093/mp/sss015. [DOI] [PubMed] [Google Scholar]
- Walley J, Xiao Y, Wang JZ, Baidoo EE, Keasling JD, Shen Z, Briggs SP, Dehesh K. Plastid-produced interorgannellar stress signal MEcPP potentiates induction of the unfolded protein response in endoplasmic reticulum. Proc Natl Acad Sci U S A. 2015;112:6212–6217. doi: 10.1073/pnas.1504828112. Available at: http://www.pnas.org/content/112/19/6212.abstract.html?etoc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walley JW, Coughlan S, Hudson ME, Covington MF, Kaspi R, Banu G, Harmer SL, Dehesh K. Mechanical stress induces biotic and abiotic stress responses via a novel cis-element. PLoS Genet. 2007;3:1800–12. doi: 10.1371/journal.pgen.0030172. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2039767&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Gao J, Zhu Z, Dong X, Wang X, Ren G, Zhou X, Kuai B. TCP transcription factors are critical for the coordinated regulation of ISOCHORISMATE SYNTHASE 1 expression in Arabidopsis thaliana. Plant J. 2015;82:151–62. doi: 10.1111/tpj.12803. Available at: http://www.ncbi.nlm.nih.gov/pubmed/25702611. [DOI] [PubMed] [Google Scholar]
- Wanichthanarak K, Fahrmann JF, Grapov D. Genomic , Proteomic , and Metabolomic Data Integration Strategies. 2015;10:1–6. doi: 10.4137/BMI.S29511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H. ggplot2: Elegant Graphics for Data Analysis. Springer-Verlag; New York: 2009. [Google Scholar]
- Wilson MH, Holman TJ, Sørensen I, et al. Multi-omics analysis identifies genes mediating the extension of cell walls in the Arabidopsis thaliana root elongation zone. Front cell Dev Biol. 2015;3:10. doi: 10.3389/fcell.2015.00010. Available at: http://journal.frontiersin.org/article/10.3389/fcell.2015.00010/abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y, Savchenko T, Baidoo EEK, et al. Retrograde signaling by the plastidial metabolite MEcPP regulates expression of nuclear stress-response genes. [Accessed September 20, 2013];Cell. 2012 149:1525–35. doi: 10.1016/j.cell.2012.04.038. Available at: http://www.ncbi.nlm.nih.gov/pubmed/22726439. [DOI] [PubMed] [Google Scholar]
- Yu X, Pasternak T, Eiblmeier M, et al. Plastid-Localized Glutathione Reductase2 – Regulated Glutathione Redox Status Is Essential for Arabidopsis Root Apical Meristem Maintenance. Plant Cell. 2013;25:1–19. doi: 10.1105/tpc.113.117028. Available at: http://www.ncbi.nlm.nih.gov/pubmed/24249834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanetti E, Zanetti E, Chang I, et al. Immunopurification of Polyribosomal Complexes of Arabidopsis for Global Analysis of Gene Expression. Plant Physiol. 2005;138:624–635. doi: 10.1104/pp.105.059477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng J, Liu Y, Liu W, et al. Integration of Transcriptome, Proteome and Metabolism Data Reveals the Alkaloids Biosynthesis in Macleaya cordata and Macleaya microcarpa. PLoS One. 2013;8 doi: 10.1371/journal.pone.0053409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Halitschke R, Yin C, Liu CJ, Gan SS. Salicylic acid 3-hydroxylase regulates Arabidopsis leaf longevity by mediating salicylic acid catabolism. Proc Natl Acad Sci U S A. 2013;110:14807–12. doi: 10.1073/pnas.1302702110. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3767541&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Xu S, Ding P, et al. Control of salicylic acid synthesis and systemic acquired resistance by two members of a plant-specific family of transcription factors. Proc Natl Acad Sci U S A. 2010;107:18220–5. doi: 10.1073/pnas.1005225107. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2964219&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Li Q, Li Z, Staswick PE, Wang M, Zhu Y, He Z. Dual regulation role of GH3.5 in salicylic acid and auxin signaling during Arabidopsis-Pseudomonas syringae interaction. Plant Physiol. 2007;145:450–64. doi: 10.1104/pp.107.106021. Available at: /pmc/articles/PMC2048736/?report=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Zhou M, Yoo H, Pruneda-Paz JL, Spivey NW, Kay Sa, Dong X. Spatial and temporal regulation of biosynthesis of the plant immune signal salicylic acid. Proc Natl Acad Sci. 2015;112:201511182. doi: 10.1073/pnas.1511182112. Available at: http://www.pnas.org/lookup/doi/10.1073/pnas.1511182112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng XY, Spivey NW, Zeng W, Liu PP, Fu ZQ, Klessig DF, He SY, Dong X. Coronatine promotes pseudomonas syringae virulence in plants by activating a signaling cascade that inhibits salicylic acid accumulation. Cell Host Microbe. 2012;11:587–596. doi: 10.1016/j.chom.2012.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information Figure S1. Metabolite accumulation in ceh1, ceh1/eds16, and eds16.
Supporting Information Figure S2. SA biosynthesis and metabolism is largely downregulated in eds16.
Supporting Information Figure S3. JA biosynthesis is largely unchanged in eds16.
Supporting Information Figure S4. Only particular aspects of glutathione biosynthesis and use are altered in MEcPP-overaccumulating genotypes.
Supporting Information Figure S5. Independent analysis confirms MEcPP effect on glutathione reduction status.
Supporting Information Table S1. Statistical identification of differentially regulated pathways in ceh1, ceh1/eds16, and eds16.
Supporting Information Table S2. Gene lists of identified KEGG pathways
Supporting Information Table S3. Metabolite peak values and comparison among genotypes.
Supporting Information Table S4. Normalized read counts and comparisons among genotypes for RNAseq data
Supporting Information Table S5. Expression of UGT glycosyltransferases with in vitro activity against benzoates.
Supporting Information Table S6. Calculated ratio, percent reduction and redox potentials for reducing equivalents
Supporting Information Table S7. MRM parameters for targeted analysis of charged hydrophilic metabolites.
Supporting Information Method S1. Metabolite extraction, profiling and analyses.


