Abstract
Purpose
To investigate the molecular basis of posterior polymorphous corneal dystrophy (PPCD) by examining the PPCD transcriptome and the effect of decreased ZEB1 expression on corneal endothelial cell (CEnC) gene expression.
Methods
Next-generation RNA sequencing (RNA-seq) analyses of corneal endothelium from two PPCD-affected individuals (one with PPCD3 and one of unknown genetic cause) compared with two age-matched controls, and primary human CEnC (pHCEnC) transfected with siRNA-mediated ZEB1 knockdown. The expression of selected differentially expressed genes was validated by quantitative polymerase chain reaction (qPCR) and/or assessed by in situ hybridization in the corneal endothelium of four independent cases of PPCD (one with PPCD3 and three of unknown genetic cause).
Results
Expression of 16% and 46% of the 104 protein-coding genes specific to ex vivo corneal endothelium was lost in the endothelium of two individuals with PPCD. Thirty-two genes associated with ZEB1 and 3 genes (BMP4, CCND1, ZEB1) associated with OVOL2 were differentially expressed in the same direction in both individuals with PPCD. Immunohistochemistry staining and RNA-seq analyses demonstrated variable expression of type IV collagens in PPCD corneas. Decreasing ZEB1 expression in pHCEnC altered expression of 711 protein-coding genes, many of which are associated with canonical pathways regulating various cellular processes.
Conclusions
Identification of the altered transcriptome in PPCD and in a cell-based model of PPCD provided insight into the molecular alterations characterizing PPCD. Further study of the differentially expressed genes associated with ZEB1 and OVOL2 is expected to identify candidate genes for individuals with PPCD and without a ZEB1 or OVOL2 mutation.
Keywords: ZEB1, posterior polymorphous corneal dystrophy, corneal endothelium, RNA-seq, COL4A3, gene expression
Posterior polymorphous corneal dystrophy (PPCD) is a rare, bilateral, autosomal dominant disorder characterized by varied corneal changes, which range from asymptomatic morphologic endothelial irregularities to significant corneal steepening, glaucoma, and/or edema.1,2 PPCD typically presents within the first decade and leads to the need for corneal transplantation in approximately 25% of affected individuals.2–4 Endothelial cells from corneas affected with PPCD have been demonstrated to exhibit epithelial-like characteristics, such as multicellular stratification and expression of epithelial cell markers.5–7
PPCD is associated with locus heterogeneity, with mutations having been reported in the ovo-like 2 (OVOL2; PPCD1) and zinc-finger E-box binding homeobox 1 (ZEB1; PPCD3) genes, which account for approximately 35% to 45% of all PPCD cases.8–18 ZEB1 is a transcription factor that plays a role in the epithelial-to-mesenchymal transition (EMT) pathway by modulating gene expression via binding to E2 box motifs within the promoter regions or target genes (e.g., CDH1).19–22 Since Krafchak and colleagues12 first reported the association of ZEB1 mutations with PPCD3 (Online Mendelian Inheritance in Man [OMIM] 609141), 37 unique heterozygous nonsense, frameshift, and copy number mutations have been reported.8–18 PPCD-associated ZEB1 truncating mutations are predicted to result in the loss of canonical ZEB1 functions and are hypothesized to lead to ZEB1 haploinsufficiency and subsequent altered corneal endothelial expression of genes regulated by ZEB1.10
OVOL2 is a transcription factor that has been shown to downregulate ZEB1 expression, thereby suppressing EMT and driving mesenchymal-to-epithelial transition (MET) instead.23 Promoter mutations in OVOL2 are associated with PPCD1 (OMIM 122000), with cell-based assays indicating that each of the three identified mutations leads to increased OVOL2 promoter activity.24,25 Thus, it is hypothesized that PPCD1-associated OVOL2 promoter region mutations cause the gain or loss of transcription factor binding sites that lead to increased OVOL2 expression and subsequent ZEB1 repression, ultimately producing corneal endothelial changes similar to PPCD3.25 As such, while the relationship between OVOL2 and ZEB1 has been described, their role in the pathomechanism of PPCD1 and PPCD3 remains to be fully elucidated.
The initial report of ZEB1 mutations as the cause of PPCD3 identified ectopic expression of collagen, type IV, alpha 3 (COL4A3) in the corneal endothelium of an affected individual, leading to a proposed theory of pathogenesis.12 Since then, additional groups have reported alterations in the expression of collagens in PPCD and in a mouse model of PPCD3.26,27 Recently, we demonstrated that COL4A3 gene expression is negatively regulated by ZEB1 binding to E2 box motifs in the COL4A3 promoter region and that a PPCD-associated ZEB1 truncating mutation caused the loss of ZEB1-dependent repression of COL4A3 expression.28 While the experimental evidence that ZEB1 can negatively regulate COL4A3 expression is convincing, the level of COL4A3 expression in PPCD varies widely, with some PPCD tissues demonstrating reduced levels compared to controls.27 Herein, we provide additional evidence that the corneal endothelial expression of COL4A3 and other type IV collagens varies widely in affected individuals. Therefore, we sought to identify molecular biomarkers of PPCD by performing transcriptomic analyses on next-generation RNA sequencing (RNA-seq) data obtained from PPCD endothelium and on a cell-based model of PPCD. We identified differentially expressed genes that are involved in canonical pathways and biological processes associated with cell proliferation, migration, adhesion, and morphology. In addition, several differentially upregulated genes, associated with ZEB1 and involved in RNA splicing, histone modifications, transcriptional activation, and cell cycle regulation, were identified and may serve as potential biomarkers and gene candidates for PPCD in the approximately 60% of affected individuals who do not demonstrate a ZEB1 or OVOL2 mutation.
Methods
Informed written consent was obtained from all subjects in this study according to the tenets of the Declaration of Helsinki. This study was approved by the Institutional Review Board at the University of California at Los Angeles (UCLA IRB no. 11–000020).
Patient Identification and DNA Isolation
Patients were examined with slit-lamp biomicroscopic imaging, and the diagnosis of PPCD was based on the presence of characteristic corneal endothelial changes that include guttae, bands, vesicular-appearing lesions, and grayish opacification in one or both eyes (Supplementary Table S1).8 After informed consent was obtained, the patients were enrolled and genomic DNA was purified from peripheral blood leukocytes using the Flexigene DNA Isolation kit (Qiagen, Valencia, CA, USA) according to the manufacturer's instructions.
PCR Amplification and Sanger Sequencing
Genomic DNA from three PPCD patients (P1, P5, and P6) was used for screening the nine exons of ZEB1, an alternative exon 1, and the 1-kb region upstream of the initiation methionine (ATG) using previously described primers and conditions.9 As a ZEB1 mutation was not identified in P6, the promoter, 5′ untranslated region (UTR), and coding regions of OVOL2 were screened in P6 using published primers, PCR conditions, and gene region parameters.29 ZEB1 promoter region sequences were compared to the RefSeqGene sequence (GenBank accession number NG_017048.1), and ZEB1 coding region nucleotide sequences (including the donor and acceptor splice sites) were read manually by comparing to the cDNA sequences for transcript variants 1 (GenBank accession number NM_001128128.2) and 2 (GenBank accession number NM_030751). OVOL2 coding region sequences were compared to the Genbank OVOL2 transcript NM_021220.3, and OVOL2 promoter region sequences were compared to RefSeqGene sequence (GenBank accession number NG_046859.1). ZEB1 coding and promoter region screening in PPCD patients P2 to P4 were previously performed and did not identify a presumed pathogenic mutation.8,9
Immunohistochemistry Imaging of Type IV Collagens
The encoded protein for each of the type IV collagens was assessed by fluorescence immunohistochemistry (F-IHC). A normal donor cornea from an individual without a history of corneal disease and corneas from PPCD-affected individuals with (P1) and without (P2–P4) a ZEB1 mutation were fixed in 10% Tris-buffered formalin and subsequently paraffin embedded. Immunodetection was performed using a standard immunohistochemistry protocol, as described previously, with antibodies directed against each type IV collagen (Supplementary Table S2).24 Images were obtained by fluorescence confocal microscopy using FluoView FV1000 software (Olympus America, Inc., Center Valley, PA, USA). Fluorescence signal quantification was performed using the Volocity 3D Image Analysis Software (PerkinElmer, Waltham, MA, USA). Final fluorescence quantities were determined by subtracting nonspecific fluorescence values, which were obtained by measuring the fluorescence in a field devoid of tissue in each image from the fluorescence in the stroma, Descemet membrane, or endothelium of the secondary antibody–only control.
In Situ Hybridization
In situ hybridization was performed using RNAscope 2.5 HD Detection Reagent Kits (Cat. no. 322350 and Cat. no. 322430; Advanced Cell Diagnostics [ACD], Newark, CA, USA) according to manufacturer's protocol. Briefly, formalin-fixed, paraffin-embedded cornea sections from an individual without a history of corneal disease and PPCD-affected individuals with (P1) and without (P2–P4) a ZEB1 mutation were baked at 60°C for 1 hour on glass slides and then deparaffinized. After treatment with hydrogen peroxidase solution, sections were incubated in target retrieval solution at 95°C to 100°C for 15 minutes, followed by treatment with protease solution for 30 minutes at 40°C. Probes targeting ZEB1 (Hs-ZEB1-C2, Cat. no. 313191-C2; ACD), CCND1 (Hs-CCND1, Cat. no. 591821; ACD), BMP4 (Hs-BMP4, Cat. no. 454301; ACD), or dapB of Bacillus subtilis as a negative control (negative control probe, Cat no. 320751; ACD) were hybridized for 2 hours at 40°C, followed by a series of signal amplification and wash cycles. Tissue sections were counterstained with hematoxylin. Hybridized probes were detected by a chromogenic reaction that produced red chromogenic punctate dots, which were visualized by brightfield microscopy using the Axio Imager 2 microscope (Zeiss, Dublin, CA, USA). The average number of probes per nucleus was calculated by quantifying the total number of red chromogenic dots residing in the nuclear regions of corneal endothelial cells and dividing it by the total number of nuclei in the corneal endothelium of each tissue sample. Background nonspecific probe counts, obtained by using the negative control probe, were subtracted from the ZEB1, CCND1, and BMP4 targeting probe counts for each tissue section.
Ex Vivo Corneal Endothelium Collection for RNA-Seq Analysis
Corneal endothelial tissue from two patients with PPCD, ages 15 (P5) and 5 (P6), were obtained when the patients underwent a Descemet membrane endothelial keratoplasty (P5) or a penetrating keratoplasty (PK) (P6). The Descemet membrane and endothelium were placed onto a piece of filter paper and submerged in TRI Reagent (Thermo Fisher, Waltham, MA, USA) for RNA isolation. Human cornea tissue from two age-matched donors, ages 6 and 17, were obtained from commercial eye banks. Descemet membrane and corneal endothelium were stripped from the donor corneas using a published technique and placed into TRI Reagent for RNA isolation.30,31
Isolation and Culturing of Primary Human Corneal Endothelial Cells
Primary cultures of human corneal endothelial cells (pHCEnC) were isolated from ex vivo donor cornea as previously described.32 Cells were plated in a 12-well tissue culture–treated, nonpyrogenic polystyrene plastic plate, and experiments were performed when the cells achieved an intact and confluent monolayer, prior to the first passage.
ZEB1 Knockdown in Cultured Primary Human Corneal Endothelial Cells
To assess the effect of decreased ZEB1 protein on gene expression in pHCEnC, confluent pHCEnC cultures were transfected with 10 nM anti-ZEB1 small interfering RNA (siRNA) (rGrGrCrCrUrArCrArArUrArArCrUrArGrCrArUrUrUrGrUTG) or scrambled siRNA (OriGene Technologies, Rockville, MD, USA) using Lipofectamine LTX (Life Technologies, Waltham, MA, USA) per the manufacturer's instructions. At 24, 48, 72, and 96 hours post transfection, the transfected cells were lysed in either Tri Reagent (Sigma-Aldrich Corp., St. Louis, MO, USA) or radioimmunoprecipitation assay (RIPA) buffer (150 mM NaCl, 100 mM Tris pH 7.6, 1 mM EDTA, 1% Triton X-100, 1% deoxycholic acid, 0.1% sodium dodecyl sulfate [SDS]) supplemented with fresh 50 mM sodium fluoride, 20 mM phenylmethylsulfonyl fluoride, and protease and phosphatase inhibitors (Life Technologies), for RNA and protein isolation, respectively. ZEB1 knockdown was confirmed by quantitative polymerase chain reaction (qPCR) and Western blotting as described below.
Total RNA Isolation
Total RNA isolated using TRI Reagent from ex vivo corneal endothelium and cultured pHCEnC was purified using the RNeasy Clean-Up Kit (Qiagen). The integrity of the purified RNA was analyzed by the Agilent 2100 Electrophoresis Bioanalyzer System (Agilent Technologies, Inc., Santa Clara, CA, USA).
Next-Generation RNA Sequencing (RNA-Seq)
Purified total RNA was submitted to the UCLA Clinical Microarray Core for library preparation and sequencing. Enrichment of poly(A)-RNA and library preparation for pHCEnC samples were performed using the KAPA Stranded mRNA-Seq Kit, with KAPA mRNA Capture Beads (KAPA Biosystems, Wilmington, MA, USA) per the manufacturer's instructions. Due to low total RNA yields from the PPCD samples, the RNA isolated from PPCD endothelium and age-matched controls was first amplified using the Ovation RNA-Seq System V2 kit (NuGEN Technologies, Inc., San Carlos, CA, USA), followed by library preparation using the KAPA Library Preparation Kit (KAPA Biosystems). Single-end 50-bp reads were generated on the Illumina Hi-seq 3000 (Illumina, Inc., San Diego, CA, USA). The generated FASTQ files and quantitative results are available from the GEO DataSets database (accession number GSE90489; National Center for Biotechnology Information [NCBI], Bethesda, MD, USA).
Next-Generation Sequencing Data Analyses
The FASTQ files containing the RNA-sequencing data were uploaded to the Partek Flow servers (Partek, Inc., Chesterfield, MO, USA). Reads were aligned to the human genome (hg38) using the TopHat2 aligner. Results of the alignment were output in BAM files, which were uploaded to the Partek Genomics Suite software, and the reads and read-depth were transformed to reads per kilobase per million (RPKM) values, a normalized quantity that accounts for gene size. Annotation was performed using the Ensembl 82 transcript database (http://sep2015.archive.ensembl.org; in the public domain, Ensembl, Hinxton, UK). The gene expression threshold level for positive detection of a transcript was calculated as previously described by Ramskold et al.33 Briefly, background RPKM values were calculated by mapping reads from each sample to intergenic regions that have been matched to have identical length distributions and were devoid of expressed sequence tags. After binning samples, false-positive and -negative rates, as a function of RPKM value, were calculated. For each bin, the RPKM value at the intersection point between the false-positive and false-negative rates was determined to be the gene expression threshold level for positive detection. Differential expression analysis was performed and P values were calculated with a 1-way ANOVA model using method of moments. Ingenuity Pathway Analysis (IPA) was used to generate gene-association lists for ZEB1 and OVOL2 and to perform canonical pathway analyses (http://www.ingenuity.com; in the public domain).34 Differentially expressed gene lists were uploaded into the Protein Analysis THrough Evolutionary Relationships (PANTHER, http://pantherdb.org; in the public domain) classification system. Based on overrepresented gene ontology (GO) terms in the gene list data, enriched biological processes and molecular functions were identified using PANTHER's Gene List Analysis software.35,36
Quantitative Polymerase Chain Reaction
Quantitative polymerase chain reaction was used to validate the level of gene expression determined by RNA-seq. Complementary DNA (cDNA) was synthesized from 1 μg total RNA isolated from pHCEnC using the SuperScript III First-Strand kit (Life Technologies). Subsequently, qPCR was performed on the LightCycler 480 System (Roche, Basel, Switzerland) using the KAPA SYBR FAST qPCR Kit (KAPA Biosystems) and transcript-specific oligonucleotide primers that were obtained from the Harvard Primer Bank database (http://pga.mgh. harvard.edu/primerbank/index.html; in the public domain) (Supplementary Table S3).37–39 Reaction conditions were as previously described.40 Relative expression was obtained by comparison to the housekeeping gene RAB7A and calculated using the comparative Ct (2−ΔΔCt) method.41 Relative expression levels were plotted as 2−ΔCt values.
Western Blotting
Western blotting was used to validate ZEB1 knockdown in pHCEnC cultures. At 24, 48, 72, and 96 hours after transfection, protein lysates from each transfected pHCEnC culture were prepared in RIPA buffer. Anti-ZEB1 Western blotting was performed as previously described.32 As a loading control, detection of the glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and alpha-tubulin proteins was performed using anti-GADPH (MAB374, 1:5,000 dilution; Millipore, Billerica, MA, USA) and anti-alpha-tubulin (DM1A, 1:50,000 dilution; Cell Signaling, Danvers, MA, USA) primary antibodies, respectively, with an anti-mouse peroxidase-coupled secondary antibody (170-5047, 1:40,000; BioRad, Hercules, CA, USA).
Results
Type IV Collagen Expression Analysis in PPCD Corneas Using F-IHC
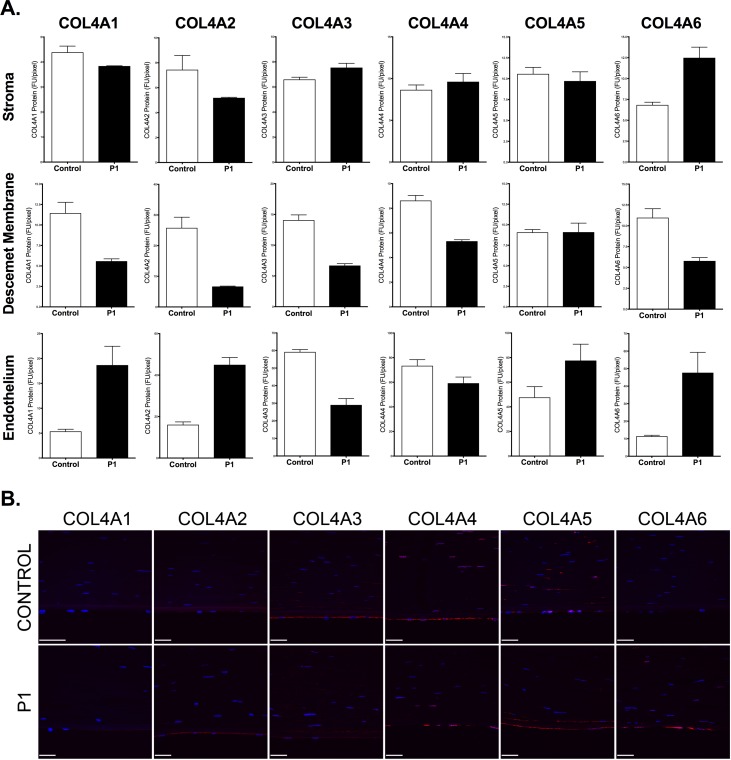
Fluorescence immunohistochemistry staining for each of the type IV collagens (COL4A1, COL4A2, COL4A3, COL4A4, COL4A5, and COL4A6) was performed on a corneal button excised from an individual with PPCD3 (P1) in whom screening of ZEB1 revealed a heterozygous truncating mutation (c.1613delC; p.(Pro538Glnfs*10)). Endothelial expression of COL4A3 was decreased while expression of COL4A1, COL4A2, and COL4A6 was increased compared to a normal cornea (Figs. 1A, 1B). While the expression of COL4A3 in Descemet membrane mirrored that in the endothelium (decreased), the expression of COL4A1, COL4A2, and COL4A6 was decreased instead of increased. The level of expression of COL4A5 in the endothelium and Descemet membrane was not markedly different in the PPCD3 cornea compared to the control cornea. In contrast to the decreased level of COL4A3 in the corneal endothelium and Descemet membrane, it was increased in the stroma in the PPCD3 cornea.
Figure 1.
Dysregulated corneal expression of type IV collagens in posterior polymorphous corneal dystrophy associated with a ZEB1 mutation. (A) Stromal (top row), Descemet membrane (DM) (middle row), and corneal endothelial (bottom row) expression of each of the six type IV collagens in a normal cornea (control) and a cornea from an individual with PPCD3 associated with the p.(Pro538Glnfs*10) mutation in ZEB1 (P1). Five independent fields of view encompassing the stroma, DM, and endothelium were captured for the control and PPCD3 corneas. FU/pixel: fluorescence units per pixel; error bars: SEM between the five independent fields. (B) Representative field of view encompassing the stroma, DM, and endothelium captured from a normal cornea and a cornea from P1. Primary antibodies directed against each of the six type IV collagens and a secondary antibody conjugated to a fluorescent moiety (Alexa Fluor 594, red) were used to detect each type IV collagen protein expression. Cornea sections were counterstained with 4′,6-diamidino-2-phenylindole (DAPI), which stained the nuclei blue.
F-IHC staining for each of the type IV collagens performed on corneal buttons from three individuals with PPCD who did not have a ZEB1 mutation (P2–P4) revealed inconsistent results. In the endothelium, none of the type IV collagens demonstrated consistently increased or decreased expression compared with control, with corneas from P2 to P4 demonstrating both increased and decreased expression of COL4A3 and COL4A5 (Figs. 2A, 2B). In Descemet membrane, the expression of two of the type IV collagens was consistently altered in corneas from P2 to P4 compared to control (COL4A3 expression was decreased and COL4A4 expression was consistently increased). In the stroma, the expression of three of the type IV collagens was consistently altered in corneas from P2 to P4 compared to control (COL4A2 expression decreased and COL4A3 and COL4A4 expression increased).
Figure 2.
Dysregulated corneal expression of type IV collagens in posterior polymorphous corneal dystrophy not associated with a ZEB1 mutation. (A) Stromal (top row), Descemet membrane (middle row), and corneal endothelial (bottom row) expression of each of the six type IV collagens in a normal cornea (control) and three corneas from individuals with PPCD not associated with a ZEB1 mutation (P2–P4). Five independent fields of view encompassing the stroma, DM, and endothelium were captured for the control and non-PPCD3 corneas. FU/pixel: fluorescence units per pixel; error bars: SEM between the five independent fields. (B) Representative field of view encompassing the stroma, DM, and endothelium captured from a normal cornea and corneas from P2 to P4. Primary antibodies directed against each of the six type IV collagens and a secondary antibody conjugated to a fluorescent moiety (Alexa Fluor 594, red) were used to detect each type IV collagen protein expression. Cornea sections were counterstained with DAPI, which stained the nuclei blue.
Type IV Collagen Expression Analysis in PPCD Endothelium Using RNA-Seq
To complement our F-IHC studies of type IV collagens in whole corneal buttons from individuals with PPCD, we performed RNA-seq analysis on corneal endothelium from two PPCD individuals, P5 and P6, who underwent endothelial keratoplasty. Screening of the promoter and coding regions of OVOL2 and ZEB1 in DNA samples from individuals P5 and P6 identified a ZEB1 truncating mutation (c.1381delinsGACGAT) in P5 and no pathogenic variant in either gene in P6. A total of 15,500,029 and 92,764,601 reads were aligned to the hg38 human genome reference for P5 and P6, respectively. For comparison, RNA-seq was performed on normal corneal endothelial cells from aged-matched donors, C5 and C6, which produced 18,118,557 and 57,325,954 aligned reads, respectively. Reads per kilobase per million values of genes expressed within the endothelium of corneas from P5 and P6, and their respective age-matched control tissues, were determined and used to derive gene expression fold changes of type IV collagen in PPCD versus normal ex vivo (ev)HCEnC. Corneal endothelium from P5 and P6 both demonstrated decreased expression of COL4A3, COL4A4, and COL4A6; increased expression of COL4A2; and divergent expression of COL4A1 and COL4A5 (Table 1).
Table 1.
RPKM Values and Fold Changes of Type IV Collagens in PPCD Versus Normal Ex Vivo Corneal Endothelial Cells

Transcriptome Analysis of PPCD Endothelium Using RNA-Seq
As neither COL4A3 nor any of the other type IV collagen subtypes demonstrated a consistent alteration in expression in whole cornea and corneal endothelial samples from individuals with PPCD, we performed a comprehensive transcriptomic analysis on the corneal endothelial samples from individuals C5, C6, P5, and P6. The threshold levels of detection of gene expression for P5 and P6 averaged approximately 0.1 RPKM, which we have designated to be the minimum RPKM value at which any gene is considered to be expressed in our samples. Based on the minimum RPKM value of 0.1 as the detection threshold of expression, the number of genes detected in each sample was P5, 16,681; C5, 18,992; P6, 24,327; and C6, 19,728.
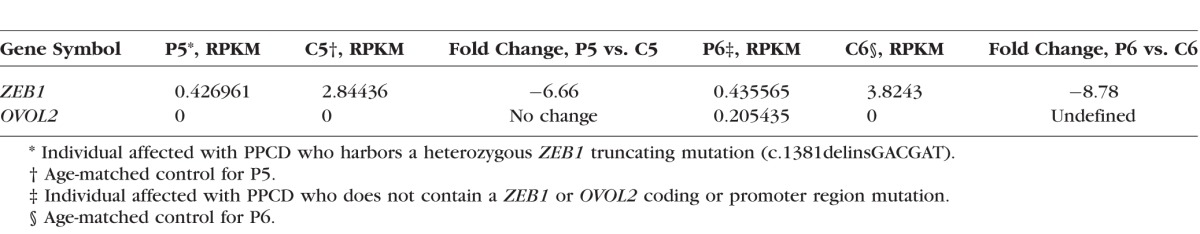
Expression of ZEB1 and OVOL2
ZEB1 expression was decreased 6.7- and 8.8-fold compared to controls in the endothelium of individuals P5 and P6, respectively. OVOL2 was undetected in the corneal endothelium of individual P5, while it was detected in the corneal endothelium of individual P6 (RPKM of 0.205). The age-matched control, C6, exhibited no OVOL2 expression (Table 2).
Table 2.
RPKM Values and Fold Changes of ZEB1 and OVOL2 in PPCD Versus Normal Ex Vivo Corneal Endothelial Cells
Expression of Genes Associated With ZEB1 and OVOL2
Corneal endothelium from individuals P5 and P6 demonstrated differential expression (RPKM value ≥ 0.1, absolute fold change ≥ 2 compared to aged-matched controls) of 5049 and 5952 protein-coding genes, respectively. Fifty-two of the 5049 and 67 of 5952 differentially expressed protein-coding genes are known to be associated with ZEB1, 36 of which were differentially expressed in the corneal endothelium of both individuals, 32 differentially expressed in the same direction (Table 3). One of these 32 genes is LAMC2, which belongs to a group of laminin proteins normally found in the subendothelial region of Descemet membrane. An approximately 16-fold increase in expression of LAMC2 was identified in P5 and a greater than 600-fold increase in expression of LAMC2 was identified in P6 compared to control (RPKM values for P5, 1.03; C5, 0.06223) (RPKM values for P6, 218.383; C6, 0.3363).
Table 3.
Differentially Expressed Genes in PPCD Known to be Associated With ZEB1

Three protein-coding genes (BMP4, CCND1, ZEB1) known to be associated with OVOL2 were differentially expressed in the corneal endothelium from both individuals P5 and P6, each in the same direction. While ZEB1 expression was decreased in both P5 and P6, BMP4 and CCND1 demonstrated increased expression compared to controls (BMP4, 2.3- and 13.1-fold increases, respectively; CCND1, 26.2- and 98.4-fold increases, respectively).
Expression of Corneal Endothelial-Specific Genes
In a previous report, we identified 138 genes, of which 104 are protein coding, considered specific to evHCEnC, as they were expressed in evHCEnC with a mean RPKM value of ≥1, had a RPKM value of <1 in ex vivo human corneal epithelial cells (evHCEpC) and ex vivo human corneal fibroblast cells (evHCFC), and had a fold change in expression of ≥5 in evHCEnC compared with evHCEpC and evHCFC.32 Using a threshold of expression of RPKM ≥ 0.1, 83.7% (87/104) and 53.8% (56/104) of the 104 protein-coding evHCEnC-specific genes were expressed in P5 and P6, respectively, as compared to 97.1% (101/104) and 95.2% (99/104) in the respective controls, C5 and C6. A total of 49 evHCEnC-specific genes were expressed in each of the two PPCD and two control HCEnC samples, while six evHCEnC-specific genes (CA10, CRB2, EPHB1, GPR158, MGAT3, and PPP1R1B) were not expressed in either P5 or P6, but were expressed in the controls (Supplementary Table S4).
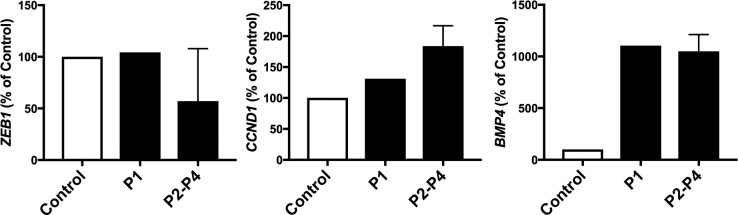
Validation of Differential Expression of ZEB1, BMP4, and CCND1 Using In Situ Hybridization
Data obtained from in situ hybridization of probes targeting ZEB1, BMP4, and CCND1 transcripts in independent corneas from individuals affected with PPCD with (P1) or without (P2–P4) a truncating ZEB1 mutation corroborated the RNA-seq data by demonstrating increased BMP4 and CCND1 transcript levels compared to control (Fig. 3). However, while RNA-seq demonstrated decreased ZEB1 transcript levels in individuals affected with PPCD compared to control, no difference was observed using in situ hybridization comparing P1, P3, and P4 to control.
Figure 3.
Detection of ZEB1, CCND1, and BMP4 transcripts in corneas from individuals with posterior polymorphous corneal dystrophy. In situ hybridization demonstrating the average number of probes targeting ZEB1, CCND1, or BMP4 transcripts per nucleus of endothelial cells in corneas of individuals affected with PPCD (P1–P4) and from an individual with no history of corneal disease (C). P1 is an individual with a ZEB1 mutation (p.(Pro538Glnfs*10)), while P2 to P4 are individuals with PPCD but without a ZEB1 or OVOL2 mutation. BMP4 and CCND1 transcript levels in PPCD are increased compared to the control while ZEB1 transcript levels are unchanged in P1 and decreased in P2 to P4 compared to the control. Error bars: SEM of probe detection between P2, P3, and P4.
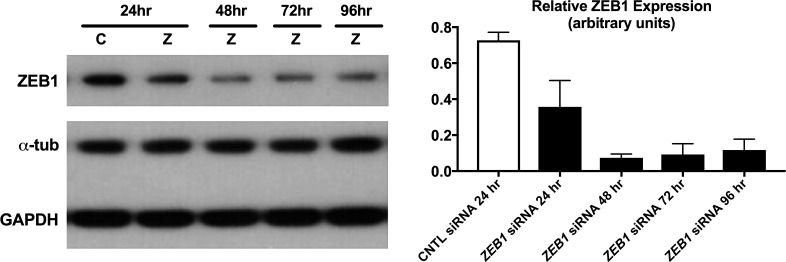
ZEB1 Knockdown in pHCEnC Results in Differentially Expressed Protein-Coding Genes
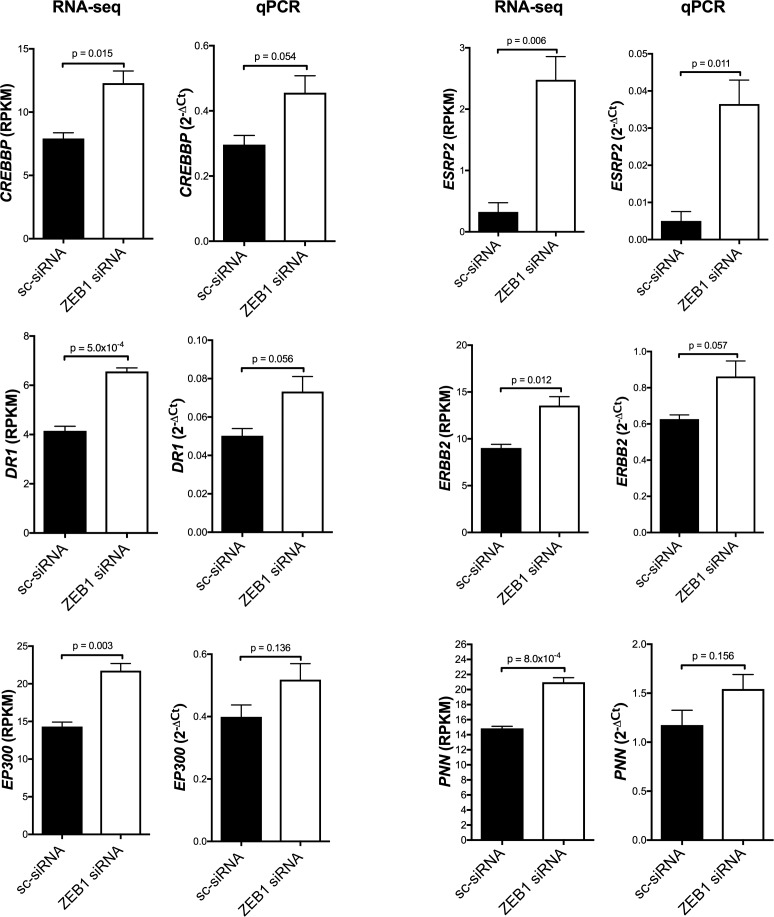
To determine the effects of reduced ZEB1 levels in HCEnC, RNA-seq analysis was performed on pHCEnC that were transfected with either anti-ZEB1 or control siRNA. Transfections were performed in triplicates from three pHCEnC cultures, each isolated from a different normal donor cornea. Western blotting performed to validate successful ZEB1 knockdown demonstrated decreases in ZEB1 protein levels compared to controls between 24 and 96 hours post transfection, with the largest decrease at 48 hours post transfection (Fig. 4). For purposes of generating the baseline for differential expression analyses and determining early expression differences, if any, between pHCEnC transfected with ZEB1 siRNA and pHCEnC transfected with control siRNA, RNA was extracted from transfected pHCEnC cultures at 24 hours post transfection. In addition, RNA was extracted 72 hours post transfection to observe the expected peak effect on genes whose expression is regulated by ZEB1, given that maximal ZEB1 knockdown is observed at 48 hours post transfection. Samples were sequenced by RNA-seq and an average of 31,152,058 aligned reads were obtained per sample. As the duration of decreased ZEB1 protein levels in transfected pHCEnC is significantly shorter than it is in HCEnC in an affected individual with PPCD3, a less stringent fold-change criterion was used for differential expression analysis in pHCEnC transfected with ZEB1 siRNA. Using filtering criteria of RPKM values ≥ 0.1, absolute fold changes ≥ 1.25, and P values < 0.05, only one gene (AL390877.1), which is noncoding, was identified as differentially expressed 24 hours after ZEB1 siRNA transfection compared to controls. Seventy-two hours after ZEB1 siRNA transfection, 889 genes were differentially expressed (RPKM values ≥ 0.1, absolute fold changes ≥ 1.25, and P values < 0.05) compared to controls, with 793 upregulated and 96 downregulated. Seven hundred eleven of the 889 genes are protein coding, of which six (DR1, ESRP2, PNN, ERBB2, CREBBP, and EP300) are known to be associated with ZEB1 and none are known to be associated with OVOL2. qPCR performed to validate differential expression revealed mean fold-change patterns consistent with the RNA-seq analyses for each of the six genes, with four genes (ESRP2, CREBBP, DR1, ERBB2) having P values of 0.057 or less (Fig. 5).
Figure 4.
Validation of ZEB1 knockdown in pHCEnC by Western blotting. ZEB1 protein levels were measured in primary HCEnC at 24, 48, 72, and 96 hours post transfection with ZEB1 siRNA (n = 3) and at 24 hours post transfection with scrambled siRNA (n = 3). This image is a representative blot from three independent samples. Alpha-tubulin and GAPDH were used as loading controls. Error bars represent SEM.
Figure 5.
ZEB1 knockdown increases transcript abundance of select ZEB1-associated protein-coding genes. Transcript levels of six genes regulated by ZEB1 were measured in primary HCEnC 72 hours after transfection with either scrambled siRNA (sc-siRNA) or ZEB1 siRNA. For each gene, ZEB1 knockdown resulted in significantly increased transcript abundance. Statistical analysis was performed using unpaired t-test (n = 3). Error bars represent mean ± SEM of RPKM (RNA-seq) or 2−ΔCt (qPCR) values.
Analysis of Differentially Expressed Protein-Coding Genes in pHCEnC Following ZEB1 Knockdown
Canonical pathway, GO, and gene list comparisons were performed on the 711 differentially expressed protein-coding genes identified in pHCEnC 72 hours after ZEB1 siRNA transfection compared to controls.
Canonical Pathways Analysis
Using Ingenuity Pathway Analysis software, we analyzed the relationships between the 711 differentially expressed protein-coding genes to determine the most impacted canonical pathways following ZEB1 reduction in pHCEnC. Associated with the 711 differentially expressed genes, a total of 38 significantly affected (P value < 0.05) canonical pathways were identified, including pathways involved in proliferation (e.g., HIPPO signaling,42 ERK/MAPK signaling,43 ephrin signaling44); cell adhesion and migration (e.g., ephrin signaling,44 epithelial adherens junction signaling,45 regulation of actin-based motility by Rho,46 cdc42 signaling47); and cell morphology (e.g., Cdc42 signaling47) (Supplementary Table S5).
Gene Ontology Analysis
Based on enriched GO terms, we grouped the 711 differentially expressed protein-coding genes into functional categories based on biological processes and molecular functions using PANTHER. In terms of biological processes, cellular processes (GO:0009987), metabolic processes (GO:0008152), and cellular component organization/biogenesis (GO:0071840) were the three most enriched with over 70% of the 711 genes falling into one or more of these three biological processes (Supplementary Table S6). In terms of molecular functions, binding (GO:0005488) and catalytic activity (GO:0003824) were the two most enriched, with 85% of the 711 genes associated with one or both of these molecular functions (Supplementary Table S7).
Comparison to the Differentially Expressed Genes in PPCD3
When comparing the 5049 differentially expressed protein-coding genes in the corneal endothelium of P5, in whom a ZEB1 truncating mutation was identified, to the 711 differentially expressed protein-coding genes in pHCEnC 72 hours post transfection with ZEB1 siRNA, 211 genes were shared, with the expression of 65 of the 211 genes changed in the same direction (Supplementary Table S8). Five of the 65 genes (PAK6, TTC22, THBS2, SLC7A5, and HIST1H2BK) demonstrated absolute fold changes > 4 based on the differential expression in pHCEnC 72 hours after ZEB1 siRNA transfection compared to controls.
Discussion
Our IHC and RNA-seq analyses of type IV collagens in PPCD are consistent with the IHC results by Merjava and colleagues,27 who demonstrated variable type IV collagen expression in 10 PPCD corneal tissues, one of which was from a patient harboring a ZEB1 truncating mutation (PPCD3).27 Interestingly, the IHC analyses performed on PPCD3 corneal tissue by our group and by Merjava et al. demonstrated overall decreases in COL4A3 expression at the Descemet membrane and/or endothelium compared to control tissue, contrary to the report of ectopic COL4A3 expression in the Descemet membrane and endothelium of a patient affected with PPCD3 using IHC.12,27 The observed variability of type IV collagen expression within Descemet membrane and endothelium of individuals with PPCD in the current study and that of Merjava and colleagues suggests that while a general dysregulation of the expression of collagens is a molecular characteristic of PPCD, the differential expression of a single type IV collagen (e.g., COL4A3) cannot be considered a biomarker of PPCD.
Along with collagen, laminin is another component of Descemet membrane that has been implicated in the pathogenesis of another corneal endothelial dystrophy, Fuchs endothelial corneal dystrophy (FECD). While laminin is normally found in the subendothelial region of Descemet membrane, it is present in increased amounts in an autosomal dominant early-onset form of FECD (FECD1, OMIM 136800)48,49 and its expression in corneal endothelial cells has been shown to be upregulated in FECD.50 In addition, a recently performed genome-wide association study using the largest number of FECD cases and controls to date identified three novel loci (in addition to the TCF4 locus) meeting genome-wide significance (P value < 5 × 10−8), one of which was rs3768617, located in an intron of LAMC1 and approximately 63 kb from LAMC2.51 Formed in different combinations of an alpha-, beta-, and gamma-chain, laminin is a trimeric glycoprotein with 15 isoforms and is involved in cellular processes such as cell signaling, differentiation, migration, and proliferation.52–55 As we have demonstrated a significant increase in LAMC2 expression in two corneas with PPCD, and as it has been previously shown that ZEB1 regulates laminin production and binds to the promoter of LAMC2, it is quite possible that ZEB1 reduction in corneal endothelial cells leads to the altered production and/or ratio of laminin isoforms, impacting corneal endothelial cell signaling and function.56
As well as exhibiting dysregulation of collagens, PPCD leads to the loss of the normal corneal endothelial transcriptome profile, as we have demonstrated that evHCEnC from two individuals with PPCD did not express 16.3% and 46.2% of the 104 protein-coding genes considered to be specific to evHCEnC.32 Therefore it is likely that the loss of expression of evHCEnC-specific genes is contributing to the alterations in corneal endothelial morphology and function that characterize PPCD. In addition to demonstrating a loss of evHCEnC-specific genes, both individuals with PPCD exhibited ectopic expression of the classic epithelial marker CDH1 while showing a greater than 6-fold decrease in ZEB1 expression (Table 3), supporting the hypothesis that PPCD is caused by MET as a result of ZEB1 inactivation. In a previous report, we demonstrated that identified ZEB1 truncating mutations can lead to decreased ZEB1 protein abundance in immortalized HCEnC and suggested, as other investigators have previously, that nonsense-mediated decay (NMD) may be playing a role in PPCD3.9,10,12,16 The observed 6.7-fold decrease in corneal endothelial ZEB1 expression in P5, in whom a heterozygous truncating ZEB1 mutation was identified, supports the hypothesis that PPCD-associated ZEB1 mutations can lead to ZEB1 haploinsufficiency by NMD.
Several OVOL2 promoter mutations have recently been linked to PPCD1 and are hypothesized to cause increased OVOL2 expression by altering regulatory binding sites.24,25 Here, we present the first patient-derived findings that support the hypothesis that increased OVOL2 expression can lead to PPCD by suppressing ZEB1 expression in corneal endothelial cells by demonstrating a nearly 9-fold decrease in ZEB1 expression in the presence of OVOL2 ectopic expression in the corneal endothelium of P6, in whom a promoter or coding region mutation was not identified in either ZEB1 or OVOL2. In addition, we have identified an approximate 2.3- and 13-fold increase of BMP4 expression in P5 and P6, respectively, compared to their respective controls. Regulated by Wnt/beta-catenin signaling, BMP4 is involved in cell differentiation and morphogenesis, and has been demonstrated to induce stratification of corneal epithelial cells during overexpression.57–60 As the expression of BMP4 was increased in evHCEnC affected with PPCD, and BMP4 is known to induce OVOL2 expression, genes involved in BMP4-dependent activation of OVOL2 expression, such as the SMAD genes that form the SMAD1/5/8 complex, should also be considered functional candidate genes for PPCD.59
While a number of possible genes and pathways involved in PPCD have been identified in our initial transcriptomic investigation of evHCEnC from two individuals affected with PPCD, statistical analyses were limited. To complement our ex vivo RNA-seq study, we performed RNA-seq on pHCEnC with transient reduction in ZEB1 levels. In our investigation of the impact of ZEB1 reduction in pHCEnC, ESRP2 demonstrated the largest fold change (a positive fold change of 7.616) among the genes known to associate with ZEB1. Interestingly, ESRP1, the highly conserved paralogue of ESRP2, was also demonstrated to be differentially upregulated in both PPCD endothelium samples, P5 and P6, compared to controls (Table 3). ESRP1 and ESRP2 globally enhance or suppress the utilization of splice sites and promote the formation of epithelial-specific transcript isoforms.61–65 Encoding the epithelial promoter factor pinin, PNN is another splice-regulating gene demonstrated to be upregulated in pHCEnC during ZEB1 reduction. Having roles in maintaining cell–cell adhesion and modulating alternative pre-mRNA splicing, pinin has been demonstrated to bind with ESRP1 to form a complex in cultured human corneal epithelial cells.66–71
In addition to having roles in modulating splicing, pinin (in a complex with other factors) is known to stabilize the nascent CCND1 transcript.72,73 Therefore, it is likely that the increased expression of PNN is contributing to the observed increased expression of CCND1 in corneal endothelial cells from individuals affected with PPCD. Encoding cyclin D1, CCND1 is a highly conserved regulator of cell cycle progression and also has roles in regulating transcriptional activity by modulating the recruitment of chromatin remodeling histone acetyltransferases, p300 and CREB-binding protein (CBP), in a gene-specific manner.73–79 The closely related p300 and CBP are transcriptional activators involved in regulating various cellular processes and are encoded by EP300 and CREBBP, respectively, both of which are upregulated in pHCEnC treated with ZEB1 siRNA.80,81 Future studies investigating the impact of upregulated ESRP1, ESRP2, PNN, CCND1, CREBBP, and EP300 on cellular processes such as alternative splicing, chromatin remodeling, and cell cycle regulation in HCEnC will likely provide additional insights into the development of PPCD and may provide useful biomarkers.
Further detection and study of differentially expressed genes associated with PPCD may identify causative mutations in individuals with PPCD who do not harbor a ZEB1 or OVOL2 mutation, which represents the majority of reported PPCD cases. Given that several splicing factors and histone modifiers associated with ZEB1 have been identified to be differentially upregulated in HCEnC during ZEB1 reduction and/or in individuals affected with PPCD, it is likely that altered splicing events and/or chromatin remodeling is associated with the development of PPCD. Therefore, genome-wide analyses of alternate transcripts and chromatin remodeling in HCEnC from individuals with PPCD and in whom ZEB1 expression has been reduced will likely provide additional insights into the pathomechanisms of PPCD and the regulatory pathways involved in the maintenance of corneal endothelial function.
Supplementary Material
Acknowledgments
Supported by National Eye Institute Grants R01 EY022082 (AJA), P30 EY000331 (Core Grant), the Walton Li Chair in Cornea and Uveitis (AJA), the Stotter Revocable Trust, and an unrestricted grant to Stein Eye Institute from Research to Prevent Blindness.
Disclosure: D.D. Chung, None; R.F. Frausto, None; B.R. Lin, None; E.M. Hanser, None; Z. Cohen, None; A.J. Aldave, None
References
- 1. Krachmer JH. Posterior polymorphous corneal dystrophy: a disease characterized by epithelial-like endothelial cells which influence management and prognosis. Trans Am Ophthalmol Soc. 1985; 83: 413–475. [PMC free article] [PubMed] [Google Scholar]
- 2. Weiss JS,, Moller HU,, Aldave AJ,, et al. IC3D classification of corneal dystrophies--edition 2. Cornea. 2015; 34: 117–159. [DOI] [PubMed] [Google Scholar]
- 3. Aldave AJ,, Ann LB,, Frausto RF,, Nguyen CK,, Yu F,, Raber IM. Classification of posterior polymorphous corneal dystrophy as a corneal ectatic disorder following confirmation of associated significant corneal steepening. JAMA Ophthalmol. 2013; 131: 1583–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Aldave AJ,, Han J,, Frausto RF. Genetics of the corneal endothelial dystrophies: an evidence-based review. Clin Genet. 2013; 84: 109–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Moroi SE,, Gokhale PA,, Schteingart MT,, et al. Clinicopathologic correlation and genetic analysis in a case of posterior polymorphous corneal dystrophy. Am J Ophthalmol. 2003; 135: 461–470. [DOI] [PubMed] [Google Scholar]
- 6. Jirsova K,, Merjava S,, Martincova R,, et al. Immunohistochemical characterization of cytokeratins in the abnormal corneal endothelium of posterior polymorphous corneal dystrophy patients. Exp Eye Res. 2007; 84: 680–686. [DOI] [PubMed] [Google Scholar]
- 7. Merjava S,, Malinova E,, Liskova P,, et al. Recurrence of posterior polymorphous corneal dystrophy is caused by the overgrowth of the original diseased host endothelium. Histochem Cell Biol. 2011; 136: 93–101. [DOI] [PubMed] [Google Scholar]
- 8. Aldave AJ,, Yellore VS,, Yu F,, et al. Posterior polymorphous corneal dystrophy is associated with TCF8 gene mutations and abdominal hernia. Am J Med Genet A. 2007; 143A: 2549–2556. [DOI] [PubMed] [Google Scholar]
- 9. Bakhtiari P,, Frausto RF,, Roldan AN,, Wang C,, Yu F,, Aldave AJ. Exclusion of pathogenic promoter region variants and identification of novel nonsense mutations in the zinc finger E-box binding homeobox 1 gene in posterior polymorphous corneal dystrophy. Mol Vis. 2013; 19: 575–580. [PMC free article] [PubMed] [Google Scholar]
- 10. Chung DW,, Frausto RF,, Ann LB,, Jang MS,, Aldave AJ. Functional impact of ZEB1 mutations associated with posterior polymorphous and Fuchs' endothelial corneal dystrophies. Invest Ophthalmol Vis Sci. 2014; 55: 6159–6166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Evans CJ,, Liskova P,, Dudakova L,, et al. Identification of six novel mutations in ZEB1 and description of the associated phenotypes in patients with posterior polymorphous corneal dystrophy 3. Ann Hum Genet. 2015; 79: 1–9. [DOI] [PubMed] [Google Scholar]
- 12. Krafchak CM,, Pawar H,, Moroi SE,, et al. Mutations in TCF8 cause posterior polymorphous corneal dystrophy and ectopic expression of COL4A3 by corneal endothelial cells. Am J Hum Genet. 2005; 77: 694–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lechner J,, Dash DP,, Muszynska D,, et al. Mutational spectrum of the ZEB1 gene in corneal dystrophies supports a genotype-phenotype correlation. Invest Ophthalmol Vis Sci. 2013; 54: 3215–3223. [DOI] [PubMed] [Google Scholar]
- 14. Liskova P,, Evans CJ,, Davidson AE,, et al. Heterozygous deletions at the ZEB1 locus verify haploinsufficiency as the mechanism of disease for posterior polymorphous corneal dystrophy type 3. Eur J Hum Genet. 2016; 24: 985–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Liskova P,, Palos M,, Hardcastle AJ,, Vincent AL. Further genetic and clinical insights of posterior polymorphous corneal dystrophy 3. JAMA Ophthalmol. 2013; 131: 1296–1303. [DOI] [PubMed] [Google Scholar]
- 16. Liskova P,, Tuft SJ,, Gwilliam R,, et al. Novel mutations in the ZEB1 gene identified in Czech and British patients with posterior polymorphous corneal dystrophy. Hum Mutat. 2007; 28: 638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nguyen DQ,, Hosseini M,, Billingsley G,, Heon E,, Churchill AJ. Clinical phenotype of posterior polymorphous corneal dystrophy in a family with a novel ZEB1 mutation. Acta Ophthalmol. 2010; 88: 695–699. [DOI] [PubMed] [Google Scholar]
- 18. Vincent AL,, Niederer RL,, Richards A,, Karolyi B,, Patel DV,, McGhee CN. Phenotypic characterisation and ZEB1 mutational analysis in posterior polymorphous corneal dystrophy in a New Zealand population. Mol Vis. 2009; 15: 2544–2553. [PMC free article] [PubMed] [Google Scholar]
- 19. Behrens J,, Lowrick O,, Klein-Hitpass L,, Birchmeier W. The E-cadherin promoter: functional analysis of a G.C-rich region and an epithelial cell-specific palindromic regulatory element. Proc Natl Acad Sci U S A. 1991; 88: 11495–11499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Giroldi LA,, Bringuier PP,, de Weijert M,, Jansen C,, van Bokhoven A,, Schalken JA. Role of E boxes in the repression of E-cadherin expression. Biochem Biophys Res Commun. 1997; 241: 453–458. [DOI] [PubMed] [Google Scholar]
- 21. Remacle JE,, Kraft H,, Lerchner W,, et al. New mode of DNA binding of multi-zinc finger transcription factors: deltaEF1 family members bind with two hands to two target sites. EMBO J. 1999; 18: 5073–5084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Vandewalle C,, Van Roy F,, Berx G. The role of the ZEB family of transcription factors in development and disease. Cell Mol Life Sci. 2009; 66: 773–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kitazawa K,, Hikichi T,, Nakamura T,, et al. OVOL2 maintains the transcriptional program of human corneal epithelium by suppressing epithelial-to-mesenchymal transition. Cell Rep. 2016; 15: 1359–1368. [DOI] [PubMed] [Google Scholar]
- 24. Le DJ,, Chung DW,, Frausto RF,, Kim MJ,, Aldave AJ. Identification of potentially pathogenic variants in the posterior polymorphous corneal dystrophy 1 locus. PLoS One. 2016; 11: e0158467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Davidson AE,, Liskova P,, Evans CJ,, et al. Autosomal-dominant corneal endothelial dystrophies CHED1 and PPCD1 are allelic disorders caused by non-coding mutations in the promoter of OVOL2. Am J Hum Genet. 2016; 98: 75–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Liu Y,, Peng X,, Tan J,, Darling DS,, Kaplan HJ,, Dean DC. Zeb1 mutant mice as a model of posterior corneal dystrophy. Invest Ophthalmol Vis Sci. 2008; 49: 1843–1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Merjava S,, Liskova P,, Jirsova K. Immunohistochemical characterization of collagen IV in control corneas and in corneas obtained from patients suffering from posterior polymorphous corneal dystrophy [in Slovak]. Cesk Slov Oftalmol. 2008; 64: 115–119. [PubMed] [Google Scholar]
- 28. Chung DW,, Frausto RF,, Chiu S,, Lin BR,, Aldave AJ. Investigating the molecular basis of PPCD3: characterization of ZEB1 regulation of COL4A3 expression. Invest Ophthalmol Vis Sci. 2016; 57: 4136–4143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chung DD,, Frausto RF,, Cervantes AE,, et al. Confirmation of the OVOL2 promoter mutation c.-307T>C in posterior polymorphous corneal dystrophy 1. PLoS One. 2017; 12: e0169215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bednarz J,, Teifel M,, Friedl P,, Engelmann K. Immortalization of human corneal endothelial cells using electroporation protocol optimized for human corneal endothelial and human retinal pigment epithelial cells. Acta Ophthalmol Scand. 2000; 78: 130–136. [DOI] [PubMed] [Google Scholar]
- 31. Price MO,, Giebel AW,, Fairchild KM,, Price FW., Jr. Descemet's membrane endothelial keratoplasty: prospective multicenter study of visual and refractive outcomes and endothelial survival. Ophthalmology. 2009; 116: 2361–2368. [DOI] [PubMed] [Google Scholar]
- 32. Frausto RF,, Le DJ,, Aldave AJ. Transcriptomic analysis of cultured corneal endothelial cells as a validation for their use in cell replacement therapy. Cell Transplant. 2016; 25: 1159–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ramskold D,, Wang ET,, Burge CB,, Sandberg R. An abundance of ubiquitously expressed genes revealed by tissue transcriptome sequence data. PLoS Comput Biol. 2009; 5: e1000598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jimenez-Marin A,, Collado-Romero M,, Ramirez-Boo M,, Arce C,, Garrido JJ. Biological pathway analysis by ArrayUnlock and Ingenuity Pathway Analysis. BMC Proc. 2009; 3 suppl 4: S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mi H,, Muruganujan A,, Casagrande JT,, Thomas PD. Large-scale gene function analysis with the PANTHER classification system. Nat Protoc. 2013; 8: 1551–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mi H,, Poudel S,, Muruganujan A,, Casagrande JT,, Thomas PD. PANTHER version 10: expanded protein families and functions, and analysis tools. Nucleic Acids Res. 2016; 44: D336–D342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Spandidos A,, Wang X,, Wang H,, Dragnev S,, Thurber T,, Seed B. A comprehensive collection of experimentally validated primers for Polymerase Chain Reaction quantitation of murine transcript abundance. BMC Genomics. 2008; 9: 633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Spandidos A,, Wang X,, Wang H,, Seed B. PrimerBank: a resource of human and mouse PCR primer pairs for gene expression detection and quantification. Nucleic Acids Res. 2010; 38: D792–D799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wang X,, Seed B. A. PCR primer bank for quantitative gene expression analysis. Nucleic Acids Res. 2003; 31: e154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kim MJ,, Frausto RF,, Rosenwasser GO,, et al. Posterior amorphous corneal dystrophy is associated with a deletion of small leucine-rich proteoglycans on chromosome 12. PLoS One. 2014; 9: e95037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Livak KJ,, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001; 25: 402–408. [DOI] [PubMed] [Google Scholar]
- 42. Ehmer U,, Sage J. Control of proliferation and cancer growth by the Hippo signaling pathway. Mol Cancer Res. 2016; 14: 127–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sun Y,, Liu WZ,, Liu T,, Feng X,, Yang N,, Zhou HF. Signaling pathway of MAPK/ERK in cell proliferation, differentiation, migration, senescence and apoptosis. J Recept Signal Transduct Res. 2015; 35: 600–604. [DOI] [PubMed] [Google Scholar]
- 44. Steinle JJ,, Meininger CJ,, Chowdhury U,, Wu G,, Granger HJ. Role of ephrin B2 in human retinal endothelial cell proliferation and migration. Cell Signal. 2003; 15: 1011–1017. [DOI] [PubMed] [Google Scholar]
- 45. Shi Y,, Tabesh M,, Sugrue SP. Role of cell adhesion-associated protein, pinin (DRS/memA), in corneal epithelial migration. Invest Ophthalmol Vis Sci. 2000; 41: 1337–1345. [PubMed] [Google Scholar]
- 46. Spiering D,, Hodgson L. Dynamics of the Rho-family small GTPases in actin regulation and motility. Cell Adh Migr. 2011; 5: 170–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kodama A,, Takaishi K,, Nakano K,, Nishioka H,, Takai Y. Involvement of Cdc42 small G protein in cell-cell adhesion, migration and morphology of MDCK cells. Oncogene. 1999; 18: 3996–4006. [DOI] [PubMed] [Google Scholar]
- 48. Gottsch JD,, Zhang C,, Sundin OH,, Bell WR,, Stark WJ,, Green WR. Fuchs corneal dystrophy: aberrant collagen distribution in an L450W mutant of the COL8A2 gene. Invest Ophthalmol Vis Sci. 2005; 46: 4504–4511. [DOI] [PubMed] [Google Scholar]
- 49. Zhang C,, Bell WR,, Sundin OH,, et al. Immunohistochemistry and electron microscopy of early-onset fuchs corneal dystrophy in three cases with the same L450W COL8A2 mutation. Trans Am Ophthalmol Soc. 2006; 104: 85–97. [PMC free article] [PubMed] [Google Scholar]
- 50. Matthaei M,, Hu J,, Kallay L,, et al. Endothelial cell microRNA expression in human late-onset Fuchs' dystrophy. Invest Ophthalmol Vis Sci. 2014; 55: 216–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Afshari NA,, Igo RP, Jr,, Morris NJ,, et al. Genome-wide association study identifies three novel loci in Fuchs endothelial corneal dystrophy. Nat Commun. 2017; 8: 14898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Aumailley M. The laminin family. Cell Adh Migr. 2013; 7: 48–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Miner JH,, Yurchenco PD. Laminin functions in tissue morphogenesis. Annu Rev Cell Dev Biol. 2004; 20: 255–284. [DOI] [PubMed] [Google Scholar]
- 54. Scheele S,, Nystrom A,, Durbeej M,, Talts JF,, Ekblom M,, Ekblom P. Laminin isoforms in development and disease. J Mol Med (Berl). 2007; 85: 825–836. [DOI] [PubMed] [Google Scholar]
- 55. Okumura N,, Kakutani K,, Numata R,, et al. Laminin-511 and -521 enable efficient in vitro expansion of human corneal endothelial cells. Invest Ophthalmol Vis Sci. 2015; 56: 2933–2942. [DOI] [PubMed] [Google Scholar]
- 56. Drake JM,, Barnes JM,, Madsen JM,, Domann FE,, Stipp CS,, Henry MD. ZEB1 coordinately regulates laminin-332 and {beta}4 integrin expression altering the invasive phenotype of prostate cancer cells. J Biol Chem. 2010; 285: 33940–33948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Baker JC,, Beddington RS,, Harland RM. Wnt signaling in Xenopus embryos inhibits bmp4 expression and activates neural development. Genes Dev. 1999; 13: 3149–3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Fujimori S,, Novak H,, Weissenbock M,, et al. Wnt/beta-catenin signaling in the dental mesenchyme regulates incisor development by regulating Bmp4. Dev Biol. 2010; 348: 97–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zhang T,, Zhu Q,, Xie Z,, et al. The zinc finger transcription factor Ovol2 acts downstream of the bone morphogenetic protein pathway to regulate the cell fate decision between neuroectoderm and mesendoderm. J Biol Chem. 2013; 288: 6166–6177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zhang Y,, Yeh LK,, Zhang S,, et al. Wnt/beta-catenin signaling modulates corneal epithelium stratification via inhibition of Bmp4 during mouse development. Development. 2015; 142: 3383–3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Dittmar KA,, Jiang P,, Park JW,, et al. Genome-wide determination of a broad ESRP-regulated posttranscriptional network by high-throughput sequencing. Mol Cell Biol. 2012; 32: 1468–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Hayakawa A,, Saitoh M,, Miyazawa K. Dual roles for epithelial splicing regulatory proteins 1 (ESRP1) and 2 (ESRP2) in cancer progression. Adv Exp Med Biol. 2017; 925: 33–40. [DOI] [PubMed] [Google Scholar]
- 63. Ishii H,, Saitoh M,, Sakamoto K,, et al. Epithelial splicing regulatory proteins 1 (ESRP1) and 2 (ESRP2) suppress cancer cell motility via different mechanisms. J Biol Chem. 2014; 289: 27386–27399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Warzecha CC,, Jiang P,, Amirikian K,, et al. An ESRP-regulated splicing programme is abrogated during the epithelial-mesenchymal transition. EMBO J. 2010; 29: 3286–3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Warzecha CC,, Shen S,, Xing Y,, Carstens RP. The epithelial splicing factors ESRP1 and ESRP2 positively and negatively regulate diverse types of alternative splicing events. RNA Biol. 2009; 6: 546–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Alpatov R,, Munguba GC,, Caton P,, et al. Nuclear speckle-associated protein Pnn/DRS binds to the transcriptional corepressor CtBP and relieves CtBP-mediated repression of the E-cadherin gene. Mol Cell Biol. 2004; 24: 10223–10235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Ouyang P,, Sugrue SP. Characterization of pinin, a novel protein associated with the desmosome-intermediate filament complex. J Cell Biol. 1996; 135: 1027–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Shi J,, Sugrue SP. Dissection of protein linkage between keratins and pinin, a protein with dual location at desmosome-intermediate filament complex and in the nucleus. J Biol Chem. 2000; 275: 14910–14915. [DOI] [PubMed] [Google Scholar]
- 69. Wang P,, Lou PJ,, Leu S,, Ouyang P. Modulation of alternative pre-mRNA splicing in vivo by pinin. Biochem Biophys Res Commun. 2002; 294: 448–455. [DOI] [PubMed] [Google Scholar]
- 70. Zimowska G,, Shi J,, Munguba G,, et al. Pinin/DRS/memA interacts with SRp75, SRm300 and SRrp130 in corneal epithelial cells. Invest Ophthalmol Vis Sci. 2003; 44: 4715–4723. [DOI] [PubMed] [Google Scholar]
- 71. Joo JH,, Correia GP,, Li JL,, Lopez MC,, Baker HV,, Sugrue SP. Transcriptomic analysis of PNN- and ESRP1-regulated alternative pre-mRNA splicing in human corneal epithelial cells. Invest Ophthalmol Vis Sci. 2013; 54: 697–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Bracken CP,, Wall SJ,, Barre B,, Panov KI,, Ajuh PM,, Perkins ND. Regulation of cyclin D1 RNA stability by SNIP1. Cancer Res. 2008; 68: 7621–7628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Witzel, II,, Koh LF,, Perkins ND. Regulation of cyclin D1 gene expression. Biochem Soc Trans. 2010; 38: 217–222. [DOI] [PubMed] [Google Scholar]
- 74. Alao JP. The regulation of cyclin D1 degradation: roles in cancer development and the potential for therapeutic invention. Mol Cancer. 2007; 6: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Qie S,, Diehl JA. Cyclin D1, cancer progression, and opportunities in cancer treatment. J Mol Med (Berl). 2016; 94: 1313–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Fu M,, Wang C,, Rao M,, et al. Cyclin D1 represses p300 transactivation through a cyclin-dependent kinase-independent mechanism. J Biol Chem. 2005; 280: 29728–29742. [DOI] [PubMed] [Google Scholar]
- 77. McMahon C,, Suthiphongchai T,, DiRenzo J,, Ewen ME. P/CAF associates with cyclin D1 and potentiates its activation of the estrogen receptor. Proc Natl Acad Sci U S A. 1999; 96: 5382–5387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Pestell RG. New roles of cyclin D1. Am J Pathol. 2013; 183: 3–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Reutens AT,, Fu M,, Wang C,, et al. Cyclin D1 binds the androgen receptor and regulates hormone-dependent signaling in a p300/CBP-associated factor (P/CAF)-dependent manner. Mol Endocrinol. 2001; 15: 797–811. [DOI] [PubMed] [Google Scholar]
- 80. Chan HM,, La Thangue NB. p300/CBP proteins: HATs for transcriptional bridges and scaffolds. J Cell Sci. 2001; 114: 2363–2373. [DOI] [PubMed] [Google Scholar]
- 81. Goodman RH,, Smolik S. CBP/p300 in cell growth, transformation, and development. Genes Dev. 2000; 14: 1553–1577. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.