Significance
Bcc bacteria are intrinsically resistant to multiple antibiotics. They are major pathogens in patients with cystic fibrosis (CF) and can cause severe necrotizing pneumonia, which is often fatal. Hopanoid biosynthesis is one of the major mechanisms involved in multiple antimicrobial resistance of Bcc pathogens. The hpnN gene of B. multivorans encodes an integral membrane protein of the HpnN family of transporters, which is responsible for shuttling hopanoids to the outer membrane. Here, we report crystal structures of B. multivorans HpnN that indicate a plausible pathway for hopanoid transport. Overall our data suggest a novel mechanism for hopanoid transport involved in cell wall remodeling, which is critical for mediating multidrug resistance in Burkholderia.
Keywords: hopanoid transport, HpnN transporter, Burkholderia multivorans, multidrug resistance, cell wall remodeling
Abstract
Strains of the Burkholderia cepacia complex (Bcc) are Gram-negative opportunisitic bacteria that are capable of causing serious diseases, mainly in immunocompromised individuals. Bcc pathogens are intrinsically resistant to multiple antibiotics, including β-lactams, aminoglycosides, fluoroquinolones, and polymyxins. They are major pathogens in patients with cystic fibrosis (CF) and can cause severe necrotizing pneumonia, which is often fatal. Hopanoid biosynthesis is one of the major mechanisms involved in multiple antimicrobial resistance of Bcc pathogens. The hpnN gene of B. multivorans encodes an integral membrane protein of the HpnN family of transporters, which is responsible for shuttling hopanoids to the outer membrane. Here, we report crystal structures of B. multivorans HpnN, revealing a dimeric molecule with an overall butterfly shape. Each subunit of the transporter contains 12 transmembrane helices and two periplasmic loops that suggest a plausible pathway for substrate transport. Further analyses indicate that HpnN is capable of shuttling hopanoid virulence factors from the outer leaflet of the inner membrane to the periplasm. Taken together, our data suggest that the HpnN transporter is critical for multidrug resistance and cell wall remodeling in Burkholderia.
The successful human pathogen Burkholderia multivorans is a member of the Burkholderia cepacia complex (Bcc) that causes pneumonia in immunocompromised individuals with underlying lung diseases, such as cystic fibrosis (CF) and chronic granulomatous disease (CGD) (1, 2). Bcc consists of a group of at least 17 closely related Gram-negative bacteria with extreme genetic capacity and metabolic diversity. All Bcc members can trigger chronic airway infections in patients with CF and have emerged as opportunistic pulmonary pathogens (3). Burkholderia cenocepacia and B. multivorans are the two most commonly isolated species (4, 5), which are threats for outbreaks. Bcc infections in patients with CF are associated with enhanced morbidity and mortality. They also have the capacity to cause rapid clinical deterioration with septicemia that leads to death. Several outbreaks of B. multivorans causing severe morbidity and mortality in both patients with CF and patients without CF have occurred (6–8). In 2013, the rapid emergence of a ceftazidime-resistant strain of B. multivorans in a patient with CF was identified in the United States (7). It was found that the resistant strain maintained dominance, resulting in an overall decline in patient health and treatment efficiency. Subsequently, a widespread outbreak of infection caused by B. multivorans was reported from the Czech Republic (8). Surprisingly, this outbreak of B. multivorans affected patients without CF with 24% mortality rate, indicating that B. multivorans is a significant and emerging threat beyond patients with CF.
Bcc pathogens are intrinsically resistant to a broad range of antimicrobials, including β-lactams, fluoroquinolones, aminoglycosides, polymyxins, and cationic peptides, creating a major challenge to the treatment of Bcc pulmonary infections (9–12). A critical line of defense against antimicrobial agents in Burkholderia species is the permeability barrier of the outer membrane. Most of these species contain a modified lipopolysaccharide, which results in polymyxin resistance (13). In addition, the permeability of the major outer membrane porin channel Omp38 appears to be low for antibiotics (14). The presence of a large number of multidrug efflux pumps, belonging to the resistance-nodulation-cell division (RND) superfamily, also plays a major role in the intrinsic resistance to a variety of antimicrobials. It has been found that some Bcc bacteria harbor between 11 and 16 hydrophobic/amphiphile efflux 1 (HAE1)-RND pumps that are responsible for drug efflux (13).
In B. multivorans, it has been reported that hopanoids play a predominant role in supporting membrane stability and barrier function, thus participating in multidrug resistance (9, 11). A mutant strain of B. multivorans lacking the ability to synthesize hopanoids exhibits hypersensitivity to polymyxin B and colistin (9). Hopanoids are pentacyclic triterpenoid lipids that are sterol analogs in prokaryotic membranes (15–17). Like cholesterols in eukaryotic membranes, hopanoids are capable of inserting in bacterial membranes and contributing to their stability and stiffness (18). Hopanoids help membranes withstand damaging stress conditions, including high temperature, low pH, and the presence of antibiotics (9, 11, 12, 18). Not all bacteria produce hopanoids, but they play a vital role in those that do make them. It has been shown that hopanoid production plays an important role in the physiology and pathogenesis of B. cenocepacia (12, 19).
Despite the importance of hopanoids in bacteria, the mechanism of intracellular hopanoid trafficking for cell wall remodeling has not been explored. A subfamily of the RND superfamily of transporters (20), termed hopanoid biosynthesis-associated RND (HpnN) transporters (21), are responsible for shuttling hopanoids from the cytoplasmic membrane to the outer membrane of Gram-negative bacteria (22). Typically, an RND efflux pump works in conjunction with a periplasmic membrane fusion protein (MFP) (23–25). However, the HpnN-subfamily transporters do not seem to associate with any of these MFPs. As an initial step to elucidate the mechanism of hopanoid transport, we here present the crystal structure of the B. multivorans HpnN transporter that is essential for cell wall biogenesis in this pathogen. A combination of the 3D structure and genetic analysis allows us to identify important residues for the function of this membrane protein.
Results
Overall Structure of B. multivorans HpnN.
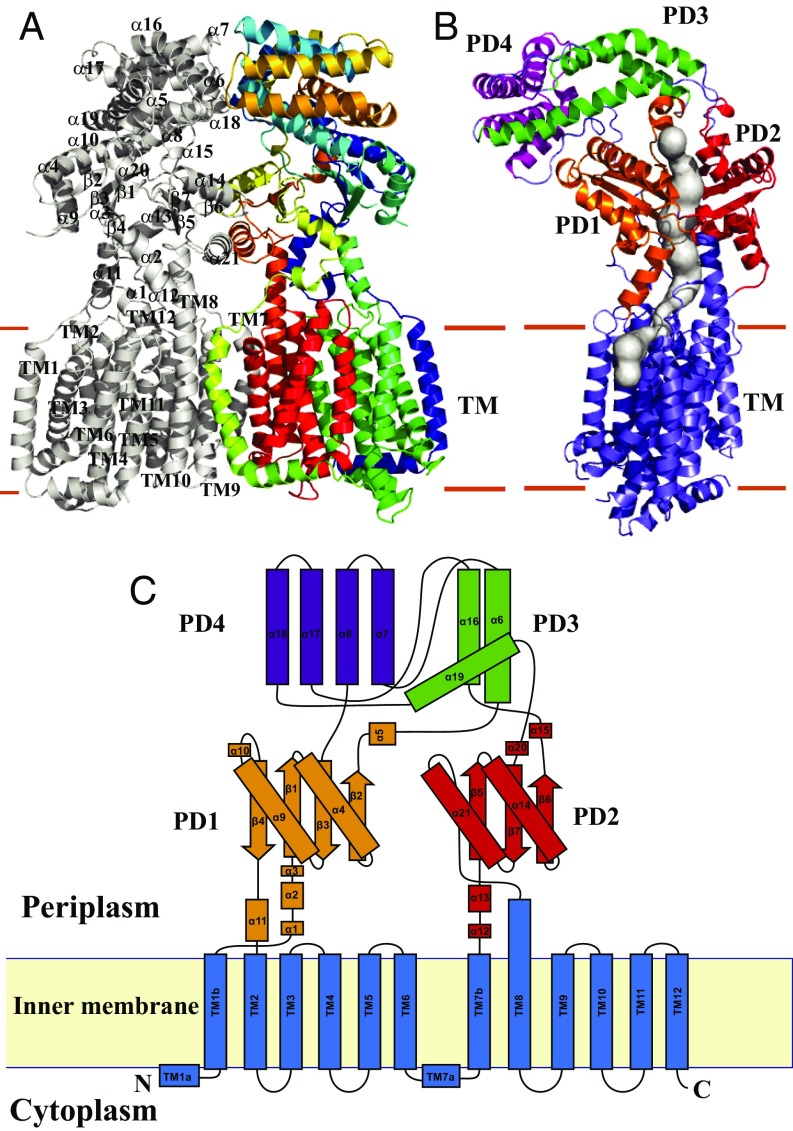
B. multivorans HpnN consists of 877 amino acids (SI Appendix, Fig. S1). Two distinct conformations of HpnN (forms I and II) were captured in two different forms of crystals (SI Appendix, Table S1). In each structure, two monomers were found in the asymmetric unit arranged as a dimer (SI Appendix, Figs. S2 and S3). The dimeric form of HpnN in the crystal lattice is in good agreement with the oligomerization state of this membrane protein in detergent solution, in which it assembles as a dimer (SI Appendix, Fig. S4). Overall, the topology of HpnN is unique. The HpnN dimer is butterfly shaped with a twofold symmetry axis perpendicular to the membrane plane (Fig. 1A). The overall structure of HpnN indicates that this membrane protein mainly constitutes the transmembrane and periplasmic domains. Viewed in parallel to the membrane, the dimer is about 110 Å tall, 100 Å wide, and 52 Å thick.
Fig. 1.
Structure of the B. mulitvorans HpnN transporter. (A) Ribbon diagram of a dimer of HpnN viewed in the membrane plane. The Right subunit of the dimer is colored using a rainbow gradient from the N terminus (blue) to the C terminus (red), whereas the Left subunit is colored gray. Overall, the HpnN dimer forms a butterfly-shaped structure. (B) Each subunit of the HpnN transport forms a channel (colored gray) spanning the outer leaflet of the inner membrane and up to the periplasmic domain. This figure depicts the Left subunit of the form I structure of the HpnN dimer. The orientation of this HpnN subunit has been rotated by 60° counterclockwise, based on the vertical C2 symmetry axis of the HpnN dimer, compared with the orientation of A. This channel was calculated using the program CAVER (loschmidt.chemi.muni.cz/caver). The transmembrane helices are colored slate. PD1–PD4 are colored orange, red, green, and magenta, respectively. (C) Secondary structural topology of the HpnN monomer. The topology was constructed based on the crystal structure of HpnN. The TM domain, PD1–PD4 are colored blue, orange, red, green, and purple, respectively.
Each protomer of HpnN in the dimer contains 12 transmembrane helices (TMs 1–12 and TMs 1′–12′, respectively). In addition, the monomer possesses a large periplasmic domain formed by two periplasmic loops between TMs 1 and 2 (loop 1), and between TMs 7 and 8 (loop 2). Loop 1 is composed of 11 α-helices and four β-strands, whereas loop 2 constitutes 10 α-helices and three β-strands. The N-terminal and C-terminal halves of the transmembrane region are assembled in a pseudosymmetrical fashion, although the periplasmic loops 1 and 2 are quite asymmetrical. Interestingly, the transmembrane domain of HpnN can be superimposed to those of AcrB (26), CusA (27), and SecDE (28) with root mean square deviation (rmsd) values of 2.8 Å, 2.7 Å, and 2.1 Å (SI Appendix, Fig. S5). However, the architectures of their periplasmic domains are completely different. The TMs are membrane embedded, but TM8 is significantly longer and protrudes into the periplasm. TM2 and TM8 directly tether the periplasmic subdomains PD1 and PD2, respectively (Fig. 1 B and C). A hairpin is formed in the middle section of each periplasmic loop (loops 1 and 2). These two α-helical hairpins contact one another through a coiled-coil interaction and form a four α-helix bundle, contributing to the hairpin subdomain PD4. PD4 is connected to PD1 and PD2 through an elongated α-helical subdomain PD3, which is composed of three α-helices (Fig. 1). Several long flexible loops are found to link each periplasmic subdomain, suggesting that the periplasmic domain of HpnN is quite flexible in nature.
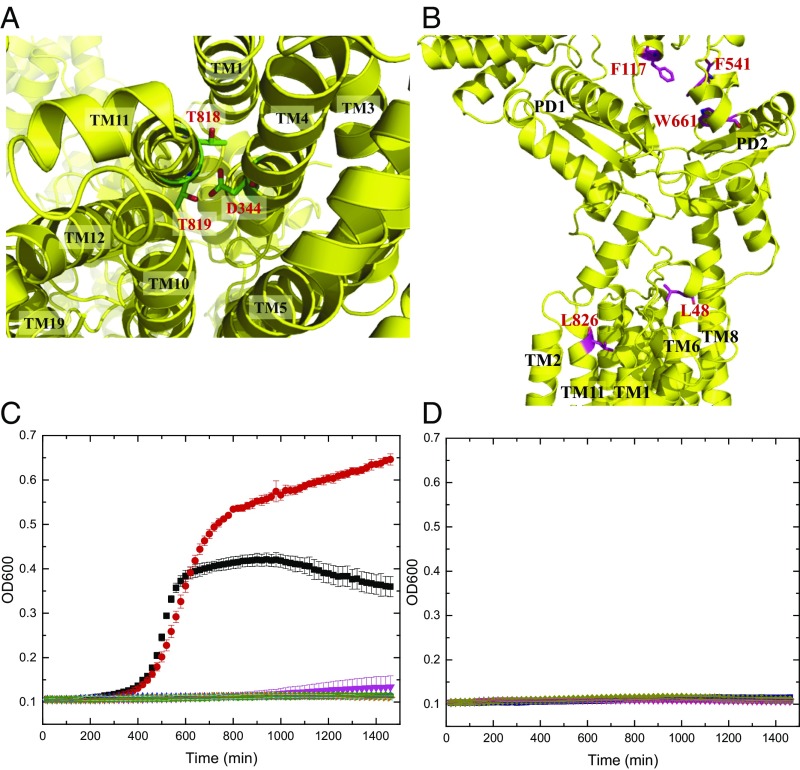
The crystal structure reveals that TMs 7–9 are involved in the formation of the dimer. In addition, α6, α14, α18, α21, and β6 contribute to form the dimer interface. Dimerization occurs mainly through van der Waals interactions, as the interaction surface is mostly hydrophobic in nature. The buried surface areas at the interface between the two monomers of form I and form II were estimated to be ∼2,610 Å2 and 2,070 Å2 (SI Appendix, Fig. S6) with the shape complementarity index scores of 0.66 and 0.65, suggesting that the oligomerization interface is extensive for this transmembrane protein (29). Surprisingly, each protomer of HpnN forms a channel spanning the outer leaflet of the inner membrane and up to the periplasmic domain (Figs. 1B and 2). We suspect that the HpnN transporter may use this channel to shuttle hopanoid molecules to the outer membrane (Fig. 2). The narrowest region of this channel is situated at the two flexible loops connecting TM1 and TM2 to the periplasmic domain. Residue L48 is positioned at this narrowest region, indicating that this conserved amino acid plays a significant functional role. A cavity is formed within TMs 2, 4, and 11, which also creates the beginning section of the channel. This cavity runs horizontally along the surface of the outer leaflet of the inner membrane and then directly connects to the vertical portion of the channel, leading to the periplasm. Potentially, this cavity may form a hopanoid binding site. Interestingly, within the vicinity of this cavity, the hydrophobic residues F270, V332, V339, and L826 are conserved (SI Appendix, Fig. S1). These residues may play a crucial role in recognizing hopanoids. The end of this channel is located at the top portion of PD1 and PD2, where the short helix α5 is also involved in forming this exit. Alignment of protein sequences indicates that several conserved aromatic residues, including F117, F541, and W661 (SI Appendix, Fig. S1), are found to line the wall of this exiting site. These conserved residues may play an important functional role for transporting hopanoids in this membrane protein.
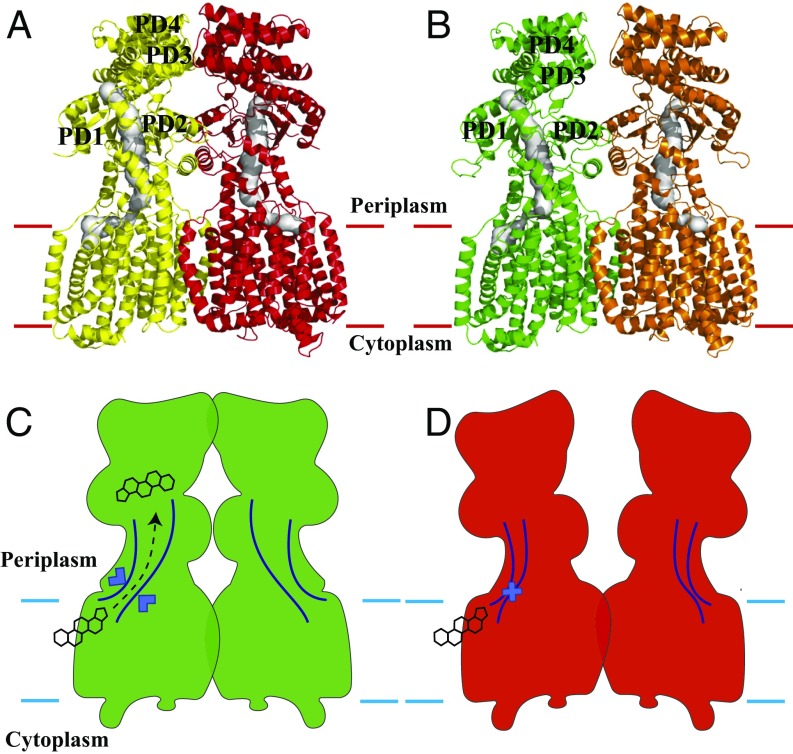
Fig. 2.
Channel in the HpnN transporter. (A) Channel formed by each subunit of the form I structure of HpnN. Each channel is colored gray. The Left and Right subunits of HpnN are colored yellow and red, respectively. (B) Channel formed by each subunit of the form II structure of HpnN. Each channel is colored gray. The Left and Right subunits of HpnN are colored green and orange, respectively. The calculations were done using the program CAVER (loschmidt.chemi.muni.cz/caver). (C) Schematic representation illustrating the channel is open in each subunit of the form I structure of HpnN. The HpnN transporter may use this open channel to shuttle hopanoid molecules to the outer membrane. (D) Schematic representation illustrating the channel is closed in each subunit of the form II structure of HpnN. In this conformation, hopanoid molecules may not be able to pass through the channel.
HpnN is a proton-motive-force (PMF)–dependent transporter (17, 30). Within the transmembrane region, we found that the conserved residues D344, T818, and T819 (SI Appendix, Fig. S1) of HpnN are in close proximity and seem to interact with each other to form a triad. It has been reported that threonine can participate in forming part of a proton translocation pathway (31). These three residues are less than 4 Å away from each other. Thus, they may be involved in creating a proton-relay network to translocate protons for energy coupling. As we crystallized HpnN at pH 3.5, residue D344 should be protonated under this acidic environment. In addition, it has been found by NMR spectroscopy that threonine can act as a proton donor or acceptor to form a hydrogen bond (32). Therefore, it is possible that one of the threonines, probably T819, may donate its hydroxyl proton to interact with the side chain oxygen of D344 to form a hydrogen bond. If this is the case, then T818 may participate in the proton-relay network as a proton acceptor to receive the acidic proton from D344 in this triad. During proton translocation, other nearby amino acids or solvent molecules may contribute to help transfer protons from the periplasm to the cytosol.
Conformational Flexibility of the Periplasmic Domain of HpnN.
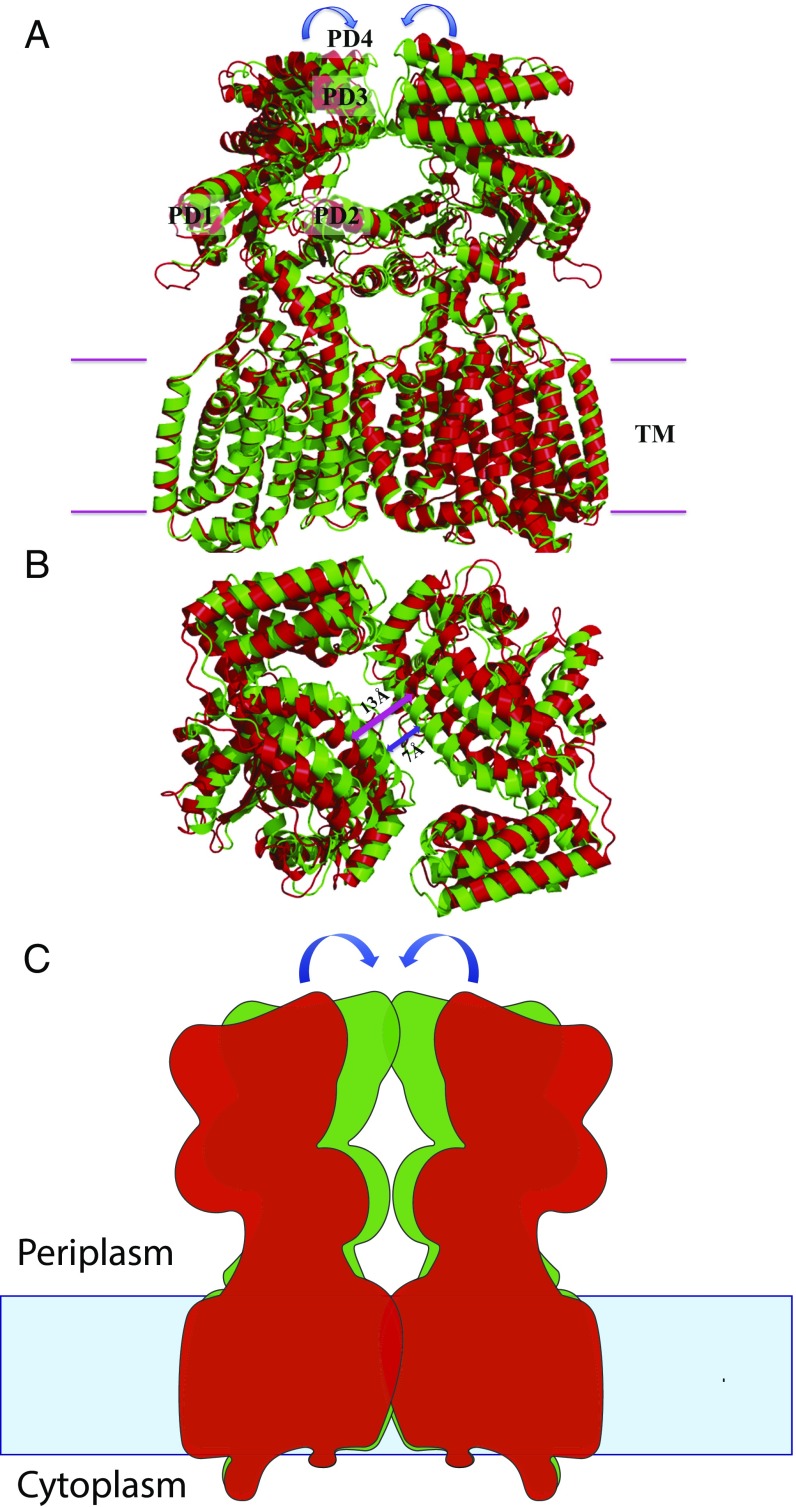
A comparison of the dimeric structures of forms I and II suggests that these two structures depict two different transient states of the transporter (Figs. 2 and 3). Superimposition of the form I and form II structures of HpnN results in an overall rmsd of 2.6 Å (Fig. 3). The major difference between these two structures is in the periplasmic domain of the transporter. This conformational change can be interpreted as a rigid-body swinging motion of the periplasmic domain with respect to the transmembrane domain (Fig. 3). Although the two conformations captured by crystallization may be caused by the difference in crystallization conditions, we suspect that this swinging motion may be related to the transport mechanism of HpnN. The two flexible loops connecting TM1 and TM2 to the periplasmic domain appear to form the hinge, which also creates the narrowest region of the elongated channel. The conserved residue L48 is found in the vicinity of the narrowest region. This residue may be important for the gating of this transporter. Based on the crystal structures of HpnN, the form II structure may represent the closed state, whereas the form I conformation may imitate the open state of the elongated channel of the transporter (Fig. 2).
Fig. 3.
Structural comparison of forms I and II of the HpnN transporter. (A) Superimposition of the dimeric structures of forms I and II (green, form I; red, form II). For clarity, only the Left subunit, PD1–PD4, is labeled. Each arrow indicates a rigid-body swinging motion of the periplasmic domain of each monomer, allowing it to come closer to the next subunit within the dimer. (B) Superimposition of the dimeric structures of forms I and II (green, form I; red, form II) viewed from the periplasmic side. This view depicts the two monomers of the form I structure as at least 6 Å closer to each other at the dimer interface compared with those of the form II structure. For clarity, the transmembrane domain of the HpnN transporter is not included in this figure. (C) Schematic representation illustrating the rigid-body swinging motion of the periplasmic domains of dimeric HpnN. Each arrow indicates a rigid body swinging motion of the periplasmic domain of each monomer. The swinging motion is capable of converting the conformations between form I (green) and form II (red).
Docking of Hopanoids to HpnN.
To elucidate whether B. multivorans HpnN has the capacity to bind hopanoids, AutoDock Vina (33) was used to calculate the potential binding modes of HpnN with a variety of hopanoids, including diploptene, 17β(H),21β(H)-hopane, 17α(H),21β(H)-hopane, and 17α(H)-22,29,30-trisnorhopan. Vina suggested that all of these ligands prefer to bind at the cavity formed within TMs 2, 4, and 11 (SI Appendix, Fig. S7). Interestingly, the conserved residue L826 located at the potential substrate-binding site is frequently involved in contacting these four hopanoid compounds. The predicted binding free energies are between −7.7 and −8.2 kcal/mol for these hopanoids.
As L826 is often found to interact with a variety of hopanoid molecules in our docking calculations, this conserved residue may be critical for recognizing hopanoids. Thus, we chose to mutate residue L826 to a phenylalanine (L826F) and investigate whether this point mutation would affect the affinity for hopanoids. Energy minimization was performed using PHENIX (34). The optimized structure of the L826F mutant was then subjected to predict the binding free energies for the hopanoid molecules diploptene, 17β(H),21β(H)-hopane, and 17α(H),21β(H)-hopane. Vina suggested that none of these ligands bind the HpnN transporter, indicating that L826 may be a crucial residue for recognizing hopanoids.
Binding of Hopanoids to HpnN.
To confirm that HpnN is directly involved in binding of hopanoids, we performed isothermal titration calorimetry (ITC) experiment to determine the binding affinity of 17β(H),21β(H)-hopane to the HpnN protein. The ITC data indicate that HpnN specifically binds 17β(H),21β(H)-hopane with an equilibrium dissociation constant (KD) of 9.1 ± 1.2 μM (SI Appendix, Fig. S8 and Table S2). We also studied the binding of 17β(H),21β(H)-hopane with the L826F HpnN mutant. We found that this mutant transporter binds 17β(H),21β(H)-hopane seven times weaker than that of the wild-type HpnN transporter (SI Appendix, Fig. S8 and Table S2), suggesting that the L826 residue is critical for recognizing hopanoids.
The hpnN Gene Is Essential for Cell Growth in the Presence of Antibiotics.
As B. multivorans is a biosafety level 2 pathogen, we turned to the biosafety level 1 null mutant strain Burkholderia thailandensis E264∆hpnN, which lacks the hpnN gene, for our mutagenesis studies and to elucidate the important function of the conserved HpnN amino acids. Alignment of protein sequences suggests that B. multivorans HpnN and B. thailandensis HpnN share 82% identity (SI Appendix, Fig. S9). We made a plasmid pHERD20ΩhpnN that contains the hpnN gene of B. thailandensis. We then transformed these E264∆hpnN cells with pHERD20ΩhpnN, expressing B. thailandensis HpnN, or the empty vector pHERD20. The knockout B. thailandensis E264∆hpnN cells, either alone or transformed with the empty vector pHERD20, were capable of growing in liquid Luria–Bertani broth (LB) in the absence of antimicrobial agents. However, they could not grow in this liquid medium in the presence of various antibiotics, including chloramphenicol, novobiocin, and polymyxin B (Fig. 4). Surprisingly, B. thailandensis E264∆hpnN cells transformed with pHERD20ΩhpnN were capable of growing in liquid LB under these damaging conditions. The experiment suggests that HpnN is critical for mediating multidrug resistance and the expression of B. thailandensis hpnN in E264∆hpnN is able to compensate for the loss of the B. thailandensis hpnN gene.
Fig. 4.
Important conserved amino acids of HpnN. (A) Ion pairs in the transmembrane domain viewed from the cytoplasmic side. Residues D344 of TM4, and T818 and T819 of TM11 that form ion pairs, which may play an important role in proton translocation, are in green sticks. (B) Side view of a protomer of HpnN that forms a channel. Residues L48, F117, F541, W661, and L826, which line the wall of the channel are in magenta sticks. (C) Time course of the growth of B. thailandensis E264∆hpnN cells harboring mutants of the proton relay network. Cells expressing the mutant transporter D344Y, T818A, or T819A could not grow in liquid LB in the presence of 5 μg/mL chloramphenicol (black, E264 cells; red, E264∆hpnN/pHERD20Ωbt_hpnN cells expressing HpnN; magneta, E264∆hpnN cells; blue, E264∆hpnN/pHERD20 cells; cyan, cells expressing D344Y; orange, T818A; and green, T819A). Error bars represent SD (n = 3). (D) Time course of the growth of B. thailandensis E264∆hpnN cells harboring mutant transporters L48F, F117R, F541R, W661H, and L826F. Growth of cells expressing these mutant transporters was severely attenuated in liquid LB supplemented with 5 μg/mL chloramphenicol (deep purple, L48F; gray, F117R; hot pink, F541R; smudge, W661H; and dark blue, L826F). Growth of E264 cells, E264∆hpnN cells, E264∆hpnN/pHERD20Ωbt_hpnN cells expressing B. thailandensis HpnN, and E264∆hpnN/pHERD20 cells carrying the empty vector is shown in A. Error bars represent SD (n = 3).
To determine whether the conserved residue D344, which forms a hydrogen-bonded triad with T818 and T819, is important for the function of the transporter, we mutated this corresponding residue in B. thailandensis HpnN to tyrosine (D344Y) (SI Appendix, Table S3). In B. multivorans HpnN, residues D344, T818, and T819 most likely establish the proton-relay network and translocate protons for energy coupling (Fig. 4 and SI Appendix, Fig. S10). We therefore also replaced the corresponding residues of T818 and T819 in B. thailandensis HpnN by alanines to create the T818A and T819A mutant transporters (SI Appendix, Table S3). We expressed these mutant HpnN transporters in B. thailandensis E264∆hpnN cells. We then monitored the growth of these cells harboring the mutant transporter D344Y, T818A, or T819A over time in liquid LB supplemented with chloramphenicol, novobiocin, or polymyxin B. Similar to the knockout E264∆hpnN cells transformed with the empty vector, growth of cells expressing these mutant transporters was severely attenuated in these liquid media (Figs. 4 and 5). The data indicate that a mutation on residue D344, T818, or T819 abolishes the function of the HpnN transporter.
Fig. 5.
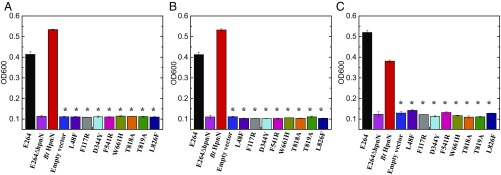
Growth of cells in the presence of antibiotics. B. thailandensis E264∆hpnN cells expressing the mutant transporter L48F, F117R, D344Y, F541R, W661H, T818A, T819A, or L826F were retarded in growth in liquid LB supplemented with (A) 5 μg/mL chloramphenicol, (B) 1 μg/mL novobiocin, or (C) 1 mg/mL polymyxin B compared with cells expressing wild-type B. thailandensis HpnN. Error bars represent SD (n = 3). *Values of E264∆hpnN/pHERD20 and E264∆hpnN cells expressing the mutant transporters that are significantly lower than that of E264∆hpnN/pHERD20Ωbt_hpnN expressing wild-type HpnN (P < 6 × 10−6 for A, P < 2 × 10−6 for B, and P < 7 × 10−5 for C; Student’s t test).
To understand whether the conserved HpnN residues L48 and L826 that line the wall of the channel are crucial for transport function, we mutated these two corresponding leucines in B. thailandensis HpnN into phenylalanines to produce the single-point mutant transporters L48F and L826F (SI Appendix, Table S3). We found that mutations on these two residues abrogate cell growth in liquid LB supplemented with chloramphenicol, novobiocin, or polymyxin B (Figs. 4 and 5), demonstrating that these two residues are necessary for the function of the transporter.
The three conserved aromatic residues F117, F541, and W661 likely create the exiting site of the channel within the transporter. Therefore, we replaced each corresponding residue in B. thailandensis HpnN by arginine or histidine to make single-point mutants F117R, F541R, and W661H (SI Appendix, Table S3). Again, B. thailandensis E264∆hpnN cells harboring these mutant transporters are retarded in growth in liquid LB supplemented with chloramphenicol, novobiocin, or polymyxin B (Figs. 4 and 5), indicating the important role of these amino acids. It should be noted that cells expressing wild-type or each mutant HpnN transporter grow similarly in liquid LB in the absence of antimicrobials (SI Appendix, Fig. S11).
In addition, we tested for the susceptibility of B. thailandensis E264∆hpnN cells carrying the wild-type, L48F, F117R, D344Y, F541R, W661H, T818A, T819A, or L826F transporter to polymyxin B, chloramphenicol, and novobiocin (SI Appendix, Table S4). We found that the B. thailandensis E264∆hpnN cells producing wild-type HpnN were eight times less sensitive to polymyxin B compared with E264∆hpnN cells harboring the L48F, F117R, D344Y, F541R, W661H, T818A, T819A, or L826F mutant. This E264∆hpnN transformant carrying wild-type HpnN was found to be fivefold more resistant to chloramphenicol in comparison with cells containing these mutant transporters. We also found that the minimum inhibitory concentration (MIC) of E264∆hpnN cells carrying wild-type HpnN was four or eight times higher than those of E264∆hpnN cells harboring the HpnN mutants (SI Appendix, Table S4). These data indeed indicate that residues L48, F117, D344, F541, W661, T818, T819, and L826 are critical for the function of the HpnN transporter.
Discussion
In this paper, we report crystal structures of the B. multivorans HpnN transporter, revealing a dimeric assembly of this membrane protein. Each subunit of HpnN creates a channel spanning the outer leaflet of the inner membrane and up to the periplasmic space. Based on our experimental data, we believe that HpnN is capable of transferring hopanoid lipids from the inner membrane to the periplasm. Presumably, the next step is that they spontaneously insert to the inner leaflet of the outer membrane, strengthening the cell envelope.
It is not surprising that B. thailandensis is resistant to polymyxins, because its lipopolysaccharide located at the outer layer of the cell envelope does not bind these polycationic drugs. However, it is not known why the defective export of hopanoids makes cells more susceptible to chloramphenicol and novobiocin, as these two drugs are not supposed to interfere with the membrane. One possible explanation is that these hydrophobic drugs are capable of using the bilayer region of outer membrane for influx. Novobiocin is too large for porin channels. Although chloramphenicol is small, its permeability through porins is low. It is likely that the outer membrane of cells carrying the HpnN mutants is much more permeable for these drugs.
We suspect that hopanoid transport is related to the rigid-body swinging motion of the periplasmic domain of HpnN with respect to its transmembrane domain. This movement may be critical for controlling the opening and closing of the tunnel formed within each subunit of the transporter. The narrowest region of the tunnel is found within the vicinity of residue L48. It is possible that this narrowest region creates a gate for the tunnel and L48 is crucial for gating. Indeed, our data show that a mutation of this leucine to an arginine results in cells hypersensitive to polymyxin B, chloramphenicol, and novobiocin. As the crystal structures indicate that HpnN is dimeric, it is likely that this transporter functions as a dimer. If this is the case, then HpnN may use a very different mechanism for substrate transport in comparison with those efflux pumps within the HAE1-RND subfamily, where they assemble as a trimer. Overall, our data suggest a mechanism for hopanoid transport involved in cell wall remodeling, which is critical for mediating multidrug resistance in Burkholderia. Exactly how HpnN export substrates must await confirmation by additional structural and functional studies of this transporter.
Methods
Crystals of the HpnN protein were obtained using sitting-drop vapor diffusion. The form I crystals were grown at room temperature in 24-well plates with the following procedures: A 2-μL protein solution containing 20 mg/mL HpnN in 20 mM Na-Hepes (pH 7.5) and 0.05% (wt/vol) n-dodecyl-β-d-maltoside (DDM) was mixed with a 2 μL of reservoir solution containing 16% PEG 2000, 0.1 M sodium citrate (pH 3.5) and 0.2 M Li2SO4. The resultant mixture was equilibrated against 500 μL of the reservoir solution at 25 °C. For the form II crystals, a 2-μL protein solution containing 20 mg/mL HpnN in 20 mM Na-Hepes (pH 7.5) and 0.05% (wt/vol) DDM was mixed with 2 μL of reservoir solution containing 15% PEG 2000, 0.1 M sodium citrate (pH 4.0), and 0.2 M (NH4)2SO4. The resultant mixture was equilibrated against 500 μL of the reservoir solution at 25 °C. The crystallization conditions for SeMet-HpnN were the same as those for crystallizing the form II crystals. Crystals of HpnN (both forms I and II) and SeMet-HpnN grew to a full size in the drops within a month. Typically, the dimensions of the crystals were 0.2 mm × 0.2 mm × 0.2 mm. Cryoprotection of these crystals was achieved by raising the glycerol concentration stepwise to 30% with a 5% increment in each step. Crystals of the tungsten cluster derivative were prepared by incubating the form I crystals in solution containing 18% PEG 2000, 0.1 M sodium citrate (pH 3.5), 0.2 M Li2SO4, 0.05% (wt/vol) DDM, and 0.5 mM (NH4)2W6(μ-O)6(μ-Cl)6Cl6 for 5 h at 25 °C. A full description of methods for protein expression and purification, X-ray data collection and analysis, docking, isothermal titration calorimetry, construction of complementing plasmid for B. thailandensis, site-directed mutagenesis, plasmid mobilization, cell growth, and drug susceptibility are described in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Dr. Miguel A. Valvano (Queen’s University Belfast) and Dr. Silvia Cardona (University of Manitoba) for providing us the plasmid pScrhaB2 and Hongwei D. Yu (Marshall University) for sharing the plasmid pHERD20T. This work is based upon research conducted at the Northeastern Collaborative Access Team beamlines of the Advanced Photon Source, supported by Award GM103403 from the National Institutes of General Medical Sciences. Use of the Advanced Photon Source is supported by the US Department of Energy, Office of Basic Energy Sciences, under Contract DE-AC02-06CH11357. This work was supported by NIH Grant R01AI114629 (to E.W.Y.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org [PDB ID code 5KHN (form I) and 5KHS (form II)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1619660114/-/DCSupplemental.
References
- 1.Mahenthiralingam E, Urban TA, Goldberg JB. The multifarious, multireplicon Burkholderia cepacia complex. Nat Rev Microbiol. 2005;3:144–156. doi: 10.1038/nrmicro1085. [DOI] [PubMed] [Google Scholar]
- 2.Sajjan U, et al. Immunolocalisation of Burkholderia cepacia in the lungs of cystic fibrosis patients. J Med Microbiol. 2001;50:535–546. doi: 10.1099/0022-1317-50-6-535. [DOI] [PubMed] [Google Scholar]
- 3.Ledson MJ, Gallagher MJ, Corkill JE, Hart CA, Walshaw MJ. Cross infection between cystic fibrosis patients colonised with Burkholderia cepacia. Thorax. 1998;53:432–436. doi: 10.1136/thx.53.5.432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zlosnik JE, et al. Burkholderia species infections in patients with cystic fibrosis in British Columbia, Canada. 30 years’ experience. Ann Am Thorac Soc. 2015;12:70–78. doi: 10.1513/AnnalsATS.201408-395OC. [DOI] [PubMed] [Google Scholar]
- 5.Drevinek P, Mahenthiralingam E. Burkholderia cenocepacia in cystic fibrosis: Epidemiology and molecular mechanisms of virulence. Clin Microbiol Infect. 2010;16:821–830. doi: 10.1111/j.1469-0691.2010.03237.x. [DOI] [PubMed] [Google Scholar]
- 6.Whiteford ML, et al. Outcome of Burkholderia (Pseudomonas) cepacia colonisation in children with cystic fibrosis following a hospital outbreak. Thorax. 1995;50:1194–1198. doi: 10.1136/thx.50.11.1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stokell JR, Gharaibeh RZ, Steck TR. Rapid emergence of a ceftazidime-resistant Burkholderia multivorans strain in a cystic fibrosis patient. J Cyst Fibros. 2013;12:812–816. doi: 10.1016/j.jcf.2013.01.009. [DOI] [PubMed] [Google Scholar]
- 8.Hanulik V, et al. An outbreak of Burkholderia multivorans beyond cystic fibrosis patients. J Hosp Infect. 2013;84:248–251. doi: 10.1016/j.jhin.2013.04.001. [DOI] [PubMed] [Google Scholar]
- 9.Malott RJ, Steen-Kinnaird BR, Lee TD, Speert DP. Identification of hopanoid biosynthesis genes involved in polymyxin resistance in Burkholderia multivorans. Antimicrob Agents Chemother. 2012;56:464–471. doi: 10.1128/AAC.00602-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aaron SD, Ferris W, Henry DA, Speert DP, Macdonald NE. Multiple combination bactericidal antibiotic testing for patients with cystic fibrosis infected with Burkholderia cepacia. Am J Respir Crit Care Med. 2000;161:1206–1212. doi: 10.1164/ajrccm.161.4.9907147. [DOI] [PubMed] [Google Scholar]
- 11.Malott RJ, et al. Fosmidomycin decreases membrane hopanoids and potentiates the effects of colistin on Burkholderia multivorans clinical isolates. Antimicrob Agents Chemother. 2014;58:5211–5219. doi: 10.1128/AAC.02705-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schmerk CL, Bernards MA, Valvano MA. Hopanoid production is required for low-pH tolerance, antimicrobial resistance, and motility in Burkholderia cenocepacia. J Bacteriol. 2011;193:6712–6723. doi: 10.1128/JB.05979-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rhodes KA, Schweizer HP. Antibiotic resistance in Burkholderia species. Drug Resist Updat. 2016;28:82–90. doi: 10.1016/j.drup.2016.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Siritapetawee J, Prinz H, Krittanai C, Suginta W. Expression and refolding of Omp38 from Burkholderia pseudomallei and Burkholderia thailandensis, and its function as a diffusion porin. Biochem J. 2004;384:609–617. doi: 10.1042/BJ20041102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ourisson G, Rohmer M, Poralla K. Prokaryotic hopanoids and other polyterpenoid sterol surrogates. Annu Rev Microbiol. 1987;41:301–333. doi: 10.1146/annurev.mi.41.100187.001505. [DOI] [PubMed] [Google Scholar]
- 16.Mahato SB, Sen S. Advances in triterpenoid research, 1990-1994. Phytochemistry. 1997;44:1185–1236. doi: 10.1016/s0031-9422(96)00639-5. [DOI] [PubMed] [Google Scholar]
- 17.Saenz JP, et al. Hopanoids as functional analogues of cholesterol in bacterial membranes. Proc Natl Acad Sci USA. 2015;112:11971–11976. doi: 10.1073/pnas.1515607112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Welander PV, et al. Hopanoids play a role in membrane integrity and pH homeostasis in Rhodopseudomonas palustris TIE-1. J Bacteriol. 2009;191:6145–6156. doi: 10.1128/JB.00460-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tseng SP, et al. The contribution of antibiotic resistance mechanisms in clinical Burkholderia cepacia complex isolates: An emphasis on efflux pump activity. PLoS One. 2014;9:e104986. doi: 10.1371/journal.pone.0104986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tseng TT, et al. The RND permease superfamily: an ancient, ubiquitous and diverse family that includes human disease and development proteins. J Mol Microbiol Biotechnol. 1999;1:107–125. [PubMed] [Google Scholar]
- 21.Doughty DM, et al. The RND-family transporter, HpnN, is required for hopanoid localization to the outer membrane of Rhodopseudomonas palustris TIE-1. Proc Natl Acad Sci USA. 2011;108:E1045–E1051. doi: 10.1073/pnas.1104209108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Daligault HE, et al. 2014. Whole-genome assemblies of 56 burkholderia species. Genome Announc 2: pii: e01106-14.
- 23.Symmons MF, Bokma E, Koronakis E, Hughes C, Koronakis V. The assembled structure of a complete tripartite bacterial multidrug efflux pump. Proc Natl Acad Sci USA. 2009;106:7173–7178. doi: 10.1073/pnas.0900693106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mikolosko J, Bobyk K, Zgurskaya HI, Ghosh P. Conformational flexibility in the multidrug efflux system protein AcrA. Structure. 2006;14:577–587. doi: 10.1016/j.str.2005.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Su CC, et al. Crystal structure of the CusBA heavy-metal efflux complex of Escherichia coli. Nature. 2011;470:558–562. doi: 10.1038/nature09743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eicher T, et al. Transport of drugs by the multidrug transporter AcrB involves an access and a deep binding pocket that are separated by a switch-loop. Proc Natl Acad Sci USA. 2012;109:5687–5692. doi: 10.1073/pnas.1114944109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Long F, et al. Crystal structures of the CusA efflux pump suggest methionine-mediated metal transport. Nature. 2010;467:484–488. doi: 10.1038/nature09395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsukazaki T, et al. Structure and function of a membrane component SecDF that enhances protein export. Nature. 2011;474:235–238. doi: 10.1038/nature09980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Duarte JM, Biyani N, Baskaran K, Capitani G. An analysis of oligomerization interfaces in transmembrane proteins. BMC Struct Biol. 2013;13:21. doi: 10.1186/1472-6807-13-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Perrin E, et al. A census of RND superfamily proteins in the Burkholderia genus. Future Microbiol. 2013;8:923–937. doi: 10.2217/fmb.13.50. [DOI] [PubMed] [Google Scholar]
- 31.Takatsuka Y, Nikaido H. Threonine-978 in the transmembrane segment of the multidrug efflux pump AcrB of Escherichia coli is crucial for drug transport as a probable component of the proton relay network. J Bacteriol. 2006;188:7284–7289. doi: 10.1128/JB.00683-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brockerman JA, Okon M, McIntosh LP. Detection and characterization of serine and threonine hydroxyl protons in Bacillus circulans xylanase by NMR spectroscopy. J Biomol NMR. 2014;58:17–25. doi: 10.1007/s10858-013-9799-6. [DOI] [PubMed] [Google Scholar]
- 33.Trott O, Olson AJ. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Adams PD, et al. PHENIX: Building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.