Significance
The class of bioactive cyclic plant natural products called orbitides was first identified nearly half a century ago. Here we describe how a single enzyme can catalyze the cyclization of a range of ribosomally synthesized linear peptides into the corresponding cyclic products of varying ring sizes. These studies may provide a means for producing large libraries of cyclic peptides without any sequence bias.
Keywords: RiPP, biosynthesis, peptide, plant, orbitide
Abstract
Enzymes that can catalyze the macrocyclization of linear peptide substrates have long been sought for the production of libraries of structurally diverse scaffolds via combinatorial gene assembly as well as to afford rapid in vivo screening methods. Orbitides are plant ribosomally synthesized and posttranslationally modified peptides (RiPPs) of various sizes and topologies, several of which are shown to be biologically active. The diversity in size and sequence of orbitides suggests that the corresponding macrocyclases may be ideal catalysts for production of cyclic peptides. Here we present the biochemical characterization and crystal structures of the plant enzyme PCY1 involved in orbitide macrocyclization. These studies demonstrate how the PCY1 S9A protease fold has been adapted for transamidation, rather than hydrolysis, of acyl-enzyme intermediates to yield cyclic products. Notably, PCY1 uses an unusual strategy in which the cleaved C-terminal follower peptide from the substrate stabilizes the enzyme in a productive conformation to facilitate macrocyclization of the N-terminal fragment. The broad substrate tolerance of PCY1 can be exploited as a biotechnological tool to generate structurally diverse arrays of macrocycles, including those with nonproteinogenic elements.
Macrocyclic peptides have become appealing targets for drug discovery efforts due to the emergence over the past decade of multiple routes for rapid synthesis and screening (1). The drug-like properties of cyclic peptides arise from their constrained rigid structure, improved bioavailability, membrane permeability relative to linear peptides, and resistance to degradation by host proteases (2–4). Organic synthesis of large peptide libraries is constrained by practical concerns, thereby limiting the diversity of sequence variants that would be necessary to truly explore scaffold space. More recent approaches for macrocyclic peptide synthesis have focused on in vivo production, in which the use of genetic templates can provide routes toward the production of libraries of diverse structures (5). Enzymatic routes for macrocycle production include the use of isolated thioesterase domains from nonribosomal peptide synthetases (6), GST (7), or split inteins (5), as well as technologies based on reprogramming the genetic code (8). However, these approaches either require the use of conjugated biomimetics or are not sufficiently substrate-tolerant to thoroughly sample the diversity of possible structures.
Ribosomally synthesized and posttranslationally modified peptides (RiPPs) represent an abundant source of chemically and structurally diverse natural products (9). RiPPs are translated by the ribosome as linear peptides that are subsequently enzymatically modified to yield compounds with a range of biological activities (10). Many RiPPs undergo enzymatic macrocyclization, as observed in several diverse classes of compounds including bottromycins (11), cyanobactins (12), lanthipeptides (13), streptides (14), lasso peptides (15), and thiopeptides (16). Understanding the biosynthetic pathways for RiPP macrocyclization continues to be important for biotechnological application, for the identification of new gene clusters, and for the production of novel RiPP derivatives (17).
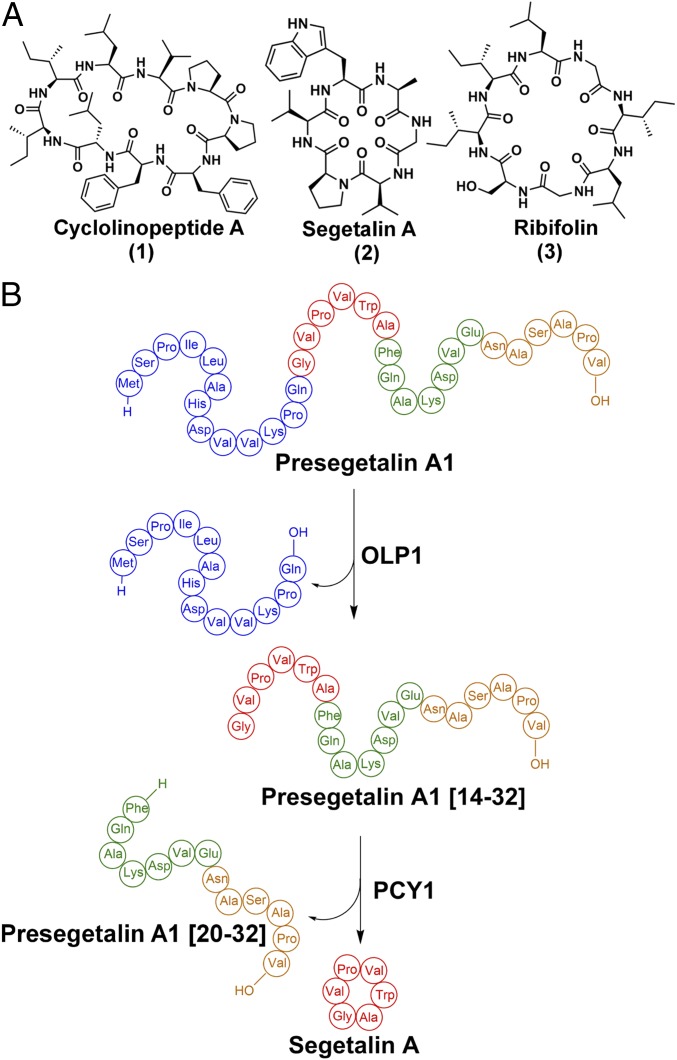
Orbitides encompass a class of plant homodetic macrocyclic natural products, usually 5–12 amino acids in size, which are characterized by the intramolecular condensation between the N and C termini of their linear precursors (10). The chemical structures of orbitides consist solely of α-amide linkages and are distinguished from other macrocyclic plant peptide natural products such as cyclotides, which are further interlocked through multiple disulfide bonds (18), and amatoxins, which contain an unusual tryptathionine moiety formed by a crosslink between a Cys and Trp residue (19). The first macrocyclic orbitide, cyclolinopeptide A, was isolated in 1959 from flaxseed (Linum usitatissimum) oil and determined to be a cyclic peptide of ILVPPFFLI (Fig. 1A) (20). Since then, over 168 Caryophyllaceae-like nonredundant orbitides have been identified, and encoding sequences have been confirmed in Annonaceae, Rutaceae, Euphorbiaceae, and Linaceae (21). Many orbitides demonstrate biological activities including antimalarial (22), vasodilatory (23), immunomodulating (24), and xenoestrogenic activities (25).
Fig. 1.
(A) Structures of the orbitides cyclolinopeptide A, segetalin A, and ribifolin. (B) Scheme showing the biosynthesis of segetalin A by S. vaccaria.
Analysis of expressed sequence tag libraries from developing seeds of Saponaria vaccaria (Vaccaria hispanica) identified genes that likely encode for peptide precursors of orbitides. Expression of candidate precursors in the roots of a nonsegetalin A-producing S. vaccaria strain resulted in the production of fully processed cyclic peptide products (26), confirming that orbitides are indeed RiPPs. Subsequent biochemical assays with seed extracts, using synthetic peptide substrates, revealed the order of posttranslational modifications that generate the orbitide segetalin A (Fig. 1 A and B) (27). Briefly, segetalin A is first produced ribosomally as a 32-aa peptide termed presegetalin A1. A serine protease (oligopeptidase 1 or OLP1) cleaves the first 15 amino acids to yield a product containing an N-terminal glycine (termed presegetalin A1 [14–32]). A second enzyme [peptide cyclase 1 (PCY1)] acts on this product to excise the linear C-terminal 13 residues (termed presegetalin A1 [20–32]), with the concomitant macrocyclization of the remaining N-terminal six residues to yield the cyclo[GVPVWA] segetalin A product (Fig. 1B). The biosynthesis of cyclotides (28, 29), amatoxins (30), and cyanobactins (31) uses similar biosynthetic strategies in which a sequence-specific peptidase catalyzes the transamidation of a linear peptide substrate to produce a macrocyclic product.
Primary sequence analysis identifies PCY1 as a member of the S9A protease family that includes several clades of prolyl oligopeptidases (POP). Proteases of the S9A class catalyze the hydrolytic cleavage of peptide substrates at the amide linkage C-terminal to Pro to yield two linear products. However, PCY1 is distinguished from canonical S9A proteases by its unique product profile, as the enzyme generates a macrocyclic product rather than the linear counterpart. In addition, PCY1 does not require a Pro in the sequence of the cyclized peptide. Analysis of the predicted primary sequences of dozens of putative orbitide precursor peptides reveals that residues within the core sequence that is cyclized are highly divergent, whereas residues in the preceding leader sequence (removed by OLP1) and the trailing follower sequence (excised as a linear peptide by PCY1) are highly conserved. Similar patterns of conservation are observed for the precursors of other cyclic peptides including cyanobactins (31) and amatoxins (19, 32). Alanine scanning mutational analysis of presegetalin A1 demonstrates that the necessary elements for substrate recognition by PCY1 reside solely in the 12-residue follower sequence (hereafter, presegetalin A1 [20–32]) that is excised during the formation of the macrocyclic product (27). Notably, PCY1 can accept a range of substrates leading to rings of different sizes as this single enzyme is capable of processing almost all of the orbitides that are observed in S. vaccaria. Consequently, PCY1 may be used as a biotechnology tool for the production of macrocycles of diverse sequences. A similar strategy has been used by employing the peptide cyclase butelase 1 to produce cyclic bacteriocins (33).
The structure of the unrelated cyanobactin transamidating protease (PatG) revealed a peptidase S8 (subtilisin-like) fold that is decorated with a “capping” domain, and biochemical studies suggest that this domain insertion likely facilitates macrocyclization of the peptide substrate (34, 35). The primary sequence of PCY1 fails to identify any such obvious insertions, relative to canonical S9A protease family members, raising the question of how an enzyme from a class of generic proteases can be adapted to produce a macrocyclic product. To address this issue, we have carried out biochemical and crystallographic characterization of PCY1.
Results
Kinetic Analysis of PCY1.
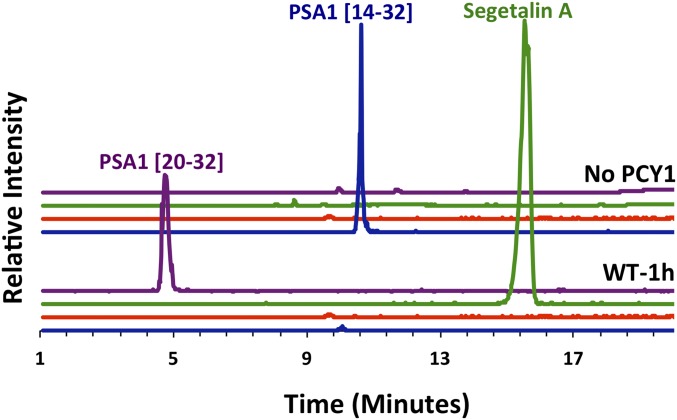
Incubation of recombinant PCY1 with a synthetic peptide corresponding to presegetalin A1 [14–32] resulted in a near-complete conversion into cyclic[GVPVWA] segetalin A (and corresponding linear follower peptide) (Fig. 2). The yield of linear core peptide was minimal, demonstrating that the enzyme favors production of macrocyclic products relative to the linear counterpart. To determine steady-state kinetic parameters, we used a LC-MS–based assay to quantify rates of product formation. Complete Michaelis–Menten kinetics were carried out for the presegetalin A1 [14–32] substrate, yielding a kcat value of 6.0 × 10−2⋅s−1 with a KM of 0.77 μM (SI Appendix, Fig. S1 and Table S3). In comparison, the asparaginyl endopeptidases involved in cyclotide macrocyclization (butelase 1 and AEP) have similar kcat values, but with KM values that are 5- to 10-fold higher (28, 29). Additionally, the transamidative protease involved in the biosynthesis of amatoxin (GmPOPB) was initially shown to have a similar KM value and a 100-fold faster rate (kcat of 5.7⋅s−1) (30), whereas more extensive kinetic analysis indicated a KM about 75-fold higher and a kcat approximately 4-fold faster than PCY1 (36).
Fig. 2.
Extracted-ion chromatograms of products derived upon incubation of recombinant PCY1 with a synthetic peptide corresponding to presegetalin A1 [14–32]. Chromatograms showing the mass to charge ratio (m/z) values of the segetalin A product (m/z = 610.5, z = 1; green trace), of the linear [GVPVWA] product (m/z = 628.5, z = 1; red trace), of the presegetalin A1 [14–32] substrate (m/z = 993.2, z = 2; blue trace), and the presegetalin A1 [20–32] by-product (m/z = 688.5, z = 2; purple trace) are shown as indicated. The wild-type PCY1 used most of the presegetalin A1 [14–32] substrate to produce almost exclusively cyclic segetalin A product after a 1-h reaction.
Structural Characterization of PCY1.
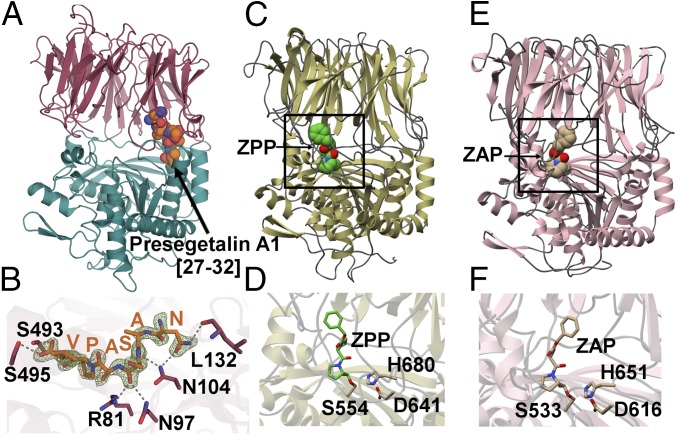
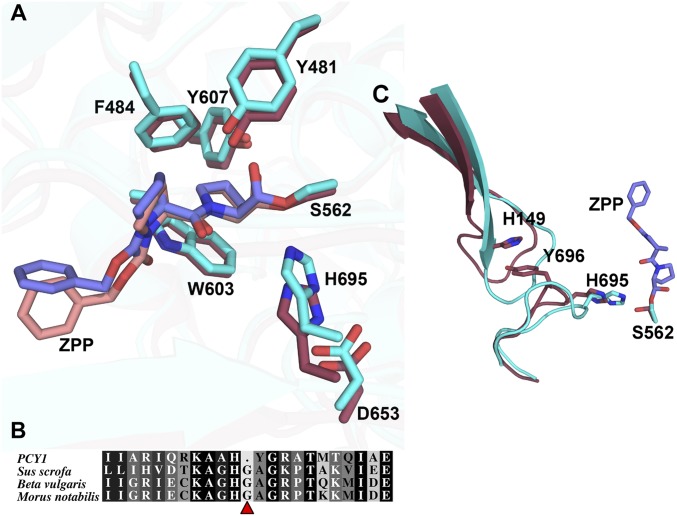
To elucidate how an S9A protease family can catalyze macrocyclization, we determined the crystal structure of PCY1 that had been incubated with the presegetalin A1 [14–32] substrate (Fig. 3 A and B) (37). The 1.9-Å resolution structure of this PCY1 complex revealed an overall architecture similar to other prolyl oligopeptidase family members, such as the eukaryotic enzyme from porcine muscle (Sus scorfa) (PDB ID code: 1QFS; rmsd of 1.6 Å over 691 aligned Cα atoms) (38, 39) and the bacterial enzyme from Myxococcus xanthus (PDB ID code: 2BKL; rmsd of 1.7 Å over 691 aligned Cα atoms) (40) (Fig. 3 C and E). The structure of PCY1 consists of an α/β hydrolase domain composed of residues Met1-Val77 and Asp437-Asp724 and a β-propeller domain consisting of residues Cys82-Glu432. A hinge region encompassing residues Asn78-Arg81 and Ser433-Pro436 links the two domains and presumably facilitates movement between an open form competent for binding substrate and a closed form where chemistry is carried out (41, 42). The PCY1–presegetalin A1 complex was crystallized in the closed form with a large internal cavity (10,317 Å3 as calculated by CASTp), which can accommodate the binding of large peptides (41, 43). A close-up view of the active sites of canonical prolyl oligopeptidases from S. scorfa and M. xanthus reveal the protrusion of various loops that significantly decrease the corresponding internal cavity (Fig. 3 D and F). In PCY1, residues Asp653, His695, and Ser562 make up the active-site catalytic triad, and a hydrophobic pocket enriched in aromatic residues forms a binding pocket for the target peptide (Fig. 4A). Finally, Tyr481 acts to stabilize the tetrahedral intermediate oxyanion that is formed following the attack of the Ser562 alkoxide on the peptide substrate.
Fig. 3.
(A) The 1.9-Å PCY1 structure with the α/β hydrolase domain shown in green and β-propeller domain shown in red. (B) The C-terminal six amino acids of the presegetalin A1 [14–32] substrate (orange) is bound near the hinge region of PCY1. A simulated annealing difference Fourier maps (Fo-Fc), calculated with the coordinates of the peptide omitted, is superimposed and contoured to 2.5σ (blue). (C) Structure of the canonical POP enzyme from S. scorfa (PDB ID code 1QFS) (38, 39) bound to covalent inhibitor ZPP, with the active site demarcated in the rectangle, and (D) a close-up view of the active site. (E) Structure of the bacterial enzyme from M. xanthus (PDB ID code 2BKL) (40) bound to covalent inhibitor Z-Ala-prolinal, with the active site demarcated, and (F) a close-up view of the active site.
Fig. 4.
(A) Active-site alignment of PCY1 (red, pink) and porcine muscle POP (cyan, blue) with covalently bound ZPP. All residues are conserved and align well except for the catalytic histidine. Residues are labeled according to PCY1’s numbering. (B) Sequence alignment of PCY1 and POP enzymes from S. scorfa [domestic pig, National Center for Biotechnology Information (NCBI): NP_001004050.1], Beta vulgaris (beet, NCBI: XP_010667346.1) and Morus notabilis (mulberry, NCBI: XP_010101294.1). (C) Changes in structures at regions distal to the active site between PCY1 (red) and S. scorfa POP (blue) indicate significant difference in the orientation of the hairpin loop. The hairpin is stabilized in PCY1 by a stacking interaction between His149 and Tyr696.
Although PCY1 crystallization was carried out using presegetalin A1 [14–32], the active site revealed unambiguous electron density only for the terminal six residues (NASAPV) of the follower peptide (Fig. 3B). The residues within the follower sequence bind to PCY1 near the hinge region and engage in hydrogen bonds with amino acids in both the α/β hydrolase and the β-propeller domains. Hydrogen-bonding interactions can be observed between PCY1 residues Arg81, Asn97, Asn104, and backbone carbonyl and amide of Ala28 and Ser29, respectively, of the follower peptide. The only sequence-specific interaction with the follower peptide occurs between its side chain of Asn27 and the side chain of Asp104 and backbone carbonyl of Leu132 of PCY1. The binding of the carboxyl terminus of the peptide is of particular interest, as it forms hydrogen bonds with PCY1 residues Ser495 and Ser493. This observation explains prior alanine-scanning mutational analysis of presegetalin A1 [14–32], which demonstrates that PCY1 is tolerant of mutation along the entirety of the primary sequence of the substrate but cannot process a substrate with the deletion of the C-terminal Val32, indicating the importance of the follower peptide C-terminal carboxylate (27).
To visualize a structure of PCY1 bound to intact presegetalin A1, we carried out cocrystallization with two different active-site variants, His695→Ala and Tyr481→Phe. In both instances, electron density could be observed only for the six-residue follower sequence, despite the presence of the presegetalin A1 [20–32] peptide in the crystals (as determined by mass spectrometry), consistent with disorder/flexibility for the remainder of the substrate (SI Appendix, Figs. S2 and S3). Hence, only the follower region of presegetalin A1 is engaged by PCY1, allowing for flexibility in the substrate N-terminal to the cleavage site. This flexibility explains how S. vaccaria can produce nine different orbitides of different ring sizes and sequence composition, as only the follower sequences are conserved in the progenitors of the macrocyclic products (SI Appendix, Fig. S4).
Most RiPP biosynthetic enzymes use a PqqD domain (RiPP precursor peptide recognition element) (44) to engage the leader precursor peptide through a limited number of hydrophobic interactions. This has been observed in at least some of the enzymes involved in the biosynthesis of pyrroloquinoline quinone (45), lantibiotics (46), lasso peptides (44), and cyanobactins (47). In contrast, the follower peptide binding observed in PCY1 does not rely on hydrophobic interactions, but rather on hydrogen-bonding interactions. Hydrogen-bonding interactions also mediate binding of leader peptides by microvirdin biosynthetic enzymes (48) and of the follower peptide by PatG (34).
PCY1-Binding Affinity Measurements.
To determine the affinity of PCY1 for various follower peptides, we carried out competition binding measurements (Table 1). A six-residue presegetalin A1 [27–32] peptide conjugated to fluorescein isothiocyanate bound to PCY1 with a dissociation constant (Kd) of 4.83 μM as determined by fluorescence polarization (SI Appendix, Fig. S5A). Competitive binding between this labeled peptide and various unlabeled peptides was used to determine the Ki values for the unlabeled peptides. These competition experiments demonstrate that unlabeled presegetalin A1 [27–32] bound to PCY1 with a Ki of 31.0 μM (Table 1 and SI Appendix, Fig. S5B). The longer, full-length linear by-product of the cyclization reaction, presegetalin A1 [20–32], binds with a much higher affinity with a Ki of 0.40 μM (Table 1 and SI Appendix, Fig. S5C). The nearly 80-fold greater binding affinity for the longer presegetalin A1 [20–32] peptide is curious given that only the last six residues of the peptide are involved in contacts with PCY1 in the cocrystal structure. The additional binding affinity, relative to presegetalin A1 [27–32], may be due to solvent exclusion or another nonsequence-dependent mechanism. The amino acids that make up the final cyclic product do not seem to contribute to binding as substrate presegetalin A1 [14–32] was measured to bind to the catalytically inhibited PCY1 H695A with a Ki of 1.10 μM, very similar to that of presegetalin A1 [20–32] (Table 1 and SI Appendix, Fig. S5D). This contrasts with binding studies completed with GmPOPB, in which the cyclized region of the peptide enhances binding affinity (36).
Table 1.
Binding constants for the follower peptide and mutants
| Ligand | IC50 (μM) | Ki (μM) |
| Presegetalin A1 [20–32] | 1.97 ± 0.34 | 0.40 ± 0.007 |
| Presegetalin A1 [14–32]* | 3.03 ± 0.61 | 1.10 ± 0.22 |
| Presegetalin A1 [27–32] | 47.4 ± 1.0 | 31.0 ± 0.65 |
| Presegetalin A1 [27–32] N27A | 197 ± 8 | 131 ± 5 |
| Presegetalin A1 [27–32] amide | >1,000 | >650 |
Presegetalin A1 [14–32] binding experiments were completed with PCY1 H695A to prevent turnover.
To verify the roles of individual interactions observed in the structure, we measured the affinity of PCY1 for variants of the presegetalin A1 [27–32] peptide. Based on the crystal structure, the side chain of Asn27 of presegetalin A1 [27–32] is engaged in specific hydrogen-bonding interactions. Consequently, the Asn27→Ala mutation in presegetalin A1 [27–32] results in a reduction in the PCY1-binding affinity (Ki of 131 μM) (Table 1 and SI Appendix, Fig. S5E). The C terminus of the follower peptide forms hydrogen bonds with PCY1 residues Ser495 and Ser493, and the binding affinity of PCY1 for the presegetalin A1 [27–32] variant with a C-terminal amide is diminished and outside the detection range of the assay (Table 1 and SI Appendix, Fig. S5F). Mutations were made to PCY1 residues Ser493 and Asn97, both of which bind the follower peptide. Ser493→Ala had minimal changes on catalytic efficiency relative to the wild type, whereas Asn97→Ala greatly diminished activity (SI Appendix, Table S3 and Fig. S6).
Follower Peptide Induces Conformational Change in PCY1.
Based on the kinetic studies, we hypothesized that the high affinity of PCY1 for the presegetalin A1 [27–32] follower peptide may ensure that the enzyme is maintained in the closed conformation throughout catalysis and is displaced only after turnover. To test this, we fluorescently labeled PCY1 with the thiol-specific probe N-1-pyrene maleimide. Addition of the presegetalin A1 [27–32] to labeled PCY1 resulted in the quenching of fluorescence compared with a buffer control (SI Appendix, Fig. S7). Conversely, addition of a control peptide that does not bind (presegetalin A1 [27–32] amide) showed no change in fluorescence. As there are no cysteines present near the binding site of the peptide (SI Appendix, Fig. S8), these changes in fluorescence have been attributed to changes in the conformation of PCY1. These findings are consistent with the notion that binding of presegetalin A1 [27–32] alone induces a conformational change in PCY1 to the closed state.
As presegetalin A1 [27–32] binding induces closure of the PCY1 structure, addition of exogenous peptide would be expected to inhibit productive turnover of the presegetalin A1 [14–32] substrate. We tested this hypothesis using a 1-h end-point reaction of PCY1 incubated with 30 μM of substrate in the presence of stoichiometric and 10-fold excess of the presegetalin A1 [20–32] linker+follower peptide product, as well as 100-fold excess of the presegetalin A1 [27–32] follower peptide (SI Appendix, Fig. S9). Although PCY1 consumed nearly all of the presegetalin A1 [14–32] substrate alone (Fig. 2), addition of the follower peptides resulted in a decrease in turnover in a concentration-dependent manner. These data are consistent with the follower peptide maintaining a closed state for PCY1. The role of the leader/follower peptide in stabilizing active conformations of biosynthetic enzymes is an emerging theme in RiPP biosynthesis. For example, during cyanobactin biosynthesis, binding of the PatE′ leader peptide to the LynD cyclodehydratase stabilizes the enzyme in a catalytically competent form (47).
Using the structural data as a guide, we generated and characterized mutants of several active-site residues to test their roles in catalysis (SI Appendix, Table S3 and Fig. S10A). As expected, both the Ser562→Ala and His695→Ala mutation of the catalytic triad residues produced inactive enzyme, confirming the necessity of these conserved residues. Finally, Tyr481 is necessary to stabilize the oxyanion of the tetrahedral intermediate as the Try481→Phe variant is catalytically inactive. We next interrogated the extent to which the enzyme could accommodate primary amines other than a peptide α-amino group for transamidation. First, we used a variant presegetalin A1 [14–32] substrate peptide that contained both the α-amino terminus and a side-chain amine (Val15→Lys). Kinetic characterization of PCY1 using the Val15→Lys presegetalin A1 [14–32] as a substrate demonstrates that the KM increased by 10-fold relative to the cognate presegetalin A1 [14–32] peptide as a substrate but the kcat also increased by almost 10-fold, resulting in a catalytic efficiency similar to that using the cognate peptide. Although PCY1 was able to completely convert this substrate to the cyclic product (SI Appendix, Table S3 and Fig. S10B), tandem mass spectrometric analysis revealed that the α-amino group was used for transamidation (SI Appendix, Fig. S11). We did not observe any products consistent with the use of the side chain of Lys15 as part of the macrocycle. We then tested whether PCY1 could generate a macrocyclic product using a nonproteinogenic amine. To this end, we used a presegetalin A1 [14–32] substrate that contained an N-terminal aminohexanoic acid (Ahx) in place of Gly14. PCY1 largely produced a linear product with this substrate, although a minor amount of cyclic [AhxVPVWA] could also be detected (SI Appendix, Table S3 and Fig. S12). These data suggest that PCY1-catalyzed macrocyclization is most effective via the use of a peptide α-amine.
Dual Function of His695 Allows for Cyclization.
Prior studies on other macrocyclases, such as the PatG protease involved in cyanobactin biosynthesis, suggest a plausible route to cyclic peptide production (34, 35). Briefly, following the formation of an acyl-enzyme intermediate during the first half protease reaction, hydrolysis of the intermediate is averted by solvent exclusion from the active site. Instead, the α-amino terminus of the bound peptide substrate is oriented back toward the catalytic Ser, and deprotonation of the amine can then facilitate a transpeptidation reaction to yield the cyclic peptide product. A comparison of the structure of PCY1 with other prolyl oligopeptidases reveals a strong conservation of overall structure and a near-identical alignment of putative active-site residues (Fig. 4A). While nearly all of these features in the PCY1 active site superimpose with the structures of other S9A proteases, a notable exception is the ∼2.4-Å displacement of His695 away from the catalytic Ser562.
Studies of hydrolytic serine proteases have established that the catalytic His moves following formation of a tetrahedral intermediate, both to tune pKa and to serve as an effective proton donor to the leaving group (49–51). To determine if the misaligned His695 is reflective of a catalytically competent active site, we determined the 3.3 Å cocrystal structure of PCY1 covalently bound to an aldehyde inhibitor that mimics a tetrahedral intermediate [Z-Proprolinal (ZPP)] (Fig. 4A). Although the structure of ZPP differs from that of the true presegetalin substrate peptide, PCY1 is sufficiently tolerant for the sequence preceding the scissile bond to accommodate this inhibitor. The cocrystal structure shows that ZPP is covalently bound to Ser562 and adopts a conformation similar to that in canonical S9A enzymes, with the Pro oriented within the aromatic hydrophobic pocket. Surprisingly, the disposition of His695 relative to the unliganded enzyme remained unchanged (0.9-Å movement of Nγ) and was outside the range of a hydrogen-bonding interaction with Ser562.
The obvious change in the orientation of the catalytic His695 suggested that it may play some role in directing the ability of PCY1 to catalyze macrocyclization. A cursory inspection of the PCY1 active site revealed a single residue deletion at a Gly that is normally adjacent to the catalytic His in canonical S9A proteases (Fig. 4B). Surmising that this deletion may force the displacement of His695 away from Ser562, we generated a Gly insertion mutant in PCY1, which would mimic the canonical S9A protease active site. If the misalignment of His695 indeed plays a role in the macrocyclase activity of PCY1, this insertion mutant should generate linear, hydrolytic products, similar to that of canonical S9A proteases. Notably, this PCY1 mutant slowly produces a significant amount of the linear, hydrolytic product (ratio of cyclic:linear product of ∼3.7:1), whereas the wild-type enzyme almost exclusively produces the macrocyclic product (Fig. 2 and SI Appendix, Table S3 and Fig. S13). These data support the assertion that the altered position of His695 plays some role in macrocycle formation by PCY1.
Although the insertion mutants showed the expected phenotype of production of both linear and cyclic products, the catalytic competency of the enzyme was severely compromised. To determine the basis for the diminished activity, we determined the 2.8-Å resolution structure of the PCY1 Gly696 insertion variant. The structure reveals significant disorder in the loop harboring His695 and could not be confidently modeled. A more detailed comparison of wild-type PCY1 against canonical S9A proteases reveals additional structural deviations near the active site, relative to canonical S9A proteases, in addition to the single Gly deletion (Fig. 4C). In particular, in canonical S9A enzymes, such as porcine prolyl oligopeptidase (PDB ID code 1QFS), the loop of an adjacent β-hairpin (encompassing Leu143 through Val156) is longer and buttresses the orientation of the catalytic His in the active site. In PCY1, the equivalent loop is one residue shorter and is displaced away from the active site. The side chain of Tyr696 (normally an Ala) occupies the volume created by this loop displacement. The Tyr696→Ala/Gly insertion double mutant had a greatly compromised activity, further underscoring that the repositioning of His695 in PCY1 involves a confluence of many local changes (SI Appendix, Fig. S14).
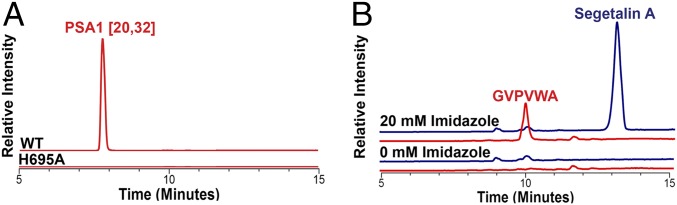
Based on the above data, we hypothesized that the His695 in PCY1 may serve two roles. It could serve to deprotonate the catalytic Ser562 (as in other serine proteases), but also may deprotonate the α-amino terminus of the peptide substrate to facilitate transamidation. To further investigate the dual functionality of the catalytic His, we analyzed the PCY1 His695→Ala variant under single turnover conditions (Fig. 5A). If His695 does play a role in the deprotonization of Ser562, this variant should still be unable to yield the hydrolytic, linear presegetalin A1 [20–32] by-product that forms after generation of the acyl-enzyme intermediate. As expected, the His695→Ala variant was completely inactive, indicating that His695 is necessary for activation of Ser562 despite the long distance between the two residues. Next, we sought to determine if His695 played a role in proton removal from the substrate α-amino group by testing the activity of PCY1 His695→Ala in the presence of imidazole. At the pH of the assay (pH = 8.5), imidazole should be deprotonated and may be able to function as an imperfect, mobile surrogate for His. Surprisingly, the addition of 20 mM imidazole resulted in a gain of function for the His695→Ala variant, which could generate both linear and cyclic [GVPVWA] segetalin A products from the presegetalin A1 [14–32] substrate (Fig. 5B). Competition between the mobile imidazole surrogate and solvent for attack on the acyl-enzyme intermediate may account for the observance of both linear and cyclic products in the His695→Ala variant. This result suggests that imidazole (and presumably His695 in the wild-type enzyme) can both deprotonate Ser562 to facilitate formation of an acyl-enzyme adduct and deprotonate the α-amino terminus of the substrate to facilitate transamidation.
Fig. 5.
(A) Formation of presegetalin A1 [20–32] was observed under single turnover conditions by LC-MS. After a 5-min incubation, wild-type PCY1 was able to catalyze formation of the acyl-enzyme intermediate, producing the presegetalin A1 [20–32] by-product. The H695A mutant did not catalyze formation of presegetalin A1 [20–32], indicating that it could not form the acyl-enzyme intermediate. (B) An 18-h incubation of H695A with 20 mM imidazole allowed for the production of both linear [GVPVWA] and cyclic segetalin A. However, the H695A variant produces a significantly greater amount of hydrolytic, linear product (ratio of cyclic:linear product of ∼3.7:1), whereas wild-type PCY1 almost exclusively produces the macrocyclic product (Fig. 2).
Discussion
Recent studies of various RiPP biosynthetic enzymes have provided several examples of biocatalysts that can generate a macrocyclic product from a linear peptide substrate (14, 28, 30, 31, 52–54). Such enzymes represent a next-generation technological utility, but many of these catalysts are often slow or require sequence constraints on substrates. The biosynthesis of orbitides, as well as other RiPPs such as cyclotides (28, 29), amatoxins (30), and cyanobactins (31), all use convergent strategies based on Ser proteases that can catalyze the formation of a cyclic product via transamidation, rather than a linear product via hydrolysis. Structural and biochemical studies of the cyanobactin macrocyclase PatG reveal that the enzyme has a helix-turn-helix insertion that sits above the active site, which presumably shields the acyl-enzyme intermediate from solvent. Full enclosure of the PatG active site upon binding of substrate is completed by the AYDG follower peptide sequence necessary for substrate recognition. The structural and biochemical data presented here on the orbitide macrocyclase PCY1 reveals a more nuanced strategy of achieving the same result. Rather than containing obvious insertions, transamidation by PCY1 is facilitated by the binding of the follower peptide, which maintains the enzyme in a closed state that precludes solvent from the active site, potentially limiting the competing hydrolysis reaction. The selectivity for substrates is maintained via recognition of the follower sequence, and the lack of appreciable affinity for the residues to be cyclized (as demonstrated by fluorescence polarization data) allows for product diversity. Specificity dictated by recognition of the follower peptide is not common in RiPP biosynthetic enzymes, and the only other known cases are of enzymes involved in the biosynthesis of cyanobactins and bottromycin (34).
The active site of PCY1 is also altered to more efficiently catalyze the cyclization reaction. The placement of His695 facilitates two roles: the canonical role of activating the Ser nucleophile to help form an acyl-enzyme intermediate and an unexpected role in deprotonating the α-amine of the peptide substrate to facilitate transamidation (SI Appendix, Fig. S15). The dual functionality for His695 is aided by its location in a mobile loop, which could accommodate the range of movements necessary for each function. Our biochemical studies provide a foundation for understanding the mechanistic basis for peptide macrocyclization in plants, an activity that is observed in growing classes of RiPP natural products.
Materials and Methods
Methods describing the cloning, expression, purification, biochemical analysis, kinetic and biochemical characterization, and crystallization of PCY1 and ligand complexes are described in detail in SI Appendix, SI Materials and Methods. Chemical data and analytical methods are also provided.
Supplementary Material
Acknowledgments
We thank the entire staff at the Life Sciences Collaborative Access Team at Argonne National Laboratory for their assistance with X-ray data collection. J.R.C. was supported in part by the Lowell P. Hager Fellowship from the Department of Biochemistry. The Bruker UltrafleXtreme mass spectrometer was purchased in part with a grant from the National Center for Research Resources, National Institutes of Health (S10 RR027109 A).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Crystallographic coordinates have been deposited in the Protein Data Bank, www.pdb.org [PDB ID codes 5UW3 (PCY1 with follower peptide), 5UW5 (PCY1 H695A variant with follower peptide), 5UW6 (PCY1 with follower peptide and covalent inhibitor ZPP), 5UW7 (PCY1 Y481F with follower peptide), and 5UZW (PCY1 G696 insertion variant with follower peptide and covalent inhibitor ZPP)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1620499114/-/DCSupplemental.
References
- 1.Passioura T, Katoh T, Goto Y, Suga H. Selection-based discovery of druglike macrocyclic peptides. Annu Rev Biochem. 2014;83:727–752. doi: 10.1146/annurev-biochem-060713-035456. [DOI] [PubMed] [Google Scholar]
- 2.Driggers EM, Hale SP, Lee J, Terrett NK. The exploration of macrocycles for drug discovery: An underexploited structural class. Nat Rev Drug Discov. 2008;7:608–624. doi: 10.1038/nrd2590. [DOI] [PubMed] [Google Scholar]
- 3.White TR, et al. On-resin N-methylation of cyclic peptides for discovery of orally bioavailable scaffolds. Nat Chem Biol. 2011;7:810–817. doi: 10.1038/nchembio.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rezai T, Yu B, Millhauser GL, Jacobson MP, Lokey RS. Testing the conformational hypothesis of passive membrane permeability using synthetic cyclic peptide diastereomers. J Am Chem Soc. 2006;128:2510–2511. doi: 10.1021/ja0563455. [DOI] [PubMed] [Google Scholar]
- 5.Scott CP, Abel-Santos E, Wall M, Wahnon DC, Benkovic SJ. Production of cyclic peptides and proteins in vivo. Proc Natl Acad Sci USA. 1999;96:13638–13643. doi: 10.1073/pnas.96.24.13638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kohli RM, Walsh CT, Burkart MD. Biomimetic synthesis and optimization of cyclic peptide antibiotics. Nature. 2002;418:658–661. doi: 10.1038/nature00907. [DOI] [PubMed] [Google Scholar]
- 7.Zhang C, Dai P, Spokoyny AM, Pentelute BL. Enzyme-catalyzed macrocyclization of long unprotected peptides. Org Lett. 2014;16:3652–3655. doi: 10.1021/ol501609y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hipolito CJ, Suga H. Ribosomal production and in vitro selection of natural product-like peptidomimetics: The FIT and RaPID systems. Curr Opin Chem Biol. 2012;16:196–203. doi: 10.1016/j.cbpa.2012.02.014. [DOI] [PubMed] [Google Scholar]
- 9.Velásquez JE, van der Donk WA. Genome mining for ribosomally synthesized natural products. Curr Opin Chem Biol. 2011;15:11–21. doi: 10.1016/j.cbpa.2010.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arnison PG, et al. Ribosomally synthesized and post-translationally modified peptide natural products: Overview and recommendations for a universal nomenclature. Nat Prod Rep. 2013;30:108–160. doi: 10.1039/c2np20085f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shimamura H, et al. Structure determination and total synthesis of bottromycin A2: A potent antibiotic against MRSA and VRE. Angew Chem Int Ed Engl. 2009;48:914–917. doi: 10.1002/anie.200804138. [DOI] [PubMed] [Google Scholar]
- 12.Sivonen K, Leikoski N, Fewer DP, Jokela J. Cyanobactins-ribosomal cyclic peptides produced by cyanobacteria. Appl Microbiol Biotechnol. 2010;86:1213–1225. doi: 10.1007/s00253-010-2482-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gross E, Morell JL. The structure of nisin. J Am Chem Soc. 1971;93:4634–4635. doi: 10.1021/ja00747a073. [DOI] [PubMed] [Google Scholar]
- 14.Schramma KR, Bushin LB, Seyedsayamdost MR. Structure and biosynthesis of a macrocyclic peptide containing an unprecedented lysine-to-tryptophan crosslink. Nat Chem. 2015;7:431–437. doi: 10.1038/nchem.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hegemann JD, Zimmermann M, Xie X, Marahiel MA. Lasso peptides: An intriguing class of bacterial natural products. Acc Chem Res. 2015;48:1909–1919. doi: 10.1021/acs.accounts.5b00156. [DOI] [PubMed] [Google Scholar]
- 16.Bagley MC, Dale JW, Merritt EA, Xiong X. Thiopeptide antibiotics. Chem Rev. 2005;105:685–714. doi: 10.1021/cr0300441. [DOI] [PubMed] [Google Scholar]
- 17.Sardar D, Lin Z, Schmidt EW. Modularity of RiPP enzymes enables designed synthesis of decorated peptides. Chem Biol. 2015;22:907–916. doi: 10.1016/j.chembiol.2015.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Craik DJ, Malik U. Cyclotide biosynthesis. Curr Opin Chem Biol. 2013;17:546–554. doi: 10.1016/j.cbpa.2013.05.033. [DOI] [PubMed] [Google Scholar]
- 19.Hallen HE, Luo H, Scott-Craig JS, Walton JD. Gene family encoding the major toxins of lethal Amanita mushrooms. Proc Natl Acad Sci USA. 2007;104:19097–19101. doi: 10.1073/pnas.0707340104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaufmann HP, Tobschirbel A. An oligopeptide from flaxseed. Chem Ber. 1959;92:2805–2809. [Google Scholar]
- 21.Gui B, et al. Identification and quantification of cyclolinopeptides in five flaxseed cultivars. J Agric Food Chem. 2012;60:8571–8579. doi: 10.1021/jf301847u. [DOI] [PubMed] [Google Scholar]
- 22.Pinto MEF, et al. Ribifolin, an orbitide from Jatropha ribifolia, and its potential antimalarial activity. J Nat Prod. 2015;78:374–380. doi: 10.1021/np5007668. [DOI] [PubMed] [Google Scholar]
- 23.Morita H, et al. Structure of a new cyclic nonapeptide, segetalin F, and vasorelaxant activity of segetalins from Vaccaria segetalis. Bioorg Med Chem Lett. 2006;16:4458–4461. doi: 10.1016/j.bmcl.2006.06.083. [DOI] [PubMed] [Google Scholar]
- 24.Gaymes TJ, Cebrat M, Siemion IZ, Kay JE. Cyclolinopeptide A (CLA) mediates its immunosuppressive activity through cyclophilin-dependent calcineurin inactivation. FEBS Lett. 1997;418:224–227. doi: 10.1016/s0014-5793(97)01345-8. [DOI] [PubMed] [Google Scholar]
- 25.Itokawa H, Yun Y, Morita H, Takeya K, Yamada K. Estrogen-like activity of cyclic peptides from Vaccaria segetalis extracts. Planta Med. 1995;61:561–562. doi: 10.1055/s-2006-959373. [DOI] [PubMed] [Google Scholar]
- 26.Condie JA, et al. The biosynthesis of Caryophyllaceae-like cyclic peptides in Saponaria vaccaria L. from DNA-encoded precursors. Plant J. 2011;67:682–690. doi: 10.1111/j.1365-313X.2011.04626.x. [DOI] [PubMed] [Google Scholar]
- 27.Barber CJS, et al. The two-step biosynthesis of cyclic peptides from linear precursors in a member of the plant family Caryophyllaceae involves cyclization by a serine protease-like enzyme. J Biol Chem. 2013;288:12500–12510. doi: 10.1074/jbc.M112.437947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nguyen GKT, et al. Butelase 1 is an Asx-specific ligase enabling peptide macrocyclization and synthesis. Nat Chem Biol. 2014;10:732–738. doi: 10.1038/nchembio.1586. [DOI] [PubMed] [Google Scholar]
- 29.Bernath-Levin K, et al. Peptide macrocyclization by a bifunctional endoprotease. Chem Biol. 2015;22:571–582. doi: 10.1016/j.chembiol.2015.04.010. [DOI] [PubMed] [Google Scholar]
- 30.Luo H, et al. Peptide macrocyclization catalyzed by a prolyl oligopeptidase involved in α-amanitin biosynthesis. Chem Biol. 2014;21:1610–1617. doi: 10.1016/j.chembiol.2014.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee J, McIntosh J, Hathaway BJ, Schmidt EW. Using marine natural products to discover a protease that catalyzes peptide macrocyclization of diverse substrates. J Am Chem Soc. 2009;131:2122–2124. doi: 10.1021/ja8092168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li P, Deng W, Li T. The molecular diversity of toxin gene families in lethal Amanita mushrooms. Toxicon. 2014;83:59–68. doi: 10.1016/j.toxicon.2014.02.020. [DOI] [PubMed] [Google Scholar]
- 33.Hemu X, Qiu Y, Nguyen GKT, Tam JP. Total synthesis of circular bacteriocins by butelase 1. J Am Chem Soc. 2016;138:6968–6971. doi: 10.1021/jacs.6b04310. [DOI] [PubMed] [Google Scholar]
- 34.Koehnke J, et al. The mechanism of patellamide macrocyclization revealed by the characterization of the PatG macrocyclase domain. Nat Struct Mol Biol. 2012;19:767–772. doi: 10.1038/nsmb.2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Agarwal V, Pierce E, McIntosh J, Schmidt EW, Nair SK. Structures of cyanobactin maturation enzymes define a family of transamidating proteases. Chem Biol. 2012;19:1411–1422. doi: 10.1016/j.chembiol.2012.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Czekster CM, Naismith JH. Kinetic landscape of a peptide bond-forming prolyl oligopeptidase. Biochemistry. 2017;56:2086–2095. doi: 10.1021/acs.biochem.7b00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tsilikounas E, Kettner CA, Bachovchin WW. Identification of serine and histidine adducts in complexes of trypsin and trypsinogen with peptide and nonpeptide boronic acid inhibitors by 1H NMR spectroscopy. Biochemistry. 1992;31:12839–12846. doi: 10.1021/bi00166a019. [DOI] [PubMed] [Google Scholar]
- 38.Holm L, Rosenström P. Dali server: Conservation mapping in 3D. Nucleic Acids Res. 2010;38:W545-9. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fülöp V, Böcskei Z, Polgár L. Prolyl oligopeptidase: An unusual beta-propeller domain regulates proteolysis. Cell. 1998;94:161–170. doi: 10.1016/s0092-8674(00)81416-6. [DOI] [PubMed] [Google Scholar]
- 40.Shan L, Mathews II, Khosla C. Structural and mechanistic analysis of two prolyl endopeptidases: Role of interdomain dynamics in catalysis and specificity. Proc Natl Acad Sci USA. 2005;102:3599–3604. doi: 10.1073/pnas.0408286102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li M, Chen C, Davies DR, Chiu TK. Induced-fit mechanism for prolyl endopeptidase. J Biol Chem. 2010;285:21487–21495. doi: 10.1074/jbc.M109.092692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Polgár L. The prolyl oligopeptidase family. Cell Mol Life Sci. 2002;59:349–362. doi: 10.1007/s00018-002-8427-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dundas J, et al. CASTp: Computed atlas of surface topography of proteins with structural and topographical mapping of functionally annotated residues. Nucleic Acids Res. 2006;34:W116-8. doi: 10.1093/nar/gkl282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Burkhart BJ, Hudson GA, Dunbar KL, Mitchell DA. A prevalent peptide-binding domain guides ribosomal natural product biosynthesis. Nat Chem Biol. 2015;11:564–570. doi: 10.1038/nchembio.1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tsai TY, Yang CY, Shih HL, Wang AHJ, Chou SH. Xanthomonas campestris PqqD in the pyrroloquinoline quinone biosynthesis operon adopts a novel saddle-like fold that possibly serves as a PQQ carrier. Proteins. 2009;76:1042–1048. doi: 10.1002/prot.22461. [DOI] [PubMed] [Google Scholar]
- 46.Ortega MA, et al. Structure and mechanism of the tRNA-dependent lantibiotic dehydratase NisB. Nature. 2015;517:509–512. doi: 10.1038/nature13888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Koehnke J, et al. Structural analysis of leader peptide binding enables leader-free cyanobactin processing. Nat Chem Biol. 2015;11:558–563. doi: 10.1038/nchembio.1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li K, Condurso HL, Li G, Ding Y, Bruner SD. Structural basis for precursor protein-directed ribosomal peptide macrocyclization. Nat Chem Biol. 2016;12:973–979. doi: 10.1038/nchembio.2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cleland WW. Low-barrier hydrogen bonds and enzymatic catalysis. Arch Biochem Biophys. 2000;382:1–5. doi: 10.1006/abbi.2000.2011. [DOI] [PubMed] [Google Scholar]
- 50.Radisky ES, Lee JM, Lu C-JK, Koshland DE., Jr Insights into the serine protease mechanism from atomic resolution structures of trypsin reaction intermediates. Proc Natl Acad Sci USA. 2006;103:6835–6840. doi: 10.1073/pnas.0601910103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Topf M, Richards WG. Theoretical studies on the deacylation step of serine protease catalysis in the gas phase, in solution, and in elastase. J Am Chem Soc. 2004;126:14631–14641. doi: 10.1021/ja047010a. [DOI] [PubMed] [Google Scholar]
- 52.Duquesne S, et al. Two enzymes catalyze the maturation of a lasso peptide in Escherichia coli. Chem Biol. 2007;14:793–803. doi: 10.1016/j.chembiol.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 53.Yan K-P, et al. Dissecting the maturation steps of the lasso peptide microcin J25 in vitro. ChemBioChem. 2012;13:1046–1052. doi: 10.1002/cbic.201200016. [DOI] [PubMed] [Google Scholar]
- 54.Li B. 2006. et al. Structure and mechanism of the lantibiotic cyclase involved in nisin biosynthesis. Science 311:1464–1467.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.