Significance
Projection neurons of the neocortex represent a diverse neuronal population that can be conveniently classified into three broad categories based on axon projection patterns: corticothalamic, subcerebral, and callosal. These neuronal subtypes are known to be specified postmitotically by a cortical derepression circuit involving the key transcription factors Tbr1, Fezf2, Satb2, and Ctip2. However, projection neuron identities are also known to be determined at the progenitor cell level, but the molecular mechanisms are poorly understood. Here we reveal that the proneural genes Neurog2 and Ascl1, which are together expressed in neocortical progenitors, cooperate to regulate the expression of components of the cortical derepression circuit to specify corticothalamic and subcerebral identities while repressing a callosal fate.
Keywords: neocortex, laminar fate specification, derepression circuit, proneural genes, temporal identity
Abstract
A derepression mode of cell-fate specification involving the transcriptional repressors Tbr1, Fezf2, Satb2, and Ctip2 operates in neocortical projection neurons to specify six layer identities in sequence. Less well understood is how laminar fate transitions are regulated in cortical progenitors. The proneural genes Neurog2 and Ascl1 cooperate in progenitors to control the temporal switch from neurogenesis to gliogenesis. Here we asked whether these proneural genes also regulate laminar fate transitions. Several defects were observed in the derepression circuit in Neurog2−/−;Ascl1−/− mutants: an inability to repress expression of Tbr1 (a deep layer VI marker) during upper-layer neurogenesis, a loss of Fezf2+/Ctip2+ layer V neurons, and precocious differentiation of normally late-born, Satb2+ layer II–IV neurons. Conversely, in stable gain-of-function transgenics, Neurog2 promoted differentiative divisions and extended the period of Tbr1+/Ctip2+ deep-layer neurogenesis while reducing Satb2+ upper-layer neurogenesis. Similarly, acute misexpression of Neurog2 in early cortical progenitors promoted Tbr1 expression, whereas both Neurog2 and Ascl1 induced Ctip2. However, Neurog2 was unable to influence the derepression circuit when misexpressed in late cortical progenitors, and Ascl1 repressed only Satb2. Nevertheless, neurons derived from late misexpression of Neurog2 and, to a lesser extent, Ascl1, extended aberrant subcortical axon projections characteristic of early-born neurons. Finally, Neurog2 and Ascl1 altered the expression of Ikaros and Foxg1, known temporal regulators. Proneural genes thus act in a context-dependent fashion as early determinants, promoting deep-layer neurogenesis in early cortical progenitors via input into the derepression circuit while also influencing other temporal regulators.
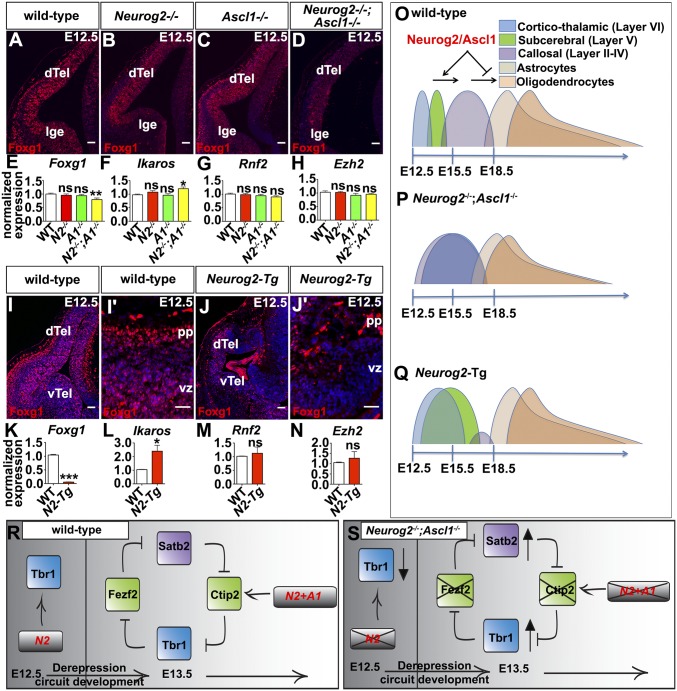
Neocortical neurons project to nearby or distant targets, depending on their molecular identity and correlating with laminar position. Deep-layer corticofugal neurons project subcortically and include layer VI corticothalamic neurons, which target the thalamus, and layer V subcerebral neurons, which project to the spinal cord, basal ganglia, and other distant targets (1). Conversely, layer IV granular neurons are the major site of thalamic input, whereas layer II/III callosal neurons form cortico–cortical connections. A cross-repressive gene-regulatory network operates in postmitotic projection neurons to specify laminar fates and to repress competing laminar identities (Fig. 1A) (2). Tbr1, a T-box transcription factor expressed in layer VI, specifies a corticothalamic neuronal fate (3) and also represses the expression of Fezf2, a zinc finger transcription factor that specifies a layer V subcerebral identity (4, 5). Fezf2 represses Tbr1 expression and a corticothalamic fate in layer V neurons (6, 7) and also represses the expression of Satb2 (8), an AT-rich DNA-binding protein that specifies a layer II–IV callosal identity (9, 10). In layer II–IV callosal neurons, Satb2 represses the expression of Ctip2/Bcl11b, a zinc finger transcription factor that controls layer V subcerebral projection patterns (11, 12). Ctip2 in turn represses Tbr1 expression in layer V, closing the negative feedback loop (2).
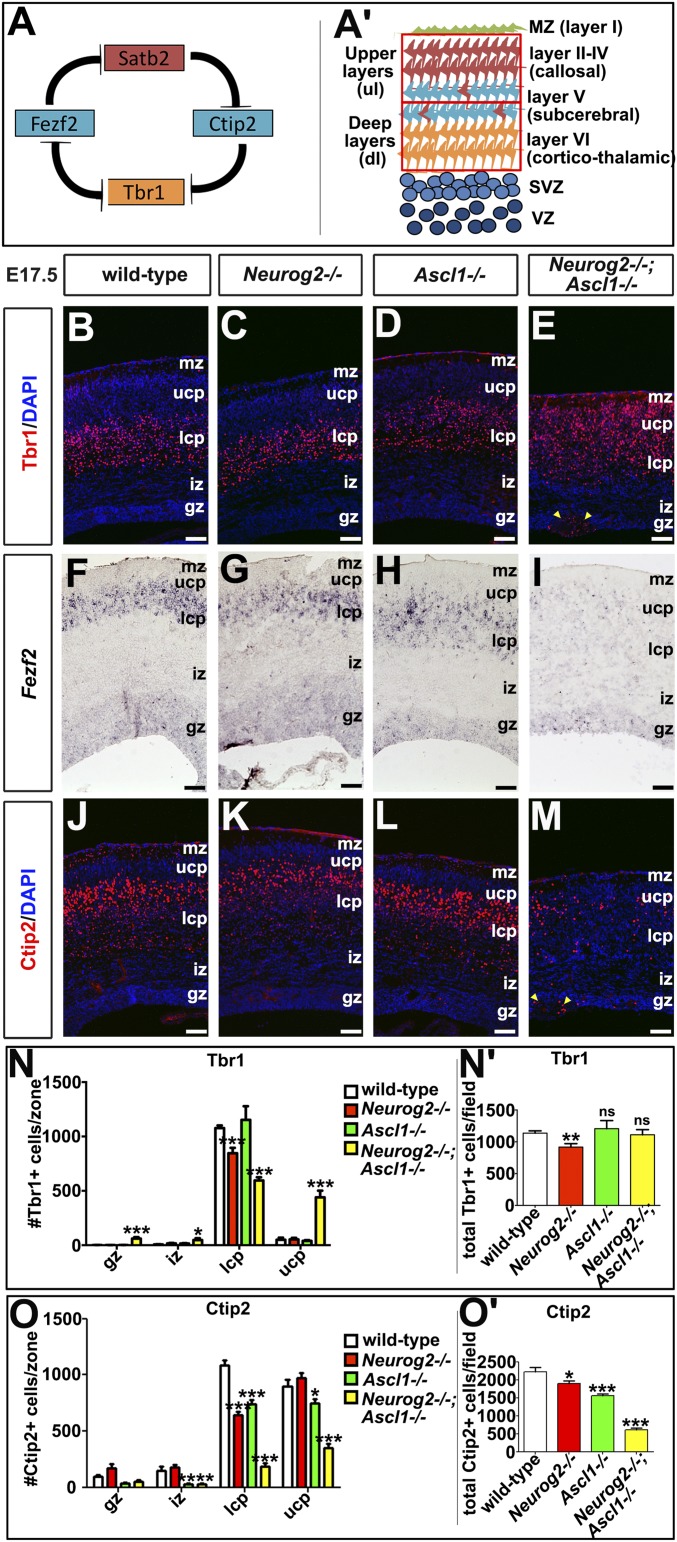
Fig. 1.
Expansion of layer VI and loss of layer V identities in Neurog2−/−;Ascl1−/− cortices. (A) Model of cross-repressive interactions among Tbr1, Fezf2, Satb2, and Ctip2. (A′) Model of the laminar organization of the neocortex, showing expression domains for Tbr1+ (orange, layer VI), Fezf2+/Ctip2+ (blue, layer V), and Satb2+ (red, layers II–IV). Red boxes mark upper and lower bins for quantitation. (B–M) Expression of Tbr1 (B–E), Fezf2 (F–I), and Ctip2 (J–M) in E17.5 wild-type, Neurog2−/−, Ascl1−/−, and Neurog2−/−;Ascl1−/− cortices. Blue is DAPI counterstain. Yellow arrowheads mark neuronal aggregates at the ventricular surface in E and M. (N–O′) Quantitation of Tbr1+ (N and N′) and Ctip2+ (O and O′) neurons in each cortical zone and in total. *P < 0.05; **P < 0.01; ***P < 0.005; ns, not significant. gz, germinal zone; iz, intermediate zone; lcp, lower cortical plate; mz, marginal zone; ucp, upper cortical plate. (Scale bars: 50 μm.)
Laminar identity is refined in projection neurons, but layer fates are initially determined at the progenitor cell level (13). The six neocortical layers arise from dorsal telencephalic (cortical) progenitor cells between days E10.5 and E17 in mouse. The earliest-born neurons form a preplate (PP) that later is split into an overlying marginal zone (layer I) and underlying subplate (transient layer VII). The remaining neuronal layers then differentiate sequentially, with layer VI neurons born next, followed by the differentiation of layer V, IV, III, and II neurons (14). Cortical neurons arise from two progenitor pools: radial glial cells (RGCs) in the ventricular zone (VZ) and intermediate neuronal progenitors (INPs) in the subventricular zone (SVZ). Cortical RGCs are multipotent, undergoing sequential fate restrictions to give rise to deep- and then upper-layer neurons (15).
Intrinsic signals that specify temporal identities within cortical progenitors include Foxg1, a winged-helix transcription factor that initially represses a layer I identity and later promotes deep-layer cortical fates (16). The subsequent switch from deep- to upper-layer cell fates also involves the repression of Tbr1 by Foxg1 (17). In addition, Ikaros/Ikzf1, a homolog of the temporal regulator hunchback in Drosophila, confers an early identity when overexpressed in the cortex (18). Finally, loss of Ezh2, part of polycomb repressive complex 2 (PRC2), confers an early identity to cortical progenitors (19). The proneural genes Neurogenin 2 (Neurog2) and Achaete scute-like 1 (Ascl1) also act in cortical progenitors to specify early-layer fates; Neurog2 is required for the generation of layer I, VI, and V early-born neurons but not late-born upper-layer neurons (20–22), and Ascl1 is required for the generation of a subset of early-born layer I neurons (23). Strikingly, Neurog2 and Ascl1 act together to regulate the timing of the switch from neurogenesis to gliogenesis, with precocious gliogenesis occurring in Neurog2−/−;Ascl1−/− cortices (24). However, the influence of these proneural genes on laminar fate transitions and on the postmitotic derepression circuit has not been assessed directly. In this study we used both loss- and gain-of-function strategies to provide evidence that Neurog2 and Ascl1 control the timing of neuronal laminar fate transitions by regulating the postmitotic derepression circuit, in addition to controlling other temporal regulators.
Results
Tbr1+ Neurons Expand into the Upper Cortical Plate in Neurog2−/−;Ascl1−/− Cortices.
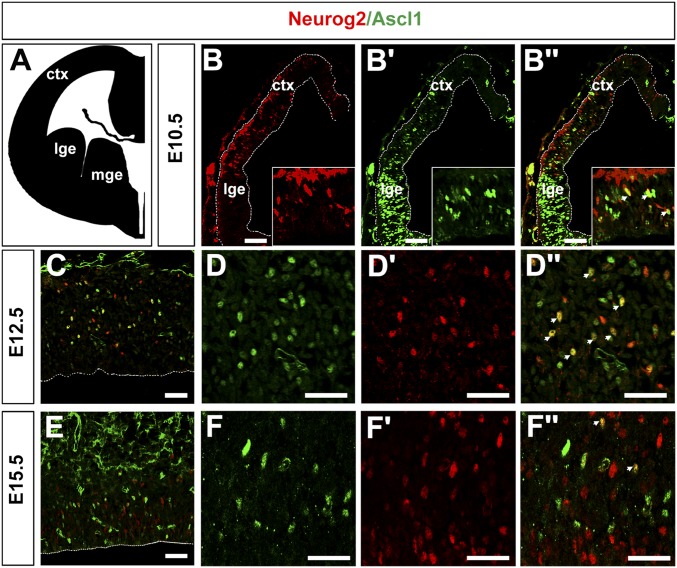
Neurog2 and Ascl1 are both expressed in cortical progenitors (25), beginning at E10.5, when neurogenesis begins, extending through E12.5, when deep-layer neurons differentiate, and continuing through (and past) E15.5, when upper-layer neurogenesis occurs (Fig. S1). We predicted that Neurog2 and Ascl1 may regulate temporal fate changes, controlling not only the switch from neurogenesis to gliogenesis (24) but also time-dependent transitions between laminar fates. To address this possibility, we examined components of the derepression circuit, Tbr1, Fezf2, Satb2, and Ctip2, in Neurog2;Ascl1 single and double mutants (Fig. 1A). We could not assess postnatal stages, when neurons have migrated to their final layers, because Neurog2;Ascl1 single and double mutants die at birth, so we conducted our studies at E17.5, when neurogenesis is complete. All analyses were conducted in dorsomedial domains, where a third proneural gene, Neurog1, is not highly expressed (20). In addition, because migration is still ongoing and layers are not clearly demarcated at E17.5, for quantitative purposes, the cortical plate (CP) was divided in half into what are hereafter called the “upper CP” and “lower CP” (schematized in Fig. 1A′). Finally, all comparisons of neuronal number were made relative to wild-type controls.
Fig. S1.
Neurog2 and Ascl1 are both expressed in a subset of cortical progenitors throughout the neurogenic period. (A) Schematic illustration of a coronal hemisection of an E12.5 mouse telencephalon, depicting the site of analysis. (B–F′′) Coimmunolabeling of E10.5 (B–B′′), E12.5 (C–D′′), and E15.5 (E–F′′) coronal sections with anti-Neurog2 (red) and anti-Ascl1 (green). Insets in B–B′′ show 4× magnification views. D–D′′ are 2× magnification images of C, and F–F′′ are 4× magnification images of E. Arrowheads mark cells that coexpress Neurog2 and Ascl1. ctx, neocortex; lge, lateral ganglionic eminence; mge, medial ganglionic eminence. (Scale bars: 50 μm.)
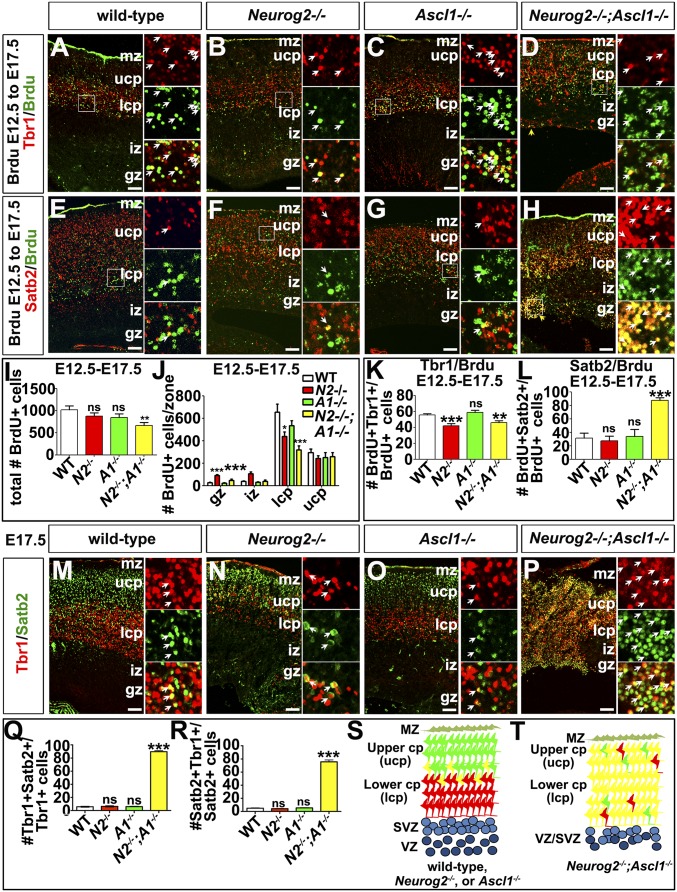
We first examined Tbr1, a factor required for the differentiation of layer VI corticothalamic neurons (3). Fewer Tbr1+ neurons were found in the lower CP (1.27-fold decrease; n = 3; P < 0.01) and overall (1.24-fold decrease; n = 3; P < 0.01) in Neurog2−/− cortices (Fig. 1 B, C, N, and N′), a finding that is consistent with the requirement for Neurog2 to specify a layer VI identity (21). In contrast, Tbr1+ neurons were at wild-type levels in Ascl1−/− cortices (Fig. 1 B, D, N, and N′). In Neurog2−/−;Ascl1−/− cortices, even fewer Tbr1+ neurons were observed in the lower CP (1.80-fold decrease; n = 3; P < 0.001), but Tbr1 was ectopically expressed in the upper CP, where this marker is not normally expressed at high levels (8.76-fold increase; n = 3; P < 0.01) (Fig. 1 B, E, N, and N′). Expansion of Tbr1 expression into the upper CP in double mutants was most evident rostrally, where all additional analyses were focused. Because Tbr1+ neurons were reduced in the lower CP and were expanded in the upper CP, the overall number of Tbr1+ neurons was unchanged in Neurog2−/−;Ascl1−/− cortices (Fig. 1N′). Finally, ectopic Tbr1+ cell clusters were also observed in the Neurog2−/−;Ascl1−/− germinal zone (GZ) (46.92-fold increase; n = 3; P < 0.001) (Fig. 1 E and N). Similar ectopic clusters of cells expressing the pan-neuronal markers Tuj1 (Fig. S2 A–D) and NeuN (Fig. S2 E–H) were evident in the Neurog2−/−;Ascl1−/− GZ, suggesting that these misplaced Tbr1+ cells were neurons.
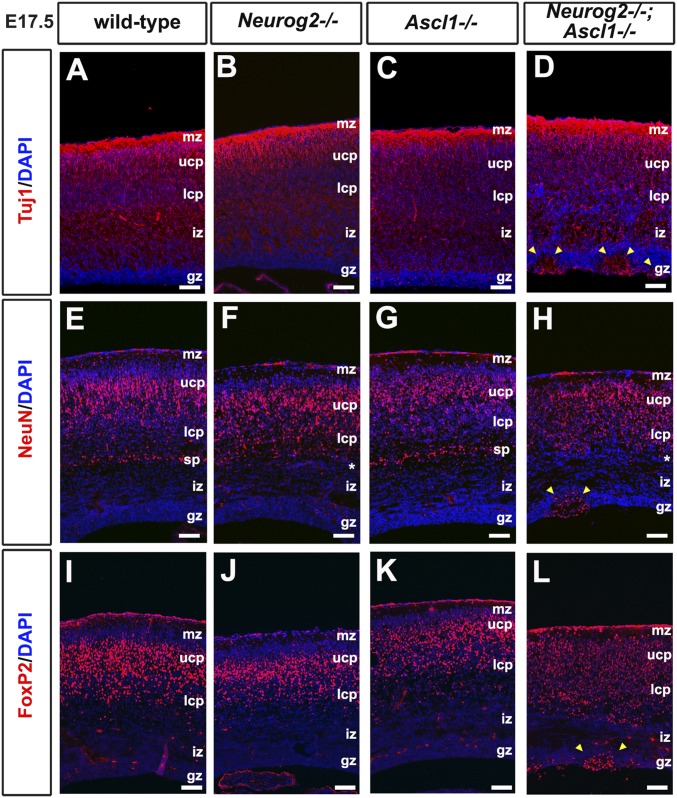
Fig. S2.
Neurons aggregate at the ventricular surface, and an expansion of layer VI neuronal identities is seen in Neurog2−/−;Ascl1−/− cortices. (A–L) Expression of Tuj1 (A–D), NeuN (E–H), and FoxP2 (I–L) in E17.5 wild-type (A, E, and I), Neurog2−/− (B, F, and J), Ascl1−/− (C, G, and K), and Neurog2−/−;Ascl1−/− (D, H, and L) cortices. Yellow arrowheads in D, H, and L mark neurons that aggregate near the ventricular surface. Asterisks mark the missing subplate in Neurog2−/− (F) and Neurog2−/−;Ascl1−/− (H) cortices. gz, germinal zone; iz, intermediate zone; lcp, lower cortical plate; mz, marginal zone; sp, subplate; ucp, upper cortical plate. (Scale bars: 50 μm.)
To confirm the expansion of deep-layer markers into the upper CP in Neurog2−/−;Ascl1−/− cortices, we also examined FoxP2, a definitive layer VI marker (26). FoxP2 was primarily expressed in deep layer VI neurons in wild-type, Neurog2−/−, and Ascl1−/− cortices but also was detected in the upper CP in Neurog2−/−;Ascl1−/− cortices (Fig. S2 I–L). Layer VI corticothalamic markers thus expand into the upper CP in the absence of Neurog2 and Ascl1, suggesting that these proneural genes together are required to extinguish a deep-layer fate during upper-layer neurogenesis.
Neurog2 and Ascl1 Are Required to Generate Fezf2+ and Ctip2+ Layer V Neurons.
In the postmitotic depression model, Tbr1 represses Fezf2 expression and a subcerebral identity (4). We observed ectopic Tbr1 expression in the upper CP in Neurog2−/−;Ascl1−/− cortices, which in our binning system contained part of presumptive layer V. We thus predicted that ectopic Tbr1 might repress Fezf2 expression in Neurog2−/−;Ascl1−/− cortices. Indeed, Fezf2 was expressed in presumptive layer V neurons in E17.5 wild-type, Neurog2−/−, and Ascl1−/− cortices, but Fezf2 transcripts were undetectable in Neurog2−/−;Ascl1−/− rostral cortices (Fig. 1 F–I).
We next assessed the expression of Ctip2, which establishes a subcerebral layer V identity in the derepression model (2). In Neurog2−/− cortices, fewer Ctip2+ neurons were found in the lower CP (1.68-fold decrease; n = 3) and overall (1.17-fold decrease; n = 3; P < 0.01; Fig. 1 J, K, O, and O′). There also was a reduction in Ctip2+ neurons in Ascl1−/− cortices in the lower CP (1.46-fold decrease; n = 3; P < 0.001), upper CP (1.21-fold decrease; P < 0.05), and overall (1.43-fold decrease; P < 0.001; Fig. 1 J, L, O, and O′). Finally, in Neurog2−/−;Ascl1−/− cortices, there was an even sharper decline in Ctip2+ neurons in the lower CP (5.96-fold; n = 3; P < 0.001), upper CP (2.56-fold; P < 0.001), and overall (3.62-fold; P < 0.001) (Fig. 1 J, M, O, and O′). Neurog2 and Ascl1 are thus partially redundant for the generation of Ctip2+ layer V neurons, with 85, 70, and 28% of the normal complement of Ctip2+ neurons generated in Neurog2−/−, Ascl1−/−, and Neurog2−/−;Ascl1−/− cortices, respectively.
Satb2+ Neurons Expand into the Lower CP in Neurog2−/−;Ascl1−/− Cortices.
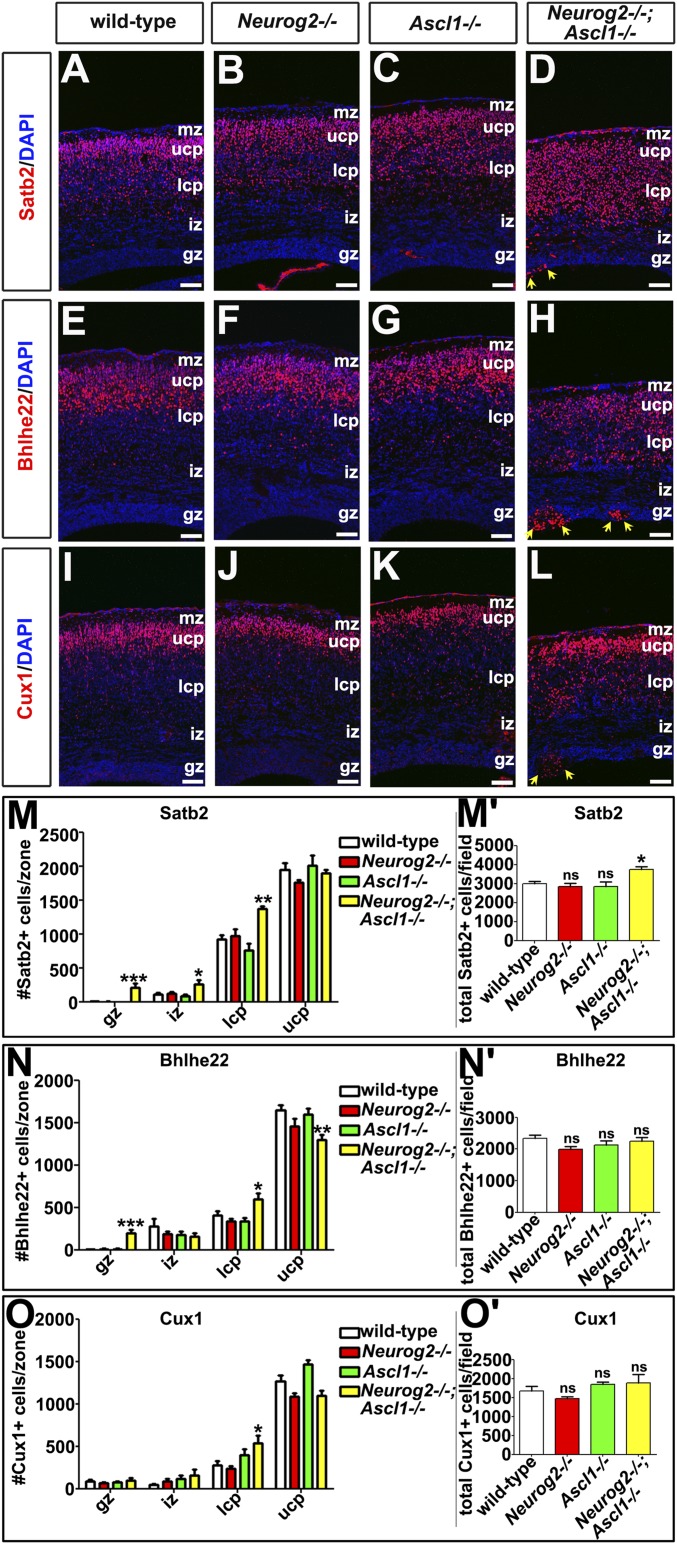
Fezf2 represses Satb2 expression, a determinant of a callosal identity, in postmitotic neurons (8). We thus predicted that the loss of Fezf2 transcripts in Neurog2−/−;Ascl1−/− cortices would increase Satb2+ neurogenesis (8). Indeed, although Satb2 was preferentially expressed in the upper CP (layers II–IV) in all genotypes (Fig. 2 A–D), Satb2 was also ectopically expressed in the Neurog2−/−;Ascl1−/− lower CP (1.48-fold increase; n = 3; P < 0.01), GZ (58.29-fold increase; P < 0.001), and intermediate zone (IZ) (2.42-fold increase; P < 0.05) (Fig. 2 D and M). Although these increases could be indicative of migratory defects, there also was an overall increase in the total number of Satb2+ neurons in Neurog2−/−;Ascl1−/− cortices (1.25-fold increase; P < 0.05) (Fig. 2M′). More neurons thus acquire a Satb2+ phenotype in the absence of Neurog2 and Ascl1.
Fig. 2.
Upper-layer neurons expand into the lower CP in Neurog2−/−;Ascl1−/− cortices. (A–L) Expression of Satb2 (A–D), Bhlhe22 (E–H), and Cux1 (I–L) in E17.5 wild-type, Neurog2−/−, Ascl1−/−, and Neurog2−/−;Ascl1−/− cortices. Blue is DAPI counterstain. Yellow arrows mark neuronal aggregates at the ventricular surface in D, H, and L. (M–O′) Quantitation of Satb2+ (M and M′), Bhlhe22+ (N and N′), and Cux1+ (O and O′) neurons in each cortical zone and in total. *P < 0.05; **P < 0.01; ***P < 0.005; ns, not significant. gz, germinal zone; iz, intermediate zone; lcp, lower cortical plate; mz, marginal zone; ucp, upper cortical plate. (Scale bars: 50 μm.)
We next examined the expression of upper-layer markers that are not known to be part of the derepression circuit, Bhlhe22 (27) and Cux1/2 (28), which are expressed in layer II–IV neurons. In Neurog2−/−;Ascl1−/− cortices, there was a 1.27-fold decrease in Bhlhe22+ neurons in the upper CP (n = 3; P < 0.05), but more Bhlhe22+ neurons were present in the lower bin (1.48-fold increase; P < 0.05) and GZ (48.49-fold increase; P < 0.001) (Fig. 2 E–H and N). Overall, the total number of Bhlhe22+ neurons was unchanged in Neurog2−/−;Ascl1−/− cortices (Fig. 2N′), with the loss of Bhlhe22+ layer V neurons in the upper bin likely masking the increase in layer VI. Similarly, more Cux1/2+ neurons were observed in the lower CP in Neurog2−/−;Ascl1−/− embryos (1.92-fold increase; n = 3; P < 0.05) (Fig. 2 I–L and O), but the overall number of Cux1/2+ neurons was similar in all genotypes (Fig. 2O′). Satb2, Cux1/2, and Bhlhe22 are thus all ectopically expressed in lower cortical layers in Neurog2−/−;Ascl1−/− cortices, suggesting that upper-layer markers are precociously expressed in deep layers and/or indicating upper-layer migratory errors, possibilities we assess below.
Satb2+ Neurons Are Generated Precociously in Neurog2−/−;Ascl1−/− Cortices.
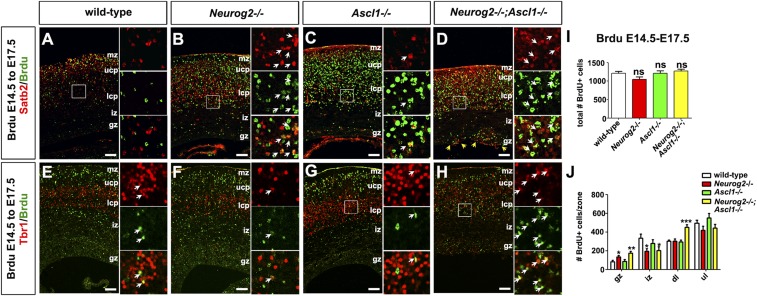
To examine migration and the timing of neurogenesis, we injected BrdU into pregnant dams at E12.5 or E14.5 to label proliferating progenitors and harvested embryos at E17.5. Only the progenitors that exited the cell cycle to undergo neurogenesis at the time of labeling retained a BrdU signal. Fewer BrdU-labeled postmitotic cells were detected in Neurog2−/−;Ascl1−/− cortices (1.54-fold reduction; n = 3; P < 0.01) (Fig. 3 A–D and I), most notably in the lower CP (2.07-fold decrease; P < 0.01) (Fig. 3J). There were no overall differences in the total number of E12.5-born neurons in Neurog2−/− or Ascl1−/− single-mutant cortices (Fig. 3 A–D and I). However, there were fewer E12.5-born neurons in the lower CP in Neurog2−/− cortices (1.49-fold; P < 0.05), and there was a corresponding increase in the GZ (3.32-fold increase; P < 0.001) and IZ (2.75-fold increase; P < 0.001) (Fig. 3J), a finding that is consistent with previously described migration defects (21). In contrast, there were no differences in overall numbers of E14.5-born neurons in wild-type or Neurog2;Ascl1 single and double mutants (Fig. S3 A–I). However, more late-born neurons were detected in the lower CP and GZ in double mutants (Fig. S3J), suggesting that migratory errors may contribute, in part, to the ectopic expression of Satb2, Cux1, and Bhlhe22.
Fig. 3.
Altered timing of deep- and upper-layer neurogenesis in Neurog2−/−;Ascl1−/− cortices. (A–H) E12.5 to E17.5 BrdU birthdating showing colabeling with BrdU and Tbr1 (A–D) or Satb2 (E–H) in wild-type, Neurog2−/−, Ascl1−/−, and Neurog2−/−;Ascl1−/− cortices. Insets at the right are 4× magnified views of the regions depicted in white boxes. White arrows mark double-positive cells. Yellow arrowheads mark ectopic BrdU+ cells at the ventricular surface of Neurog2−/−;Ascl1−/− cortices in D and H. (I and J) Quantitation of the total number of BrdU+ cells (I) and the number of BrdU+ cells per cortical zone (J). (K and L) Quantitation of the percentage of BrdU+ cells labeled at E12.5 that coexpress Tbr1 (K) and Satb2 (L). (M–P) Coexpression of Tbr1 and Satb2 in E17.5 wild-type, Neurog2−/−, Ascl1−/−, and Neurog2−/−;Ascl1−/− cortices. White arrows mark double-positive cells in the 4× magnification panels on the right. (Q and R) Quantitation of the number of Tbr1+ (Q) and Satb2+ (R) neurons as a ratio of total Tbr1+ and Satb2+ neurons, respectively. (S and T) Depiction of laminar distribution in wild-type, Neurog2−/−, and Ascl1−/− (S) and Neurog2−/−;Ascl1−/− (T) cortices. *P < 0.05; **P < 0.01; ***P < 0.005; ns, not significant. gz, germinal zone; iz, intermediate zone; lcp, lower cortical plate; mz, marginal zone; ucp, upper cortical plate. (Scale bars: 50 μm.)
Fig. S3.
Altered migration patterns of late-born neurons in Neurog2−/− and Neurog2−/−;Ascl1−/− cortices. (A–H) E14.5 to E17.5 BrdU birthdating showing colabeling of BrdU with Satb2 (A–D) and Tbr1 (E–H) in wild-type (A and E), Neurog2−/− (B and F), Ascl1−/− (C and G), and Neurog2−/−;Ascl1−/− (D and H) cortices. Insets to the right show 4× magnified views of the regions in white boxes. White arrows mark double-positive cells. Yellow arrowheads mark ectopic BrdU+ cells at the ventricular surface of Neurog2−/−;Ascl1−/− (D) cortices. (I and J) Quantitation of the total number of BrdU+ cells (I) and BrdU+ cells per cortical zone (J). *P < 0.05; **P < 0.01; ***P < 0.005; ns, not significant. gz, germinal zone; iz, intermediate zone; lcp, lower cortical plate; mz, marginal zone; ucp, upper cortical plate. (Scale bars: 50 μm.)
To assess the timing of neurogenesis, we combined layer markers with E12.5→E17.5 BrdU birthdating. Colabeling of BrdU and Tbr1 revealed that there were fewer E12.5-born Tbr1+ neurons in E17.5 Neurog2−/− (1.32-fold decrease; P < 0.001) and Neurog2−/−;Ascl1−/− (1.21-fold decrease; P < 0.01) cortices (Fig. 3 A–D and K), a finding that is consistent with the decreased number of Tbr1+ neurons in the lower CP of these embryos (Fig. 1N). The reduction in Tbr1+ neurons in Neurog2−/− cortices likely reflects the misspecification of early-born neurons to a GABAergic fate, as previously shown (20). However, GABAergic neurons are not ectopically generated in Neurog2−/−;Ascl1−/− cortices (20, 21). Instead, there was a striking increase in the number of Satb2+ neurons born at E12.5 in double mutants, as is consistent with the precocious differentiation of these normally late-born neurons (2.79-fold increase; P < 0.05; Fig. 3 E–H and L).
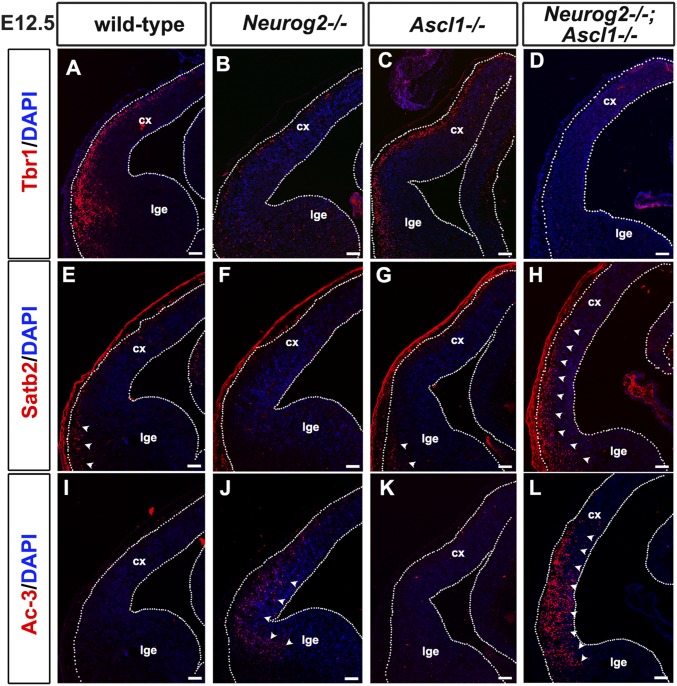
To confirm that Satb2+ neurons were born prematurely in double mutants, we examined laminar marker expression at E12.5, when only deep-layer Tbr1+ neurons are normally present. Compared with wild-type and Ascl1−/− cortices, a reduction but not a complete loss of Tbr1+ neurons was observed in E12.5 Neurog2−/− and Neurog2−/−;Ascl1−/− cortices (Fig. S4 A–D). Conversely, Satb2, which marks upper-layer cortical neurons that are born after E14.5 and which is normally expressed only in the very lateral cortex at E12.5, was precociously expressed in Neurog2−/−;Ascl1−/− cortices (Fig. S4 E–H), suggestive of a Tbr1+-to-Satb2+ fate switch. Finally, apoptosis was elevated in the lateral PP in Neurog2−/−;Ascl1−/− and to a lesser extent in Neurog2−/− cortices, as assessed by the expression of activated caspase 3 (Fig. S4 I–L). Satb2+ neurons thus differentiate precociously in the absence of Neurog2 and Ascl1, and some of these misspecified, early-born neurons undergo cell death.
Fig. S4.
Upper-layer markers are precociously expressed in E12.5 Neurog2−/−;Ascl1−/− cortices. (A–L) Expression of Tbr1 (A–D), Satb2 (E–H), and activated caspase 3 (Ac-3) (I–L) in E12.5 wild-type (A, E, and I), Neurog2−/− (B, F, and J), Ascl1−/− (C, G, and K), and Neurog2−/−;Ascl1−/− (D, H, and L) cortices. Arrowheads in E, G, and H mark extent of Satb2 expression domain. Note that apparent red labeling outside the white outline of the cortex in E–H is background pial staining. Arrowheads in J and L mark apoptotic cells. cx, neocortex; lge, lateral ganglionic eminence. (Scale bars: 100 μm.)
Neurog2−/−;Ascl1−/− Cortical Neurons Have a Mixed Tbr1+/Satb2+ Identity and Aberrant Axonal Projections.
BrdU birthdating showed that both Satb2+ and Tbr1+ neurons were born in an overlapping fashion at E12.5 in Neurog2−/−;Ascl1−/− cortices, suggesting that these markers could be coexpressed within the same cells. In wild-type cortices, Tbr1 and Satb2 are coexpressed in a small proportion of wild-type neurons during early corticogenesis, but by E15.5 double-positive cells largely resolve into single-positive cells (9). We similarly found that a small number of cortical neurons coexpressed Tbr1 and Satb2 in E17.5 wild-type, Neurog2−/−, and Ascl1−/− cortices, but in Neurog2−/−;Ascl1−/− cortices almost all Tbr1+ neurons coexpressed Satb2 (15.52-fold increase vs. wild-type; P < 0.0001), and most Satb2+ neurons coexpressed Tbr1 (15.35-fold increase; P < 0.0001) (Fig. 3 M–R). Thus, in the absence of Neurog2 and Ascl1, cortical neurons acquire a mixed identity that is reminiscent of an earlier developmental stage (Fig. 3 S and T).
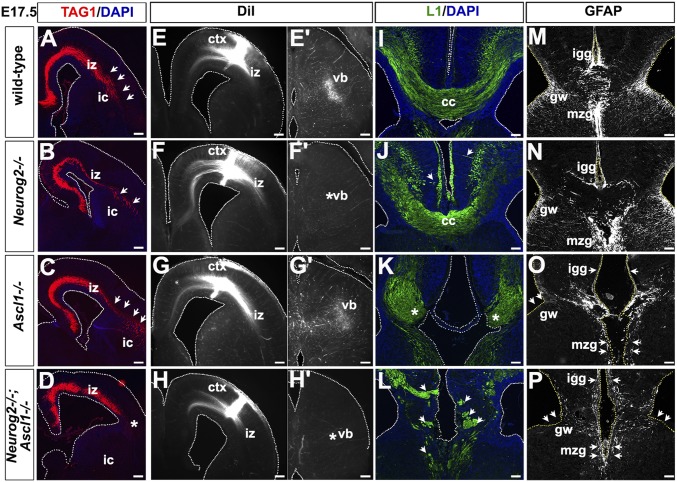
Specification of appropriate laminar identities is essential for the development of callosal and corticofugal projections. Accordingly, although transient axonal glycoprotein 1 (TAG1)-labeled axons projected laterally to enter the ventral telencephalon in Neurog2 and Ascl1 single mutants, they failed to do so in Neurog2−/−;Ascl1−/− cortices, instead truncating prematurely in the cortical IZ (Fig. 4 A–D). The further lateral extension of corticothalamic projections into the ventral telencephalon was also disrupted in E17.5 Neurog2−/− mutants, as previously reported (21, 29) and as shown by TAG1 immunostaining (Fig. 4 A–D). In addition, anterograde labeling of corticofugal axons with a 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindocarbocyanine perchlorate (DiI) injection in the rostral cortex (Fig. 4 E–H) labeled projections to the ventrobasal nucleus in the thalamus in wild-type and Ascl1 mutants but not in Neurog2−/− and Neurog2−/−;Ascl1−/− cortices (Fig. 4 E′–H′).
Fig. 4.
Influence of Neurog2 and Ascl1 on corticofugal and callosal projections. (A–D) Expression of TAG1 in E17.5 wild-type (A), Neurog2−/− (B), Ascl1−/− (C), and Neurog2−/−;Ascl1−/− (D) cortices. Blue is DAPI counterstain. Arrows in A–C mark corticofugal axons entering the ventral telencephalon. The asterisk in D marks premature truncation of corticofugal axons. (E–H′) Anterograde labeling of corticofugal axons with DiI injection in the rostral cortex (E, F, G, and H) and traced into the ventrobasal nucleus (E′, F′, G′, and H′) in E17.5 wild-type (E and E′), Neurog2−/− (F and F′), Ascl1−/− (G and G′), and Neurog2−/−;Ascl1−/− (H and H′) cortices. Asterisks mark the absence of anterograde labeling of the thalamus in Neurog2−/− (F′) and Neurog2−/−;Ascl1−/− (H′) cortices. (I–L) Expression of L1 in E17.5 wild-type (I), Neurog2−/− (J), Ascl1−/− (K), and Neurog2−/−;Ascl1−/− (L) cortices. Blue is DAPI counterstain. Asterisks in K mark Probst bundles. Arrowheads in J and L mark misguided, defasciculated axon tracts. (M–P) Expression of GFAP in E17.5 wild-type (M), Neurog2−/− (N), Ascl1−/− (O), and Neurog2−/−;Ascl1−/− (P) cortices. Arrowheads in O and P mark the loss of midline glia. ca, callosal axons; ctx, neocortex; gw, glial wedge; ic, internal capsule; igg, indusium griseum glia; iz, intermediate zone; mzg, midline zipper glia; vb, ventrobasal complex. (Scale bars: 250 μm in A–H and E′–H′ and 100 μm in I–P.)
L1 labeling revealed mild, incompletely penetrant defects in the corpus callosum in E17.5 Neurog2−/− cortices, with some axons defasciculating and projecting aberrantly instead of targeting the midline (Fig. 4 I and J), as reported previously (29). In contrast, very severe defects were observed in the midline in E17.5 Ascl1−/− cortices: The two hemispheres were not properly fused, the corpus callosum was missing, and L1-labeled callosal axons did not cross the midline, instead forming aberrant Probst bundles (Fig. 4K), as previously documented (30). However, an even more striking phenotype was observed in the midline of E17.5 Neurog2−/−;Ascl1−/− cortices, with L1+ axons undergoing widespread defasciculation to form multiple, misdirected axonal bundles (Fig. 4L).
We asked whether the failure of callosal axons to extend across the midline in E17.5 Ascl1−/− and Neurog2−/−;Ascl1−/− cortices arose because of defects in midline glia, which serve as a substrate for callosal axons and are required for corpus callosum formation (31). In E17.5 wild-type and Neurog2−/− cortices, GFAP marked all midline glia populations, including indusium griseum glia and midline zipper glia, which attract callosal axons to cross the midline, and the glial wedge, which prevents callosal axons from entering the ventral septum (Fig. 4 M and N). In contrast, in E17.5 Ascl1−/− and Neurog2−/−;Ascl1−/− cortices, a pronounced reduction in GFAP expression was observed in the midline (Fig. 4 O and P). The inability of callosal neurons to cross the midline in Ascl1−/− and Neurog2−/−;Ascl1−/− cortices thus is likely caused, at least in part, by defects in midline glia. However, the misspecification of laminar fates in double mutants may contribute to the more severe defects in both subcortical and callosal axonal targeting.
Generation of a Conditional Neurog2 Gain-of-Function Allele.
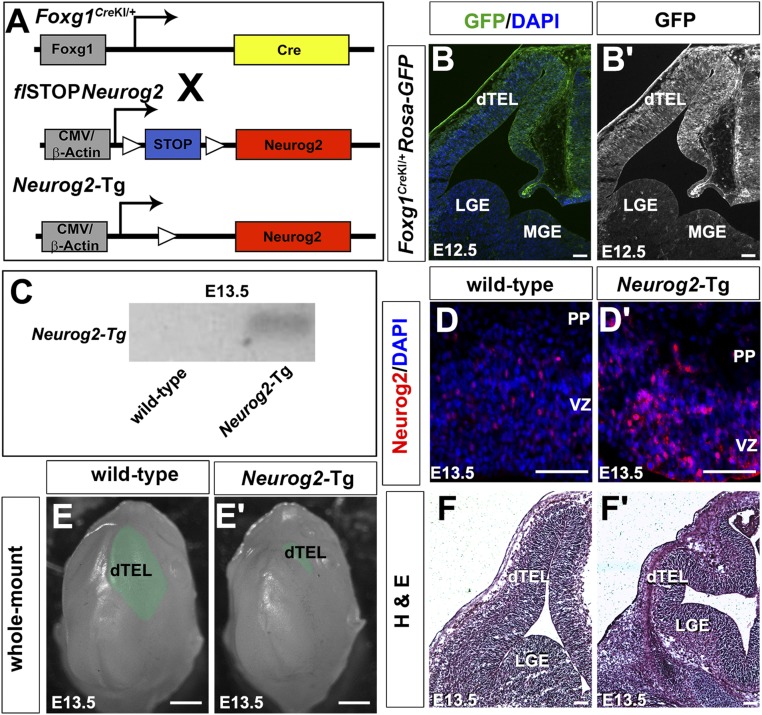
Given the loss of Tbr1+ neurons in deep layers in Neurog2−/− and Neurog2−/−;Ascl1−/− cortices and of Ctip2+ neurons in Neurog2−/−, Ascl1−/−, and Neurog2−/−;Ascl1−/− cortices, we asked whether these proneural genes were sufficient to specify deep-layer identities. We first focused on Neurog2, generating a conditional gain-of-function transgenic line to assess its laminar fate-specification properties. We did not generate Ascl1 transgenics, because Ascl1 promotes the ectopic expression of both neuronal and glial markers, complicating analyses in vivo (32, 33). A conditional Neurog2 gain-of-function allele was generated using a transgene carrying a floxed “stop” cassette and the Neurog2 coding sequence (flSTOP::Neurog2) (Fig. S5A). flSTOP::Neurog2 and Foxg1CreKI mice were crossed to generate double transgenics hereafter referred to as “Neurog2-Tgs.” Foxg1 is expressed throughout the telencephalon from E9.5; however, the genetic background influences Cre tissue specificity in Foxg1CreKI mice (34). In E12.5 Foxg1CreKI;Rosa-GFP mice, GFP was expressed throughout the dorsal but not the ventral telencephalon (Fig. S5 B and B′), suggesting that in our CD1 background this driver is preferentially active in cortical lineages. Accordingly, we amplified a transgene-specific transcript from E13.5 cortical tissue using primers anchored in the 5′ region of Neurog2 and the unique 3′ UTR (Fig. S5C). Moreover, immunostaining of E13.5 sections revealed that more cortical progenitors expressed Neurog2 protein in Neurog2-Tg than in wild-type cortices (Fig. S5 D and D′). We thus have generated a conditional Neurog2 gain-of-function transgenic line that drives Neurog2 expression in a sustained manner rather than in its normal oscillatory pattern of expression (35).
Fig. S5.
Generation of a conditional Neurog2 gain-of-function mouse line. (A) Strategy to generate Neurog2-Tg. (B and B′) Spatially restricted Foxg1-cre activity at E12.5, monitored with Rosa-GFP. (C) Amplification of Neurog2-Tg transcript. (D and D′) Neurog2 expression in E13.5 wild-type (D) and Neurog2-Tg (D′) cortices. (E and E′) Whole-mount views of E13.5 wild-type (E) and Neurog2-Tg (E′) heads (telencephalons are false-colored green). (F and F′) H&E staining of E13.5 wild-type (F) and Neurog2-Tg (F′) cortices. (Scale bars: 50 μm in D and D′; 250 μm in E and E′; 300 μm in B, B′, F, and F′.) dTEL, dorsal telencephalon; LGE, lateral ganglionic eminence; MGE, medial ganglionic eminence; PP, preplate; VZ, ventricular zone.
More Early-Born Neurons and Fewer Late-Born Neurons Are Produced in Early–Mid Corticogenesis in Neurog2-Tgs.
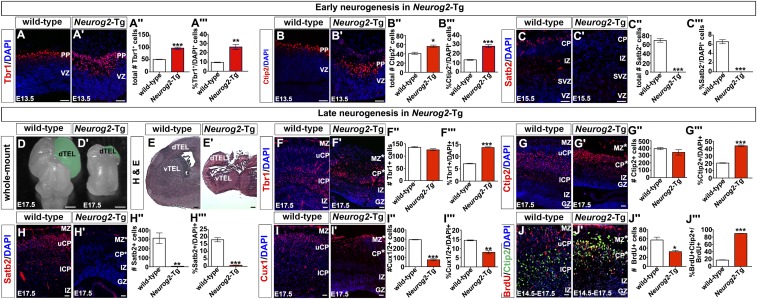
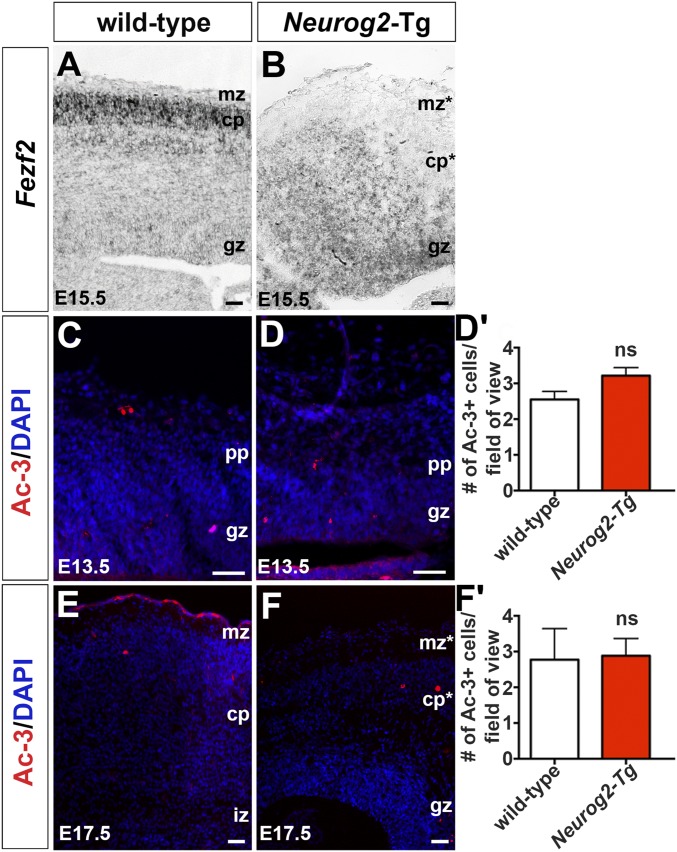
We reasoned that if Neurog2 instructed early identities, Neurog2-Tgs should generate more Tbr1+ and Ctip2+ neurons at the expense of Satb2+ neurons. At E13.5, a reduction in overall cortical size was evident in Neurog2-Tg brains (Fig. S5 E and E′), and this finding was confirmed in H&E-stained E13.5 coronal sections (Fig. S5 F and F′). Nevertheless, more Tbr1+ neurons were generated in Neurog2-Tg cortices, both overall (Tbr1+; 1.90-fold increase; n = 3; P < 0.001) and in proportional representation (Tbr1+/DAPI+; 2.58-fold increase; P < 0.01) (Fig. 5 A–A′′′). The same pattern was observed for Ctip2+ neurons in Neurog2-Tgs, with increases in both total number (Ctip2+; 1.37-fold increase; n = 3; P < 0.05) and proportional representation (Ctip2+/DAPI+; 2.11-fold increase; n = 3; P < 0.001) (Fig. 5 B–B′′′). Collectively, these data suggested that a greater fraction of Tbr1+ and Ctip2+ neurons occupied the E13.5 Neurog2-Tg cortex. As a corollary, we anticipated a decrease in the production of Satb2+ neurons. Indeed, although 6.01 ± 0.18% of DAPI+ cells in the CP expressed Satb2 in E15.5 wild-type cortices, no Satb2+ neurons were observed in E15.5 Neurog2-Tg cortices (P < 0.001) (Fig. 5 C–C′′′). Conversely, Fezf2, a Satb2 repressor, was ectopically expressed in the CP as well as the GZ in E15.5 Neurog2-Tg cortices (Fig. S6 A and B), the latter reminiscent of the early (E12.5) expression of Fezf2 in wild-type cortical progenitors (36). Thus there is a striking bias toward the production of layer V and VI neuronal phenotypes in Neurog2-Tgs, along with an apparent repression of Satb2+ upper-layer identities.
Fig. 5.
Increased early neurogenesis and decreased late neurogenesis in E17.5 Neurog2-Tg cortices. (A–C) Early neurogenesis in Neurog2-Tgs. Expression of Tbr1 (A and A′) and Ctip2 (B and B′) at E13.5 and Satb2 (C and C′) at E15.5 in wild-type and Neurog2-Tg cortices. Quantitation of Tbr1+ (A′′ and A′′′), Ctip2+ (B′′ and B′′′), and Satb2+ (C′′ and C′′′) neurons in total and as the percent of marker+/DAPI+ cells. (D–J) Late neurogenesis in Neurog2-Tgs. (D and D′) Whole-mount views of E17.5 wild-type (D) and Neurog2-Tg (D′) brains (telencephalons false-colored green). (E and E′) H&E staining of E17.5 wild-type (E) and Neurog2-Tg (E′) telencephalons. (F–J′) Expression of Tbr1 (F and F′), Ctip2 (G and G′), Satb2 (H and H′), Cux1/2 (I and I′), and Brdu/Ctip2 (J and J′) in E17.5 cortices. (F′′–I′′′) Quantitation of Tbr1+ (F′′ and F′′′), Ctip2+ (G′′ and G′′′), Satb2+ (H′′ and H′′′), and Cux1/2+ (I′′ and I′′′) cells in total and as the percent of marker+/DAPI+ cells. (J′′ and J′′′) Quantitation of BrdU+ (J′′) and BrdU+Ctip2+/BrdU+ (J′′′) cells. The asterisks in the images mark aberrant marginal zones and cortical plates. *P < 0.05; **P < 0.01; ***P < 0.005. CP, cortical plate; dTEL, dorsal telencephalon; GZ, germinal zone; lCP, lower cortical plate; IZ, intermediate zone; MZ, marginal zone; PP, preplate; SVZ, subventricular zone; uCP, upper cortical plate; vTEL, ventral telencephalon; VZ, ventricular zone. (Scale bars: 50 μm in A–C′ and F–J′, 250 μm in E and E′, and 400 μm in D and D′.)
Fig. S6.
Expression of Fezf2 and activated caspase 3 in Neurog2-Tgs. (A and B) Expression of Fezf2 in E15.5 wild-type (A) and Neurog2-Tg (B) cortices. (C–F) Expression of activated caspase-3 (Ac-3) in E13.5 (C and D) and E17.5 (E and F) wild-type and Neurog2-Tg cortices. (D′ and F′) Quantification of Ac-3+ cells at E13.5 (D′) and E17.5 (F′). Asterisks denote aberrant marginal zone and CP in Neurog2-Tgs. cp, cortical plate; gz, germinal zone; iz, intermediate zone; mz, marginal zone; ns, not significant; pp, preplate. (Scale bars: 50 μm.)
More Early-Born and Fewer Late-Born Neurons in Late Corticogenesis in Neurog2-Tgs.
If Neurog2 was indeed a true determinant of an early identity, we reasoned that the bias toward deep-layer marker expression would be maintained in E17.5 Neurog2-Tgs, when cortical neurogenesis is complete. At E17.5, Neurog2-Tg brains were dramatically reduced in size in whole-mount (Fig. 5 D and D′) and in H&E-stained sections (Fig. 5 E and E′). Analyses of laminar fates in dorsomedial domains, where mutant studies were performed, revealed that the total number of Tbr1+ neurons was similar in wild-type and Neurog2-Tg cortices (Fig. 5 F–F′′). However, given the reduced size of the cortex, there was an increase in the proportion of Tbr1+/DAPI+ cells (1.92-fold increase; P < 0.001) (Fig. 5F′′′). Similarly, although the overall number of Ctip2+ neurons was unchanged in Neurog2-Tg cortices, the proportion of Ctip2+/DAPI+ cells was increased (2.14-fold increase; n = 3; P < 0.001) (Fig. 5 G–G′′′). In contrast, very few Satb2+ neurons were generated in Neurog2-Tg cortices, both in total (50.85-fold decrease; n = 3; P < 0.01) and with respect to DAPI+ cells (22.59-fold decrease; n = 3; P < 0.001) (Fig. 5 H–H′′′). Similarly, in E17.5 Neurog2-Tg cortices, both the total number (3.89-fold decrease; n = 3; P < 0.001) and proportional distribution of Cux1/2+/DAPI+ cells (1.82-fold decrease; P < 0.01) were significantly lower in Neurog2-Tgs (Fig. 5 I–I′′′).
The increased production of early-born Tbr1+ and Ctip2+ neurons as a proportion of total cell number in Neurog2-Tgs might arise from a general increase in cell cycle exit and depletion of the progenitor pool rather than from a specification function of Neurog2. We used BrdU birthdating to investigate whether Neurog2 could specify early-born fates outside the normal early window. Pregnant dams were injected with BrdU at E14.5, when primarily Satb2+ cells are normally being generated, and were killed at E17.5. The proportion of BrdU+ cells that coexpressed Ctip2 was greatly increased in Neurog2-Tg compared with wild-type cortices (5.17-fold increase; n = 3; P < 0.0001) (Fig. 5 J–J′′′). Thus, Neurog2 can increase the time of deep-layer neurogenesis. However, this lengthening could either indicate a role for Neurog2 in fate specification or be an indirect consequence of the depletion of the pool of cortical progenitors that are restricted to upper-layer phenotypes (37). We addressed these possibilities in more detail as discussed below.
Altered Progenitor Cell Dynamics in Neurog2-Tg Cortices.
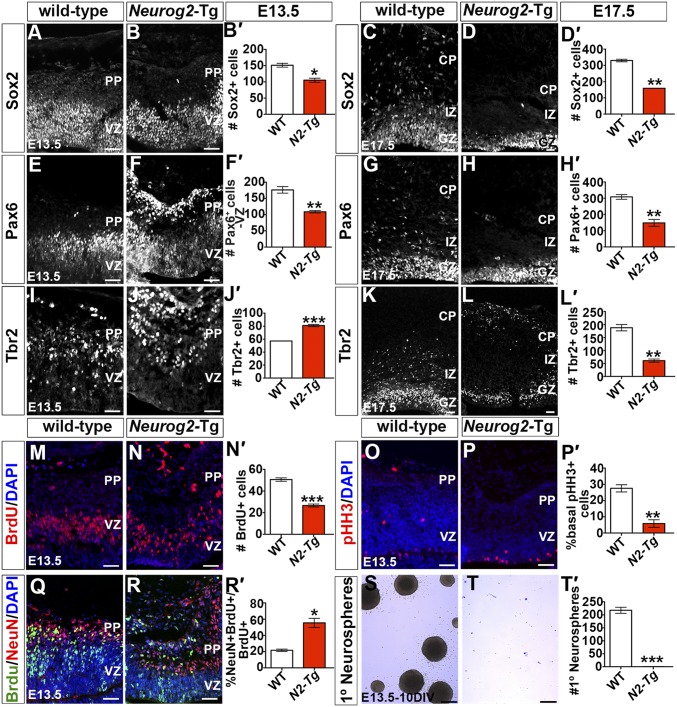
To ask whether Neurog2 influenced neuronal production by altering progenitor cell dynamics, we used Sox2 and Pax6 to label RGCs and Tbr2 to label INPs. Fewer Sox2+ RGCs were detected in the VZ of Neurog2-Tgs at both E13.5 (1.44-fold decrease; n = 3; P < 0.05) (Fig. 6 A–B′) and E17.5 (2.07-fold decrease; n = 3; P < 0.01) (Fig. 6 C–D′). Similarly, fewer Pax6+ VZ progenitors were detected in E13.5 (1.62-fold decrease; n = 3; P < 0.01) (Fig. 6 E–F′) and E17.5 (2.08-fold decrease; n = 3; P < 0.01) (Fig. 6 G–H′) Neurog2-Tg cortices, confirming that the RGC pool was indeed reduced in size. In contrast, although more Tbr2+ INPs were detected in the E13.5 Neurog2-Tg GZ (1.42-fold increase; P < 0.001) (Fig. 6 I–J′), this number was reduced by E17.5 (3.07-fold decrease; n = 3; P < 0.001) (Fig. 6 K–L′).
Fig. 6.
Altered progenitor cell dynamics in Neurog2-Tgs. (A–L) Expression of Sox2 (A–D), Pax6 (E–H), and Tbr2 (I–L) in E13.5 (A, B, E, F, I, and J) and E17.5 (C, D, G, H, K, and L) wild-type and Neurog2-Tg cortices. (B′, D′, F′, H′, J′, and L′) Quantitation of total number of Sox2+ (B′ and D′), Pax6+ (F′ and H′), and Tbr2+ (J′ and L′) cells at E13.5 and E17.5. (M–P) Expression of BrdU (M and N) and pHH3 (O and P) in E13.5 wild-type and Neurog2-Tg cortices. (N′ and P′) Quantitation of total BrdU+ cells (N′) and the percent of pHH3+ cells in basal domains (P′). (Q and R) E13.5 wild-type and Neurog2-Tg cortices injected with BrdU at E12.5, and labeled with NeuN and BrdU. (R′) Quantitation of the percent of NeuN+BrdU+/BrdU+ cells. (S–T) Primary neurospheres from E13.5 wild-type and Neurog2-Tg cortical progenitors. (T′) Quantitation of the percent of primary neurospheres from E13.5 wild-type and Neurog2-Tg cortical progenitors. *P < 0.05; **P < 0.01; ***P < 0.005; ns, not significant. CP, cortical plate; GZ, germinal zone; IZ, intermediate zone; PP, preplate; VZ, ventricular zone. (Scale bars: 50 μm.)
Pax6 and Tbr2 were also ectopically expressed in the Neurog2-Tg PP/CP, which are nonproliferative layers (e.g., see lack of BrdU incorporation in Fig. 6N). Tbr2 is a direct Neurog2 target gene (38), so its ectopic expression was anticipated. In contrast, ectopic Pax6 expression was unexpected, given that the same phenotype is observed in Neurog2−/− cortices (39). Thus, both the loss of Neurog2 (39) and its sustained expression, as achieved in Neurog2-Tg cortices, as opposed to its normal oscillatory expression (35), prevent Neurog2 from carrying out its customary role in turning off Pax6 expression.
To conclude, Neurog2 not only promotes the excessive production of early-born, deep-layer Ctip2+/Tbr1+ neurons, possibly with an aberrant identity but also promotes RGC-to-INP progression and the precocious depletion of INPs, as is in line with previous acute gain-of-function assays (39). The decline in progenitor cell number was not caused by cell death, because apoptosis was not increased in Neurog2-Tgs at E13.5 (n = 3) (Fig. S6 C, D, and D′) or E17.5 (n = 3) (Fig. S6 E, F, and F′), as assessed by the expression of activated caspase-3.
The reduction in progenitor pool size in Neurog2-Tgs suggested that progenitors were undergoing differentiation at the expense of proliferation. Indeed, labeling of S-phase progenitors with a 30-min BrdU pulse revealed that there were fewer dividing cells in Neurog2-Tg cortices (1.90-fold reduction: n = 3; P < 0.001) (Fig. 6 M–N′). The reduction in proliferating cells was particularly evident in the basal progenitor pool, where fewer pHH3+ cells were detected in E13.5 Neurog2-Tgs (4.67-fold reduction; P < 0.01) (Fig. 6 O–P′). In further support of the idea that Neurog2-Tg cortical progenitors preferentially differentiated rather than proliferated, we calculated the leaving fraction, labeling E13.5 S-phase progenitors with BrdU and then assessing the percentage of labeled cells that differentiated 24 h later by costaining with NeuN. Neurog2-Tg cortices displayed a significant increase in number of BrdU+NeuN+ neurons exiting the cell cycle at E13.5 (2.55-fold increase; n = 3; P < 0.05) (Fig. 6 Q–R′). Finally, cortical cells isolated from Neurog2-Tgs displayed an almost complete inability to form primary neurospheres, indicating a lack of proliferative and self-renewal capacity (290-fold decrease; n = 4; P < 0.001) (Fig. 6 S–T′). Neurog2-overexpressing RGCs thus either undergo deep-layer neurogenesis directly or give rise to basal progenitors, which themselves differentiate rapidly, all at the expense of proliferation and self-renewal.
Neurog2 and Ascl1 Specify Early Laminar Fates in Early Cortical Progenitors.
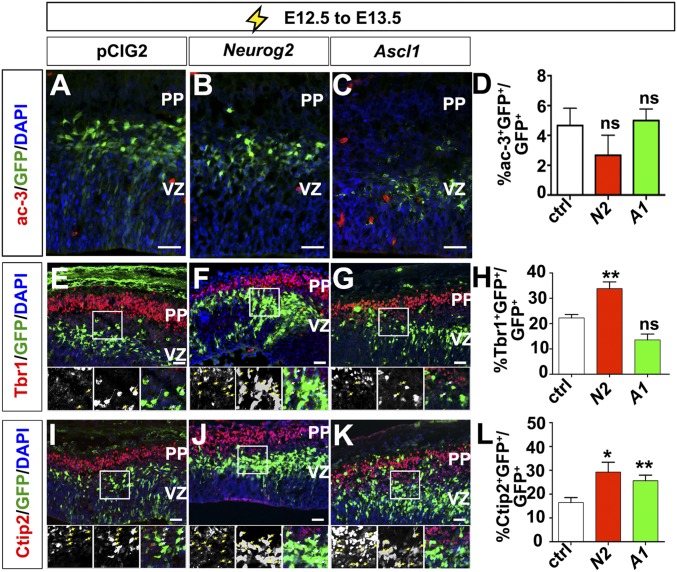
To test whether Neurog2 specifies a deep-layer fate without the confounding issue of progenitor cell depletion, and also to examine the specification properties of Ascl1, we misexpressed these genes both within and outside the normal window of deep-layer neurogenesis using a transient gain-of-function approach. Bicistronic expression vectors containing an internal ribosome entry site (IRES)-EGFP cassette were electroporated into E12.5 cortical progenitors in utero (40). Notably, activated caspase 3 was not induced in E12.5→E13.5 cortical electroporations, allowing us to assess fate decisions without the concern of apoptosis (Fig. S7 A–D). In E12.5→E14.5 electroporations, Neurog2 induced a 2.39-fold increase in the number of GFP+ cells that expressed Tbr1 relative to pCIG2 controls (n = 3; P < 0.05), as demonstrated previously (39), whereas Ascl1 did not alter the numbers of Tbr1+GFP+ cells (Fig. 7 A–D). In contrast, Neurog2 (2.97-fold increase vs. pCIG2; n = 3; P < 0.01) and Ascl1 (2.01-fold increase; n = 3; P < 0.05) had a similar capacity to induce the generation of Ctip2+GFP+ cells (Fig. 7 E–H). The induction of laminar markers was also seen within 24 h, with Neurog2 inducing 1.7- and 1.8-fold increases in Tbr1 and Ctip2 expression (n = 3; P < 0.05), respectively, in E12.5 to E13.5 electroporations, whereas Ascl1 induced a 1.6-fold increase in Ctip2 expression (n = 3; P < 0.05) (Fig. S7 E–L).
Fig. S7.
Overexpression of proneural genes increases early-born neuronal fates. (A–C) Expression of activated caspase-3 (Ac-3) in E12.5→E13.5 electroporations of cortices transfected with pCIG2 (A), Neurog2 (B), and Ascl1 (C). (D) Quantification of GFP+Ac-3+ cells. (E–K) Expression of Tbr1 (red, E–G) and Ctip2 (red, I–K) in cortices electroporated with pCIG2 (E and I), Neurog2 (F and J), and Ascl1 (G and K) at E12.5 and dissected at E13.5. Lower panels are 1.5× magnification images of the boxed area in the larger panel. (H and L) Quantification of ectopic Tbr1 (H) and Ctip2 (L) expression in GFP+ electroporated cortical cells. *P < 0.05; **P < 0.01; ns, not significant. PP, preplate; VZ, ventricular zone. (Scale bars: 50 μm.)
Fig. 7.
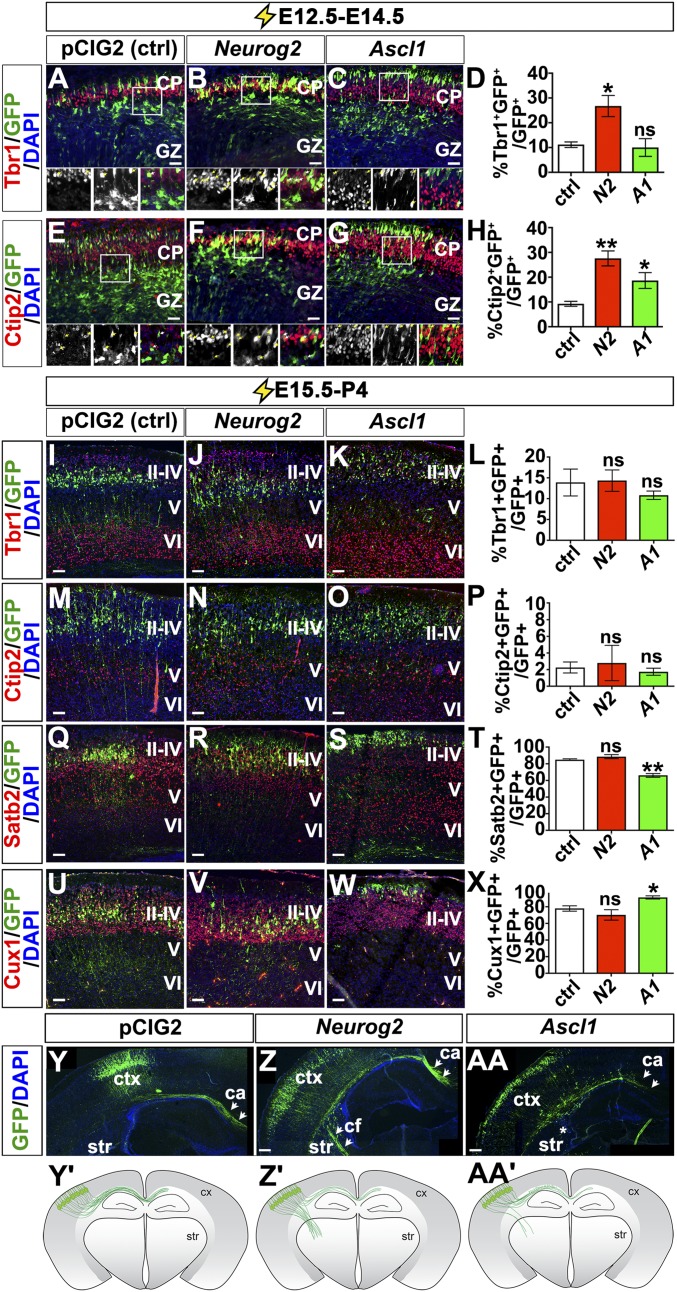
Neurog2 and Ascl1 promote corticofugal molecular identities in early but not in late cortical progenitors. (A–H) Expression of Tbr1 (red, A–C) and Ctip2 (red, E–G) in cortices electroporated with pCIG2 (A and E), Neurog2 (B and F), and Ascl1 (C and G) at E12.5 and dissected at E14.5. (D and H) Quantitation of Tbr1 (D) and Ctip2 (H) expression in GFP+ electroporated cortical cells. (I–Z and AA) E15.5 to P4 electroporations of pCIG2 (I, M, Q, U, and Y), Neurog2 (J, N, R, V, and Z), and Ascl1 (K, O, S, W, and AA) costained with GFP and Tbr1 (red, I–K), Ctip2 (red, M–O), Satb2 (red, Q–S), or Cux1 (red, U–W) or showing GFP+ axonal tracts (green, Y, Z, and AA). (L, P, T, and X) Quantitation of Tbr1 (L), Ctip2 (P), Satb2 (T), and Cux1 (X) expression in GFP+ electroporated cortical cells. (Y′, Z′, and AA′) Schematic illustrations of axonal tracts in pCIG2 (Y′), Neurog2 (Z′), and Ascl1 (AA′) misexpressing neurons from E15.5 to P4 electroporations. Blue is DAPI counterstain in all panels. Y, Z, and AA show multiple merged images to cover the entire hemisphere. Arrowheads mark callosal and corticofugal projections. *P < 0.05; **P < 0.01; ns, not significant. ca, callosal axons; cf, corticofugal; CP, cortical plate; ctx, cortex; GZ, germinal zone; str, striatum. (Scale bars: 50 μm in A–C and E–G; 100 μm in I–K, M–O, Q–S, and U–W; and 200 μm in Y, Z, and AA.)
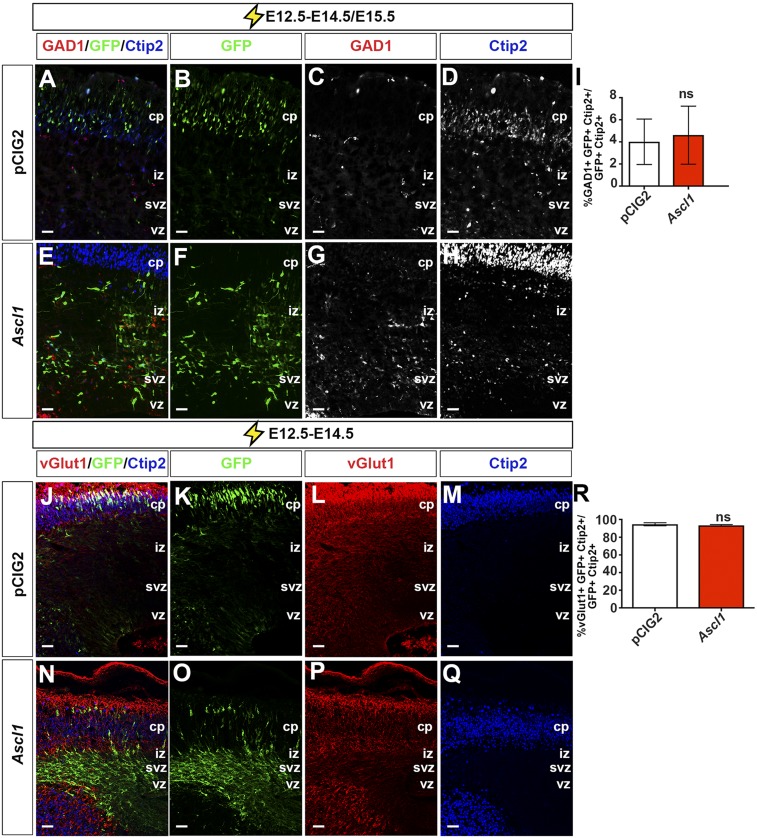
Ctip2 marks not only glutamatergic, subcerebral layer V neurons but also a subset of GABAergic interneurons (41). Ascl1 is also necessary and sufficient to specify a GABAergic fate (20, 25, 30), prompting us to assess the neurotransmitter phenotype of Ctip2+ cells generated by Ascl1 misexpression. Of the Ctip2+ neurons derived from E12.5 electroporations of pCIG2 or Ascl1, less than 5% coexpressed GAD1 (Fig. S8 A–I), but more than 93% coexpressed vGlut1 (n = 3) (Fig. S8 J–R). Neurog2 and Ascl1 are thus both sufficient to promote the acquisition of features of a glutamatergic, layer V subcerebral cell identity, whereas only Neurog2 promotes a Tbr1+ layer VI fate.
Fig. S8.
Overexpression of proneural genes does not alter neurotransmitter phenotypes. (A–H) Expression of GAD1 (red) in E12.5 to E14.5/E15.5 cortical electroporations of pCIG2 (A–D) and Ascl1 (E–H). (I) Quantification of the number of GAD1+GFP+Ctip2+ cells. (J–Q) Expression of vGlut1 in E12.5 to E14.5 cortical electroporations of pCIG2 (J–M) and Ascl1 (N–Q). (R) Quantification of the number of vGlut1+GFP+Ctip2+ cells. cp, cortical plate; iz, intermediate zone; ns, not significant; svz, subventricular zone; vz, ventricular zone. (Scale bars: 50 μm.)
Neurog2 Has a Limited Capacity to Specify Early Neuronal Features in Late Cortical Progenitors.
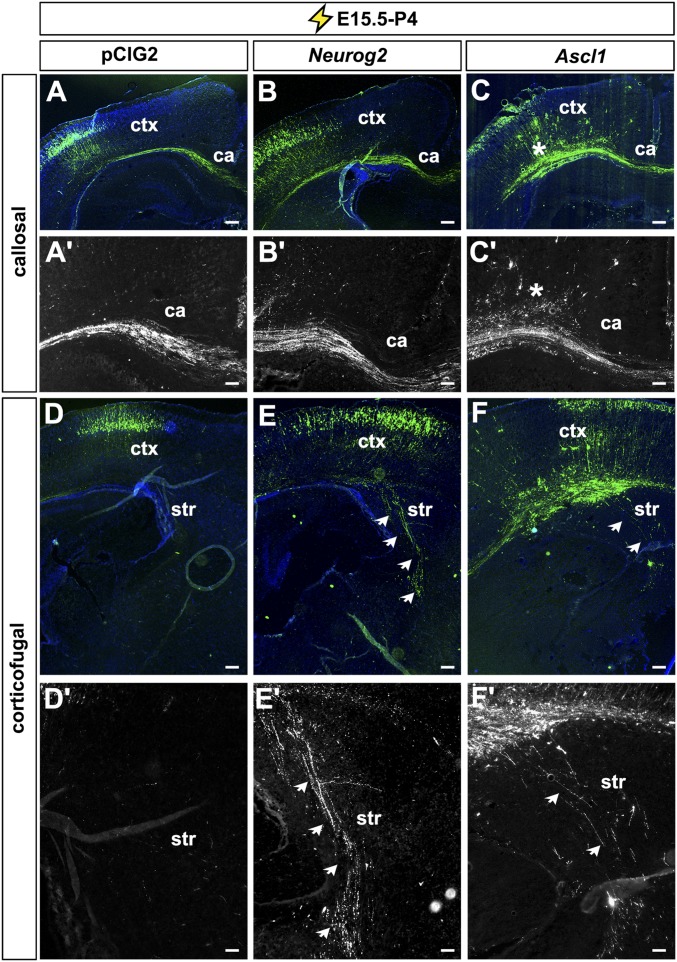
Highlighting the instructive properties of the proneural genes, Neurog2 promotes the ectopic expression of glutamatergic neuronal markers, including Tbr1, in ectopic sites in the ventral telencephalon (42), whereas Ascl1 induces the expression of GABAergic markers in dorsal domains (25, 33). To test whether Neurog2 and Ascl1 were sufficient to induce a deep-layer neuronal identity outside the normal temporal window, we performed E15.5→P4 cortical electroporations, targeting progenitors that give rise to upper layer neurons. Strikingly, in E15.5→P4 electroporations, Neurog2 did not induce the ectopic expression of Tbr1 (Fig. 7 I–L), in agreement with our previous report (42), nor did it induce Ctip2 (Fig. 7 M–P), Satb2 (Fig. 7 Q–T), or Cux1 (Fig. 7 U–X). However, although pCIG2-transfected E15.5 progenitors primarily gave rise to P4 neurons that projected their axons medially (i.e., callosal projections) (Fig. 7 Y and Y′), a subset of neurons derived from Neurog2-overexpressing E15.5 progenitors aberrantly projected their axons laterally to target the ventral telencephalon, a characteristic of earlier-born corticofugal neurons (Fig. 7 Z and Z′ and Fig. S9). These data suggest that Neurog2 is sufficient, as well as required (21), to form subcortical projections and furthermore suggest that Neurog2 has some capacity to confer characteristics of deep-layer identities onto late progenitors, although these effects are unlikely to be mediated by components of the derepression circuit.
Fig. S9.
Neurog2 and Ascl1 overexpression in late cortical progenitors perturbs axonal trajectories. (A–F). E15.5 to P4 in utero electroporation of pCIG2 (A, A′, D, and D′), Neurog2 (B, B′, E, and E′), and Ascl1 (C, C′, F, and F′). The asterisks in C and C′ mark defasciculated axons in the midline. Arrowheads in E, E′, F, and F′ mark aberrant corticofugal projections. (Scale bars: 200 μm in A–C; 100 μm in A′–C′; 150 μm in D–F; and 75 μm in D′–F′.)
Ascl1 misexpression at E15.5 was also not sufficient to alter the expression of Tbr1 (Fig. 7 I–L) or Ctip2 (Fig. 7 M–P) by postnatal day (P)4. However, Ascl1 promoted a 1.27-fold decrease in Satb2+ neurons (n = 3; P < 0.05) (Fig. 7 Q–T) and a corresponding 1.17-fold increase in Cux1/2+ neurons (n = 3; P < 0.05) (Fig. 7 U–X). Cux1 expression is more prominent than Satb2 in the more superficial cortical layers (9) and is required for the formation of callosal projections (43). P4 cortical neurons derived from Ascl1-overexpressing E15.5 progenitors primarily projected their axons medially across the corpus callosum, but the axon tracts were less tightly fasciculated than control axons (Fig. 7 AA and AA′), and some misdirected axons projected laterally into the ventral telencephalon and dorsally in the midline (Fig. S9).
Therefore Neurog2 and Ascl1 are, for the most part, not sufficient to alter the cortical derepression circuit at E15.5, with the exception of Ascl1 inhibiting Satb2 and an upper-layer fate. However, their misexpression induces axon-guidance defects, including the generation of a subset of neurons with corticofugal projections that are characteristic of earlier-born neurons.
Neurog2 and Ascl1 Regulate the Expression of Known Temporal Identity Determinants.
Finally, to assess whether Neurog2 and Ascl1 confer early cortical fates via known temporal determinants, we profiled the expression of Foxg1 (16, 17), Ikaros (18), and PRC1 (Rnf2) and PRC2 (Ezh2) components (19) in Neurog2;Ascl1 mutants. At E12.5, Foxg1, which is required to repress Tbr1 during the transition to upper-layer neurogenesis (17), was expressed in the VZ and in early-born PP neurons (Fig. 8 A–D), as previously reported (16). A reduction in Foxg1 expression was evident in Neurog2−/−;Ascl1−/− cortices, both by immunostaining (Fig. 8D) and by RT-qPCR (1.26-fold decrease; n = 3; P < 0.001) (Fig. 8E). Conversely, expression of Ikaros, the overexpression of which promotes an early deep-layer identity (18), was elevated 1.23-fold in E12.5 Neurog2−/−;Ascl1−/− cortices (n = 3; P < 0.001) (Fig. 8F). In contrast, no changes were observed in the expression of Rnf2 or Ezh2 (Fig. 8 G and H), suggesting that Neurog2 and Ascl1 do not regulate laminar fates by controlling PRC complexes.
Fig. 8.
Neurog2 and Ascl1 regulate the expression of temporal fate determinants. (A–D) Foxg1 expression in E12.5 wild-type (A), Neurog2−/− (B), Ascl1−/− (C), and Neurog2−/−;Ascl1−/− (D) cortices. (E–H) RT-qPCR analysis of Foxg1 (E), Ikzf4 (F), Rnf2 (G), and Ezh2 (H). (I–J′) Foxg1 expression in E12.5 wild-type (I and I′) and Neurog2-Tg (J and J′) cortices. I′ and J′ are 4× magnification images of I and J. (K–N) RT-qPCR analysis of Foxg1 (K), Ikaros (L), Rnf2 (M), and Ezh2 (N). (O–Q) Schematic of laminar fate transitions in wild-type (O), Neurog2−/−;Ascl1−/− (P), and Neurog2-Tg (Q) cortices. (R and S) Tbr1–Fezf2–Satb2–Ctip2 negative feedback loop in wild-type (R) and Neurog2−/−;Ascl1−/− (S) cortices. *P < 0.05; **P < 0.01; ***P < 0.005; ns, not significant. dTel, dorsal telencephalon; lge, lateral ganglionic eminence; pp, preplate; vTel, ventral telencephalon; vz, ventricular zone. (Scale bars: 100 μm in A–D, I, and J; 50 μm in I′ and J′.)
We next assessed whether Neurog2 was sufficient to regulate this set of temporal determinants in Neurog2-Tg cortices. Foxg1 expression was almost undetectable in the dorsal telencephalon of Neurog2-Tgs (Fig. 8 I, I′, J, and J′), and accordingly, there was an 18.68-fold reduction in Foxg1 transcripts in E13.5 Neurog2-Tg cortices by RT-qPCR (n = 3; P < 0.0001) (Fig. 8K). Conversely, we found that Ikaros was up-regulated 2.29-fold in Neurog2-Tg cortices (n = 3, P < 0.05) (Fig. 8L), a finding that also is consistent with the increase in Tbr1+ and Ctip2+ neurons in these transgenics. Finally, as is consistent with the loss-of-function data, transcript levels for the PRC complex components Rnf2 and Ezh2 were not altered in E12.5 Neurog2-Tgs (Fig. 8 M and N).
Discussion
In an instructive mode of cell-fate specification, genetic switches turn one set of genes on while repressing genes in competing lineages (39). Alternatively, cell fates are specified by a derepression circuit of transcriptional repressors, as seen during the specification of laminar fates in the neocortex (1) and of dorsoventral identities in the spinal cord (44). In Neurog2−/−;Ascl1−/− cortices, we observed several defects in the derepression circuit, including a failure to extinguish deep-layer Tbr1 expression in late-born, upper-layer neurons and the precocious expression of Satb2, an upper-layer marker, in early-born, deep-layer neurons (Fig. 8 O and P). Using gain-of-function assays, we found that Neurog2 is sufficient to specify both Ctip2+ and Tbr1+ early-born neuronal identities while preventing the differentiation of Satb2+ late-born neurons, but only when misexpressed in early, but not late, cortical progenitors (Fig. 8Q). Similarly, Ascl1 is sufficient to specify a Ctip2+ identity only in early, and not in late, progenitors, although it can suppress a Satb2+ fate at later stages. Thus the fate-specification properties of the proneural genes are temporally regulated, as discussed further below.
Our data suggest that the cortical derepression circuit is not yet operational at E12.5, when only deep-layer Tbr1+ neurons are being generated (Fig. 8 R and S). If the derepression circuit were active at E12.5, the loss of Tbr1+ cells in both Neurog2−/−;Ascl1−/− and Neurog2−/− cortices (this study and ref. 21) should lead to ectopic Fezf2 and Ctip2 expression, which we did not observe. The main disruption of the derepression circuit in Neurog2−/−;Ascl1−/− cortices is at the level of Fezf2/Ctip2 (Fig. 8 Q–S). The loss of Ctip2 removes the inhibitory signals blocking Tbr1 expression in upper layers, whereas the loss of Fezf2 removes the inhibition on Satb2, resulting in Neurog2−/−;Ascl1−/− cortical neurons taking on a mixed Tbr1+/Satb2+ identity. Consistent with this model, both Neurog2 and Ascl1 gain of function increased the number of Ctip2+ cells, and in Neurog2-Tgs, Ctip2+ cells were born at E14.5, outside their normal temporal window. In addition, Fezf2 was ectopically expressed in Neurog2-Tg cortices, including in the CP and GZ. Fezf2 is normally expressed in both zones at E12.5 but becomes restricted to the CP by E15.5 (36). Prolonging GZ expression of Fezf2 up to E15.5 in Neurog2-Tgs may expand the temporal window for subcerebral neurogenesis, consistent with our finding that Ctip2+ cells were ectopically generated at E15.5, outside their normal time frame. Although we do not yet know whether the interactions between the proneural genes and Fezf2/Ctip2 are direct, the ability of Neurog2 to regulate Fezf2 expression in progenitors suggests that these two factors form a feedback loop, because Fezf2 also indirectly regulates Neurog2 expression by repressing Hes5 (45).
Our data suggest that Ascl1 is also a laminar fate determinant, rather than simply promoting cell-cycle exit and generic neurogenesis. Indeed, misexpression of Ascl1 in E12.5 progenitors promotes the formation of Ctip2+ neurons that are glutamatergic and not GABAergic. Ascl1 also promotes a glutamatergic fate in other contexts, including in E10.5 cortical progenitors [Cajal–Retzius neurons (23)] and in direct neuronal reprogramming studies in fibroblasts (46). Ascl1 thus is promiscuous in its choice of target genes, promoting the formation of Ctip2+ glutamatergic projection neurons in early cortical progenitors (this study) as well as GABAergic interneurons (25) and proliferative Sox9+ glioblasts (33) and Cux1+ neurons in later progenitors (this study). How Ascl1 selects its downstream transcriptional targets to promote different cell fates is not entirely clear, although RAS/ERK signaling has been implicated in biasing Ascl1-expressing progenitors toward glioblast lineages (33).
The fate-specification properties of Neurog2 and Asc1l are temporally regulated, suggesting that they may not be instructive determinants or that the competence window for their functions may be short. For example, misexpression of Neurog2 in E15.5 cortical progenitors outside the normal temporal window for deep-layer neurogenesis did not induce ectopic Tbr1 expression (this study and ref. 42). However, in other contexts Neurog2 is instructive for a Tbr1+ neuronal fate, promoting the ectopic generation of Tbr1+ neurons when misexpressed in the embryonic ventral telencephalon (39) and in adult neural stem cells (47). One reason for Neurog2’s altered activity at E15.5 may be that Neurog2 becomes phosphorylated by GSK3 at this stage, limiting its proneural functions and altering target gene selection (48). In addition, chromatin becomes more compacted as corticogenesis proceeds, so Neurog2 target genes that are accessible at E12.5 may no longer be accessible at E15.5 (49).
One striking feature of the E15.5→P4 Neurog2 electroporations was that, even though Neurog2 did not influence the derepression circuit, it altered the axonal projection patterns of resultant neurons, some of which elaborated aberrant corticofugal projections that are not normally made at this time point. These data are consistent with previous reports that Neurog2 is required for the formation of subcortical (21, 29), mitral cell (22), and thalamic (50) axonal projections, highlighting the importance of Neurog2 in regulating axon guidance. These data also argue that Tbr1, which is not induced by Neurog2 misexpression at E15.5, may not be required for the formation of corticofugal projections. Indeed, Tbr1−/− deep-layer neurons project into the internal capsule, although there are defects in innervation of the thalamus (51). In contrast, Fezf2 and Ctip2, which also are not induced by Neurog2 at E15.5, are required for the formation of subcerebral projections (5, 11, 12). Neurog2 therefore must induce the expression of other factors that alter the axonal projection patterns of E15.5-born neurons.
Finally, we found that by overexpressing or knocking out Neurog2 and Ascl1 we altered the timing of fate specification, increasing deep-layer identities in both cases, but for different reasons. In gain-of-function assays, Neurog2 induced Tbr1 expression, whereas Neurog2 and Ascl1 induced Ctip2 expression, with Neurog2 expanding the temporal window in which these early-born neurons are generated. In loss-of-function assays, upper-layer neurons are generated precociously (Satb2) in Neurog2−/−;Ascl1−/− cortices, but at the same time, because the derepression circuit is altered, Tbr1, which marks a deep-layer fate, is ectopically expressed in later-born, upper-layer cortical neurons. Neurog2 is required both to turn on and to turn off inappropriate gene-expression programs to regulate lineage expression (39), so the inability to turn off Tbr1 in double mutants suggests that some aspects of an early identity are expressed outside their normal temporal window. Thus Tbr1 is expressed ectopically in both Neurog2;Ascl1 and Neurog2-Tg animals, and, accordingly, Foxg1, which is required to repress Tbr1 (17), is down-regulated. Conversely, the expression of Ikaros, which promotes an early identity (18), is up-regulated in both loss- and gain-of-function models.
Materials and Methods
Animals.
Animal procedures were approved by the University of Calgary (AC11-0053) and Sunnybrook Research Institute (16-604) Animal Care Committees in compliance with the Canadian Council of Animal Care. Neurog2GFPKI (25), Ascl1GFPKI (012881; Jackson Laboratories) and Foxg1CreKI (34) mice were previously described, and Neurog2-Tg mice are described in SI Materials and Methods. All mutations were maintained on a CD1 background. Embryos of either sex were used throughout the study.
In Utero Electroporation.
cDNA expression vectors were introduced into dorsal telencephalic (cortical) progenitors using in utero electroporation as previously described (23, 48). pCIG2-Neurog2 (48) and pCIG2-Ascl1 (23) expression vectors were described previously.
Immunohistochemistry, RNA in Situ Hybridization.
Brains were dissected in PBS and fixed in 4% paraformaldehyde (PFA)/1× PBS overnight at 4 °C. Brains were washed three times in 1× PBS, cryoprotected in 20% sucrose/1× PBS overnight at 4 °C, blocked in optimum cutting temperature (OCT) compound, and cryosectioned at 10 μm. RNA in situ hybridization (33, 48) and immunohistochemistry (33) were performed as described previously.
BrdU Labeling.
BrdU (Sigma) was injected at 100 μg/g body weight either 30 min or 24 h before mice were killed. BrdU-immunolabeled sections were treated with 2N HCl for 25 min at 37 °C before immunostaining as described above.
Axonal Tracing.
Brains were dissected and fixed overnight in 4% PFA. Corticofugal projections were labeled by placing a DiI crystal in the dorsal cortex as described (50).
Neurosphere Assay.
Dissociated cortical cells were seeded in 24-well plates at a density of 6,000 cells per well and were cultured in proliferation medium in the presence of mitogens FGF and EGF for 7–9 d [DMEM/F12 (3:1), 2% B27 supplement, 40 ng/mL FGF, 20 ng/mL EGF, 50 U/mL penicillin/streptomycin, 40 µg/mL Fungizone]. After 10 d of culture, neurospheres were counted in each well.
RT-qPCR.
Tissue for RNA extraction was collected at E12.5 by microdissecting dorsal telencephalic tissue. RNA was isolated with TRIzol reagent (15596-026; Thermo Fisher Scientific) using the manufacturer’s protocol. Reverse transcriptase cDNA synthesis was performed with a RT2 First Strand Kit (330401; Qiagen), and qPCR was performed with RT2 SYBR Green (330500; Qiagen) using the manufacturer’s protocols.
Quantitation.
Quantitation was performed on at least three independent brains (n = 3) and on at least three sections per brain. Cell counts were performed rostral to the hippocampus. Cortical zones were delineated by density of DAPI+ nuclei; GZ and CP are cell dense and are separated by a cell-sparse IZ. The upper and lower CP were divided in half for cell counts. Values were compared using a two-tailed Student’s t test (to compare two values) or ANOVA and Tukey’s multiple-comparison test (to compare more than two values) using Prism software (GraphPad). Data are represented as mean ± SEM; P values are denoted as follows: *P < 0.05; **P < 0.01; ***P < 0.005.
SI Materials and Methods
Imaging.
Images were captured with a QImaging RETIGA 2000R or QImaging RETIGA EX digital camera and a Leica DMRXA2 optical microscope using OpenLab5 software (Improvision).
Pronuclear Injection.
To generate flSTOP::Neurog2 mice, Neurog2 was PCR cloned into a pCCALL2 construct, and linearized DNA for pronuclear injection was isolated with StuI and XmnI digestion. The products of the restriction digest were separated using agarose gel electrophoresis, and the appropriate fragment was excised and extracted from the gel using a column-based QIAquick Gel Extraction Kit (Qiagen). The transgenic fragments were eluted off the columns with filtered (0.2-μm pore size) Tris-EDTA (TE) buffer (10 mM Tris HCl, 0.1 mM EDTA, pH 7.4) and diluted to a concentration of 15 ng/μL in TE buffer. Microinjections and transfer of manipulated fertilized ova into the oviduct of recipient pseudopregnant females were carried out using routine procedures by the Clara Christie Centre for Mouse Genomics, University of Calgary. Two independent lines were generated.
Genotyping Information.
The following PCR primers and conditions were used for genotyping: Neurog2GFPKI: 35 cycles of 98 °C for 1 s and 60 °C for 30 s using primers for wild-type (Neurog2*F and Neurog2*R) and mutant (VD187 and ZF92) alleles. Neurog2*F: 5′-TAGACGCAGTGACTTCTGTGACCG-3′; Neurog2*R: 5′-ACCTCCTCTTCCTC CTTCAACTCC-3′; VD187: 5′-GGACATTCCCGGACACACAC-3′; ZF92: 5′-GCATCACCTTCACCCTCTCC-3′; Ascl1GFPKI: 35 cycles of 94 °C for 30 s, 64 °C for 30 s, and 72 °C for 30 s using primers for wild-type (Ascl1*WT_F and Ascl1*WT_R) and mutant (Ascl1*MT_F and Ascl1*MT_R) alleles. Ascl1*WT_F: 5′-TCCAACGACTTGAACTCTATGG-3′; Ascl1*WT_R: 5′-CCAGGACTCAATACGCA GGG-3′; Ascl1*MT_F: 5′- AACTTTCCTCCGGGGCTCGT TTC-3′; Ascl1*MT_R: 5′-TGGCTGTTGTAGTTGTACTCCAGC-3′; Foxg1CreKI: 35 cycles of 95 °C for 1 min, 60 °C for 1 min; and 72 °C for 130 min for Foxg1CreKI allele (BF1-U and BF1-L). BF1-U: GGGCAACAACCACTCCTTCTCCAC; BF1-L: GACCCCTGATTTTGATGTGTGAAA; flSTOP::Neurog2: 40 cycles of 95 °C for 1 min, 60 °C for 1 min, and 72 °C for 1.5 min with Neurog2 (CGGAGAAGCATC GTTATGCG) and pCCALL2 vector (GCAGGGGGCTGTTTCATATACTG) primers.
Antibodies.
Primary antibodies used in this study included rabbit anti-Neurog2 (gift of Masato Nakafuku, Cincinnati Children's Hospital, Cincinnati, mouse anti-Ascl1 (1:500) (BD Biosciences), rabbit anti-Tbr1 (1:800) (Chemicon), mouse anti-NeuN (1:500) (Chemicon), mouse anti-Tuj1 (neuronal III β−tubulin, 1:500) (Covance), rabbit anti-GFP (1:500) (Chemicon), rabbit anti-Cux1/2 (Cutl1, 1:100) (Abcam), rabbit anti-Ctip2 (1:100) (Abcam), mouse anti-Satb2 (1:350) (Abcam), rat anti-L1 (1:500) (Chemicon), mouse anti-FoxP2 (1:500) (Abcam), rabbit anti-Sox2 (1:500) (Cell Signaling Technology), rabbit anti-Pax6 (1:250) (Covance), rabbit anti-Tbr2 (1:500) (Abcam), rabbit anti–phospho-histone H3 (pHH3, 1:500) (Millipore Biotechnology), rabbit anti-active caspase 3 (1:500) (Abcam), rabbit anti-GFAP (1:500) (DakoCytomation), rat anti-BrdU (1:50) (Serotec), mouse anti-TAG1 (1:100) (Developmental Studies Hybridoma Bank), mouse anti-Gad65 (1:100) (BD Pharmingen), and rabbit anti-Vglut1 (1:100) (SYnaptic Systems). Secondary antibodies were conjugated to Cy3 (Jackson ImmunoResearch), Alexa488, or Alexa568 diluted 1:500 (Molecular Probes).
In Utero Electroporation.
Briefly, DNA (3 μg/μL) mixed with Fast Green FCF dye (1:200) was injected into the telencephalic (i.e., lateral) ventricles at defined embryonic stages using pulled borosilicate needles and a FemtoJet microinjector (Eppendorf). Next, seven pulses of 45–55 mV were applied within a 7-s interval to the uterus surrounding the head of the embryo using a BTX electroporator (Harvard Apparatus). The uterus was replaced in the body cavity, the peritoneum was sutured, skin was stapled, and embryos were allowed to develop until the designated stage of analysis. To generate expression constructs, cDNAs were cloned into pCIG2, an expression vector that promotes the transcription of a bicistronic mRNA EGFP and a second mRNA of interest from an IRES.
RT-qPCR.
RT2 qPCR primers were from Qiagen: Gapdh (PPM02946E), B2m (PPM03562A), Hrpt (PPM03559F), Foxg1 (PPM04640A), Ikaros/IKZF1 (PPM05292E), Ring1b/Rnf2 (PPM27885A), and Ezh2 (PPM05645A). qPCR was performed with three biological replicates (RNA from three embryos), with the exception of Asc1−/− (RNA from two embryos) and with three technical replicates for each biological replicate. Gene-expression analysis was determined using the ΔΔCt method standardizing relative to the geometric average of three housekeeping genes (Gapdh, B2M, and Hrpt) and wild-type control.
Acknowledgments
We thank N. Klenin and D. Zinyk for technical support; C. Hanashima, P. Mattar, and M. Cayouette for reagents; and P. Mattar for critical review of an earlier version of the manuscript. This work was supported by Canadian Institutes of Health Research (CIHR) Grant MOP-44094 (to C.S.), Studentships from an Alberta Children’s Hospital Research Institute/CIHR Training Grant (to G.W., S.L., and L.A.), Alberta Innovates-Health Solutions (G.W.), and a CIHR Canada Graduate Scholarship (to G.W.). C.S. holds the Dixon Family Chair in Ophthalmology Research at Sunnybrook Research Institute.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1701495114/-/DCSupplemental.
References
- 1.Toma K, Hanashima C. Switching modes in corticogenesis: Mechanisms of neuronal subtype transitions and integration in the cerebral cortex. Front Neurosci. 2015;9:274. doi: 10.3389/fnins.2015.00274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Srinivasan K, et al. A network of genetic repression and derepression specifies projection fates in the developing neocortex. Proc Natl Acad Sci USA. 2012;109:19071–19078. doi: 10.1073/pnas.1216793109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hevner RF, et al. Tbr1 regulates differentiation of the preplate and layer 6. Neuron. 2001;29:353–366. doi: 10.1016/s0896-6273(01)00211-2. [DOI] [PubMed] [Google Scholar]
- 4.McKenna WL, et al. Tbr1 and Fezf2 regulate alternate corticofugal neuronal identities during neocortical development. J Neurosci. 2011;31:549–564. doi: 10.1523/JNEUROSCI.4131-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Han W, et al. TBR1 directly represses Fezf2 to control the laminar origin and development of the corticospinal tract. Proc Natl Acad Sci USA. 2011;108:3041–3046. doi: 10.1073/pnas.1016723108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Molyneaux BJ, Arlotta P, Hirata T, Hibi M, Macklis JD. Fezl is required for the birth and specification of corticospinal motor neurons. Neuron. 2005;47:817–831. doi: 10.1016/j.neuron.2005.08.030. [DOI] [PubMed] [Google Scholar]
- 7.Chen B, Schaevitz LR, McConnell SK. Fezl regulates the differentiation and axon targeting of layer 5 subcortical projection neurons in cerebral cortex. Proc Natl Acad Sci USA. 2005;102:17184–17189. doi: 10.1073/pnas.0508732102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McKenna WL, et al. Mutual regulation between Satb2 and Fezf2 promotes subcerebral projection neuron identity in the developing cerebral cortex. Proc Natl Acad Sci USA. 2015;112:11702–11707. doi: 10.1073/pnas.1504144112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alcamo EA, et al. Satb2 regulates callosal projection neuron identity in the developing cerebral cortex. Neuron. 2008;57:364–377. doi: 10.1016/j.neuron.2007.12.012. [DOI] [PubMed] [Google Scholar]
- 10.Britanova O, et al. Satb2 is a postmitotic determinant for upper-layer neuron specification in the neocortex. Neuron. 2008;57:378–392. doi: 10.1016/j.neuron.2007.12.028. [DOI] [PubMed] [Google Scholar]
- 11.Chen B, et al. The Fezf2-Ctip2 genetic pathway regulates the fate choice of subcortical projection neurons in the developing cerebral cortex. Proc Natl Acad Sci USA. 2008;105:11382–11387. doi: 10.1073/pnas.0804918105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arlotta P, et al. Neuronal subtype-specific genes that control corticospinal motor neuron development in vivo. Neuron. 2005;45:207–221. doi: 10.1016/j.neuron.2004.12.036. [DOI] [PubMed] [Google Scholar]
- 13.Shen Q, et al. The timing of cortical neurogenesis is encoded within lineages of individual progenitor cells. Nat Neurosci. 2006;9:743–751. doi: 10.1038/nn1694. [DOI] [PubMed] [Google Scholar]
- 14.Caviness VS., Jr Neocortical histogenesis in normal and reeler mice: A developmental study based upon [3H]thymidine autoradiography. Brain Res. 1982;256:293–302. doi: 10.1016/0165-3806(82)90141-9. [DOI] [PubMed] [Google Scholar]
- 15.McConnell SK, Kaznowski CE. Cell cycle dependence of laminar determination in developing neocortex. Science. 1991;254:282–285. doi: 10.1126/science.254.5029.282. [DOI] [PubMed] [Google Scholar]
- 16.Kumamoto T, et al. Foxg1 coordinates the switch from nonradially to radially migrating glutamatergic subtypes in the neocortex through spatiotemporal repression. Cell Reports. 2013;3:931–945. doi: 10.1016/j.celrep.2013.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Toma K, Kumamoto T, Hanashima C. The timing of upper-layer neurogenesis is conferred by sequential derepression and negative feedback from deep-layer neurons. J Neurosci. 2014;34:13259–13276. doi: 10.1523/JNEUROSCI.2334-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alsiö JM, Tarchini B, Cayouette M, Livesey FJ. Ikaros promotes early-born neuronal fates in the cerebral cortex. Proc Natl Acad Sci USA. 2013;110:E716–E725. doi: 10.1073/pnas.1215707110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pereira JD, et al. Ezh2, the histone methyltransferase of PRC2, regulates the balance between self-renewal and differentiation in the cerebral cortex. Proc Natl Acad Sci USA. 2010;107:15957–15962. doi: 10.1073/pnas.1002530107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fode C, et al. A role for neural determination genes in specifying the dorsoventral identity of telencephalic neurons. Genes Dev. 2000;14:67–80. [PMC free article] [PubMed] [Google Scholar]
- 21.Schuurmans C, et al. Sequential phases of cortical specification involve Neurogenin-dependent and -independent pathways. EMBO J. 2004;23:2892–2902. doi: 10.1038/sj.emboj.7600278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dixit R, et al. Neurog1 and Neurog2 control two waves of neuronal differentiation in the piriform cortex. J Neurosci. 2014;34:539–553. doi: 10.1523/JNEUROSCI.0614-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dixit R, et al. Ascl1 participates in Cajal-Retzius cell development in the neocortex. Cereb Cortex. 2011;21:2599–2611. doi: 10.1093/cercor/bhr046. [DOI] [PubMed] [Google Scholar]
- 24.Nieto M, Schuurmans C, Britz O, Guillemot F. Neural bHLH genes control the neuronal versus glial fate decision in cortical progenitors. Neuron. 2001;29:401–413. doi: 10.1016/s0896-6273(01)00214-8. [DOI] [PubMed] [Google Scholar]
- 25.Britz O, et al. A role for proneural genes in the maturation of cortical progenitor cells. Cereb Cortex. 2006;16:i138–i151. doi: 10.1093/cercor/bhj168. [DOI] [PubMed] [Google Scholar]
- 26.Ferland RJ, Cherry TJ, Preware PO, Morrisey EE, Walsh CA. Characterization of Foxp2 and Foxp1 mRNA and protein in the developing and mature brain. J Comp Neurol. 2003;460:266–279. doi: 10.1002/cne.10654. [DOI] [PubMed] [Google Scholar]
- 27.Joshi PS, et al. Bhlhb5 regulates the postmitotic acquisition of area identities in layers II-V of the developing neocortex. Neuron. 2008;60:258–272. doi: 10.1016/j.neuron.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nieto M, et al. Expression of Cux-1 and Cux-2 in the subventricular zone and upper layers II-IV of the cerebral cortex. J Comp Neurol. 2004;479:168–180. doi: 10.1002/cne.20322. [DOI] [PubMed] [Google Scholar]
- 29.Hand R, Polleux F. Neurogenin2 regulates the initial axon guidance of cortical pyramidal neurons projecting medially to the corpus callosum. Neural Dev. 2011;6:30. doi: 10.1186/1749-8104-6-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Casarosa S, Fode C, Guillemot F. Mash1 regulates neurogenesis in the ventral telencephalon. Development. 1999;126:525–534. doi: 10.1242/dev.126.3.525. [DOI] [PubMed] [Google Scholar]
- 31.Gobius I, et al. Astroglial-mediated remodeling of the interhemispheric midline is required for the formation of the corpus callosum. Cell Reports. 2016;17:735–747. doi: 10.1016/j.celrep.2016.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parras CM, et al. The proneural gene Mash1 specifies an early population of telencephalic oligodendrocytes. J Neurosci. 2007;27:4233–4242. doi: 10.1523/JNEUROSCI.0126-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li S, et al. RAS/ERK signaling controls proneural genetic programs in cortical development and gliomagenesis. J Neurosci. 2014;34:2169–2190. doi: 10.1523/JNEUROSCI.4077-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hébert JM, McConnell SK. Targeting of cre to the Foxg1 (BF-1) locus mediates loxP recombination in the telencephalon and other developing head structures. Dev Biol. 2000;222:296–306. doi: 10.1006/dbio.2000.9732. [DOI] [PubMed] [Google Scholar]
- 35.Shimojo H, Ohtsuka T, Kageyama R. Oscillations in notch signaling regulate maintenance of neural progenitors. Neuron. 2008;58:52–64. doi: 10.1016/j.neuron.2008.02.014. [DOI] [PubMed] [Google Scholar]
- 36.Eckler MJ, et al. Multiple conserved regulatory domains promote Fezf2 expression in the developing cerebral cortex. Neural Dev. 2014;9:6. doi: 10.1186/1749-8104-9-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Franco SJ, et al. Fate-restricted neural progenitors in the mammalian cerebral cortex. Science. 2012;337:746–749. doi: 10.1126/science.1223616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ochiai W, et al. Periventricular notch activation and asymmetric Ngn2 and Tbr2 expression in pair-generated neocortical daughter cells. Mol Cell Neurosci. 2009;40:225–233. doi: 10.1016/j.mcn.2008.10.007. [DOI] [PubMed] [Google Scholar]
- 39.Kovach C, et al. Neurog2 simultaneously activates and represses alternative gene expression programs in the developing neocortex. Cereb Cortex. 2013;23:1884–1900. doi: 10.1093/cercor/bhs176. [DOI] [PubMed] [Google Scholar]
- 40.Dixit R, et al. Efficient gene delivery into multiple CNS territories using in utero electroporation. J Vis Exp. 2011 doi: 10.3791/2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nikouei K, Muñoz-Manchado AB, Hjerling-Leffler J. BCL11B/CTIP2 is highly expressed in GABAergic interneurons of the mouse somatosensory cortex. J Chem Neuroanat. 2016;71:1–5. doi: 10.1016/j.jchemneu.2015.12.004. [DOI] [PubMed] [Google Scholar]
- 42.Mattar P, et al. Basic helix-loop-helix transcription factors cooperate to specify a cortical projection neuron identity. Mol Cell Biol. 2008;28:1456–1469. doi: 10.1128/MCB.01510-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rodríguez-Tornos FM, et al. Cux1 enables interhemispheric connections of layer II/III neurons by regulating Kv1-dependent firing. Neuron. 2016;89:494–506. doi: 10.1016/j.neuron.2015.12.020. [DOI] [PubMed] [Google Scholar]
- 44.Lee SK, Pfaff SL. Transcriptional networks regulating neuronal identity in the developing spinal cord. Nat Neurosci. 2001;4:1183–1191. doi: 10.1038/nn750. [DOI] [PubMed] [Google Scholar]
- 45.Shimizu T, et al. Zinc finger genes Fezf1 and Fezf2 control neuronal differentiation by repressing Hes5 expression in the forebrain. Development. 2010;137:1875–1885. doi: 10.1242/dev.047167. [DOI] [PubMed] [Google Scholar]
- 46.Vierbuchen T, et al. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463:1035–1041. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Berninger B, Guillemot F, Götz M. Directing neurotransmitter identity of neurones derived from expanded adult neural stem cells. Eur J Neurosci. 2007;25:2581–2590. doi: 10.1111/j.1460-9568.2007.05509.x. [DOI] [PubMed] [Google Scholar]
- 48.Li S, et al. GSK3 temporally regulates neurogenin 2 proneural activity in the neocortex. J Neurosci. 2012;32:7791–7805. doi: 10.1523/JNEUROSCI.1309-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kishi Y, Fujii Y, Hirabayashi Y, Gotoh Y. HMGA regulates the global chromatin state and neurogenic potential in neocortical precursor cells. Nat Neurosci. 2012;15:1127–1133. doi: 10.1038/nn.3165. [DOI] [PubMed] [Google Scholar]
- 50.Seibt J, et al. Neurogenin2 specifies the connectivity of thalamic neurons by controlling axon responsiveness to intermediate target cues. Neuron. 2003;39:439–452. doi: 10.1016/s0896-6273(03)00435-5. [DOI] [PubMed] [Google Scholar]
- 51.Hevner RF, Miyashita-Lin E, Rubenstein JL. Cortical and thalamic axon pathfinding defects in Tbr1, Gbx2, and Pax6 mutant mice: Evidence that cortical and thalamic axons interact and guide each other. J Comp Neurol. 2002;447:8–17. doi: 10.1002/cne.10219. [DOI] [PubMed] [Google Scholar]