Abstract
The development of agricultural societies, one of the most transformative events in human and ecological history, was made possible by plant and animal domestication. Plant domestication began 12,000–10,000 y ago in a number of major world areas, including the New World tropics, Southwest Asia, and China, during a period of profound global environmental perturbations as the Pleistocene epoch ended and transitioned into the Holocene. Domestication is at its heart an evolutionary process, and for many prehistorians evolutionary theory has been foundational in investigating agricultural origins. Similarly, geneticists working largely with modern crops and their living wild progenitors have documented some of the mechanisms that underwrote phenotypic transformations from wild to domesticated species. Ever-improving analytic methods for retrieval of empirical data from archaeological sites, together with advances in genetic, genomic, epigenetic, and experimental research on living crop plants and wild progenitors, suggest that three fields of study currently little applied to plant domestication processes may be necessary to understand these transformations across a range of species important in early prehistoric agriculture. These fields are phenotypic (developmental) plasticity, niche construction theory, and epigenetics with transgenerational epigenetic inheritance. All are central in a controversy about whether an Extended Evolutionary Synthesis is needed to reconceptualize how evolutionary change occurs. An exploration of their present and potential utility in domestication study shows that all three fields have considerable promise in elucidating important issues in plant domestication and in agricultural origin and dispersal research and should be increasingly applied to these issues.
Keywords: plant domestication, agricultural origins, agricultural dispersals, extended evolutionary synthesis
The development of agricultural societies, one of the most transformative events in human and ecological history, was made possible by plant and animal domestication. The origins of plant domestication can be traced in a number of world areas, including southwest Asia, northern and southern China, and the lowland tropics of Mesoamerica and South America, to 12,000–10,000 y ago, a time of profound global environmental perturbations as the Pleistocene epoch ended and transitioned into the Holocene (Fig. S1) (1–3). Domestication is, at its heart, an evolutionary process. Indeed, it, together with plant breeding, “… are ongoing 10,000-year-old evolutionary experiments that have radically altered wild species to meet human needs” (ref. 4, p. 808). Thus, for many prehistorians evolutionary theory has been foundational for investigating when, why, why not, and how foragers became farmers (e.g., refs. 5–13).
Fig. S1.
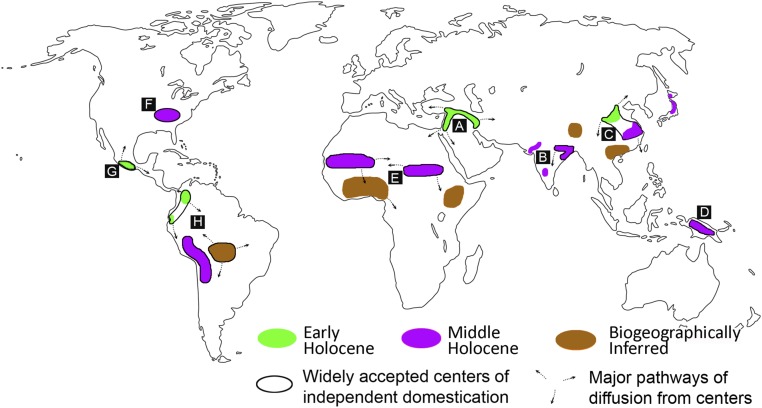
A map depicting known or likely centers of plant domestication. Black outlines surround the most widely accepted independent centers of domestication, and sources of major diffusions of domesticates are indicated by arrows. Green and purple regions are those where the domestication process took place during the late Pleistocene to early Holocene transition (12,000–8,200 y B.P.) and in the middle Holocene (8,200–4,200 y B.P.), respectively. Brown regions represent areas where, at present, the evidence for domestication is interpreted based on the presence of domestic forms indigenous to these regions found outside their native distributions. The letters A–H correspond to the following: A, Southwest Asia (wheat, barley, lentil, pea, chickpea); B, India [rice (indica), millets, mungbean]; C, China [broomcorn millet, foxtail millet, rice (japonica), soybean, melon]; D, New Guinea (banana, taro, yam); E, Africa (date palm, sorghum, pearl millet, African rice, oil palm); F, Eastern North America (acorn and spaghetti squash, sunflower, sumpweed, goosefoot); G, Mexico (maize, pumpkin squash, common and lima beans, avocado, chili pepper); H, South America [chili peppers, peanut, cotton, squashes (butternut and Hubbard), common and lima beans, manioc, sweet potato, white potato, yam, quinoa]. Reproduced from ref. 2.
During the last decade a group of evolutionary biologists and ecologists has argued that there is an urgent need to broaden the traditional Modern Synthesis (MS) to address questions they consider to be underemphasized or beyond the scope of the MS, calling this new approach an “Extended Evolutionary Synthesis” (EES). Its elements are well described (14–16) and prominently include (i) phenotypic (developmental) plasticity; (ii) transgenerational epigenetic inheritance (TEI); and (iii) niche construction theory (NCT). The elements conceptualize as critical factors in evolutionary change the preadult-hood developmental processes of organisms that give rise to the body plans and traits on which natural selection subsequently acts. They envelop controversial issues such as phenotypic before genotypic change; inheritance mechanisms outside of genes, including ecological inheritance; macro- vs. microevolution; and levels of selection. Many biologists, however, question whether there is much “new” in the EES, arguing the traditional MS adequately incorporates consideration of EES elements, and there has been considerable debate around these issues (14–16). Some question whether EES additions and modifications, such as developmental plasticity and TEI, have been shown to have much evolutionary importance (14, 16). An emerging body of literature is clarifying some of these issues (17–19).
With regard to plant domestication, it has long been known that experiments in domestication by the earliest farmers resulted in a constellation of traits found in crop species and not or rarely found in their wild progenitors; collectively, these traits are termed the “domestication syndrome” (20–22). Common among them are larger seed, fruit, and root/tuber size; nonshattering seed heads; nonbitter fruits and underground organs representing decreased natural physical defense mechanisms; decreased seed dormancy; and increased starch content along with different starch qualities. Archaeobotanical records and genetic studies of living plants are revealing much about the genetic mechanisms and human selection strategies that underwrote and drove these phenotypic transformations (e.g., refs. 1–3, 20–22). Ever-improving analytic methods for retrieving hard, empirical data from archaeological sites coupled with advances in genetic, genomic, epigenetic, and experimental research on both living and ancient plant specimens are also revising the traditional understandings of the processes and are introducing new mechanisms for them.
For example, genetic research shows that once-emphasized conventional assumptions about morphological change—e.g., that the change was driven mainly by human selection for rare mutants of a few single genes that were deleterious in wild plants and favorable in field environments or by selection for new, advantageous mutations that appeared postcultivation—have, for some major traits, been supplanted by different and/or more complex processes. These processes include (i) regulatory changes that targeted diverse developmental pathways and led to changes in gene expression (e.g., how, when, and to what degree existing genes are expressed through changes in the amount of mRNA during transcription); (ii) extensive rewiring of transcriptomic and coexpression networks; (iii) in an increasing number of wild progenitors, the presence and availability to the first cultivators of preexisting, nondeleterious genetic components for major domestication traits (known as “cryptic genetic variation”) that induce trait variation only under specific environmental or genetic conditions; and (iv) deviations from simple Mendelian expectations (e.g., refs. 4, 20, 21, 23–26).
Explored recently by archaeobotanists are phenotypic plasticity in crops and wild ancestors and how well-documented natural- and human-caused environmental changes that occurred pre- and postcultivation may have directly caused phenotypic change by inducing plasticity (27, 28). Many of these genetic and other factors mediate or result from the developmental processes and environmental contexts of organisms emphasized by the EES to account for morphological variability and change in evolution, potentially articulating with developmental plasticity, NCT, and epigenetics with TEI (14, 16). At present, however, these fields are little applied to plant domestication research by archaeologists and geneticists.
My purpose here is to examine the basic concepts, recent applications, less understood aspects, and future challenges of the three fields with regard to their utility in domestication research so that, going forward, they may be better understood by the multidisciplinary communities of investigators directly engaged in or interested in such research, as well as by scholars interested in evolutionary questions more broadly. I use examples from archaeological, paleoenvironmental, genetic, ancient DNA, and experimental research.
Phenotypic (Developmental) Plasticity
Developmental biology was not a significant part of the MS because some of the principal founders of the MS believed population genetics and microevolution could largely explain evolutionary change and that no genetic and therefore heritable basis existed for the different, preadult developmental trajectories organisms could take (see ref. 15 for discussion). As a result many developmental biologists refused to join the MS. An exception was Conrad Waddington (29, 30), who offered a view of phenotypic change and evolutionary significance rooted in an organism’s early development, now called “developmental plasticity,” that for many years was relegated to the fringe of evolutionary thinking. The emergence of the field of evolutionary developmental biology in the 1970s with the discovery of regulatory genes and emphasis on where, when, and to what degree these highly conserved genes are activated in different taxa, began to change previous attitudes (see ref. 18 for review). Some prominent evolutionary biologists still question the importance of developmental plasticity in evolutionary change (16), and some of its aspects remain controversial, but a number of its concepts are now well established in evolutionary thinking and practice (17).
Developmental plasticity is defined as the inherent capacity of a single genotype to rapidly exhibit more than one phenotype through one of several available preadult developmental pathways in direct response to environmental perturbations and stress factors (17, 31–33). New phenotypic variation is introduced into a population rapidly, without a corresponding genetic change (e.g., without the appearance or spread of a new mutation), in part through the presence of cryptic (preexisting) genetic variation, which does not normally contribute to an organism’s phenotype but may be uncovered and released upon exposure to certain environmental or genetic cues (e.g., refs. 34, 35). A capacity for plasticity should be particularly important in plants, which cannot simply get up and move to another place more to their liking when physical and biotic conditions become less favorable. Indeed, numerous examples demonstrate how diverse environmental clues, ranging from temperature, to light, to atmospheric CO2, can directly trigger phenotypic variability and change in a single generation (e.g., refs. 15, 31, 36–38).
With regard to major questions that arose about the influence of phenotypic plasticity in evolution, a number of studies now demonstrate that (i) phenotypic plasticity can be adaptive, maladaptive, or neutral; (ii) it may either speed up or slow evolutionary change, leading to more focused questions about when it is likely to be adaptive; and (iii) it may enhance the colonization of new environments or adaptations to local environmental change (see refs. 17–19 for reviews). The molecular mechanisms underwriting developmental plasticity are not well understood. However, it is increasingly shown that new phenotypes often emerge through changes in gene expression, which is known to be highly responsive to environmental perturbation (e.g., refs. 15, 32).
A fundamental and still controversial question about the importance of plasticity in evolutionary change is whether and how plastic forms can be transmitted through multiple generations if they do not initially arise from DNA sequence change. However, there is increasing evidence that plastic phenotypes can be inherited and then stabilized in all environments through a process that Waddington called “genetic assimilation” (GA) (e.g., refs. 15, 29, 30, 32, 33, 37, 39) (Here and elsewhere I differentiate GA from the more general term “genetic accommodation,” the former leading to loss of plasticity and the latter fine-tuning it.) In first demonstrating GA, Waddington (29, 30) carried out laboratory experiments with Drosophila and showed that GA can take place if plastic forms are exposed for a sufficient time to the conditions that initially induced them, so that a gradual accumulation in the gene pool from generation to generation takes the place of the alleles—representing preexisting genetic variation—that contribute to the phenotype. After a number of generations of enrichment, a threshold is reached whereby the alleles are fixed for the constitutive expression of the phenotype in any environment, and plasticity is lost. This loss of plasticity is called “canalization” or “robustness” of the phenotype. Although its frequency and importance in the natural world are still debated, recent field and laboratory research has revived its credibility, because it has been demonstrated or strongly inferred in a variety of organisms. Some examples are terrestrial locomotion in early tetrapods; lower rates of leaf stomatal conductance in a grass from the Southwestern United States; flower sexual expression in Solanum; heat-shock response in nematodes; and early life-history strategies in Homo (e.g., refs. 40–43). Despite protestations to the contrary, including claims the mechanism is Lamarckian and hence inapplicable to evolutionary change, the genetic process following the phenotypic change is Darwinian and can be integrated into conventional evolutionary theory.
Maize as a Possible Model Plant for the Study of Developmental Plasticity
Maize (Zea mays L.) was domesticated in the low-lying seasonal tropical forest of the Balsas River Valley of present-day southwestern Mexico (hereafter, simply “Mexico”) by 9,000 y B.P. (44–47) (all ages in the paper are in calendar years before present, B.P.). There, as in the lowland Neotropics generally, paleoecological evidence indicates that CO2 levels rose from 220 to 260 ppm (these were globally relevant levels), temperature increased by 4–7 °C, and precipitation rose by 20–50% during the Late Pleistocene to early Holocene period, ca. 14,000–10,000 y B.P., when foragers first collected and farmers then began to cultivate its wild ancestor teosinte (Zea mays ssp. parviglumis H.H. Iltis and Doebley) (48–52). Although these global climatic perturbations may have offered strong inducing conditions for plasticity, the way in which they may have influenced the phenotypes encountered by foragers and farmers and the dramatic phenotypic transitions undergone by domesticated plants is a neglected area of domestication research.
Because of maize’s economic importance and status as a genetic model organism, the genetics underlying maize domestication have received considerable attention. Nonetheless, many questions remain. A major quantitative trait locus (QTL), teosinte branched 1 (tb1), has been identified that in part underwrites important domestication traits such as vegetative architecture (branching and tillering) and inflorescence sexuality through a change in gene expression which it mediates during plant development (53–55). Subsequently, another QTL, grassy tillers 1 (gt1), was identified as an additional mediator of branching and tillering in maize that was selected during domestication and has been shown to be responsive to environmental cues such as light and to be dependent on the activity of tb1 (56). The investigators (ref. 56, p. E511) noted that their work “suggests that maize domestication involved modification of a developmental pathway that integrates environmental cues.” Other research revealed cryptic genetic variation in teosinte associated with major domestication traits such as ear disarticulation (24), and recent genome-wide scans found evidence of human selection during domestication at many more loci than previously identified, suggesting that as much as 5% of the genome may have played a functional role in domestication (4, 57). Furthermore, many of the transcription and coexpression networks of maize have been substantially modified during domestication (26), and a number of genes showing evidence of selection show directional changes in gene expression (4). The demonstrated importance of human selection on regulatory elements and gene expression (e.g., no fixed amino acid differences in the tb1 protein were found between maize and teosinte) together with known environmental effects on major domestication QTLs invite research into the effects of changing environmental conditions on Zea phenotypic change during the timeframe of maize domestication.
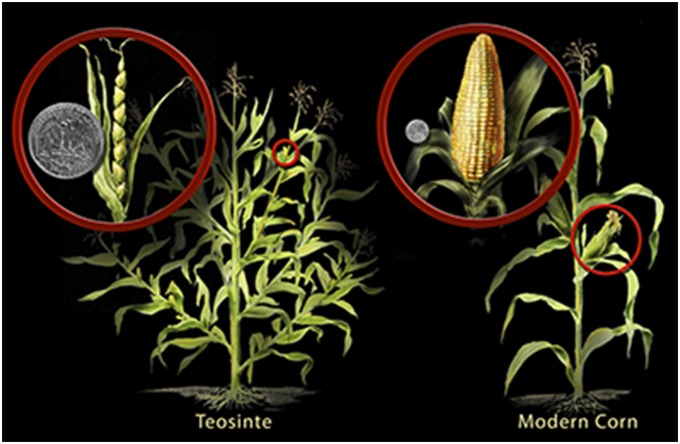
Indeed, in a paper that was ahead of its time, Hugh Iltis (58) first proposed the importance of cryptic genetic variation, environmental influences on development and phenotype, and GA in plant domestication with a theory he called a “catastrophic sexual transmutation” of teosinte to maize. He focused on the profound differences in vegetative architecture and inflorescence sexuality between wild and domesticated Zea (Fig. 1), transformations so great that scientists spent much of the 20th century debating maize’s ancestry. As discussed above, the differences were subsequently shown to be underwritten in part by a change in gene expression at tb1. Iltis proposed that environmental factors induced a rapid, macroevolutionary transformation from teosinte- to maize-type branching and inflorescence sexuality without human involvement which subsequently was fixed through GA under human selection. Without the knowledge (which would be revealed later) of the considerable Late Pleistocene cooling in maize’s homeland but drawing on previous experimental work by others on Zea responses to temperature, he suggested abnormally cool growing seasons as a possible trigger for the maize-like transformations of teosinte. Those changes were very important, because maize plants, with a single main stalk and female ears located on short branches, are examples of apical dominance, in which available nutrients are directed efficiently to the stalk, making possible the compact, large female ears with many large seeds (54). His theory didn't gain much traction at the time.
Fig. 1.
The differences between teosinte and maize today in branching architecture and inflorescence sexuality. Teosinte has several long primary lateral branches terminated by tassels and secondary lateral branching, where female ears are located. Maize has a single main stem terminated with a solitary tassel. There are a few, very short primary lateral branches and no secondary branching. The cobs are located at the ends of the branches on the main stem in the positions occupied by tassels in teosinte. Reproduced from ref. 27.
An important factor that also was unknown to Iltis concerns a developmentally plastic response in vegetative architecture and inflorescence sexuality that takes place today in teosinte and adapts plants to their local environments. In good growing conditions (adequate sunlight, deep soils), the plant has the branching and inflorescence sexuality characteristics normally associated with maize’s wild ancestor both today and in the past (Fig. 1, Left). However, stressful or less optimal habitats (shade, shallow soils, crowding) induce plasticity in gene expression, possibly from tb1 actions, resulting in plants with maize-like vegetative and floral sexuality attributes (Fig. S2) (55, 56).
Fig. S2.
Maize’s wild ancestor teosinte in its natural habitat in Guerrero, Mexico growing in poor, shallow soils. It responded with plasticity to produce maize traits in vegetative architecture and inflorescence sexuality as described in the legend of Fig. 1 caption. Image courtesy of Anthony J. Ranere.
Experimental Research on Developmental Plasticity, GA, and Maize Domestication
In a first effort to investigate empirically the environmental influences on wild Zea phenotypes during the time of domestication, an experiment was undertaken in which seeds from natural living populations of teosinte were grown in conditions that simulated the lower-than-today’s atmospheric CO2 and temperature of the Late Pleistocene (LP) [ca. 215 parts per million volume (ppmv), 21–22 °C] and early Holocene (EH) (ca. 260 ppmv, 23 °C) periods (ca. 15,000 to 10,000–9000 B.P.) (27). In a single generation, both LP and EH conditions repeatedly induced phenotypic changes uncharacteristic of modern teosinte, including the maize-like vegetative architecture and inflorescence sexuality responses predicted by Iltis, a more uniform seed maturation, and low plant height, biomass, and seed yield (Fig. 2A) (27). Plants grown in modern control conditions were uniformly of the normal teosinte-type seen today (Fig. 2B). These results indicate that the people who collected and then cultivated teosinte may have worked with phenotypes that already had important maize-like traits and were considerably different in other ways from phenotypes presently used as the baseline in research on domestication, with plasticity being a major factor accounting for some of the differences. Because suboptimal growing conditions today induce plasticity in the same vegetative and inflorescence traits, it should not be surprising that less favorable conditions in the past, such as low CO2 and temperature, would have the same effects.
Fig. 2.
(A) A maize-like phenotype plant from the environmental chamber that was adjusted to Late Pleistocene conditions. Like maize, it has a single tassel that terminates the main stem, female ears at the main stem (arrows) that terminate a few, very short lateral branches, and no secondary branching. The Inset shows a close-up view of one of the female ears, which do not differ from those seen normally in teosinte today. (B) Teosinte in the modern control chamber. As in modern natural populations, it has many long, primary lateral branches (example, upper white arrow) terminated by tassels (black arrow). Female ears, not yet developed, would be on secondary lateral branches at the location of the two lower white arrows.
Gene expression is increasingly understood to be an important molecular mechanism underlying plasticity responses; therefore RNAseq (whole-transcriptome) gene expression analysis was undertaken of the teosinte plants grown in early Holocene vs. modern conditions (59). Numerous genes demonstrated differences in gene expression [i.e., up-regulation (the gene is more active) or down-regulation (the the gene is less active)] in the two environments. These differently expressed (DE) genes include a number of genes previously shown to have undergone selection in maize during domestication, diversification, or improvement and which mediate diverse key traits such as starch synthesis and properties [sugary 1 (su1)], biomass and seed yield (nitrate reductase), and flowering (various transcription factors) (SI Text, Teosinte Gene Expression for other genes of importance that were differentially expressed and that probably contributed to phenotypic differences observed; see ref. 59 for a complete list of genes). Interestingly, ancient DNA work shows that su1, which influences starch-pasting properties (e.g., it would be difficult to make tortillas without the maize su1 gene), was not fixed in 5,300-y-old maize from present-day Mexico or in 2,000-y-old maize from the present-day Southwestern United States (60–62). The combined data open the possibility that su1 remained plastic in maize for thousands of years after initial cultivation and human fixation of other domestication traits. Other aspects of su1 plasticity that intersect with important archaeological questions and may be investigated with future DNA work on later Holocene varieties of maize in Mexico and the Southwestern United States, concern how su1 evolution was associated with regional trends in the development of ceramic and stone tool food-processing technology, leading to more productive varieties of maize that supported larger and more complex populations.
The gene expression data provide a molecular basis for the remarkable phenotypic differences observed in the experimental past vs. modern environments (e.g., vegetative architecture, inflorescence sexuality, height, biomass, seed yield). They contribute to our understanding of how teosinte and other grasses acclimatized and possibly adapted to past environments with lower CO2 and temperature. For plastic forms to have importance in evolutionary change, including by influencing domestication, they must be passed from generation to generation and then stabilized, so that new phenotypes will occur in any environment and not simply in the inducing one. As discussed above, such a process has been a controversial issue, with GA proposed as a major mechanism for the stabilization of plastic forms. Our transcriptomic analysis comparing teosinte and maize (see also refs. 63, 64) indeed showed that a substantial loss of plasticity occurred during maize domestication, because numerous genes that were differentially expressed in teosinte were not differentially expressed in maize; we inferred that these loci, including 83 with previous evidence of selection, represent GA in maize (e.g., refs. 4, 26) (SI Text, Studying GA in Maize Domestication for further details). This set of transcriptomic gene expression data provides evidence of a loss of plasticity linked to domestication and a role of GA in crop plant evolution.
In summary, the combined results indicate that ancestral biological characteristics of crop plant progenitors are not always predictable from examples growing in the modern environment and that, because of developmental plasticity, some important maize traits were likely present at initial human exploitation. If true, then early farmers would have selected for gene expression changes on those traits. (It is also possible that human movement of plants outside their natural habitats induced some phenotypic changes, as discussed in detail below in the section on niche construction.) Plastic responses could also have been important substrates for adjustments to new environments and culinary practices during maize dispersals.
SI Text
Teosinte Gene Expression.
In teosinte, other genes that were differentially expressed in early Holocene vs. modern conditions and not previously identified as domestication candidates nonetheless have functions known or suggested in maize and other grasses to mediate the following key trait differences observed in the experimental grow-outs (see refs. 27, 59 for further details): (i) vegetative architecture: phytohormone genes such as auxins were differentially expressed, and they are known to influence this trait, possibly through interactions with the tb1 gene; (ii) inflorescence sexuality: recently discovered hormones called “Brassinosteroids” that now are known to control this trait partially in maize were differentially expressed; (iii) plant height: phytohormones, including gibberellins, and gibberellin regulators called “DELLA” that modulate plant growth and responses to abiotic stressors, such as cold, in maize were differentially expressed.
Studying GA in Maize Domestication.
Approaches increasingly used by investigators to study possible incidences of GA use a comparative transcriptome analysis that correlates changes in phenotype with possible differential expression of genes in the inducing and noninducing environment (63, 64). Here we had the advantage of being able to work with both the ancestral and derived plant, because the former presumably would exhibit more plasticity than the latter, and selected traits can be placed in an evolutionary context (e.g., refs. 19, 33). Domestication studies typically will provide this beneficial circumstance, because wild progenitors of many important crops have been identified and are still found on modern landscapes. Therefore, to study GA, we grew traditional land races of Mexican maize in the same early Holocene and modern conditions of atmospheric CO2 and temperature used for teosinte. RNAseq analysis was carried out on the maize to determine if DE genes in teosinte were invariant in expression in maize in the contrasting environments; we inferred such invariability would represent GA at those loci. The comparative gene expression results do suggest that a substantial loss of plasticity occurred during maize domestication, because numerous genes that were differentially expressed in teosinte were not differentially expressed in maize, including 83 with previous evidence of selection and that mediate diverse traits. Moreover, a number of genes that were differentially expressed in teosinte (discussed in part above), that were not previously identified as targets of selection but that nonetheless have relationships to vegetative architecture, inflorescence sexuality, plant height, and yield, were invariant in maize, thus also suggesting GA.
Other Current Examples of Crop Plant Plasticity.
The plants discussed thus far exemplify examples of trait plasticity that was lost during domestication; it may turn out that plasticity was lost in a majority of cases, because farmers would be expected to decrease environmental sensitivity in their crops. However, there were instances in which plasticity was maintained in the domesticated species, reducing fitness in some environmental contexts. For example, manioc (Manihot esculenta Crantz) retained plasticity from its wild ancestor in growth form (shrub or liana depending on savanna or forested environment), but stem brittleness, which apparently was favored by farmers for ease of harvesting, limits growth of the liana form in abandoned fields (109). Another less well-documented example of plasticity in traditional cultivars concerns fruit bitterness, considered a premier wild trait in the Cucurbitaceae, a family that gave rise to many Old and New World domesticates (squashes, gourds, cucumbers, and others) including some of the earliest crops of the Americas. However, recent evidence indicates the trait is plastic in some varieties of domesticated cucumber (Cucumis sativus L.), because nonbitter fruits become bitter when exposed to cold temperature stress, a process controlled in part by a newly discovered transcription factor (110). Bitterness in all economic cucurbits is conferred by the triterpenoids cucurbitacins. Therefore, a possible association of plasticity and temperature variability in the evolution of nonbitter fruits would lend itself to study in other Cucurbitaceae, including Cucurbita spp. squashes, which in Mesoamerica and South America were domesticated at the beginning of the Holocene.
More Subjects for Research in Developmental Plasticity in Domestication
Given the little that is known about the influences of plasticity on domestication, we might expect that future research will provide divergences from conventional knowledge in a number of crops and their wild progenitors, as now seen in teosinte and maize. The limited information available already points to a number of avenues for study. For example, Chenopodium berlandieri Moq. ssp. jonesianum Smith and Funk (goosefoot) is a domesticate native to the eastern United States that first appears in archaeological assemblages about 3,800 y B.P. as part of the premaize agricultural complex (e.g., ref. 65). For phenotypic purposes, the most useful domestication indicators in goosefoot involve a complex of traits for a reduction in the thickness of the outermost seed coat or testa that weaken dormancy and favor rapid germination in field environments. However, it was reported that domesticated-like, thin-testa fruits occur in small proportions in wild populations in the region and that these morphs could be dominant in certain environmental contexts and probably were environmentally triggered (66). The morphology of some domesticated seed assemblages found in prehistoric contexts is the same as the domesticated-like forms found in wild populations in the region (67). The archaeobotanical record also indicates substantial variation through time in the testa morphs, with no evidence for a directional trend for reduced seed coat thickness during the first few thousand years of cultivation (67). Therefore it is reasonable to propose a rapid establishment of thin-testa forms in cultivated fields as a reaction to environmental cues in those contexts (7, 67). [Again, the obvious connection with human niche construction (NC) activities is discussed in detail in the next section.] Environmental influences on seed dormancy are also well known from both older and more recent literature in the closely related South American domesticate Chenopodium quinoa Willd. Seeds are more dormant at high temperatures and long photoperiods than in the converse conditions, and premature sprouting is a problem outside its region of origin (68).
Another member of the eastern North American crop plant complex, Polygonum erectum L. (erect knotweed), was cultivated about 2,500 y B.P. and is now extinct. In proposing and describing characteristics of the domestication syndrome for this plant, Mueller (28) discusses how seasonally controlled differences in fruit morphology among wild species result in distinctly different ratios of two different morphs, called “smooth” and “tubercled.” This apparent plasticity, possibly induced by differences in sunlight, rainfall, and/or temperature, is hypothesized to be a form of bet-hedging on the part of the species, because the smooth form tends to germinate quickly regardless of the conditions, and the tubercled form tends to have stronger dormancy mechanisms (28). Archaeological examples identified as cultivated and/or domesticated knotweed produce achenes of a uniform smooth morphology, and it is possible the plant evolved and lost plasticity in the stable, predictable cultivated field environments afforded by humans. Another potential example of an association between plasticity and an important domestication trait is the increase in seed size in Old World cereals, which is suggested to have been a plastic response to the enriched soils of early cultivation (69). (For a summary and for additional examples of known plasticity in crops, see SI Text, Other Current Examples of Crop Plant Plasticity and Table S1.)
Table S1.
Some known and potential examples of plasticity for important domestication traits in domesticated plants and wild ancestors
| Species | Plastic change | Environmental factor (refs.) |
| Zea mays ssp parviglumis (wild maize) | Vegetative architecture, flower sexuality | Shade, poor soils, little moisture (55, 58 ) Past low CO2 and temperature (experimental) (27) |
| Triticum monococcum (einkorn wheat) | Seed size increase | Enriched soils of early cultivation? (69) |
| Hordeum vulgare (barley) | Seed size increase | Enriched soils of early cultivation? (69) |
| Cucumis sativa (cucumber) | Fruit bitterness | Temperature (110) |
| Chenopodium berlandieri (goosefoot) | Seed dormancy traits | Age at flowering related to soil moisture? (66, 67) |
| Polygonum erectum (erect knotweed) | Seed morphology | Unspecified growing conditions (28) |
| Manihot esculenta ssp. flabellifolia (Pohl) Cifferi (wild manioc) | Growth form (shrub vs. liana) | Savanna vs. forest (109) |
| Manihot esculenta ssp. esculenta (domesticated manioc) | Growth form (shrub vs. liana) | Cultivated field vs. forest (109) |
As has been noted (7, 28), examples of plasticity from eastern North America and elsewhere invite experimental research with living plants to investigate more deeply issues such as the way the involved taxa responded to different growing conditions and cultural practices in prehistoric fields, whether plasticity worked against eventual domestication, and whether the loss of plasticity and the production of uniform forms fitting the characteristics of domesticates was accomplished without generations of artificial selection. These questions, in addition to issues of heritability, are among the key questions plasticity studies in domestication will need to address with empirical data going forward. Gene-expression studies often will be warranted in investigating them.
Niche Construction Theory
Within the last few years, niche construction theory (NCT) (70) has come under active discussion in archaeology with regard to domestication and agricultural origin research (71–76). Ecologists and evolutionary biologists debate whether NC, “the process whereby organisms, through their metabolism, their activities and their choices, modify their own and/or each other's niches” (ref. 70, p. 419), represents a separate force of evolution or instead is a proximate mechanism already subsumed in standard evolutionary theory (SET) (16, 77). Examples of disagreement between EES advocates and SET skeptics also center on whether NC systematically adapts organisms to their environments, as does natural selection, because both positive and negative NC are possible, and the evolutionary importance of one over the other can be difficult to sort out. NCT advocates respond that the developmental processes it emphasizes are by now well-understood to drive the direction and rate of adaptive evolutionary change methodically (e.g., ref. 77). One thing clear to all is that, for an organism’s impact on an environment to beget evolutionary change and be subject to study with a NCT, the impact must be more than just environmental modification and must itself change environmental selection pressures on the niche constructor’s descendants or other organisms occupying the niche, a second process of inheritance called “ecological inheritance” (70). When humans are studied, cultural inheritance becomes a third process involved in transgenerational evolutionary change resulting from NC.
It has been long known that human environmental modification of many types (e.g., fire, vegetation clearing, depression of preferred prey through overexploitation) has a deep history around the world, and these interactions appear to have intensified in some regions shortly before and during early agricultural periods (78, 79). Because most societies since ancient times have arguably modified environments to some degree, it is both especially important and probably more complicated, when using NCT in human research, to specify and test for the resulting selection pressures and evolutionary change for NC recipients—the ecological inheritances—that make NC a separate evolutionary process and not simply an example of ecological engineering (73, 80).
An NCT-based emphasis on dynamic interactions and feedbacks in natural and social systems with regard to domestication and agricultural origins, termed “cultural niche construction” (CNC), is obviously well placed (e.g., refs. 8, 9, 71–76). It emphasizes the importance of human agency in addition to natural environmental change in major cultural evolutionary transitions, also taking us back to Braidwood’s fundamental writings on the importance of cultural knowledge systems in agricultural origins (e.g., ref. 81). Whether, as some have argued recently (74, 75), CNC is conceptually broad or powerful enough to provide an overarching explanatory framework for those origins is under active discussion (e.g., refs. 8, 9, 76, 82–85). Some archaeologists also point out that CNC speaks little to why and when humans may choose to modify environments, questions that potentially complementary fields of study in human behavioral ecology, such as optimal foraging theory, may address better (e.g., refs. 8, 11, 82, 84, 85).
Turning to the key question of what in particular CNC theory can provide to domestication research, some insights and applications are evident. First, as discussed above, bringing plants from their natural habitats into human-created field niches may, in addition to natural environmental changes, have induced phenotypic plasticity. Stabilization of preferred phenotypes could then have resulted from human selection in those durable niches. Subsequent dispersals of domesticates outside their areas of origin into different floristic associations, photoperiods, and other abiotic conditions may have induced another set of phenotypes associated with cultural preferences or crop improvement, as perhaps demonstrated, in part, by the numerous, traditional land races of maize and other crops. Odling-Smee (ref. 86, p. 182), in fact, emphasized the close relationship between NC and developmental biology as illustrated by developmental plasticity and TEI, viewing the latter two as “different components of niche inheritance,” as opposed to genetic inheritance.
Second, gene–culture coevolution (GCC), a branch of theoretical population genetics that stresses how culturally derived traits influence transgenerational gene transmission, has been recast in the context of CNC with suggestions that it can provide a broad conceptual framework for studying transformational developments in prehistory, including agriculture (9). For example, it has been posited that although GCC existed throughout human evolution, the advent of agriculture may have driven uniquely strong selection on human genes because a variety of types of CNC rise to particular prominence at that time (9, 87). Ancient DNA studies of early European farming populations indeed show changes in a number of genes presumably associated with the increased carbohydrate content of domesticated foodstuffs along with agricultural settlement and demographic patterns, including those associated with celiac disease risk, immunity, vitamin D levels, light skin color, and lactose digestion (88). An integrated CNC–GCC approach also prompts further discussions about whether CNC, which is typically practiced at a group level, should result in group selection, as has been frequently assumed, or the converse (9).
EES advocates assume NC will, on average, increase fitness in the short term, so that it and its ecological inheritances are considered directional forces for adaptation similar to natural selection (77). However, especially because the niche constructors under discussion are humans with an enormous capacity not only to modify but also to damage environments, utilization of CNC theory in domestication and other cultural research should come with the acute recognition that it may have both positive and negative effects. The former, such as enhancement of abundance and/or predictability of human-preferred economic and other resources, have thus far been emphasized by some investigators (74, 75), but CNC may not increase fitness. Human-caused depression of valued prey shortly before and/or at domestication origins is well known and influenced the constituents of the plant and animal complexes chosen for attention by the last foragers and first cultivators, along with the timing of domestication origins (e.g., refs. 6, 8, 76, 89–91). The ecological contexts of early farming could have resulted from either positive or negative CNC, the latter in a context of resource limitation.
Furthermore, plasticity induced by new field niches may have slowed or even stopped the domestication process, a possibility that finds accord with the slow pace of phenotypic change under cultivation demonstrated in archaeological records for traits in maize, barley, wheat, and rice (2, 22, 92). Negative NC can also have unintended long-term positive effects, so that, especially with time frames in the hundreds of years and more to which archaeological data are most sensitive, which of the processes most influenced a particular outcome could be ambiguous. In summary, NCT clearly has considerable value in domestication research. To test NCT vs. SET arguments regarding environmental modifications and their effects adequately, their dynamic nature and numerous feedbacks need to be explored and, when possible, disentangled, with falsifiable predictions tested against empirical data with attention to and reconstruction of as many proxies of human environmental impacts as possible (see ref. 73 for an extended discussion).
Epigenetics and Transgenerational Epigenetic Inheritance
A third major element that EES advocates wish to see considered as a substrate for phenotypic and evolutionary change, and which thus far has been little investigated in plant domestication, is transgenerational epigenetic inheritance (TEI), also known as “soft inheritance” and “inclusive inheritance” (15, 16, 93, 94). Epigenetics, meaning “above or on top of genetics,” signifies organismal change without alteration of the DNA sequence through gene expression. In their most general sense then, epigenetic processes occur when effects originating from sources both internal and external to organisms and their genomes induce phenotypic variations, that, if heritable, are so through a non-Mendelian fashion. As such, the term “epigenetics” has sometimes referred to (i) mechanisms such as developmental plasticity (Waddington originated the term along with “epigenetic landscapes” in this sense); (ii) cultural transmission of traits; and (iii) influences of chemical modifications along with types of nonmessenger RNAs on DNA and gene activity. Here, epigenetics is used in its strict sense to refer to the third processes. They have not so far found a role in plant domestication research, despite being amply documented in modern plants and some domesticated animals (e.g., ref. 95).
Epigenetic research is in a remarkable period of discovery and progress, with new revelations that broaden known epigenetic mechanisms or add considerable knowledge to little-understood ones (96). Some of the best-studied epigenetic processes in diverse taxa of higher plants emanate from the action of chemicals such as methyl groups that bind to and mark DNA, or from the methylation or acetylation of histone proteins, that modify chromatin (chromosomal) structure, along with the actions of small, noncoding RNAs that often participate. All may alter gene expression by silencing or activating genes. The effects may be reversible and highly unstable in transgenerational contexts, reducing their chances for heritability. However, more stability than once thought possible is being documented (e.g., ref. 97). Many examples of epigenetic processes are found in plants (as discussed below) and are little disputed. DNA methylation, which largely silences genes, is the easiest to recognize and probably is the best understood at present. However, at this writing the heritability of epigenetic activity in plants and in animals across generations—TEI—is one of the most contentious elements of the EES. Some believe “there is little evidence for the role of inherited epigenetic modification … in adaptation: we know of no case in which a new trait has been shown to have a strictly epigenetic basis divorced from gene sequence” (ref. 16, p. 164). Other scholars argue the opposite position just as strongly (93, 94).
With regard to plants, however, there is little disagreement that DNA methylation and histone-related epigenetic actions with their often-mediating genetic influences, such as those from transposable elements (DNA sequences that move from one location to another), are common between and within species and may produce heritable changes through germ lines (98, 99). Therefore, for EES skeptics and others, the key in demonstrating what they consider to be truly TEI would be ruling out those genetic elements well known to influence epigenetic processes frequently. However, it is not clear to this author whether most EES advocates support a strict definition of TEI in any context or would accept a broader definition whereby DNA sequence and purely epigenetic influences interact to produce the outcome. In any case, TEI as strictly defined does appear to be empirically demonstrated with examples from plants, albeit, at this point, infrequently (99, 100). Prominent examples notably include maize (101) and canola (Brassica napus L.) (97). Moreover, methylation and histone epigenetic effects have been clearly demonstrated to modulate phenotypic variability for agronomically important traits, such as seed yield in canola (in addition to heritability, epigenetic factors also were shown to be amenable to manipulation through artificial selection), fruit ripening (with tomato as the model plant), plant height, and flowering time (high heritability was shown in the latter), and also, interestingly to cause an unusual frequency of dramatic phenotypic changes in hybrids as a result of novel regulatory interactions (e.g., refs. 97–99, 102–104). These findings invite more attention by geneticists and archaeobotanists in their considerations of genetic and phenotypic change during domestication.
Recent investigations of a possible influence of epigenetic actions, notably DNA methylation, in the domestication process show promise. For example, methylation differences were demonstrated in diverse lines of maize and between teosinte and maize, including in DNA regions known to have undergone selection (105). Much, although not all, of the variability in DNA methylation was associated with transposable elements and DNA sequence variation. Analyses of inheritance showed that much of the methylation variability was stably inherited in offspring, and some of the heritability appears to have been driven by purely epigenetic processes. Although the extent to which the differences in methylation influenced important phenotypic traits in the teosinte-to-maize transition is unclear, the work indicates that DNA methylation was altered during domestication, and, as with developmental plasticity, may implicate a human targeting of gene expression differences. Tomatoes have been the subject of epigenetic inquiry for a number of years (102), and these investigations show considerable promise. Also of note is a recent DNA study of 2,800- to 500-y-old barley grains from Egypt that demonstrated methylation action from stress caused by viral infection (106). The study also documented differences in methylation signatures resulting from DNA degradation over time in the warm Egyptian environment. When possible, future ancient DNA work should target methylation as well as changes in DNA sequences.
Summary
This paper has explored some important components of an EES with regard to the study of plant domestication. It argues that each component has considerable potential to inform how phenotypic and genetic change occurred during the domestication process. Recent findings from experimental research on teosinte phenotypic and gene expression responses to simulated past environments and comparative gene expression work with maize point to ways in which studies on plasticity in other wild progenitor/crop plant pairs may provide significant new information. Moreover, it is possible that phenotypic changes resulting from plasticity also involved changes in gene expression caused by epigenetic mechanisms, perhaps necessitating broader approaches to explicate the issues fully. More examples of plasticity, together with its proclivity for inheritance, need to be demonstrated empirically in crops and wild progenitors to assess further their overall importance in domestication.
Similarly, EES skeptics, as well as other investigators, want to see more empirical demonstrations of TEI (in its strictest sense) before acknowledging its importance in evolutionary change. It is likely that, as epigenetic research intensifies, more examples of true TEI will be found in plants. At this time, few studies have been carried out that assess the role of epigenetic factors in the domestication process, but results thus far on maize and the tomato provide intriguing evidence for research going forward. Investigating processes such as DNA methylation is more complex and expensive than DNA-sequence generation, and to this point the investigations are not often combined. Clearly, both developmental plasticity and epigenetics may be of considerable importance in breeding and crop improvement work and should provide informed projections of plant phenotypic and molecular responses to future climate change and the new ecological niches it creates.
With regard to NCT, CNC effectively bridges developmental plasticity and epigenetics. For example, CNC may prove particularly powerful for demonstrating what were viable ecological contexts and opportunities for phenotypic transformations from wild to domesticated species via the type of CNC in which early farmers created what, at the time, were ecologically unique field niches and transferred the wild progenitors of crops to them. This aspect of CNC is particularly amenable to field and experimental research with living crops and their extant wild ancestors. Broader questions regarding the efficacy of NCT in explaining evolutionary change, such as whether it is equivalent to natural selection, whether ecological inheritance is demonstrated, or whether adaptive NC can be shown to be more prominent than negative NC—all of which could complicated to pin down when using archaeological or paleoecological data—may be evaluated more straightforwardly with this particular focus of CNC.
Future investigations of all three fields of study will prominently include experimental research with living crops and their wild ancestors, either in simulated ancient field conditions in modern open-air environments or in simulated past natural environments in growing chambers (see also ref. 107 for a recent example of experimental work in the Old World). Experimental efforts become necessary and particularly valuable in many regions of the world because archaeological sites dating to the periods surrounding domestication origins are few in number and, when found, the macrofossil remains (seeds, fruits, stems) that can best address some of the questions are often poorly preserved. Even when early plant remains are well preserved, addressing plasticity and other questions directly is difficult.
Neglected in the broader domain of this paper are evolutionary perspectives from human behavioral ecology, which already have contributed significantly to questions about agricultural origins (e.g., refs. 3, 6–8, 11, 12, 85, 89–91) and can fit holistically with the topics considered here. Finally, Pigliucci and Finkleman (108) discuss how some who are directly involved in the SET vs. EES debate or who have offered their views of it take a middle-ground position, i.e., that ever since Darwin, evolutionary theory and practice have constantly added new concepts and approaches to then-practiced SET, while setting aside or modifying others. Arguably, this debate in domestication research and, more broadly, in evolutionary theory will proceed in a similar manner. It can be expected that future work in domestication will be accelerated, with a consequent increase in knowledge and understanding and with ever-increasing engagement by archaeologists and anthropologists.
Footnotes
The author declares no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1703658114/-/DCSupplemental.
References
- 1.Price TD, Bar-Yosef O. The origins of agriculture: New data, new ideas. Curr Anthropol. 2011;52(Suppl 4):S163–S174. [Google Scholar]
- 2.Larson G, et al. Current perspectives and the future of domestication studies. Proc Natl Acad Sci USA. 2014;111:6139–6146. doi: 10.1073/pnas.1323964111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Piperno DR. The origins of plant cultivation and domestication in the New World tropics. Curr Anthropol. 2011;52:S453–S470. [Google Scholar]
- 4.Hufford MB, et al. Comparative population genomics of maize domestication and improvement. Nat Genet. 2012;44:808–811. doi: 10.1038/ng.2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rindos D. The Origins of Agriculture an Evolutionary Perspective. Academic; New York: 1984. [Google Scholar]
- 6.Piperno DR, Pearsall DM. The Origins of Agriculture in the Lowland Neotropics. Academic; San Diego, CA: 1998. [Google Scholar]
- 7.Gremillion KJ, Piperno DR. Human behavioral ecology, phenotypic (developmental) plasticity, and agricultural origins: Insights from the emerging evolutionary synthesis. Curr Anthropol. 2009;50:615–619. doi: 10.1086/605360. [DOI] [PubMed] [Google Scholar]
- 8.Gremillion KJ, Barton L, Piperno DR. Particularism and the retreat from theory in the archaeology of agricultural origins. Proc Natl Acad Sci USA. 2014;111:6171–6177. doi: 10.1073/pnas.1308938110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O’Brien MJ, Laland KN. Genes, culture, and agriculture: An example of human niche construction. Curr Anthropol. 2012;53:434–470. [Google Scholar]
- 10.Zeder M. Evolutionary biology and the emergence of agriculture: The value of co-opted models of evolution in the study of culture change. In: Prentice A, Kuijt I, Chatters JC, editors. Macroevolution in Human Prehistory. Springer; New York: 2009. pp. 157–210. [Google Scholar]
- 11.Kennett DJ, Winterhalder B, editors. Behavioral Ecology and the Transition to Agriculture. Univ of California Press; Berkeley, CA: 2006. [Google Scholar]
- 12.Bettinger RL. Orderly Anarchy: Sociopolitical Evolution in Aboriginal California. Univ of California Press; Oakland, CA: 2015. [Google Scholar]
- 13.Richerson P, Boyd R, Bettinger RL. Was agriculture impossible during the pleistocene but mandatory during the holocene? A climate change hypothesis. Am Antiq. 2001;66:387–411. [Google Scholar]
- 14.Pigliucci M, Muller GB, editors. Evolution: The Extended Synthesis. MIT Press; Cambridge, MA: 2010. [Google Scholar]
- 15.Gilbert SF, Epel D. Ecological Developmental Biology: Integrating Epigenetics, Medicine and Evolution. Sinauer Associates, Inc.; Sunderland, MA: 2009. [Google Scholar]
- 16.Laland K, et al. Does evolutionary theory need a rethink? Nature. 2014;514:161–164. doi: 10.1038/514161a. [DOI] [PubMed] [Google Scholar]
- 17.Moczek AP. Developmental plasticity and evolution–quo vadis? Heredity (Edinb) 2015;115:302–305. doi: 10.1038/hdy.2015.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moczek AP, et al. The significance and scope of evolutionary developmental biology: A vision for the 21st century. Evol Dev. 2015;17:198–219. doi: 10.1111/ede.12125. [DOI] [PubMed] [Google Scholar]
- 19.Hendry AP. Key questions on the role of phenotypic plasticity in eco-evolutionary dynamics. J Hered. 2016;107:25–41. doi: 10.1093/jhered/esv060. [DOI] [PubMed] [Google Scholar]
- 20.Meyer RS, Purugganan MD. Evolution of crop species: Genetics of domestication and diversification. Nat Rev Genet. 2013;14:840–852. doi: 10.1038/nrg3605. [DOI] [PubMed] [Google Scholar]
- 21.Olsen KM, Wendel JF. A bountiful harvest: Genomic insights into crop domestication phenotypes. Annu Rev Plant Biol. 2013;64:47–70. doi: 10.1146/annurev-arplant-050312-120048. [DOI] [PubMed] [Google Scholar]
- 22.Martínez-Ainsworth NE, Tenaillon MI. Superheroes and masterminds of plant domestication. C R Biol. 2016;339:268–273. doi: 10.1016/j.crvi.2016.05.005. [DOI] [PubMed] [Google Scholar]
- 23.Nesbitt TC, Tanksley SD. Comparative sequencing in the genus Lycopersicon. Implications for the evolution of fruit size in the domestication of cultivated tomatoes. Genetics. 2002;162:365–379. doi: 10.1093/genetics/162.1.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lauter N, Doebley J. Genetic variation for phenotypically invariant traits detected in teosinte: Implications for the evolution of novel forms. Genetics. 2002;160:333–342. doi: 10.1093/genetics/160.1.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Doebley JF, Gaut BS, Smith BD. The molecular genetics of crop domestication. Cell. 2006;127:1309–1321. doi: 10.1016/j.cell.2006.12.006. [DOI] [PubMed] [Google Scholar]
- 26.Swanson-Wagner R, et al. Reshaping of the maize transcriptome by domestication. Proc Natl Acad Sci USA. 2012;109:11878–11883. doi: 10.1073/pnas.1201961109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Piperno DR, Holst I, Winter K, McMillan O. Teosinte before domestication: Experimental study of growth and phenotypic variability in late Pleistocene and early Holocene environments. Quat Int. 2015;363:65–77. [Google Scholar]
- 28.Mueller NG. Documenting domestication in a lost crop (Polygonum erectum L.): Evolutionary bet-hedgers under cultivation. Veg Hist Archaeobot. 2017;26:313–327. [Google Scholar]
- 29.Waddington CH. Selection of the genetic basis for an acquired character. Nature. 1952;169:625–626. doi: 10.1038/169625b0. [DOI] [PubMed] [Google Scholar]
- 30.Waddington CH. Genetic assimilation of an acquired character. Evolution. 1953;7:118–126. [Google Scholar]
- 31.West-Eberhard MJ. Developmental Plasticity and Evolution. Oxford Univ Press; New York: 2003. [Google Scholar]
- 32.Beldade P, Mateus ARA, Keller RA. Evolution and molecular mechanisms of adaptive developmental plasticity. Mol Ecol. 2011;20:1347–1363. doi: 10.1111/j.1365-294X.2011.05016.x. [DOI] [PubMed] [Google Scholar]
- 33.Moczek AP, et al. The role of developmental plasticity in evolutionary innovation. Proc Biol Sci. 2011;278:2705–2713. doi: 10.1098/rspb.2011.0971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schlichting CD. Hidden reaction norms, cryptic genetic variation, and evolvability. Ann N Y Acad Sci. 2008;1133:187–203. doi: 10.1196/annals.1438.010. [DOI] [PubMed] [Google Scholar]
- 35.Paaby AB, Rockman MV. Cryptic genetic variation: Evolution’s hidden substrate. Nat Rev Genet. 2014;15:247–258. doi: 10.1038/nrg3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sultan SE. Plant developmental responses to the environment: eco-devo insights. Curr Opin Plant Biol. 2010;13:96–101. doi: 10.1016/j.pbi.2009.09.021. [DOI] [PubMed] [Google Scholar]
- 37.Palmer CM, Bush SM, Maloof JN. Encyclopedia of Life Sciences. John Wiley & Sons, Ltd; Chichester, UK: 2012. Phenotypic and developmental plasticity in plants. [DOI] [Google Scholar]
- 38.Gratani L. 2014. Plant phenotypic plasticity in response to environmental factors Adv Bot Res, 10.1155/2014/208747.
- 39.Pigliucci M, Murren CJ, Schlichting CD. Phenotypic plasticity and evolution by genetic assimilation. J Exp Biol. 2006;209:2362–2367. doi: 10.1242/jeb.02070. [DOI] [PubMed] [Google Scholar]
- 40.Kuzawa CW, Bragg JM. Plasticity in human life history strategy. Curr Anthropol. 2012;53:S369–S382. [Google Scholar]
- 41.Diggle PK, Miller JS. Developmental plasticity, genetic assimilation, and the evolutionary diversification of sexual expression in Solanum. Am J Bot. 2013;100:1050–1060. doi: 10.3732/ajb.1200647. [DOI] [PubMed] [Google Scholar]
- 42.Standen EM, Du TY, Larsson HCE. Developmental plasticity and the origin of tetrapods. Nature. 2014;513:54–58. doi: 10.1038/nature13708. [DOI] [PubMed] [Google Scholar]
- 43.Grossman JD, Rice KJ. Contemporary evolution of an invasive grass in response to elevated atmospheric CO(2) at a Mojave Desert FACE site. Ecol Lett. 2014;17:710–716. doi: 10.1111/ele.12274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Matsuoka Y, et al. A single domestication for maize shown by multilocus microsatellite genotyping. Proc Natl Acad Sci USA. 2002;99:6080–6084. doi: 10.1073/pnas.052125199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Heerwaarden J, et al. Genetic signals of origin, spread, and introgression in a large sample of maize landraces. Proc Natl Acad Sci USA. 2011;108:1088–1092. doi: 10.1073/pnas.1013011108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Piperno DR, Ranere AJ, Holst I, Iriarte J, Dickau R. Starch grain and phytolith evidence for early ninth millennium B.P. maize from the Central Balsas River Valley, Mexico. Proc Natl Acad Sci USA. 2009;106:5019–5024. doi: 10.1073/pnas.0812525106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ranere AJ, Piperno DR, Holst I, Dickau R, Iriarte J. The cultural and chronological context of early Holocene maize and squash domestication in the Central Balsas River Valley, Mexico. Proc Natl Acad Sci USA. 2009;106:5014–5018. doi: 10.1073/pnas.0812590106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ahn J, et al. A record of atmospheric CO2 during the last 40,000 years from the Siple Dome, Antarctica ice core. J Geophys Res. 2004;109:8–13. [Google Scholar]
- 49.Piperno DR, et al. Late Pleistocene and Holocene environmental history of the Iguala Valley, central Balsas watershed of Mexico. Proc Natl Acad Sci USA. 2007;104:11874–11881. doi: 10.1073/pnas.0703442104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hodell DA, et al. An 85-ka record of climate change in lowland Central America. Quat Sci Rev. 2008;27:1152–1165. [Google Scholar]
- 51.Bush MB, et al. Re-evaluation of climate change in lowland Central America during the Last Glacial Maximum using new sediment cores from Lake Peten Itza, Guatemala. In: Vimeux F, Sylvestre F, Khodri M, editors. Past Climate Variability in South America and Surrounding Regions. Springer; Dordrecht, The Netherlands: 2009. pp. 113–128. [Google Scholar]
- 52.Correa-Metrio A, et al. Rapid climate change and no-analog vegetation in lowland Central America during the last 86,000 years. Quat Sci Rev. 2012;38:63–75. [Google Scholar]
- 53.Doebley J, Stec A, Gustus C. teosinte branched1 and the origin of maize: Evidence for epistasis and the evolution of dominance. Genetics. 1995;141:333–346. doi: 10.1093/genetics/141.1.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Doebley J, Stec A, Hubbard L. The evolution of apical dominance in maize. Nature. 1997;386:485–488. doi: 10.1038/386485a0. [DOI] [PubMed] [Google Scholar]
- 55.Hubbard L, McSteen P, Doebley J, Hake S. Expression patterns and mutant phenotype of teosinte branched1 correlate with growth suppression in maize and teosinte. Genetics. 2002;162:1927–1935. doi: 10.1093/genetics/162.4.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Whipple CJ, et al. grassy tillers1 promotes apical dominance in maize and responds to shade signals in the grasses. Proc Natl Acad Sci USA. 2011;108:E506–E512. doi: 10.1073/pnas.1102819108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wright SI, et al. The effects of artificial selection on the maize genome. Science. 2005;308:1310–1314. doi: 10.1126/science.1107891. [DOI] [PubMed] [Google Scholar]
- 58.Iltis HH. From teosinte to maize: The catastrophic sexual transmutation. Science. 1983;222:886–894. doi: 10.1126/science.222.4626.886. [DOI] [PubMed] [Google Scholar]
- 59.Lorant A, et al. 2017 The potential role of genetic assimilation during maize domestication. bioRxiv preprint, Available at biorxiv.org/content/early/2017/02/03/105940. Accessed February 3, 2017.
- 60.Jaenicke-Després V, et al. Early allelic selection in maize as revealed by ancient DNA. Science. 2003;302:1206–1208. doi: 10.1126/science.1089056. [DOI] [PubMed] [Google Scholar]
- 61.Vallebueno-Estrada M, et al. The earliest maize from San Marcos Tehuacán is a partial domesticate with genomic evidence of inbreeding. Proc Natl Acad Sci USA. 2016;113:14151–14156. doi: 10.1073/pnas.1609701113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ramos-Madrigal J, et al. Genome sequence of a 5,310-year-old maize cob provides insights into the early stages of maize domestication. Curr Biol. 2016;26:3195–3201. doi: 10.1016/j.cub.2016.09.036. [DOI] [PubMed] [Google Scholar]
- 63.Sikkink KL, Reynolds RM, Ituarte CM, Cresko WA, Phillips PC. Rapid evolution of phenotypic plasticity and shifting thresholds of genetic assimilation in the nematode Caenorhabditis remanei. G3 (Bethesda) 2014;4:1103–1112. doi: 10.1534/g3.114.010553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.DeBiasse MB, Kelly MW. Plastic and evolved responses to global change: What can we learn from comparative transcriptomics? J Hered. 2016;107:71–81. doi: 10.1093/jhered/esv073. [DOI] [PubMed] [Google Scholar]
- 65.Gremillion KJ. Seed processing and the origins of food production in eastern North America. Am Antiq. 2004;69:215–234. [Google Scholar]
- 66.Asch D, Asch NE. Chenopod as cultigen: A reevaluation of some prehistoric collections from eastern North America. Midcontinental J Archaeol. 1977;2:3–45. [Google Scholar]
- 67.Gremillion KJ. The evolution of seed morphology in domesticated Chenopodium: An archaeological case study. J Ethnobiol. 1993;13:149–169. [Google Scholar]
- 68.Ceccato D, Bertero D, Batilla D, Galati B. Structural aspects of dormancy in quinoa (Chenopodium quinoa): Importance and possible action mechanisms of the seed coat. Seed Sci Res. 2015;25:1–9. [Google Scholar]
- 69.Willcox G. Measuring grain size and identifying Near Eastern cereal domestication: Evidence from the Euphrates valley. J Archaeol Sci. 2004;31:145–150. [Google Scholar]
- 70.Odling-Smee FJ, Laland KN, Feldman MW. Niche Construction. The Neglected Process in Evolution. Princeton Univ Press; Princeton, NJ: 2003. [Google Scholar]
- 71.Smith BD. Niche construction and the behavioral context of plant and animal domestication. Evol Anthropol. 2007;165:188–199. [Google Scholar]
- 72.Freeman J, Peeples MA, Anderies JM. Toward a theory of non-linear transitions from foraging to farming. J Anthro Arch. 2015;40:109–122. [Google Scholar]
- 73.Brock WA, O’Brien MJ, Bentley RA. Validating niche-construction theory through path analysis. Archaeol Anthropol Sci. 2016;8:819–837. [Google Scholar]
- 74.Smith BD. Neo-Darwinism, niche construction theory, and the initial domestication of plants and animals. Evol Ecol. 2016;30:307–324. [Google Scholar]
- 75.Zeder MA. Domestication as a model system for niche construction theory. Evol Ecol. 2016;30:325–348. [Google Scholar]
- 76.Piperno DR, Ranere AJ, Dickau R, Aceituno F. Niche construction and optimal foraging theory in Neotropical agricultural origins: A re-evaluation in consideration of the empirical evidence. J Archaeol Sci. 2017;78:214–220. [Google Scholar]
- 77.Scott-Phillips TC, Laland KN, Shuker DM, Dickins TE, West SA. The niche construction perspective: A critical appraisal. Evolution. 2014;68:1231–1243. doi: 10.1111/evo.12332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ellis EC, et al. Used planet: A global history. Proc Natl Acad Sci USA. 2013;110:7978–7985. doi: 10.1073/pnas.1217241110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Boivin NL, et al. Ecological consequences of human niche construction: Examining long-term anthropogenic shaping of global species distributions. Proc Natl Acad Sci USA. 2016;113:6388–6396. doi: 10.1073/pnas.1525200113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ellis EC, et al. Evolving the human niche. Proc Natl Acad Sci USA. 2016;113:E4436. doi: 10.1073/pnas.1609425113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Braidwood RJ. The agricultural revolution. Sci Am. 1960;203:131–148. [PubMed] [Google Scholar]
- 82.Bird DW, Bliege Bird R, Codding BF. Pyrodiversity and the anthropocene: The role of fire in the broad spectrum revolution. Evol Anthropol. 2016;25:105–116. doi: 10.1002/evan.21482. [DOI] [PubMed] [Google Scholar]
- 83.Wallach E. Niche construction theory as an explanatory framework for human phenomena. Synthase. 2016;193:2595–2618. [Google Scholar]
- 84.Zeanah D. Foraging models, niche construction, and the eastern agricultural complex. Am Antiq. 2017;82:3–24. [Google Scholar]
- 85.Broughton JM, Cannon MD, Bartelink EJ. Evolutionary ecology, resource depression, and niche construction theory: Applications to central California hunter-gatherers and Mimbres-Mogollon agriculturalists. J Archaeol Method Theory. 2010;17:371–421. [Google Scholar]
- 86.Odling-Smee J. Niche inheritance. In: Pigliucci M, Muller GB, editors. Evolution: The Extended Synthesis. MIT Press; Cambridge, MA: 2010. pp. 175–207. [Google Scholar]
- 87.Laland KN, Odling-Smee J, Myles S. How culture shaped the human genome: Bringing genetics and the human sciences together. Nat Rev Genet. 2010;11:137–148. doi: 10.1038/nrg2734. [DOI] [PubMed] [Google Scholar]
- 88.Mathieson I, et al. Genome-wide patterns of selection in 230 ancient Eurasians. Nature. 2015;528:499–503. doi: 10.1038/nature16152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Munro ND. Small game, the younger Dryas, and the transition to agriculture in the southern Levant. Mitteilungen der Gesellschaft für Urgeschichte. 2003;12:47–71. [Google Scholar]
- 90.Codding BF, Bird DW. Behavioral ecology and the future of archaeological science. J Archaeol Sci. 2015;56:9–20. [Google Scholar]
- 91.Stutz AJ, Munro ND, Bar-Oz G. Increasing the resolution of the broad spectrum revolution in the Southern Levantine Epipaleolithic (19-12 ka) J Hum Evol. 2009;56:294–306. doi: 10.1016/j.jhevol.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 92.Purugganan MD, Fuller DQ. Archaeological data reveal slow rates of evolution during plant domestication. Evolution. 2011;65:171–183. doi: 10.1111/j.1558-5646.2010.01093.x. [DOI] [PubMed] [Google Scholar]
- 93.Jablonka E, Raz G. Transgenerational epigenetic inheritance: Prevalence, mechanisms, and implications for the study of heredity and evolution. Q Rev Biol. 2009;84:131–176. doi: 10.1086/598822. [DOI] [PubMed] [Google Scholar]
- 94.Jablonka E, Lamb MJ. Transgenerational epigenetic inheritance. In: Pigliucci M, Muller GB, editors. Evolution: The Extended Synthesis. MIT Press; Cambridge, MA: 2010. pp. 138–174. [Google Scholar]
- 95.Feeney A, Nilsson E, Skinner MK. Epigenetics and transgenerational inheritance in domesticated farm animals. J Anim Sci Biotechnol. 2014;5:48–55. doi: 10.1186/2049-1891-5-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Willyard C. An epigenetics gold rush: New controls for gene expression. Nature. 2017;542:406–408. doi: 10.1038/542406a. [DOI] [PubMed] [Google Scholar]
- 97.Hauben M, et al. Energy use efficiency is characterized by an epigenetic component that can be directed through artificial selection to increase yield. Proc Natl Acad Sci USA. 2009;106:20109–20114. doi: 10.1073/pnas.0908755106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Grant-Downton RT, Dickinson HG. The Epigenetic Network in Plants Epigenetics and its implications for plant biology. 1. The epigenetic network in plants. Ann Bot (Lond) 2005;96:1143–1164. doi: 10.1093/aob/mci273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Heard E, Martienssen RA. Transgenerational epigenetic inheritance: Myths and mechanisms. Cell. 2014;157:95–109. doi: 10.1016/j.cell.2014.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Cortijo S, et al. Mapping the epigenetic basis of complex traits. Science. 2014;343:1145–1148. doi: 10.1126/science.1248127. [DOI] [PubMed] [Google Scholar]
- 101.Eichten SR, et al. Heritable epigenetic variation among maize inbreds. PLoS Genet. 2011;7:e1002372. doi: 10.1371/journal.pgen.1002372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Gallusci P, Hodgman C, Teyssier E, Seymour CB. DNA methylation and chromatin regulation during fleshy fruit development and ripening. Fron Plant Sci. 2016;7:807. doi: 10.3389/fpls.2016.00807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Johannes F, et al. Assessing the impact of transgenerational epigenetic variation on complex traits. PLoS Genet. 2009;5:e1000530. doi: 10.1371/journal.pgen.1000530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Meyer P. Epigenetic variation and environmental change. J Exp Bot. 2015;66:3541–3548. doi: 10.1093/jxb/eru502. [DOI] [PubMed] [Google Scholar]
- 105.Eichten SR, et al. Epigenetic and genetic influences on DNA methylation variation in maize populations. Plant Cell. 2013;25:2783–2797. doi: 10.1105/tpc.113.114793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Smith O, et al. Genomic methylation patterns in archaeological barley show de-methylation as a time-dependent diagenetic process. Sci Rep. 2014;4:5559. doi: 10.1038/srep05559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Cubero i Corpas C, Ollich i Castanyer I, de Rocafiguera i Espona M, Ocana I Subirana M. From the granary to the field: Archaeobotany and experimental archaeology at l’Esquerda (Catalonia, Spain) Veg Hist Archaeobot. 2008;17:85–92. [Google Scholar]
- 108.Pigliucci M, Finkleman L. The extended (evolutionary) synthesis debate: Where science meets philosophy. Bioscience. 2014;64:511–516. [Google Scholar]
- 109.Ménard L, McKey D, Mühlen GS, Clair B, Rowe NP. The evolutionary fate of phenotypic plasticity and functional traits under domestication in manioc: Changes in stem biomechanics and the appearance of stem brittleness. PLoS One. 2013;8:e74727. doi: 10.1371/journal.pone.0074727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Shang Y, et al. Plant science. Biosynthesis, regulation, and domestication of bitterness in cucumber. Science. 2014;346:1084–1088. doi: 10.1126/science.1259215. [DOI] [PubMed] [Google Scholar]