Significance
Both plants and animals make decisions in response to the environment to maximize their fitness. Plants use dormancy in seeds to move through time and space, and timing of the transition to germination is influenced by external cues, including temperature. Here, we report the presence of a decision-making center within the root tip of dormant seeds and demonstrate that it shares a similar configuration as some systems within the human brain. Unlike in humans, where this spatial structure is used to filter out noisy inputs from the environment, seeds use this arrangement to harness fluctuating temperatures and stimulate the termination of dormancy. Variable inputs therefore act as an instructive signal for seeds, enhancing the accuracy with which plants are established in ecosystems.
Keywords: seed, dormancy, signal integration, distributed control, variability
Abstract
Plants perceive and integrate information from the environment to time critical transitions in their life cycle. Some mechanisms underlying this quantitative signal processing have been described, whereas others await discovery. Seeds have evolved a mechanism to integrate environmental information by regulating the abundance of the antagonistically acting hormones abscisic acid (ABA) and gibberellin (GA). Here, we show that hormone metabolic interactions and their feedbacks are sufficient to create a bistable developmental fate switch in Arabidopsis seeds. A digital single-cell atlas mapping the distribution of hormone metabolic and response components revealed their enrichment within the embryonic radicle, identifying the presence of a decision-making center within dormant seeds. The responses to both GA and ABA were found to occur within distinct cell types, suggesting cross-talk occurs at the level of hormone transport between these signaling centers. We describe theoretically, and demonstrate experimentally, that this spatial separation within the decision-making center is required to process variable temperature inputs from the environment to promote the breaking of dormancy. In contrast to other noise-filtering systems, including human neurons, the functional role of this spatial embedding is to leverage variability in temperature to transduce a fate-switching signal within this biological system. Fluctuating inputs therefore act as an instructive signal for seeds, enhancing the accuracy with which plants are established in ecosystems, and distributed computation within the radicle underlies this signal integration mechanism.
Plant development is guided by the perception of diverse environmental cues and their integration into key transitions (1). One major decision in the life cycle of plants is when to commence flowering (2, 3). The other major decision is when to initiate a new plant (4). This decision is achieved through seed dormancy, an adaptive trait that determines where and when plants are established, and the entry and exit of plants into and out of ecosystems (4). The germination of seeds also represents the starting point for the vast majority of world agriculture, having great industrial, economic, and societal significance (5).
During seed development, dormancy level is established in response to the environment experienced by the mother plant (6). This control is achieved through the quantitative regulation of genetically encoded regulatory factors, including the DOG1 locus (7, 8), and hormone abundance and sensitivity (9, 10). Following their release from the mother plant, the control of dormancy in seeds was proposed to be mediated by the activity of antagonistically acting factors (11). Later work identified this endogenous signal integration mechanism to consist of the antagonistically acting hormone abscisic acid (ABA) promoting dormancy and gibberellin (GA) promoting germination (9, 12). The relative abundance of these hormones constitutes a metabolic thresholding mechanism that regulates the developmental fate of this system (10, 13). Mutual inhibition is sufficient to generate a one-way developmental fate switch (14) and has been described as a mechanism used to regulate cell fate (15). ABA and GA, however, do not directly interact to antagonize one another, but rather modulate their own and one another’s abundance through a complex series of feedbacks onto hormone synthesis and degradation gene expression (16). This study seeks to understand how the endogenous hormone integration system in seeds acts to integrate information from the environment into developmental fate decisions.
Results
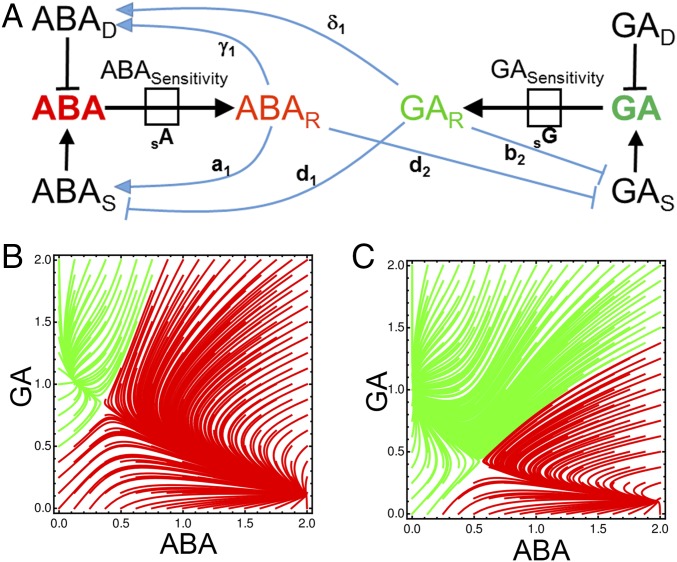
We explored hormone metabolic interactions underlying the control of seed dormancy using a mathematical modeling approach. Feedbacks between GA and ABA responses onto both hormone synthesis and degradation gene expression were identified using publicly available microarray data generated from whole seeds (17, 18) (Fig. 1A and SI Appendix, Supplementary Fig. 1A). These relationships were encoded using ordinary differential equations (ODEs) to capture the dynamic metabolic interactions governing these hormone levels (SI Appendix, Supplementary Figs. 1 and 2). Terms describing the sensitivity to each hormone, as defined by the ability to respond to these hormone levels, were also included (Fig. 1A). The model predicts how the metabolic poise of cells will change, given an initial condition, and the predicted dynamics can be visualized as attractor basins (Fig. 1 B and C) describing how a seed’s state evolves from a given condition. These attractor basins are represented as 2D plots with the relative abundance of ABA and GA on the axes. Indicated within these plots are lines that indicate how the system will shift when it is in any given state (Fig. 1 B and C). These lines ultimately lead toward convergence points that represent a stable resting state of the system. The shape of these lines, including the position of the convergence points and threshold at which the system flips from one state to the other, is determined by the underlying ODEs and their parameterization.
Fig. 1.
Mathematical model of hormone metabolic interactions underlying fate switching in Arabidopsis seeds. (A) Schematic outlining the relationships between the components of the hormone metabolic model. The hormone ABA is shown in red and bold, and the hormone GA is shown in green and bold. The response to each of these hormones, each comprising expression of several genes, is denoted by the subscript R (ABAR and GAR). The degradation of each hormone is indicated by the subscript D (ABAD and GAD), synthesis is indicated by the subscript S (ABAS and GAS), and sensitivity is written in full (ABASensitivity and GASensitivity). The directions of the arrows in the model are defined using microarray data describing ABA and GA application to Arabidopsis seeds (17, 47) and the associated gene expression changes for components representing each component (SI Appendix, Supplementary Fig. 1). GA degrading gene expression (GA 2-oxidase) was not detected at significant levels in germinating or dormant seeds. Attractor basins describing the dynamics of metabolic poise under a dynamic model parameterized by observations for deeply dormant seeds (B) and less dormant seeds (C) are shown. Axes indicate ABA and GA hormone abundance, and trajectories give the dynamics of the system starting from a given point and converging on one of two stable states.
Effective parameter sets for this model were identified harnessing a range of biological observations, and using approximate Bayesian computation with the criteria driving parameter selection was based on the similarity between simulated model behavior and observed biological behaviors (19) (SI Appendix, Supplementary Materials and Methods). The basin structure from inferred effective parameters yields a bifurcating output of relative hormone abundance in seeds, underlying bistable fate switching in this system (Fig. 1 B and C). These two outcomes are indicated by the presence of two different convergence points corresponding to a high-ABA, low-GA state and a high-GA, low-ABA state, representing the dormant and germinating states, respectively. Both deeply dormant (Fig. 1B) and less dormant (Fig. 1C) attractor basins are supported by the experimentally supported model. This shift to reduced dormancy is achieved by increasing the sensitivity of the system to GA (SI Appendix, Supplementary Fig. 2). The increase in GA sensitivity and GID1 GA receptor abundance has been associated with the progressive loss of dormancy, such as during after ripening (20) or low-temperature treatment (18) (SI Appendix, Supplementary Fig. 2). The parameterized model also captures further observed responses to perturbations. Adding the ABA synthesis inhibitor norflurazon, or applying GA, is sufficient to stimulate germination in a portion of a dormant seed population (10) (SI Appendix, Supplementary Fig. 3), and these effects increase with the progressive loss of the depth of dormancy (21). These features are present in the less dormant model (Fig. 1C) and not the highly dormant model (Fig. 1B), whereby the removal of ABA or addition of GA is sufficient to drive the system into the high-GA, low-ABA germination state. This dynamic model thus captures the bistable developmental fate switch in seeds, and recapitulates fate-switching observations following physiological and pharmacological interventions.
We next sought to understand how the spatial context of this decision-making module within the multicellular embryo body plan allows the plant to process environmental information. Genetically encoded components of GA and ABA synthesis, degradation, sensitivity, and response have been identified previously (4) (SI Appendix, Supplementary Fig. 1 and Supplementary Table 1). It has been established that germination is ultimately initiated within the Arabidopsis embryo and that this process is informed by the endosperm (10, 12, 21, 22). The cellular locations underlying this decision-making process remain unknown, however. Using 3D digital single-cell analysis, we localized ABA and GA hormone synthesis, degradation, and response reporter constructs in primary dormant embryos (23, 24) to reveal the cellular basis of decision making in seeds (Fig. 2 A–I and SI Appendix, Supplementary Figs. 4–6). All reporters investigated were enriched within the embryo radicle in primary dormant Arabidopsis seeds (SI Appendix, Supplementary Figs. 4 and 5). This result indicates that the subset of cells constituting the radicle constitutes a decision-making center within primary dormant Arabidopsis seeds.
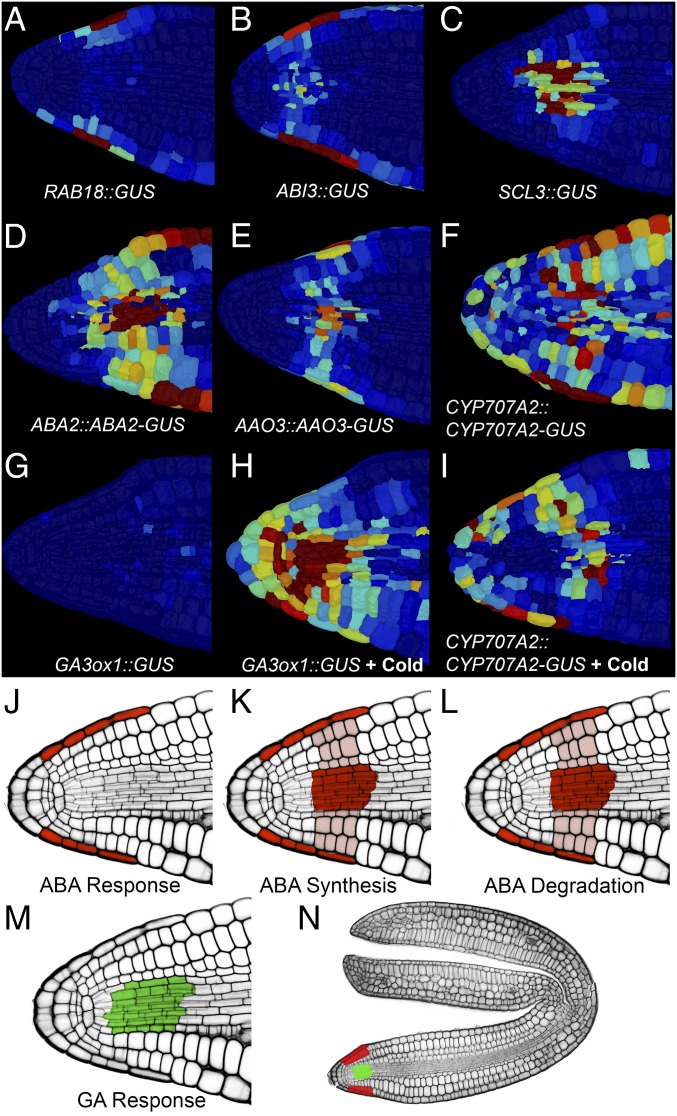
Fig. 2.
Cellular basis for developmental fate switching in primary dormant Arabidopsis seeds. The single-cell distribution of the ABA-responsive RAB18 promoter (A), the promoter of the ABA response-promoting transcription factor ABI3 (B), the GA-response proxy SCL3 promoter (C), the ABA synthesis proteins ABA2 (D) and AAO3 (E), the ABA degrading protein CYP707A2 (F), and the promoter activity of the GA synthesis enzyme GA3ox1 (G) in primary dormant embryos are shown. (H) GA3ox1 promoter activity following 1 d at 4 °C. (I) Same as H for CYP707A2 protein abundance. Schematics indicating ABA response (J), ABA synthesis (K), ABA degradation (L), and GA response (M) in the Arabidopsis radicle are shown. (N) Whole-embryo highlighting of the subset of cells enriched for ABA response (red) and GA response (green).
The activity of the ABA-responsive RAB18 promoter demonstrated the transcriptional response to this hormone to be localized to the outer cell layers of the embryo radicle, principally the root cap and epidermis (Fig. 2A). The promoter of the ABA response-promoting transcription factor ABI3 was also found to be principally within the cells of the embryonic radicle (Fig. 2B), providing a spatial overlap between this upstream regulator and the final transcriptional output of the ABA pathway.
The cellular site of GA response was identified by characterizing the localization of the activity of the promoter of SCL3. This transcription factor stimulates GA responses, and the cellular localization of this promoter activity correlates with the accumulation of GA (25) and response to GA (26) in roots. The activity of the SCL3 promoter was highly enriched within the vascular cells of the radicle (Fig. 2C), together with the GA receptors GID1A and GID1C, as well as DELLA proteins GAI and RGA (SI Appendix, Supplementary Figs. 4 and 5). These observations indicate that ABA and GA responses in nongerminating seeds occur within distinct cell types of the radicle, with the ABA response being enriched in the outer cells and the GA response within the inner cells, including the vasculature. This spatial separation of hormone responses suggests that cross-talk between ABA and GA is non–cell-autonomous and is controlled at the level of hormone movement between spatially separated signaling centers.
To understand how these distinct hormone response centers are spatially defined, we applied exogenous ABA and GA to primary dormant RAB18::GUS and SCL3::GUS seeds, respectively, to establish whether their patterns of activity were due to local hormone concentrations. Neither of these reporters showed appreciable ectopic induction in response to hormone application (SI Appendix, Supplementary Fig. 7), indicating that hormone response, and not local hormone abundance, defines the site of these signaling centers.
The spatial relationship between hormone metabolism and signaling was examined by localizing reporters to key synthesis and degradation components of these hormones. The penultimate step of ABA synthesis is catalyzed by ABA2, and the final step by AAO3. Both of these proteins were enriched within the outer cells of the radicle and within the vasculature of the radicle (Fig. 2 D and E). CYP707A2 is the primary enzyme responsible for both ABA catabolism and seed dormancy breaking in Arabidopsis seeds (27). This protein is present as well within the root cap, epidermis, and vascular cells of the radicle in dormant embryos (Fig. 2F and SI Appendix, Supplementary Figs. 4 and 5). These localizations indicate that both ABA synthesis and degradation overlap with the distinct cellular response centers to ABA and GA.
The final step of GA synthesis is catalyzed by GA 3-oxidase, and GA degradation is mediated by GA 2-oxidase. Genes encoding each of these proteins are not expressed at high levels in nongerminating Arabidopsis seeds (28, 29) (Fig. 2G and SI Appendix, Supplementary Figs. 4 and 5). The GA synthesis gene GA3ox1 is necessary for Arabidopsis dormancy breaking in response to low temperature (18) and is transcriptionally induced in response to this treatment. The spatial induction of this promoter activity in response to the germination-stimulating treatment of cold is present across diverse cell types of the radicle (Fig. 2H). The control of GA synthetic gene expression therefore overlaps with ABA and GA signaling centers. Cold treatment sustains the abundance of the CYP707A2 protein in the root cap and vasculature, with the same cells displaying high GA and ABA responses (Fig. 2I), also providing spatial overlap in the targeted control of ABA removal.
These results suggest that the abundance of ABA and GA is mediated by the modulation of hormone metabolic components within spatially separated signaling centers and that the movement of hormones between these centers mediates the cross-talk between these pathways. Supporting this model is the presence of key transporters. The AIT1 protein has been reported to transport both ABA and GA (30), and is broadly expressed at low levels throughout the primary dormant Arabidopsis embryo (SI Appendix, Supplementary Fig. 5Y). The NPF3 transporter is also a dual ABA and GA transporter (31). Despite the activity of this promoter not being detected in primary dormant seeds (SI Appendix, Supplementary Fig. 5Z), the transcript is abundant in this developmental state (28).
To determine when this distribution of decision-making components in the radicle is established, the localization of key reporters during seed development was examined. The radicle-enriched localization of decision-making components occurred during late seed development, coincident with the temporal induction of dormancy in Arabidopsis seeds during seed maturation (SI Appendix, Supplementary Fig. 8).
The radicle represents a decision-making center within the complex embryo, deconstructing signaling processing and integration at a cellular level within the context of the multicellular plant embryo body plan (Fig. 2 J–N). This site spatially overlaps with the cellular location where germination is initiated in Arabidopsis (12, 32), and where the hormone strigolactone first acts to stimulate the germination of Striga seeds (33). Fate switching in the radicle is informed by the surrounding endosperm, where all decision-making components are also present (SI Appendix, Supplementary Fig. 6), and GA synthesis is conditionally up-regulated in response to cold (18) (SI Appendix, Supplementary Figs. 5 and 6). Collectively, these observations indicate that the antagonism between ABA and GA occurs at the level of hormone metabolic feedbacks, which occur in distinct cellular response centers, and the movement of these hormones between these centers (31).
The spatial separation of antagonistically acting signaling centers imbues processing power to decision making in the human brain through the basal ganglia-cortex loop (34). This physical distance is proposed to filter variable inputs by imposing a time delay in communication between signaling centers and smoothing external signal fluctuations (35). The observation that this topological configuration is also present in a complex plant organ raises the intriguing possibility that similar topological principles are being used by humans and plants to optimize their decision making in response to variable input information (36).
We tested this possibility of a conserved role for spatial separation in processing noisy inputs by applying sets of both continuous and variable environmental inputs to primary dormant Arabidopsis seeds to establish how temperature is integrated by seeds, and whether the timing in the flipping of the fate switch is affected by fluctuations in the input signal. Strikingly, variable temperature inputs were more effective at breaking seed dormancy than continuous cold for equivalent time periods (37) (Fig. 3A). The nonlinear utilization of low-temperature signals by seeds may contrast how cold is integrated in the control of flowering time, which appears to be linear and quantitative (38). Fluctuating temperatures are therefore instructive in promoting the seed-to-seedling transition in Arabidopsis, being sought after, rather than filtered out, by the system. This result led us to pursue an understanding of the mechanism by which variable inputs can lead to the enhanced developmental fate switching in dormant seeds using our mathematical model.
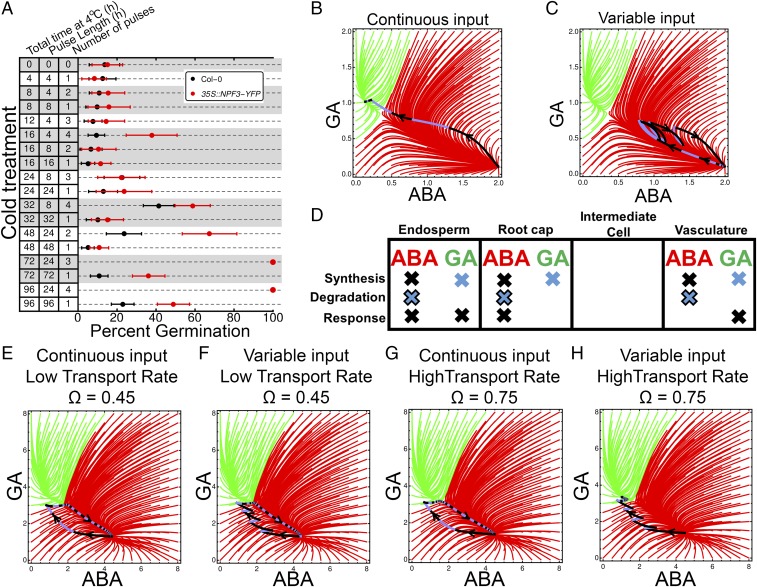
Fig. 3.
Influence of interrupted cold treatment on developmental fate switching in dormant Arabidopsis seeds. (A) Percentage germination of wild-type Col-0 and 35S::NPF3-YFP seeds with continuous or interrupted cold (4 °C) for varying lengths of time, each counted 16 d after the start of the experiment. Single-region models with continuous virtual cold input (B) and interrupted cold input (C) are shown. (D) Schematic depicting the modeled distribution of metabolic components across the four-region model, motivated by our results on spatial localization of these components. Dark crosses indicate constitutively present components, pure blue crosses indicate components induced in response to cold (GA synthesis), and blue crosses with a dark outline represent constitutively present ABA degradation that is further induced in response to the cold (18). Dynamic behavior of the four-region model with continuous input and a low hormone transport rate (0.45) (E), variable input with a low hormone transport rate (F), continuous input and a high hormone transport rate (0.75) (G), and variable input and a high hormone transport rate (H) is illustrated. For the same total cold exposure, only a varying environment (sensed by communicating, separate compartments) breaks dormancy. Intervals of temperature treatment time in B, C, and E–H are indicated by different colors on the line drawn on the attractor basin, which depicts the trajectory of the system, alternating between black and blue, with each representing a single unit of time. Each continuous temperature and alternating temperature time unit is indicated using this same color scheme. Error bars in A indicate the 95% confidence interval.
In response to cold stratification (4 °C), Arabidopsis seeds both lower their ABA levels through the further induction of CYP707A2 and increase GA levels through the initiation of GA3ox1 expression (18) (SI Appendix, Supplementary Figs. 5 and 6). We linked the abundance of these hormone metabolic nodes to the virtual application of temperature inputs to the model. We applied either four continuous units of virtual cold treatment or four cold treatments separated by an equivalent interval of time at control temperature. The trajectory the system followed is shown with a blue and black line drawn on the attractor basins in Fig. 3, with changes in the color of the line indicating the units of time of applied temperature stimulus.
The model indicated that if all processes are embedded in a single cell, continuous cold treatment is more effective than variable cold treatment in flipping the developmental fate switch following an equivalent length of low-temperature stimulus (Fig. 3 B and C). To account for our observed spatial distribution of ABA and GA metabolic and response components across the various cell types of the radicle and the endosperm of dormant seeds, we extended our model to a multicell system, where four interconnected regions are endowed with the patterns of hormone synthesis, degradation, and response components established by microscopy (Fig. 3D) and signals are exchanged between these regions through a transport rate. A bistable switch is present within the four-compartment model as in the single-cell model (Fig. 3E and SI Appendix, Supplementary Materials and Methods and Supplementary Fig. 9). In light of the microarray data used to characterize interactions between hormone response and metabolic feedbacks being generated using whole seeds, these relationships were still sufficient to generate the same output when spatially embedded. This model was also able to capture seed behavior dynamics following progressive dormancy loss through increases in GA sensitivity as observed in the single-cell model (SI Appendix, Supplementary Fig. 9), supporting the capacity of the multicell model to reflect observed seed behavior. Modifying hormone transport rates between different compartments did not drastically impact overall attractor basin architecture (SI Appendix, Supplementary Fig. 10).
The visual representation of this multidimensional system consisting of four cells and two hormones is limited in a single attractor basin 2D plot, which loses information. We note that, in some instances, this averaging can give the visual impression that a system has crossed over the threshold from one state to the other when, in the full phase space of the system, the threshold has not been passed.
When cell-to-cell communication is weak (lower transport rate), neither continuous (Fig. 3E) nor variable (Fig. 3F) cold stimuli were capable of flipping the dormancy fate switch. When cells communicated more readily (higher transport rates), continuous cold was incapable of flipping the switch (Fig. 3G), but, notably, a variable input did lead to the breaking of dormancy (Fig. 3H).
Discussion
Here, the inferred nonlinear structure of interactions means that pulses of low-temperature stimulus and relaxation are more efficient at crossing the effective ABA/GA dynamic landscape than a single stimulus of the same total length. This behavior is analogous to the increased efficiency of evolutionary progress on complex landscapes under varying environments (39).
These observations indicate that the spatial separation of hormone metabolic interactions between communicating biological compartments provides the plant with a more efficient means of integrating variable temperature inputs into the flipping of the developmental fate switch. Additionally, increasing hormone transport rates further sensitizes the system to change fate in response to fluctuating inputs. To test this theory, we made use of a transgenic line overexpressing NPF3, a dual ABA and GA transporter (31), to examine what role increasing transport rates has on integrating variable inputs leading to fate switching in primary dormant Arabidopsis seeds. The 35S::NPF3 seeds showed a greater propensity to break dormancy in response to variable inputs than the wild-type control, as predicted by the model to a greater extent than in the response to continuous cold (Fig. 3A). The prediction of our model that increased hormone transport rates further sensitize seeds to low-temperature oscillations is thus verified within Arabidopsis seeds.
We have described a distributed signal and response system to ABA and GA in dormant seeds that contributes to sensing and responding to temperature variation. This spatial embedding of molecular components within multicellular architectures can therefore provide information-processing capacity that is absent without spatial structure. This observation is consistent with the increased computational capacities of cellular consortia over their unicellular counterparts (40), and can be likened to the principles of distributed computation. The joint exploitation of the geometry of a dynamical system and its nonlinear interactions supports decision making and the stepping of variable inputs toward effecting the flipping of a developmental fate switch in seeds. Although the topological configuration at the cellular level for decision making in Arabidopsis seeds is similar to the topological configuration at the cellular level for decision making in the human brain, the exploitation of this architecture is fundamentally different in the harnessing or filtering of inputs, respectively. The recognition of oscillations in temperature associated with soil depth and changing seasons may represent an important instructive clue toward ensuring accuracy in the timing and positioning of seedling establishment (41). Temperature oscillations in the soil are greatest at the surface and dampened with increased depth, and in the case of the small-seeded species Arabidopsis, the positioning of germination close to the top of the soil is as critical as timing it across the year. The short period of the temperature oscillations and responsiveness to these oscillations suggest that such daily stimuli plays an important role in this context. This mathematical and spatially embedded model provides a cellular and molecular template to understand and reengineer seed behavior in response to a changing environment.
Materials and Methods
Plant Growth Conditions.
To produce primary dormant seeds, seeds were germinated on 0.8% 1/2 MS agar, pH 6.2, after sterilization for 5 min in 10% bleach and rinsing with water. Once germinated, seedlings were transplanted to compost and grown in greenhouse conditions with 16-h days until plants had bolted. Plants were then transferred to a growth cabinet at a constant 12 °C, with 16-h white light cycles to produce primary dormant seeds (6).
Generation of Transgenic Reporter Lines.
C-terminal GUS translational fusion lines for SLEEPY, RGA, GAI, and RGL2 were created by PCR amplification of the genomic region spanning from 2 kb upstream of the transcriptional start site until the final codon before the stop. The thymine residue of the stop codon was retained to keep the fragment in frame with the downstream protein fusion. This fragment was cloned into the GATEWAY entry vector pDNR221 and subcloned into GWB433, which contains a C-terminal GUS (42). The translational fusion of CYP707A2::CYP707A2-GUS-3′ was obtained by overlap PCR using three fragments as templates. The first fragment is the CYP707A2 genomic region, including the −1,887-bp upstream promoter region and the genomic region (excluding the stop codon); the second fragment is the GUS gene; and the third fragment is the 3′ region covering the 1,989-bp genomic region downstream of the stop codon of CYP707A2. The overlap PCR product was cloned into entry vector pDONR207 and subcloned into binary vector pMDC99 (43).
Seed Dormancy Sampling Conditions.
Primary dormant seeds were placed in 90-mm Petri dishes with three sheets of Fisherbrand filter paper (QL 100; Fisher) with sterile deionized water. Plates were placed at 22 °C in continuous white light. After 7 d, samples were dissected using a stereo binocular microscope, and embryo and endosperm samples were stained for GUS activity as described below.
Hormone Application Treatment.
For hormone application with either GA or ABA (SI Appendix, Supplementary Fig. 7), primary dormant seeds were imbibed on water for 7 d and then transferred to plates containing 50 μM GA or 50 μM ABA. Samples were then collected, stained, and imaged at the times indicated.
Dormancy Breaking Using GA and Norflurazon.
Primary dormant seeds were imbibed directly to 50 μM GA or 50 μM norflurazon, and their germination was counted after 13 d, either with or without the application of an initial cold treatment for 4 d at 4 °C (SI Appendix, Supplementary Fig. 3).
Continuous and Interrupted Cold Treatment of Seeds.
Primary dormant seeds were imbibed on water and subjected to cold treatment at 4 °C for either continuous or interrupted intervals. Each cold interval took place within the duration of a 24-h time frame, either as 24 h of continuous cold, 4 h of cold, and 20 h at 22 °C or as 8 h of cold and 16 h at 22 °C. Cold treatment lasting 24 h was followed by a 24-h period at 22 °C. The total amount of time from when seeds were first imbibed until their final germination was counted was equal (16 d) for all samples.
Statistical Analysis of Germination Data.
The 95% confidence intervals for the percentage of seeds germinated was calculated using
where is the proportion of germinated seeds, is the quantile of the standard normal distribution, and is the number of seeds tested.
Staining of Samples for Microscopy.
Dissected embryo samples were GUS-stained in X-Gluc solution consisting of 0.1 M sodium phosphate buffer (pH 7.0), 2 mM 0.1% Triton X-100, and X-Gluc for a variable time depending on the reporter used (32). Once the required level of GUS staining was attained, samples were fixed in a 3:1 solution of ethanol/acetic acid plus 500:1 DMSO, Tween 20, and Nonidet P-40 for at least 24 h. Following this treatment, cells were lysed in 0.2 N of sodium hydroxide + 1% SDS solution and kept at room temperature on a gyrotary platform until embryos turned translucent. If embryos did not turn fully translucent after this treatment, the fixing and lysis were repeated. After clearing, the embryos were incubated overnight at 37 °C in amylase to remove starch granules. After starch removal, aldehydes were produced by treating the embryos with 1% (wt/vol) periodic acid for 40 min, and cell walls could then be stained using 100 mM sodium metabisulfite in 0.15 N of HCl, mixed 9:1 with propidium iodide (44) until samples turned pink. After this final treatment, samples were cleared for at least 1 wk at a ratio of 4:1:2 by weight solution of chloral hydrate/glycerol/water.
Endosperm samples were fixed in the same way as embryos and then bleached using 25% (vol/vol) hypochlorite for 2 h to remove seed coat pigments. Samples were then mounted in Hoyer’s medium on microscope slides and imaged on a Leica ICC50HD light microscope.
Imaging and Computational Analysis of 3D Images.
Z-stacks of prepared samples were taken using an inverted Zeiss LSM710 laser-scanning confocal microscope. Samples were mounted in chloral hydrate solution (as described in the previous section) in glass-bottomed microscopy cell culture dishes (Greiner Bio One) using a 25× oil-immersion objective (LD LCI Plan-Apochromat 25×/0.8; Zeiss) and excited using an argon-ion laser. Cell wall (propidium iodide) signal was collected between 530-nm and 735-nm wavelengths, and GUS signal was collected as reflectance at between 483-nm and 493-nm wavelengths.
LSM files were then converted into tiled resulting tagged image file format (TIFF) images using Fiji (45), with separate stacks for the propidium iodide and GUS channels. The resulting TIFF images were then imported into MorphoGraphX (MGX) (23). Samples were subjected to a Gaussian image blur using a radius of 0.5, and then segmented using the auto-seeded watershed algorithm of the Insight Toolkit image-processing library. A typical threshold for the watershed was around 500, aiming to have no undersegmentation of any cells in the stack. Oversegmentation was then corrected manually, and the edited segmented stack was then used to generate a 3D mesh using the marching cubes algorithm in MGX, using a cube size of two and seven smoothing passes at the time of meshing. This mesh was then used for all downstream analyses.
For quantitative reporter concentration data, the stack containing the GUS channel was loaded into MGX along with the mesh. Using MGX’s heat map function, GUS concentration data could be obtained by reporting to a spreadsheet the “signal average” of the volumetric, “interior signal” data calculated from the mesh combined with the GUS reporter stack. For whole-axis heat maps, the same process was used. Here, images of the mesh with the heat map overlaid were taken from within MGX and exported as JPEG images. Heat maps were scaled such that the intensity of all visible cells ranged from zero (blue) to dark red.
Two-dimensional images were taken using the same Zeiss LSM710 setup, but limited to single optical sections. These images were used to highlight the distribution of reporters across the entire embryo.
Lines Used and Quantification of GUS Activity.
The lines used in this study are listed in SI Appendix, Supplementary Table 1. GUS activity was quantified using the fluorometric assays described by Stamm et al. (46).
Supplementary Material
Acknowledgments
We thank Harriet Davies for technical support. G.W.B. and A.T.T. were supported by Biotechnology and Biological Sciences Research Council (BBSRC) Grant BB/L010232/1, and G.W.B. was supported by BBSRC Grants BB/J017604/1 and BB/N009754/1. I.G.J. was supported by a Birmingham Fellowship. D.Y. and E.N. were funded by Natural Sciences and Engineering Research Council Discovery Grant RGPIN-2014-03621.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1704745114/-/DCSupplemental.
References
- 1.Domagalska MA, Leyser O. Signal integration in the control of shoot branching. Nat Rev Mol Cell Biol. 2011;12:211–221. doi: 10.1038/nrm3088. [DOI] [PubMed] [Google Scholar]
- 2.Angel A, Song J, Dean C, Howard M. A Polycomb-based switch underlying quantitative epigenetic memory. Nature. 2011;476:105–108. doi: 10.1038/nature10241. [DOI] [PubMed] [Google Scholar]
- 3.Boss PK, Bastow RM, Mylne JS, Dean C. Multiple pathways in the decision to flower: Enabling, promoting, and resetting. Plant Cell. 2004;16(Suppl):S18–S31. doi: 10.1105/tpc.015958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Finch-Savage WE, Leubner-Metzger G. Seed dormancy and the control of germination. New Phytol. 2006;171:501–523. doi: 10.1111/j.1469-8137.2006.01787.x. [DOI] [PubMed] [Google Scholar]
- 5.Finch-Savage WE, Bassel GW. Seed vigour and crop establishment: Extending performance beyond adaptation. J Exp Bot. 2016;67:567–591. doi: 10.1093/jxb/erv490. [DOI] [PubMed] [Google Scholar]
- 6.Chiang GC, Barua D, Kramer EM, Amasino RM, Donohue K. Major flowering time gene, flowering locus C, regulates seed germination in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2009;106:11661–11666. doi: 10.1073/pnas.0901367106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bentsink L, Jowett J, Hanhart CJ, Koornneef M. Cloning of DOG1, a quantitative trait locus controlling seed dormancy in Arabidopsis. Proc Natl Acad Sci USA. 2006;103:17042–17047. doi: 10.1073/pnas.0607877103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Footitt S, Douterelo-Soler I, Clay H, Finch-Savage WE. Dormancy cycling in Arabidopsis seeds is controlled by seasonally distinct hormone-signaling pathways. Proc Natl Acad Sci USA. 2011;108:20236–20241. doi: 10.1073/pnas.1116325108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karssen CM, Brinkhorst-van der Swan DL, Breekland AE, Koornneef M. Induction of dormancy during seed development by endogenous abscisic acid: Studies on abscisic acid deficient genotypes of Arabidopsis thaliana (L.) Heynh. Planta. 1983;157:158–165. doi: 10.1007/BF00393650. [DOI] [PubMed] [Google Scholar]
- 10.Bradford KJ, Trewavas AJ. Sensitivity thresholds and variable time scales in plant hormone action. Plant Physiol. 1994;105:1029–1036. doi: 10.1104/pp.105.4.1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luckwill L. Studies of fruit development in relation to plant hormones: I. Hormone production by the developing apple seed in relation to fruit drop. J Hortic Sci. 1953;28:14–24. [Google Scholar]
- 12.Bassel GW. To grow or not to grow? Trends Plant Sci. 2016;21:498–505. doi: 10.1016/j.tplants.2016.02.001. [DOI] [PubMed] [Google Scholar]
- 13.Karssen C, Lacka E. 1986. A revision of the hormone balance theory of seed dormancy: Studies on gibberellin and/or abscisic acid-deficient mutants of Arabidopsis thaliana. Plant Growth Substances 1985, Proceedings in Life Sciences, ed Bopp M, (Springer, Berlin), pp 315–323.
- 14.Tyson JJ, Chen KC, Novak B. Sniffers, buzzers, toggles and blinkers: Dynamics of regulatory and signaling pathways in the cell. Curr Opin Cell Biol. 2003;15:221–231. doi: 10.1016/s0955-0674(03)00017-6. [DOI] [PubMed] [Google Scholar]
- 15.Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: Cell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- 16.Seo M, et al. Regulation of hormone metabolism in Arabidopsis seeds: Phytochrome regulation of abscisic acid metabolism and abscisic acid regulation of gibberellin metabolism. Plant J. 2006;48:354–366. doi: 10.1111/j.1365-313X.2006.02881.x. [DOI] [PubMed] [Google Scholar]
- 17.Ogawa M, et al. Gibberellin biosynthesis and response during Arabidopsis seed germination. Plant Cell. 2003;15:1591–1604. doi: 10.1105/tpc.011650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yamauchi Y, et al. Activation of gibberellin biosynthesis and response pathways by low temperature during imbibition of Arabidopsis thaliana seeds. Plant Cell. 2004;16:367–378. doi: 10.1105/tpc.018143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Toni T, Stumpf MP. Simulation-based model selection for dynamical systems in systems and population biology. Bioinformatics. 2010;26:104–110. doi: 10.1093/bioinformatics/btp619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hauvermale AL, Tuttle KM, Takebayashi Y, Seo M, Steber CM. Loss of Arabidopsis thaliana seed dormancy is associated with increased accumulation of the GID1 GA hormone receptors. Plant Cell Physiol. 2015;56:1773–1785. doi: 10.1093/pcp/pcv084. [DOI] [PubMed] [Google Scholar]
- 21.Debeaujon I, Koornneef M. Gibberellin requirement for Arabidopsis seed germination is determined both by testa characteristics and embryonic abscisic acid. Plant Physiol. 2000;122:415–424. doi: 10.1104/pp.122.2.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee KP, Piskurewicz U, Turecková V, Strnad M, Lopez-Molina L. A seed coat bedding assay shows that RGL2-dependent release of abscisic acid by the endosperm controls embryo growth in Arabidopsis dormant seeds. Proc Natl Acad Sci USA. 2010;107:19108–19113. doi: 10.1073/pnas.1012896107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barbier de Reuille P, et al. MorphoGraphX: A platform for quantifying morphogenesis in 4D. eLife. 2015;4:05864. doi: 10.7554/eLife.05864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Montenegro-Johnson TD, et al. Digital single-cell analysis of plant organ development using 3DCellAtlas. Plant Cell. 2015;27:1018–1033. doi: 10.1105/tpc.15.00175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shani E, et al. Gibberellins accumulate in the elongating endodermal cells of Arabidopsis root. Proc Natl Acad Sci USA. 2013;110:4834–4839. doi: 10.1073/pnas.1300436110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ubeda-Tomás S, et al. Root growth in Arabidopsis requires gibberellin/DELLA signalling in the endodermis. Nat Cell Biol. 2008;10:625–628. doi: 10.1038/ncb1726. [DOI] [PubMed] [Google Scholar]
- 27.Okamoto M, et al. CYP707A1 and CYP707A2, which encode abscisic acid 8′-hydroxylases, are indispensable for proper control of seed dormancy and germination in Arabidopsis. Plant Physiol. 2006;141:97–107. doi: 10.1104/pp.106.079475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cadman CS, Toorop PE, Hilhorst HW, Finch-Savage WE. Gene expression profiles of Arabidopsis Cvi seeds during dormancy cycling indicate a common underlying dormancy control mechanism. Plant J. 2006;46:805–822. doi: 10.1111/j.1365-313X.2006.02738.x. [DOI] [PubMed] [Google Scholar]
- 29.Bassel GW, et al. Elucidating the germination transcriptional program using small molecules. Plant Physiol. 2008;147:143–155. doi: 10.1104/pp.107.110841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kanno Y, et al. Identification of an abscisic acid transporter by functional screening using the receptor complex as a sensor. Proc Natl Acad Sci USA. 2012;109:9653–9658. doi: 10.1073/pnas.1203567109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tal I, et al. The Arabidopsis NPF3 protein is a GA transporter. Nat Commun. 2016;7:11486. doi: 10.1038/ncomms11486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bassel GW, et al. Mechanical constraints imposed by 3D cellular geometry and arrangement modulate growth patterns in the Arabidopsis embryo. Proc Natl Acad Sci USA. 2014;111:8685–8690. doi: 10.1073/pnas.1404616111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsuchiya Y, et al. PARASITIC PLANTS. Probing strigolactone receptors in Striga hermonthica with fluorescence. Science. 2015;349:864–868. doi: 10.1126/science.aab3831. [DOI] [PubMed] [Google Scholar]
- 34.Bogacz R, Gurney K. The basal ganglia and cortex implement optimal decision making between alternative actions. Neural Comput. 2007;19:442–477. doi: 10.1162/neco.2007.19.2.442. [DOI] [PubMed] [Google Scholar]
- 35.Bogacz R. Optimal decision-making theories: Linking neurobiology with behaviour. Trends Cogn Sci. 2007;11:118–125. doi: 10.1016/j.tics.2006.12.006. [DOI] [PubMed] [Google Scholar]
- 36.Fitts PM. The information capacity of the human motor system in controlling the amplitude of movement. J Exp Psychol. 1954;47:381–391. [PubMed] [Google Scholar]
- 37.Morinaga T. Effect of alternating temperatures upon the germination of seeds. Am J Bot. 1926;13:141–158. [Google Scholar]
- 38.Angel A, et al. Vernalizing cold is registered digitally at FLC. Proc Natl Acad Sci USA. 2015;112:4146–4151. doi: 10.1073/pnas.1503100112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kashtan N, Alon U. Spontaneous evolution of modularity and network motifs. Proc Natl Acad Sci USA. 2005;102:13773–13778. doi: 10.1073/pnas.0503610102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Regot S, et al. Distributed biological computation with multicellular engineered networks. Nature. 2011;469:207–211. doi: 10.1038/nature09679. [DOI] [PubMed] [Google Scholar]
- 41.Thompson K, Grime JP, Mason G. Seed germination in response to diurnal fluctuations of temperature. Nature. 1977;267:147–149. doi: 10.1038/267147a0. [DOI] [PubMed] [Google Scholar]
- 42.Nakagawa T, et al. Improved Gateway binary vectors: High-performance vectors for creation of fusion constructs in transgenic analysis of plants. Biosci Biotechnol Biochem. 2007;71:2095–2100. doi: 10.1271/bbb.70216. [DOI] [PubMed] [Google Scholar]
- 43.Curtis MD, Grossniklaus U. A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 2003;133:462–469. doi: 10.1104/pp.103.027979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Truernit E, et al. High-resolution whole-mount imaging of three-dimensional tissue organization and gene expression enables the study of Phloem development and structure in Arabidopsis. Plant Cell. 2008;20:1494–1503. doi: 10.1105/tpc.107.056069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schindelin J, et al. Fiji: An open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stamm P, et al. The transcription factor ATHB5 affects GA-mediated plasticity in hypocotyl cell growth during seed germination. Plant Physiol. 2017;173:907–917. doi: 10.1104/pp.16.01099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nakabayashi K, Okamoto M, Koshiba T, Kamiya Y, Nambara E. Genome-wide profiling of stored mRNA in Arabidopsis thaliana seed germination: Epigenetic and genetic regulation of transcription in seed. Plant J. 2005;41:697–709. doi: 10.1111/j.1365-313X.2005.02337.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.