Abstract
Osteosarcoma survival rate has not improved over the past three decades, and the debilitating side effects of the surgical treatment suggest the need for alternative local control approaches. Radiotherapy is largely ineffective in osteosarcoma, indicating a potential role for radiosensitizers. Blocking DNA repair, particularly by inhibiting the catalytic subunit of DNA-dependent protein kinase (DNA-PKCS), is an attractive option for the radiosensitization of osteosarcoma. In this study, the expression of DNA-PKCS in osteosarcoma tissue specimens and cell lines was examined. Moreover, the small molecule DNA-PKCS inhibitor, KU60648, was investigated as a radiosensitizing strategy for osteosarcoma cells in vitro. DNA-PKCS was consistently expressed in the osteosarcoma tissue specimens and cell lines studied. Additionally, KU60648 effectively sensitized two of those osteosarcoma cell lines (143B cells by 1.5-fold and U2OS cells by 2.5-fold). KU60648 co-treatment also altered cell cycle distribution and enhanced DNA damage. Cell accumulation at the G2/M transition point increased by 55% and 45%, while the percentage of cells with >20 γH2AX foci were enhanced by 59% and 107% for 143B and U2OS cells, respectively. These results indicate that the DNA-PKCS inhibitor, KU60648, is a promising radiosensitizing agent for osteosarcoma.
Keywords: DNA-PK, DNA-PKcs, osteosarcoma, radiotherapy, radiosensitizers, KU60648
Introduction
Osteosarcoma (OS) is the most common malignant bone tumor, and is treated with surgery and chemotherapy [1]. The debilitating side effects of surgical treatment suggest that adding radiotherapy could potentially improve patient outcomes [2]. One approach to amplify the effects of ionizing radiation (IR) is through radiosensitizers. Sulforaphane, zoledronic acid and parthenolide are naturally occurring agents that enhanced the effects of IR in OS [3–5]. In addition, targeting molecular pathways by inhibiting RAD51, WEE1 or histone deacetylases (HDAC) have shown promise in radiosensitizing OS [6–8].
DNA repair is critical for recovery of IR induced damage and disrupting repair can increase cell killing [9]. IR induces DNA double-strand breaks (DSB), which are primarily repaired by non-homologous end joining (NHEJ) [10]. The catalytic subunit of DNA-dependent protein kinase (DNA-PKCS), which belongs to the phosphatidylinositol 3-kinase-related kinases (PIKKs) family, is an essential component of NHEJ [11]. Mutations in PRKDC, the gene that codes for DNA-PKCS, result in defects in NHEJ causing hypersensitivity to IR [12]. Hence, small molecule inhibitors of DNA-PKCS could be promising radiosensitizers for human OS cells [13].
Knocking down DNA-PKCS in OS cells enhances the response to IR [14]. While some studies have explored the use of DNA-PKCS inhibitors to sensitize OS cells to chemotherapy [15, 16], the role of these inhibitors in sensitizing OS cells to IR remains to be investigated. KU60648 is a recently developed potent small molecule DNA-PKCS inhibitor [17]. In this study, the potential of KU60648 to sensitize OS cells to IR is explored.
Materials and Methods
Next Generation RNA Sequencing
The RNASeq analysis was performed on OS tissue and cell lines and their DNA-PKCS mRNA was compared with expression in other bone tumors (chondrosarcoma, Ewing’s sarcoma, chordoma and chondroblastoma). All human specimens were collected under protocols approved by the Mayo Clinic Institutional Review Board (IRB). Each specimen was evaluated by a board-certified musculoskeletal pathologist. Once collected, the tumor specimens were immediately snap-frozen using liquid nitrogen, and later ground into a powder using a hammer. RNA from three OS and four Ewing’s sarcoma cell lines were also evaluated. For both tissues and cell lines, total RNA was harvested using the miRNeasy minikit (Qiagen, Hilden, Germany). Total RNA was quantified using the NanoDrop 2000 spectrophotometer (Thermo Fischer Scientific, Wilmington, DE). RNA sequencing and bioinformatics analysis were performed as previously described [18, 19]. Gene expression, normalized to one million reads and corrected for gene length, was reported in terms of reads per kilobase per million mapped reads (RPKM).
Cell Culture, Irradiation and Compound
Human OS cells (143B, U2OS, Saos-2 and Hos) as well as human osteoblast (HOB) cells were cultured in Dulbecco’s Advanced Modified Eagles Media – F12 (DMEM-F12) with 10% fetal bovine serum (FBS), 100 units/ml of penicillin/streptomycin and kept at 37°C under 5 % CO2. The OS cells were obtained from ATCC, while the HOB cells were established from cancellous bone obtained as surgical waste from orthopedic procedures following a protocol approved by the Mayo Clinic IRB as previously described [20]. All IR treatments were performed using a cesium 137 (137Cs) irradiator. Cells in six-well plates were exposed to specified dose of IR. The DNA-PKCS inhibitor, KU60648, was kindly provided by KuDOS Pharmaceuticals and was dissolved in DMSO as a 1 mM stock solution.
Western Blotting
Cytoplasmic protein extracts were loaded onto 3 – 8% Criterion™ Tris-acetate gels (BioRad, Hercules, CA) and subsequently transferred to PVDF membranes. High molecular weight marker, HiMark™ Protein Standard (ThermoFischer Scientific, Waltham, MA) was used for identifying DNA-PKCS. Antibodies used included anti-DNA-PKCS (NA57, Calbiochem, EMD Biosciences, Billerica, MA), phospho-S2056-DNA-PKCS (ab18192, Abcam, Cambridge, MA) and vinculin (E1E9V, Cell Signaling Technology, Beverly, MA). Inhibition of DNA-PKCS autophosphorylation was determined in cells cultured in the presence of graded concentrations of KU60648 one hour before treatment with IR (10 Gy) followed by cell lysis 30 minutes later. DNA-PKCS autophosphorylation at Ser2056 and total DNA-PKCS levels were assessed by western blotting as previously described [17].
Clonogenic Assay
Appropriate numbers of cells were counted and plated in six-well plates, and treated with vehicle control or 300 nM KU60648 one hour prior to IR treatment. Cells were then incubated with and without the drug for an additional 24 hours, rinsed with phosphate-buffered saline (PBS) and incubated in fresh media for 7–12 days to allow for colony formation. After staining with crystal violet, colonies with ≥ 50 cells were quantified and survival analysis was performed.
Cell Cycle Analysis
Cells plated in six-well plates were cultured overnight prior to adding vehicle control or 100 nM KU60648, and were treated with IR one hour later. The cells were then incubated for 24 hours and subsequently trypsinized, washed with ice-cold PBS, cell pellets were suspended in equal volume of chilled 95% ethanol and stored at 4°C until further processing. Cells were centrifuged, washed twice to remove ethanol content, and subsequently suspended in PBS containing RNase A (1.5 mg/ml; Sigma-Aldrich, St. Louis, MO). After incubation at 37°C for 15–30 minutes, propidium iodide (100 μg/ml; Roche, Indianapolis, IN) was added and cell cycle distribution was measured using fluorescence-activated cell sorting (FACS) by the Mayo Clinic Flow Cytometry Core Lab and data were analyzed using the ModFit software.
Immunocytochemistry
Cells were cultured overnight on coverslips (placed in six-well plates) prior to treatment with vehicle control or 100 nM KU60648 and irradiated one hour later. After 24 hours, cells were rinsed with PBS, fixed with 4% paraformaldehyde, permeabilized with 0.5% Triton X-100 and stained with anti-γH2AX (phospho-Ser139, 2577, Cell Signaling Technology) in 1% bovine serum albumin (BSA) at 4°C overnight. Cells were then washed with PBS and incubated with goat anti-rabbit Alexa Fluor 488 (A-11008, ThermoFisher Scientific). Finally, stained cells were mounted with DAPI containing mounting medium (H-1500, Vector Labs, Burlingame, CA) and imaged by confocal microscopy (Zeiss LSM510). Nuclei positive for γH2AX foci were quantified. For each group, a total of >150 and >90 nuclei were counted for 143B and U2OS cells, respectively.
Statistical Analysis
Unless stated otherwise, data presented are mean ± standard deviation (SD) from three or more experiments. Two-tailed Student’s t-tests were used for statistical analysis and p<0.05 was considered significant. Fitting clonogenic survival data to linear-quadratic (LQ) model was performed using CS-Cal software from German Cancer Research Center (http://angiogenesis.dkfz.de/oncoexpress/software/cs-cal/index.htm). Survival fraction (SF) is related to dose d (Gy) by SF = e−(αd+βd2), where α and β are experimentally derived parameters.
Results
DNA-PKCS Expression and Inhibition in OS
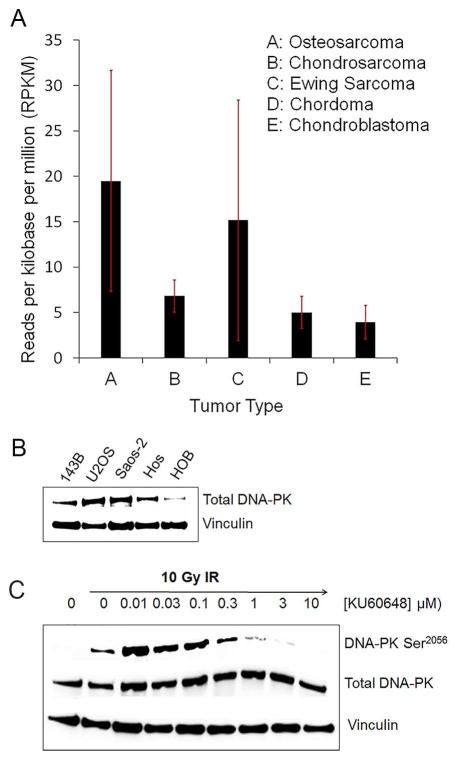
RNASeq analysis showed that DNA-PKCS mRNA was expressed at levels greater than 1 RPKM in all tumor specimens studied. OS specimens had the highest level of expression, while specimens for chondroblastoma, a benign bone tumor, had the lowest level of expression (Fig. 1A). Additionally, there was a higher level of DNA-PKCS protein expression in all OS cells compared to the non-cancerous HOB cells (Fig. 1B). The autophosphorylation induced in response to IR was greatly reduced by KU60648 treatment starting at 300 nM (Fig. 1C). This suggests that KU60648 is effective at inhibiting DNA-PKCS in OS cells.
Fig. 1. DNA-PKCS expression and inhibition in human OS tissue and cells.
A) RNASeq analysis of DNA-PKCS mRNA in primary bone tumor specimens and cell lines. Specimens included four chondrosarcomas, eight chondroblastomas, five chordoma, five Ewing’s sarcoma (one tissue and four cell lines) and four OS (one tissue and three cell lines). B) Total DNA-PKCS protein levels in OS cell lines compared with HOB cells. C) Levels of DNA-PKCS autophosphorylation at Ser2056 induced with IR (10Gy) and with graded concentration of KU60648, in 143B cells. Results are representative of three independent experiments.
KU60648 Sensitizes Human OS Cells to IR
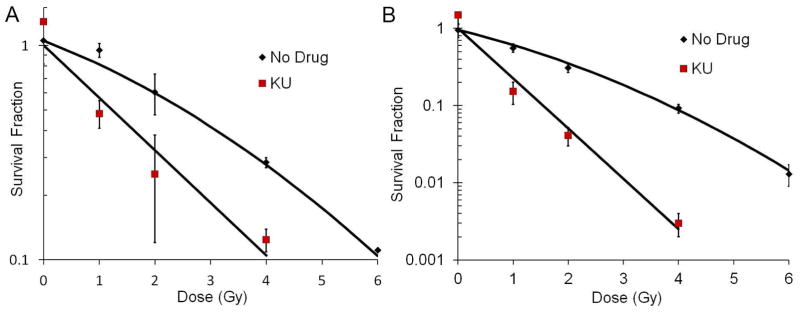
Treatment of human OS cells, 143B and U2OS, with KU60648 sensitized them to IR (Fig. 2A and B). Fitting the curves to the LQ model, α and β values for 143B and U2OS cells were α = 0.230, β = 0.256 (α/β ratio = 8.9) and α = 0.39, β = 0.05 (α/β ratio = 7.8), respectively. With KU60648 co-treatment, α values increased to 0.56 (2.4-fold) and 1.5 (3.8-fold) for 143B and U2OS cells, respectively. The survival curves with KU60648 co-treatment did not exhibit a shoulder, and hence β values approach zero and could not be accurately determined. Additionally, sensitization enhancement ratio at 10% survival (SER10) was calculated as the ratio of LD10 (lethal dose at 10% survival) without drug to LD10 with drug. With KU60648 co-treatment, the SER10 was 1.5 and 2.5 for 143B and U2OS cells, respectively. Similarly, KU60648 led to a 2.4-fold and 7.8-fold reduction in survival (at 2 Gy) for 143B and U2OS cells, respectively. KU60648 treatment alone was similar to vehicle control (normalized to 1 in the clonogenic curves). These results indicate that KU60648 greatly potentiates IR induced killing of OS cells in vitro.
Fig. 2. KU60648 sensitizes 143B and U2OS cells to IR.
Clonogenic survival for 143B cells (A) and U2OS cells (B) treated with vehicle control or 300 nM KU60648 for one hour pre-IR and 24 hours post-IR, rinsed with PBS, media changed and incubated for colony formation. Results are the mean ± SD of three independent experiments fitted to the LQ model.
Combining KU60648 with IR Alters Cell Cycle Distribution in Human OS cells
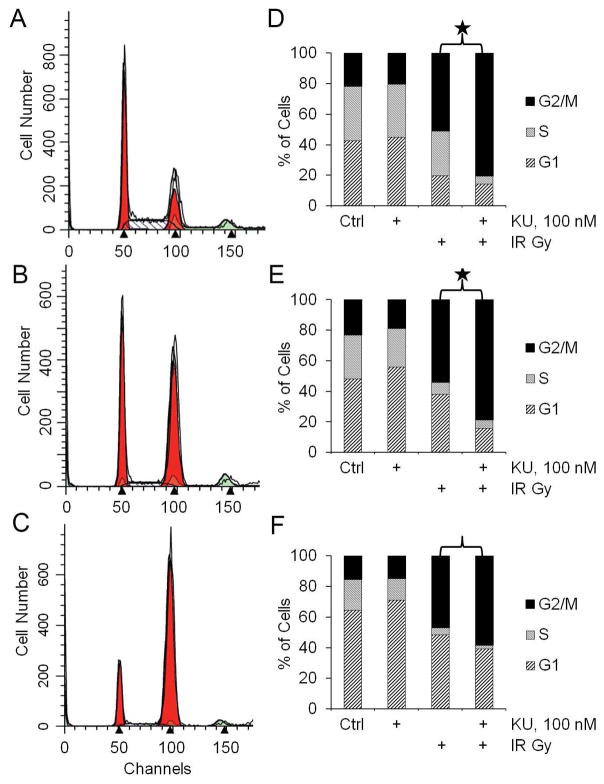
Treatment with KU60648 caused changes in cell cycle distribution when combined with IR. In the representative histograms in Fig. 3A–C (for U2OS cells), IR alone caused an increase in cell population at the G2/M transition point (Fig. 3B), which was further enhanced with KU60648 treatment (Fig. 3C). KU60648 treatment alone was similar to vehicle control. Comprehensive analysis of the results shown in Figs. 3D–F suggest that KU60648 co-treatment led to a statistically significant (P < 0.05) percentage increase in G2/M accumulation (55% and 45% in 143B and U2OS cells, respectively) compared to IR alone (Figs. 3D and E). The percentage increase of G2/M accumulation in HOB cells (Fig. 3F) was not statistically significant (P = 0.08).
Fig. 3. KU60648 enhances G2/M accumulation when combined with IR in human OS cells.
A–C) FACS histograms for U2OS cells treated with vehicle control (A), 5 Gy (B), and 5 Gy plus 100 nM KU60648 (C). Results are representative of three independent experiments. D–F) The summary of the cell cycle analyses for 143B cells (D), U2OS cells (E) and HOB cells (F). Results are mean ± SD of three or more independent experiments. (* P < 0.05)
Combining KU60648 with IR Increases DNA Damage in Human OS cells
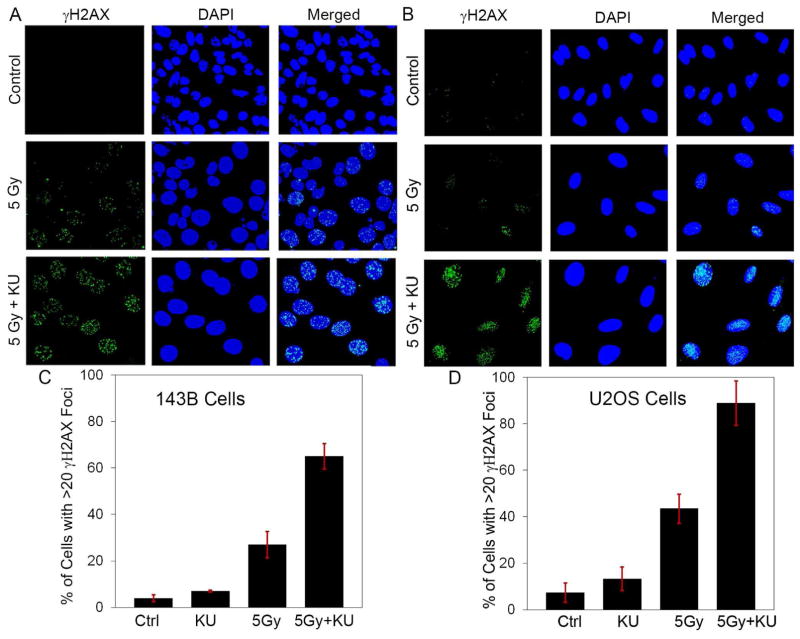
IR treatment led to increased levels of γH2AX foci, which was further enhanced by co-treatment with KU60648. Treatment with KU60648 one hour before IR enhanced the extent of γH2AX foci observed 24 hours after IR treatment (Figs. 4A and B). KU60648 co-treatment increased the percentage of cells with > 20 γH2AX foci from 27.0 ± 5.6 to 65.0 ± 5.5, for 143B cells (Fig. 4C), and from 43.5 ± 6.2 to 88.8 ± 9.6, for U2OS cells (Fig. 4D), compared to IR treatment alone. KU60648 treatment alone was similar to vehicle control. This increase in the fraction of cells with persistent γH2AX foci with KU60648 co-treatment indicates that KU60648 potentiates the DNA damage induced by IR by inhibiting DNA repair.
Fig. 4. KU60648 enhances DNA damage when combined with IR in human OS cells.
Representative confocal microscopy images of γH2AX foci 24 hours post-IR in cells treated with vehicle control, 5 Gy or 5 Gy plus 100 nM KU60648 (added 1 hour before IR) in 143B cells (A) and U2OS cells (B). Quantification of percentage of cells with > 20 γH2AX foci in cells treated with vehicle control, 100 nM KU60648 alone, 5 Gy or 5 Gy plus 100 nM KU60648 in 143B cells (C) and U2OS cells (D). Results are mean ± SD from 2 independent experiments. For each treatment group, >150 and >90 nuclei were counted for 143B and U2OS cells, respectively.
Discussion
The 5-year overall survival for extremity OS is 65–70%, while it is lower for pelvic and spinal OS [21]. For pelvic and spinal OS, surgery is more likely to lead to debilitating morbidity and higher local failure rates [22]. Radiosensitizers could potentially enhance the efficacy of radiotherapy in OS as an adjunct to surgery. Inhibiting key regulators of DNA repair has become an attractive approach to enhance the cytotoxic effects of IR [23]. DNA-PKCS is an essential component of NHEJ, the major DNA repair mechanism in IR-induced DSB [24]. Hence, inhibiting DNA-PKCS has been explored in sensitizing various cancer cells to IR [25]. In the current study, DNA-PKCS was shown to be consistently expressed in OS and the small molecule DNA-PKCS inhibitor, KU60648, effectively sensitized OS cells to IR. KU60648 co-treatment with IR also led to increased G2/M accumulation and DNA damage.
Co-treatment with KU60648 suppressed IR-induced DNA-PKCS autophosphorylation in 143B cells (Fig. 1C), consistent with the potential of the drug to radiosensitize OS cells. Moreover, RNASeq analysis indicated that OS specimens had higher levels of DNA-PKCS mRNA expression compared to the benign tumor chondroblastoma (Fig. 1A). Interestingly, specimens from Ewing’s sarcoma (ES), an IR-responsive primary bone tumor, also had high levels of DNA-PKCS mRNA expression, suggesting that DNA-PKCS inhibitors may play a role in ES treatment. Previously, Feng et al have suggested that DNA-PKCS associates with ETS gene fusions, providing a rationale for treating ES with DNA-PKCS inhibitors [26]. Taken together, the expression data in Fig. 1 indicate that DNA-PKCS can be targeted in OS treatment, but it is not entirely clear whether the level of expression correlates with the level of IR sensitivity.
The LQ model (SF = e−(αd+βd2)) is a standard method to model the dose-response relationship of clonogenic survival data from IR experiments [27]. The α and β parameters, which represent the linear and quadratic components of the curve, can provide insight into the mechanism of radiosensitization [28]. Franken et al. showed that radiosensitizers could increase α, β or both, and hence affect the α/β ratio, depending on whether they influence direct lethal damage, potentially lethal damage (PLD) or sublethal damage (SLD) [28]. Inhibiting DNA-PKcs is expected to mainly increase the β component based on the importance of DNA-PKcs on the repair of SLD [29]. As expected, at the IR doses used with KU60648 co-treatment (≤4 Gy), the survival curves did not exhibit a shoulder and β approached zero. The increase in the α value observed with KU60648 co-treatment (2.4 and 3.8-fold for 143B and U2OS cells, respectively), indicates there might also be enhanced PLD at low radiation doses. While β cannot be accurately calculated in this context, the α/β ratio becomes quite large. These results are all consistent with a radiosensitizing effect associated with the inhibition of DNA repair capacity.
Radiosensitization effects were also analyzed by comparing SER10 and survival fraction at 2 Gy. While both OS cell lines studied were effectively sensitized by KU60648, U2OS cells were sensitized to a higher degree (SER10=2.5) than 143B cells (SER10=1.5). The survival fraction at 2 Gy was also reduced to a higher degree for U2OS cells (7.5-fold) compared to 143B cells (2.4-fold). The difference might be attributed to the higher level of DNA-PKCS expression seen in U2OS cells compared to 143B cells (Fig. 1B). The other difference between the two cell lines is that U2OS is wild type for the p53 tumor suppressor gene (TP53) with active p53 proteins, while 143B is p53 mutant [30–32]. There are multiple studies to suggest that p53 wild-type cells may be more sensitive given the role of p53 in regulating DNA repair, cell cycle arrest and cell survival [33, 34]. Additional differences in DNA repair capacity due to the other arm of the DNA repair machinery, i.e. homologous recombination (HR), could also possibly explain the difference in the two cell lines.
Similar to the findings with other DNA-PKCS inhibitors [35], the radiosensitizing effect of KU60648 in OS cells is accompanied by a change in cell cycle distribution and a rise in DNA damage. While the increase in G2/M accumulation is comparable in the two cell lines (55% vs 45%), the increase in γH2AX foci is more pronounced in U2OS cells (107%) compared to 143B cells (59%). This agrees very well with the relative difference discussed above in the radiosensitization effects, and could be explained in a similar way. Moreover, the significant difference in the G2/M accumulation for the two OS cell lines compared with the control HOB cells indicates that the effect of DNA-PKCS inhibition could be more specific to tumor cells.
In conclusion, KU60648, a potent DNA-PKCS inhibitor, is an effective radiosensitizing agent for OS at much lower concentrations than those reported for other DNA-PKCS inhibitors [35]. However, given that DNA-PKcs is expressed at some level in all normal cells such as the gut and skin, future work should explore the use of drug delivery vehicles such as nanoparticles in the clinical translation of KU60648 for osteosarcoma radiotherapy.
Supplementary Material
Highlights.
DNA-PKcs is consistently expressed in human osteosarcoma tissue and cell lines
The DNA-PKcs inhibitor, KU60648, effectively radiosensitizes osteosarcoma cells
Combining KU60648 with radiation increases G2/M accumulation and DNA damage
Acknowledgments
We thank Drs. Matthew Callstrom, Scott Kaufmann, Zhenkun Lou, Krzysztof Matyjaszewski and Joel Reid for their thoughtful advice. In addition, we thank members of our laboratories, including Dr. Gasper Kitange, Dr. Lichun Lu and Dr. Lee Miller, Jim Herrick, Brian Waletzki, Sofia Jerez and Hector Araya for their support. We also thank the Mayo Clinic Flow Cytometry Core for help with FACS and cell cycle analysis.
Funding: This work was supported by the National Institutes of Health [1F31CA206388-01A1, 2016–2019; 5T32AR56950-7, 2015, T32 GM072474-10, 2015], and the John and Posy Krehbiel Endowed Professorship of Orthopedic Surgery and Biomedical Engineering [MJY, 2004-present].
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bishop MW, Janeway KA, Gorlick R. Future directions in the treatment of osteosarcoma. Curr Opin Pediatr. 2016;28:26–33. doi: 10.1097/MOP.0000000000000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.DeLaney TF, Park L, Goldberg SI, Hug EB, Liebsch NJ, Munzenrider JE, Suit HD. Radiotherapy for local control of osteosarcoma. Int J Radiat Oncol Biol Phys. 2005;61:492–498. doi: 10.1016/j.ijrobp.2004.05.051. [DOI] [PubMed] [Google Scholar]
- 3.Sawai Y, Murata H, Horii M, Koto K, Matsui T, Horie N, Tsuji Y, Ashihara E, Maekawa T, Kubo T, Fushiki S. Effectiveness of sulforaphane as a radiosensitizer for murine osteosarcoma cells. Oncol Rep. 2013;29:941–945. doi: 10.3892/or.2012.2195. [DOI] [PubMed] [Google Scholar]
- 4.Zuch D, Giang AH, Shapovalov Y, Schwarz E, Rosier R, O’Keefe R, Eliseev RA. Targeting radioresistant osteosarcoma cells with parthenolide. J Cell Biochem. 2012;113:1282–1291. doi: 10.1002/jcb.24002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim EH, Kim MS, Lee KH, Koh JS, Jung WG, Kong CB. Zoledronic acid is an effective radiosensitizer in the treatment of osteosarcoma. Oncotarget. 2016 doi: 10.18632/oncotarget.12281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Du LQ, Wang Y, Wang H, Cao J, Liu Q, Fan FY. Knockdown of Rad51 expression induces radiation- and chemo-sensitivity in osteosarcoma cells. Med Oncol. 2011;28:1481–1487. doi: 10.1007/s12032-010-9605-1. [DOI] [PubMed] [Google Scholar]
- 7.PosthumaDeBoer J, Wurdinger T, Graat HC, van Beusechem VW, Helder MN, van Royen BJ, Kaspers GJ. WEE1 inhibition sensitizes osteosarcoma to radiotherapy. BMC Cancer. 2011;11:156. doi: 10.1186/1471-2407-11-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blattmann C, Thiemann M, Stenzinger A, Christmann A, Roth E, Ehemann V, Debus J, Kulozik AE, Weichert W, Huber PE, Oertel S, Abdollahi A. Radiosensitization by histone deacetylase inhibition in an osteosarcoma mouse model. Strahlenther Onkol. 2013;189:957–966. doi: 10.1007/s00066-013-0372-8. [DOI] [PubMed] [Google Scholar]
- 9.Matt S, Hofmann TG. The DNA damage-induced cell death response: a roadmap to kill cancer cells. Cell Mol Life Sci. 2016;73:2829–2850. doi: 10.1007/s00018-016-2130-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mladenov E, Magin S, Soni A, Iliakis G. DNA double-strand break repair as determinant of cellular radiosensitivity to killing and target in radiation therapy. Front Oncol. 2013;3:113. doi: 10.3389/fonc.2013.00113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goodwin JF, Knudsen KE. Beyond DNA repair: DNA-PK function in cancer. Cancer Discov. 2014;4:1126–1139. doi: 10.1158/2159-8290.CD-14-0358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lees-Miller SP, Godbout R, Chan DW, Weinfeld M, Day RS, 3rd, Barron GM, Allalunis-Turner J. Absence of p350 subunit of DNA-activated protein kinase from a radiosensitive human cell line. Science. 1995;267:1183–1185. doi: 10.1126/science.7855602. [DOI] [PubMed] [Google Scholar]
- 13.Davidson D, Amrein L, Panasci L, Aloyz R. Small Molecules, Inhibitors of DNA-PK, Targeting DNA Repair, and Beyond. Front Pharmacol. 2013;4:5. doi: 10.3389/fphar.2013.00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tang X, Yuan F, Guo K. Repair of radiation damage of U2OS osteosarcoma cells is related to DNA-dependent protein kinase catalytic subunit (DNA-PKcs) activity. Mol Cell Biochem. 2014;390:51–59. doi: 10.1007/s11010-013-1955-5. [DOI] [PubMed] [Google Scholar]
- 15.Li X, Tian J, Bo Q, Li K, Wang H, Liu T, Li J. Targeting DNA-PKcs increased anticancer drug sensitivity by suppressing DNA damage repair in osteosarcoma cell line MG63. Tumour Biol. 2015;36:9365–9372. doi: 10.1007/s13277-015-3642-5. [DOI] [PubMed] [Google Scholar]
- 16.Zhen YF, Li ST, Zhu YR, Wang XD, Zhou XZ, Zhu LQ. Identification of DNA-PKcs as a primary resistance factor of salinomycin in osteosarcoma cells. Oncotarget. 2016 doi: 10.18632/oncotarget.12712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Munck JM, Batey MA, Zhao Y, Jenkins H, Richardson CJ, Cano C, Tavecchio M, Barbeau J, Bardos J, Cornell L, Griffin RJ, Menear K, Slade A, Thommes P, Martin NM, Newell DR, Smith GC, Curtin NJ. Chemosensitization of cancer cells by KU-0060648, a dual inhibitor of DNA-PK and PI-3K. Mol Cancer Ther. 2012;11:1789–1798. doi: 10.1158/1535-7163.MCT-11-0535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin Y, Lewallen EA, Camilleri ET, Bonin CA, Jones DL, Dudakovic A, Galeano-Garces C, Wang W, Karperien MJ, Larson AN, Dahm DL, Stuart MJ, Levy BA, Smith J, Ryssman DB, Westendorf JJ, Im HJ, van Wijnen AJ, Riester SM, Krych AJ. RNA-seq analysis of clinical-grade osteochondral allografts reveals activation of early response genes. J Orthop Res. 2016;34:1950–1959. doi: 10.1002/jor.23209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kalari KR, Nair AA, Bhavsar JD, O’Brien DR, Davila JI, Bockol MA, Nie J, Tang X, Baheti S, Doughty JB, Middha S, Sicotte H, Thompson AE, Asmann YW, Kocher JP. MAP-RSeq: Mayo Analysis Pipeline for RNA sequencing. BMC Bioinformatics. 2014;15:224. doi: 10.1186/1471-2105-15-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wimbauer F, Yang C, Shogren KL, Zhang M, Goyal R, Riester SM, Yaszemski MJ, Maran A. Regulation of interferon pathway in 2-methoxyestradiol-treated osteosarcoma cells. BMC Cancer. 2012;12:93. doi: 10.1186/1471-2407-12-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kansara M, Teng MW, Smyth MJ, Thomas DM. Translational biology of osteosarcoma. Nat Rev Cancer. 2014;14:722–735. doi: 10.1038/nrc3838. [DOI] [PubMed] [Google Scholar]
- 22.Isakoff MS, Barkauskas DA, Ebb D, Morris C, Letson GD. Poor survival for osteosarcoma of the pelvis: a report from the Children’s Oncology Group. Clin Orthop Relat Res. 2012;470:2007–2013. doi: 10.1007/s11999-012-2284-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morgan MA, Lawrence TS. Molecular Pathways: Overcoming Radiation Resistance by Targeting DNA Damage Response Pathways. Clin Cancer Res. 2015;21:2898–2904. doi: 10.1158/1078-0432.CCR-13-3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuhne C, Tjornhammar ML, Pongor S, Banks L, Simoncsits A. Repair of a minimal DNA double-strand break by NHEJ requires DNA-PKcs and is controlled by the ATM/ATR checkpoint. Nucleic Acids Res. 2003;31:7227–7237. doi: 10.1093/nar/gkg937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Novotna E, Tichy A, Pejchal J, Lukasova E, Salovska B, Vavrova J. DNA-dependent protein kinase and its inhibition in support of radiotherapy. Int J Radiat Biol. 2013;89:416–423. doi: 10.3109/09553002.2013.767993. [DOI] [PubMed] [Google Scholar]
- 26.Feng FY, Brenner JC, Hussain M, Chinnaiyan AM. Molecular pathways: targeting ETS gene fusions in cancer. Clin Cancer Res. 2014;20:4442–4448. doi: 10.1158/1078-0432.CCR-13-0275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bromley R, Oliver L, Davey R, Harvie R, Baldock C. Predicting the clonogenic survival of A549 cells after modulated x-ray irradiation using the linear quadratic model. Phys Med Biol. 2009;54:187–206. doi: 10.1088/0031-9155/54/2/002. [DOI] [PubMed] [Google Scholar]
- 28.Franken NA, Oei AL, Kok HP, Rodermond HM, Sminia P, Crezee J, Stalpers LJ, Barendsen GW. Cell survival and radiosensitisation: modulation of the linear and quadratic parameters of the LQ model (Review) Int J Oncol. 2013;42:1501–1515. doi: 10.3892/ijo.2013.1857. [DOI] [PubMed] [Google Scholar]
- 29.Polischouk AG, Cedervall B, Ljungquist S, Flygare J, Hellgren D, Grenman R, Lewensohn R. DNA double-strand break repair, DNA-PK, and DNA ligases in two human squamous carcinoma cell lines with different radiosensitivity. Int J Radiat Oncol Biol Phys. 1999;43:191–198. doi: 10.1016/s0360-3016(98)00362-9. [DOI] [PubMed] [Google Scholar]
- 30.Ohnstad HO, Paulsen EB, Noordhuis P, Berg M, Lothe RA, Vassilev LT, Myklebost O. MDM2 antagonist Nutlin-3a potentiates antitumour activity of cytotoxic drugs in sarcoma cell lines. BMC Cancer. 2011;11:211:211–211. doi: 10.1186/1471-2407-11-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tovar C, Rosinski J, Filipovic Z, Higgins B, Kolinsky K, Hilton H, Zhao X, Vu BT, Qing W, Packman K, Myklebost O, Heimbrook DC, Vassilev LT. Small-molecule MDM2 antagonists reveal aberrant p53 signaling in cancer: implications for therapy. Proc Natl Acad Sci U S A. 2006;103:1888–1893. doi: 10.1073/pnas.0507493103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ottaviano L, Schaefer KL, Gajewski M, Huckenbeck W, Baldus S, Rogel U, Mackintosh C, de Alava E, Myklebost O, Kresse SH, Meza-Zepeda LA, Serra M, Cleton-Jansen AM, Hogendoorn PC, Buerger H, Aigner T, Gabbert HE, Poremba C. Molecular characterization of commonly used cell lines for bone tumor research: a trans-European EuroBoNet effort. Genes Chromosomes Cancer. 2010;49:40–51. doi: 10.1002/gcc.20717. [DOI] [PubMed] [Google Scholar]
- 33.Sinthupibulyakit C, Grimes KR, Domann FE, Xu Y, Fang F, Ittarat W, St Clair DK, St Clair W. p53 is an important factor for the radiosensitization effect of 2-deoxy-D-glucose. Int J Oncol. 2009;35:609–615. doi: 10.3892/ijo_00000372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Robinson BW, Shewach DS. Radiosensitization by gemcitabine in p53 wild-type and mutant MCF-7 breast carcinoma cell lines. Clin Cancer Res. 2001;7:2581–2589. [PubMed] [Google Scholar]
- 35.Zhao Y, Thomas HD, Batey MA, Cowell IG, Richardson CJ, Griffin RJ, Calvert AH, Newell DR, Smith GC, Curtin NJ. Preclinical evaluation of a potent novel DNA-dependent protein kinase inhibitor NU7441. Cancer Res. 2006;66:5354–5362. doi: 10.1158/0008-5472.CAN-05-4275. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.