Abstract
Background
Colorectal cancer (CRC) is the third most prevalent cancer type and the third leading cause of cancer-related deaths worldwide, it is urgently needed to discover a new marker for the progress of CRC. Many long noncoding RNAs (lncRNAs) have been reported to be abnormally expressed in CRC, and may be feasible as effective biomarkers and prognostic factors. The aim of this study was to identify the prognostic value of various lncRNAs in CRC.
Methods
Pubmed, Web of Science, Embase and Cochrane Library were searched for potentially related studies. A total of 34 eligible studies including 30 on overall survival (OS), 7 on disease-free survival (DFS), 1 on relapse-free survival (RFS), 2 on disease-specific survival (DSS) and 29 on clinicopathological features were qualified from the databases.
Results
The results showed that the expression levels of lncRNAs were significantly associated with poor OS (hazard ratio (HR) = 2.08, 95% confidence interval (CI) = 1.68–2.57, P<0.001, I2 = 70%), DFS (HR = 1.79, 95% CI = 1.54–2.08, P<0.001, I2 = 6%) and DSS (HR = 0.11, 95% CI = 0.02–0.54, P = 0.007, I2 = 14%). Subgroup analysis further showed that lncRNA transcription level was significantly associated with tumor differentiation (odds ratio (OR) = 0.51, 95% CI = 0.34–0.77, P = 0.001), lymph node metastasis (OR = 1.63, 95% CI = 1.23–2.17, P = 0.0007), distant metastasis (OR = 2.06, 95% CI = 1.29–3.30, P = 0.002), TNM stage (OR = 0.44, 95% CI = 0.32–0.62, P<0.001), tumor invasion depth (OR = 0.48, 95% CI = 0.39–0.60, P<0.001).
Conclusions
The meta-analysis demonstrated that abnormal lncRNA transcription level may serve as a promising indicator for prognostic of patients with CRC.
Introduction
Colorectal cancer (CRC) is one of the most common malignancies, which ranks the third in the cancer morbidity and the second in the cancer mortality worldwide, with an annual 1.3 million new CRC cancer cases and 694,000 deaths according to the GLOBOCAN estimations[1, 2]. Although advancements have been made regarding the available treatment strategies, the overall survival rate of CRC patients has not improved dramatically[3, 4]. Relapse and metastasis are major factors for the poor outcome of CRC patients[5]. The occurrence of CRC involves multi-factorial and complex steps in which the abnormal gene expression plays a vital role[6]. Therefore, it is emergently necessary to identify novel molecular markers for the early detection, prognosis prediction and therapy evaluation for CRC.
Long non-coding RNAs (lncRNAs), which are defined as RNA molecules of larger than 200 nucleotides in length without protein-coding capacity, regulate gene expression at the epigenetic, transcriptional or posttranscriptional level and once were considered to be transcriptional noise[7–9]. Recently, accumulating evidences showed that lncRNAs played pivotal roles in various cancers and were associated with tumor cell proliferation, apoptosis, invasion and metastasis[10, 11]. For example, plasmacytoma variant translocation 1 gene (PVT1) could inhibit the apoptosis of breast cancer cells [12]. LncRNA H19 was significantly up-regulated in the plasma of Gastric Cancer (GC) patients, and could be a potential non-invasive diagnostic biomarker in GC[13]. In 2016 Huang et al. [14] reported that lncRNA DGCR5 was down-regulated in hepatocellular carcinoma and correlated with poor prognosis. In non-small cell lung cancer, upregulation of long non-coding RNA ATB, a TGF-β-activated lncRNA, indicated a poor prognosis by regulating cell proliferation and metastasis[15]. Recent mounting studies have shown that lncRNAs are potential diagnostic and prognostic biomarkers of CRC.
Up to now, some lncRNAs have been shown to be expressed aberrantly in CRC, such as SNHG20[16], TUG1[17], and 91H[18]. In 2015, Chen et al. [19] found that FEZF1 antisense RNA1 (FEZF1-AS1) was upregulated in CRC tissues and could serve as a potential therapeutic target in CRC. Colon cancer-associated transcript 1 (CCAT1) activated by c-Myc, plays an oncogenic role in CRC development and metastasis[20]. Promoter of CDKN 1A antisense DNA damage activated RNA (PANDAR) could affect epithelial–mesenchymal transition through inhibiting N-cadherin, vimentin, β-catenin, Snail and Twist expression and increasing the expression levels of E-cadherin. The results indicted that PANDAR could be a biomarker for poor prognosis of CRC[21]. Many studies were performed to investigate the prognostic value of lncRNAs in CRC. However, single study may be inaccurate and insufficient, Thus, studies should be analyzed systematically to gain a better insight into the potential clinical values of lncRNAs in CRC. Although some reviews have reported evaluation of the clinical values of multiple lncRNAs in CRC, meta-analysis of lncRNAs in CRC has not yet to be performed. Therefore, relevant articles were collected to evaluate the relationship between lncRNAs expression and clinical outcomes in CRC.
Materials and methods
Publication search
We retrieved Pubmed, Web of Science, Embase and Cochrane Library to obtain all relevant articles. The literature search was limited to the English language and ended in January 22, 2017. The search strategy used both MeSH terms and free-text words to increase the sensitivity of the search. The search terms included: (“Long non-coding RNA”, “lncRNA”, “LincRNA”, “Long ncRNA”, “Long intergenic non-coding RNA”) AND (“CRC”, “colorectal cancer”, “colorectal neoplasm”, “colorectal tumor”, “rectal neoplasm”, “rectal cancer”, “rectal tumor”, “colon neoplasm”, “colon cancer”, “colon tumor”). Relevant articles were also reviewed manually in case of the omission of any potentially relevant literature.
Inclusion and exclusion criteria
The eligible studies met the following criteria: patients were diagnosed with colorectal cancer; relationship between lncRNAs and colorectal cancer was investigated; the prognostic value of lncRNAs was evaluated; the association between lncRNA expression and survival (OS, disease-free survival [DFS], and disease-specific survival [DSS]) was performed; and the survival curve or sufficient relevant data was provided to obtain hazard ratios (HR) for survival rates and their 95% confidence intervals (95% CI). Exclusion criteria were as follows: duplicate studies; sample population consisted of less than 40 cases; non-English papers; letters; review articles; case reports; lack of original data; non-human studies.
Data quality assessment and extraction
Two investigators (JEW and SLD) extracted and reviewed the essential information of each eligible study independently according to pre-specified inclusion and exclusion criteria. The following data were extracted: the first author’s name, year of publication, country, sample size, tumor type, cutoff value, detection method, outcome, analysis type and quality score. The quality of the included studies was assessed with the Newcastle-Ottawa Scale (NOS) criteria for cohort studies [22]. HRs and their 95% CIs were extracted directly from the original articles or calculated from Kaplan-Meier survival curve by HR digitizer software Engauge 4.1 as described by Thierny et al[23]. Any discrepancies on data extraction and quality assessment were resolved through discussion with a third reviewer (MXY). The quality of all the included studies was assessed by The Newcastle-Ottawa Scale (NOS) method. The NOS scores ranged from 0 to 9, and a study with the NOS score more than 6 was regarded as high quality.
Statistical analysis
Statistical analysis was conducted with Review Manager5.2 (The Cochrane Collaboration, Software Update, Oxford, UK) and stata12.0 (STATA Corporation, College Station, TX, USA). HRs and its 95% CI were used to evaluate the association between lncRNAs and survival in CRC. HR > 1 indicated that the patients with high lncRNAs expression had a poor prognosis. Conversely, HR<1 implied the patients with low lncRNAs expression had a good prognosis. ORs and 95% CIs were used to assess the association between lncRNAs and clinical features in patients of CRC. The features included gender, tumor size, tumor differentiation, distant metastasis, lymph node status, TNM stage and tumor invasion depth. Heterogeneity among the eligible studies was assessed with the Q test and I2 statistic, and the I2 value indicated the degree of heterogeneity. A p-value<0.05 or I2>50% indicated significant heterogeneity, in which case a random-effects model was used, if not, a fixed-effects model was used. Publication bias was evaluated by Begg’s test and P>0.05 indicated no significant bias among studies. Sensitivity analyses were performed to access the stability of the meta-analysis results. All the P values were determined by two-sided tests.
Results
Study characteristics
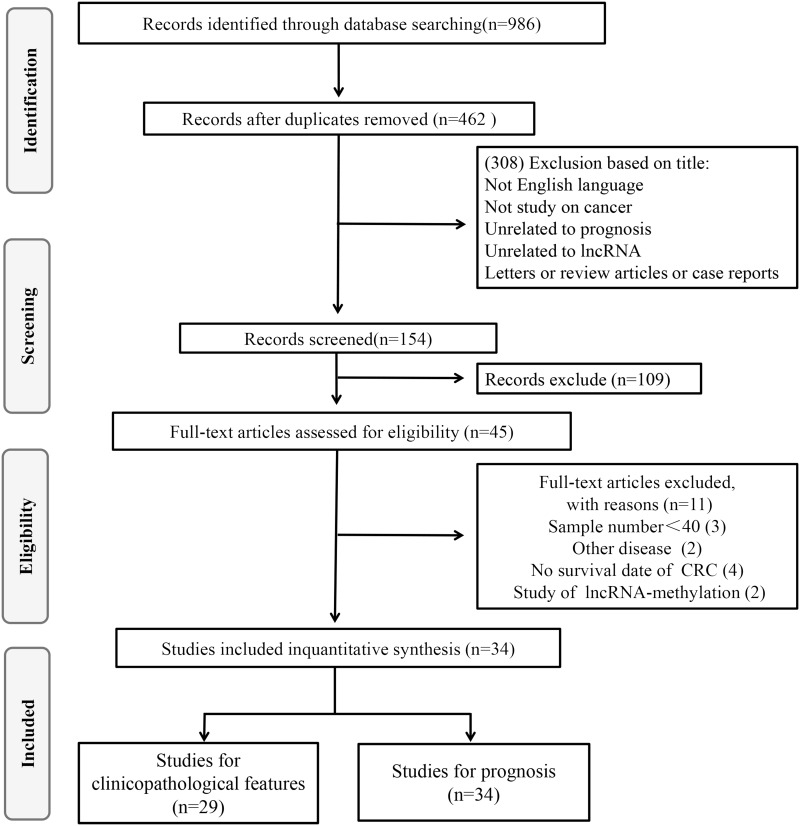
As shown in the flow diagram (Fig 1), a total of 986 records were retrieved from Pubmed, Web of Science, Embase and Cochrane Library. 524 duplicate reports were excluded. After the titles and abstracts were reviewed, 308 irrelevant articles were excluded. Subsequently, the 154 remaining full-text articles were assessed, 120 were found to be ineligible due to lack of sufficient data for further analysis, 34 studies were eligible for the current meta-analysis, including 30 on OS[16–20, 24–48], 7 on DFS[19, 25, 34, 36, 37, 42, 49], 1 on RFS[24], 2 on DSS[50, 51], and 29 on clinicopathological features[16–20, 24–29, 31, 32, 34–37, 39–46, 48–51].
Fig 1. The flow diagram of this meta-analysis in CRC.
Among these 34 studies, a total of 3653 patients were represented. The mean size of patient sample was 111 (range from 48 to 239). The most recent publication date was January 22, 2017. Among the eligible articles, 31 were from China, 3 from Japan and 1 from Czech Republic. In the including 34 studies, 29 articles analyzed the relationship between the expression of lncRNAs and gender[16–20, 24–29, 31, 32, 34–37, 39–46, 48–51], 7 studies estimated the association between lncRNAs and tumor size[19, 24, 25, 27, 41, 42, 48], 16 studies estimated the association between lncRNAs and tumor differentiation[16, 17, 19, 24, 25, 28, 29, 34, 36, 37, 39, 40, 43, 45, 46, 48], 24 studies were about lymph node metastasis(LNM)[16–20, 26, 27, 29–32, 34–37, 39, 40, 42–46, 48, 50], and 20 were about distant metastasis[16, 18–20, 25, 27–29, 35–37, 39, 40, 42–46, 48, 50], 18 studies demonstrated that lncRNAs were correlated with TNM stage[18, 20, 25, 27, 29, 35–39, 41, 42, 44, 46, 48, 50–52], 17 studies reported the association between lncRNAs and tumor invasion depth[16, 18, 19, 26–28, 31, 36, 39, 41, 42, 44–46, 48, 50, 51]. The main information and data were summarized in Table 1.
Table 1. Characteristics of studies included in this meta-analysis.
| Author Year of publication | Country | LncRNAs | Sample size (high/low) | Tumor type | Cutoff | Detection method | Outcome | Analysis type | Quality score |
|---|---|---|---|---|---|---|---|---|---|
| Qi2013 | China | LOC285194 | 33/48 | CRC | mean | qRT-PCR | DSS | Multivariate | 8 |
| Shi2014 | China | RP11-462C24.1 | 32/54 | CRC | mean | qRT-PCR | DSS | Multivariate | 8 |
| Li2016 | China | SNHG20 | 54/53 | CRC | 2.86-fold | qRT-PCR | OS | Multivariate | 7 |
| SunJ2016 | China | TUG1 | 38/23 | CRC | fivefold | qRT-PCR | OS | Kaplan-Meiercurves | 7 |
| SunY2016 | China | ANRIL | 53/44 | CRC | RE1.5 | RT-qPCR | OS | Kaplan-Meiercurves | 7 |
| Lu2016 | China | PANDAR | 62/62 | CRC | median | qRT-PCR | OS | Multivariate | 8 |
| Ge2013 | China | PCAT-1 | 50/58 | CRC | ROC | qRT-PCR | OS | Multivariate | 7 |
| Yin2014 | China | GAS5 | 33/33 | CRC | mean | qRT-PCR | OS | Multivariate | 6 |
| Yin2015 | China | MEG3 | 31/31 | CRC | mean | qRT-PCR | OS | Multivariate | 8 |
| Ye2015 | China | CLMAT3 | 45/45 | CRC | RE | qRT-PCR | OS | Multivariate | 8 |
| Takahashi2013 | Japan | PVT-1 | 131/33 | CRC | MYC expression | qRT–PCR | OS | Multivariate | 7 |
| Svoboda2014 | Czech Republic | HOTAIR | 36/37 | CRC | ROC | RT-qPCR | OS | Multivariate | 8 |
| WangW2016 | China | ZFAS1 | 79/80 | CRC | median | qRT-PCR | RFS/OS | Multivariate | 7 |
| WangF2016 | China | AFAP1-AS1 | 26/26 | CRC | median | qRT-PCR | DFS/OS | Multivariate | 8 |
| Zheng2014 | China | MALAT1 | 73/73 | CRC | RE 6.15 | qRT–PCR | DFS/OS | Multivariate | 7 |
| Iguchi2015 | Japan | lncRNA-ATB | 62/62 | CRC | median | RT-PCR | DFS | Kaplan-Meiercurves | 8 |
| Ni2015 | China | UCA1 | 27/27 | CRC | median | RT-qPCR | OS | Kaplan-Meiercurves | 8 |
| LiY2015 | China | NEAT1 | 110/129 | CRC | 2-fold | RT-PCR | DFS/OS | Multivariate | 8 |
| Han2016 | China | H19 | 48/35 | CRC | 3-fold | qRT-PCR | DFS/OS | Multivariate | 7 |
| Liu2016 | China | CRNDE-h | 71/71 | CRC | median | qRT-PCR | OS | Multivariate | 7 |
| Deng2014 | China | 91H | 30/42 | CRC | RE2.86 | qRT-PCR | OS | Multivariate | 6 |
| Wu2014 | China | HOTAIR | 40/80 | CC | 5-fold | qRT-PCR | OS | Multivariate | 8 |
| Kogo2011 | Japan | HOTAIR | 20/80 | CRC | RE 0.273 | qRT-PCR | OS | Kaplan-Meiercurves | 7 |
| Han2014 | China | UCA1 | 37/43 | CRC | mean | RT-qPCR | OS | Kaplan-Meiercurves | 7 |
| He2014 | China | CCAT1 | 24/24 | CC | median | qPCR | OS | Kaplan-Meiercurves | 8 |
| Liu2015 | China | DANCR | 52/52 | CRC | median | qRT-PCR | DFS/OS | Multivariate | 8 |
| Guo2015 | China | FTX | 75/112 | CRC | median | qRT-PCR | OS | Multivariate | 7 |
| Ren2015 | China | HOTTIP | 77/79 | CRC | median | qRT-PCR | OS | Multivariate | 6 |
| Chen2016 | China | FEZF1-AS1 | 89/64 | CRC | - | RT-PCR | OS | Multivariate | 7 |
| Chen2016 | China | FEZF1-AS1 | 89/64 | CRC | - | RT-PCR | DFS | Kaplan-Meiercurves | 8 |
| Cao2016 | China | SPRY4-IT1 | 36/48 | CRC | 2.87-fold | qRT-PCR | OS | Multivariate | 8 |
| Jiang2016 | China | UCA1 | 61/60 | CRC | median | qRT-PCR | OS | Multivariate | 7 |
| Bian2015 | China | UCA1 | 45/45 | CRC | 2-fold | qRT-PCR | OS | Multivariate | 7 |
| Qiu2015 | China | LINC01296 | 80/80 | CRC | GAPDH | GEO | OS | Multivariate | 6 |
Abbreviations: LncRNA: Long-coding RNA; CRC: Colorectal Cancer; CC: Colon Cancer; RE: Relative expression; RT-PCR: reverse transcription -polymerase chain reaction; qPCR: Real-time-PCR; qRT-PCR: Quantities reverse transcription-PCR; OS: Overall survival; DFS: Disease-free survival; DSS: Disease-specific survival; NA:Not available.
Global analysis between lncRNA transcription level and CRC survival
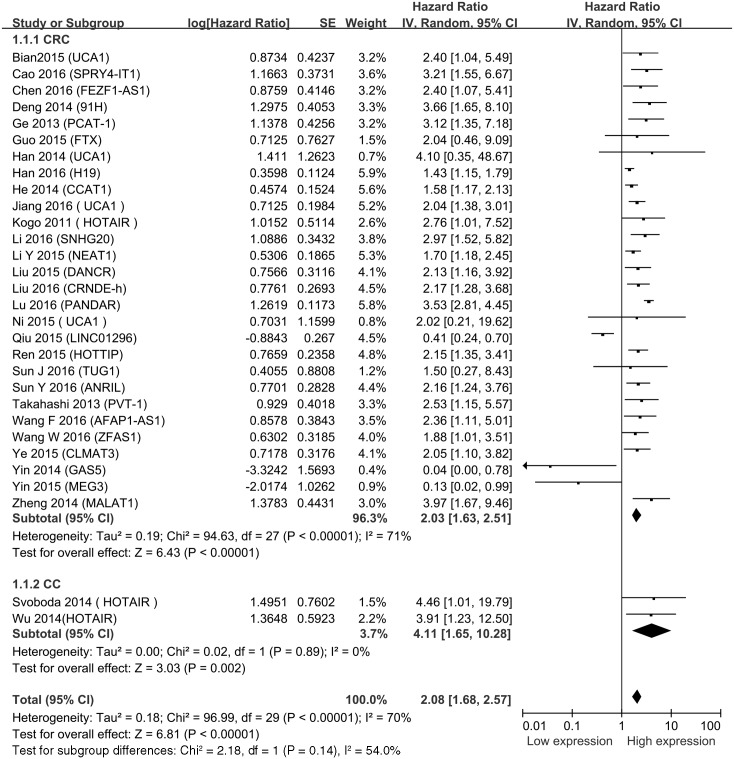
A total of 30 studies of 3361 patients reported OS of CRC. HRs and corresponding 95% CIs of OS were extracted from the included studies. The estimated pooled HR showed a significant association between lncRNAs and OS in CRC patients (HR = 2.08, 95% CI = 1.68–2.57, P<0.001, I2 = 70%) (Fig 2). There are two articles [20, 39] that reported the relationship between the expression level of lncRNAs and OS of colon cancer (HR = 4.11, 95% CI = 1.65–10.28, P = 0.002, I2 = 0%). The increased expressions of SPRY4-IT1, FEZF1-AS1, 91H, PCAT-1, H19, CCAT1, UCA1, HOTAIR, SNHG20, NEAT1, DANCR, CRNDE-h, PANDAR, HOTTIP, TUG1, ANRIL, PVT-1, AFAP1-AS1, ZFAS1 and CLMAT3 were associated with poor prognosis by promoting the proliferation and metastasis of CRC. Meanwhile, the decreased expressions of GAS5, LINC01296, LOC285194, RP11-462C24.1 and MEG3 were related to poor prognosis.
Fig 2. Forest plot for the association between lncRNAs expression levels with overall survival in CRC.
For OS, a significant degree of heterogeneity (I2 = 70%, p<0.001) was determined. Meta-regression analysis and subgroup analysis by sample size, analysis type, cut off values, and NOS score were also performed (Table 2). The cutoff value of mean indicated no statistical significance (HR = 0.296, 95% CI = 0.02–4.05). However, other factors including sample size, analysis type, NOS scores, and weight did not change the significant prognostic impact of high lncRNAs expression level on OS. Subgroup analysis and meta-regression analysis failed to reveal the source of heterogeneity (S1 Fig).
Table 2. Subgroup meta-analysis of pooled HRs for OS.
| Categories | No. of studies | No. of patients | HR (95% CI) for OS | Meta-regression P-value | Heterogeneity | |
|---|---|---|---|---|---|---|
| I2 (%) | Ph | |||||
| [1] OS | 30 | 3361 | 2.08 (1.68–2.57) | 70 | <0.001 | |
| [2]No.of patients | 0.715 | |||||
| ≥100 | 17 | 2500 | 2.16(1.62–2.88) | 74 | <0.001 | |
| <100 | 13 | 851 | 1.96 (1.46–2.63) | 53 | 0.01 | |
| [3] Analysis type | 0.863 | |||||
| Multivariate | 24 | 2862 | 2.09(1.64–2.68) | 75 | <0.001 | |
| Survival curves | 6 | 951 | 1.76(1.37–2.26) | 0 | 0.81 | |
| [4]Cut-off values | 0.243 | |||||
| Mean | 3 | 298 | 0.29(0.02–4.05) | 70 | 0.03 | |
| Median | 10 | 1237 | 2.22(1.70–2.90) | 54 | 0.02 | |
| Others | 16 | 1943 | 2.17(1.60–2.95) | 71 | <0.001 | |
| [5] NOS score | 0.853 | |||||
| >7 | 11 | 1243 | 2.35(2.03–2.73) | 68 | 0.0005 | |
| ≤7 | 19 | 2298 | 2.06 (1.57–2.70) | 69 | <0.001 | |
| [6] Weight | 0.726 | |||||
| ≥5% | 5 | 615 | 1.95(1.33–2.86) | 89 | <0.001 | |
| <5% | 24 | 2746 | 2.13(1.63–2.80) | 61 | <0.001 | |
CRC: Colorectal Cancer; TNM: Tumor node metastasis; I2>50% with the random-effects model; I2<50% with the fixed-effects model.
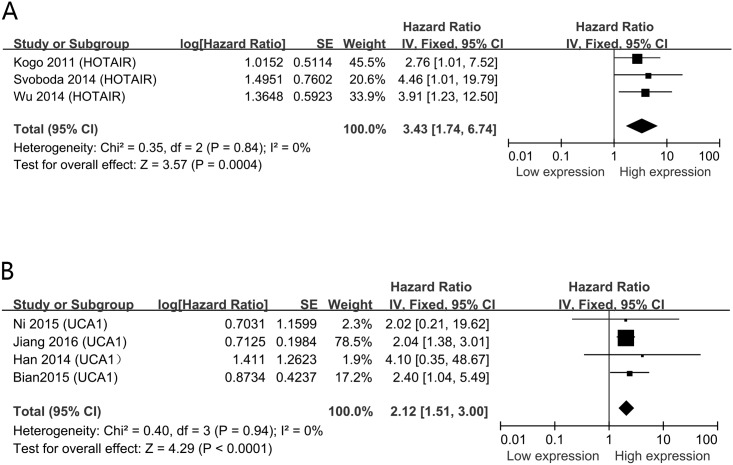
In the including studies, HOTAIR and UCA1 were investigated in three and four studies respectively. The other lncRNAs were performed in single study. We then conducted a meta-analysis on the relationship between HOTAIR/ UCA1 expression and OS of CRC patients. We found that high HOTAIR expression could predict short OS (HR = 3.43, 95% CI = 1.74–6.74, P = 0.0004, I2 = 0%) (Fig 3A). Besides, a poor prognosis in CRC was found in the upregulated levels of UCA1(HR = 2.12, 95% CI = 1.51–3.00, P<0.001, I2 = 0%) (Fig 3B).
Fig 3. Forest plots of studies evaluating hazard ratios of up-regulated lncRNAs and the overall survival of CRC patients.
A. HOTAIR; B. UCA1.
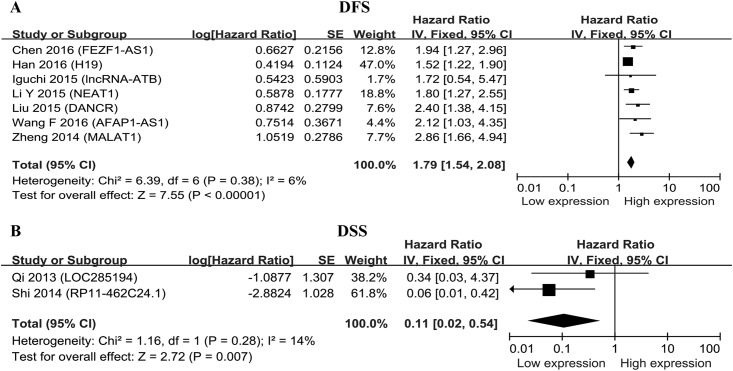
The prognostic significance of lncRNAs in DFS was evaluated in seven studies[19, 25, 34, 36, 37, 42, 49] with 901 patients and that in DSS was examined in two studies [50, 51] with 167 patients (Table 1). There was only one study showed the association between lncRNAs expression level and RFS [24] and therefore was ruled out from the meta-analysis. The up-regulated expression of lncRNAs were significantly associated with DFS (HR = 1.79, 95% CI = 1.54–2.08, P<0.001, I2 = 6%) (Fig 4A), meanwhile, the down-regulated expression of lncRNAs was significantly correlated with DSS in CRC (HR = 0.01, 95% CI = 0.02–0.54, P = 0.007, I2 = 14%) (Fig 4B).
Fig 4. Meta-analysis of the pooled HRs of DFS and DSS for CRC patients.
A.DFS; B. DSS.
Correlation of lncRNAs with clinicopathological characteristics
In order to determine whether the expression of lncRNA was related to the clinical pathological characteristics, the clinicopathological data was collected for the meta-analysis. As shown in Table 3, OR>1 implied that high levels of lncRNAs might be a risk factor in the features. In CRC, lncRNA transcription level was significantly associated with tumor differentiation (OR = 0.51, 95% CI = 0.34–0.77, P = 0.001, random-effect model), lymph node metastasis (OR = 1.63, 95% CI = 1.23–2.17, P = 0.0007, random-effect model), distant metastasis (OR = 2.06, 95% CI = 1.29–3.30, P = 0.002, random-effect model), TNM stage (OR = 0.44, 95% CI = 0.32–0.62, P<0.001, random-effect model), tumor invasion depth (OR = 0.48, 95% CI = 0.39–0.60, P<0.001, fixed-effect model), and tumor size (OR = 0.52, 95% CI = 0.31–0.88, P = 0.02, random-effect model). However, no significant correlation was found with gender (OR = 0.88, 95% CI = 0.76–1.02, P = 0.08, fixed-effect model) (S2 Fig).
Table 3. Association between high levels of lncRNAs and characteristics of patients with CRC.
| Clinicopathological Parameters | Studies | Number of patients | Relative risk of higher lncRNAs OR (95% CI) | Significant Z | Test p-value | Heterogeneity I2 (%) | Test p-value | Model |
|---|---|---|---|---|---|---|---|---|
| Gender (Female vs. male) | 29 | 3125 | 0.88(0.76–1.02) | 1.74 | 0.08 | 0 | 0.66 | Fixed effects |
| Tumor size (<5 vs ≥5) | 7 | 672 | 0.52(0.31–0.88) | 2.43 | 0.02 | 65 | 0.009 | Random effects |
| Tumor differentiation (Moderate/well vs. poor) | 16 | 1845 | 0.51(0.34–0.77) | 3.24 | 0.001 | 70 | <0.001 | Random effects |
| Lymph node metastasis (Positive vs. negative) | 24 | 2748 | 1.63(1.23–2.17) | 3.38 | 0.0007 | 66 | <0.001 | Random effects |
| Distant metastasis (Positive vs. negative) | 20 | 1998 | 2.06(1.29–3.30) | 3.03 | 0.002 | 68 | <0.001 | Random effects |
| TNM stage (I–II vs. III–IV) | 18 | 1770 | 0.44(0.32–0.62) | 4.84 | <0.001 | 62 | 0.0002 | Random effects |
| Tumor invasion depth (T1-T2 vs. T3-T4) | 17 | 1822 | 0.48(0.39–0.60) | 6.66 | <0.001 | 39 | 0.05 | Fixed effects |
Abbreviations: CRC: Colorectal Cancer; TNM: Tumor node metastasis; I2>50% with the random-effects model; I2<50% with the fixed-effects model.
Sensitivity analyses
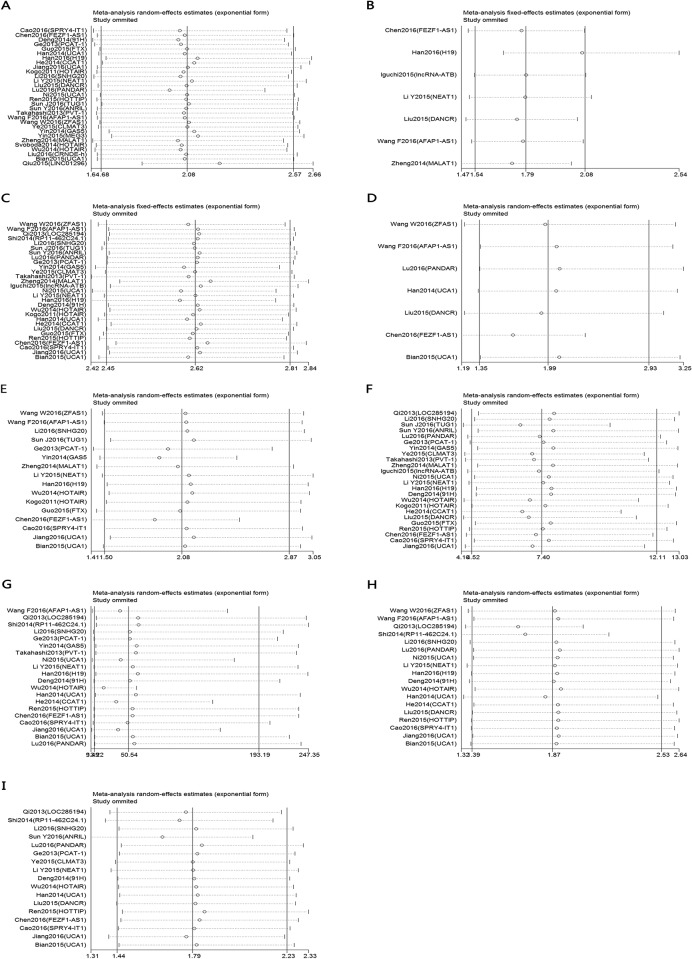
To assess whether a single study might significantly affect the overall results, we performed a sensitivity analysis using Stata12.0 software. In the current study, removing any of the included studies had no significant influence on the estimated pooled results, which demonstrated that our analyses were relatively stable and credible (Fig 5). Because of the small number of studies, the sensitivity analysis was not analyzed for DSS.
Fig 5. Sensitivity analyses of the studies.
A. overall survival; B. disease-free survival; C. gender; D. tumor size (<5 vs ≥5); E. tumor differentiation; F. lymph node metastasis; G. distant metastasis; H. TNM stage; I. Tumor invasion depth.
Publication bias
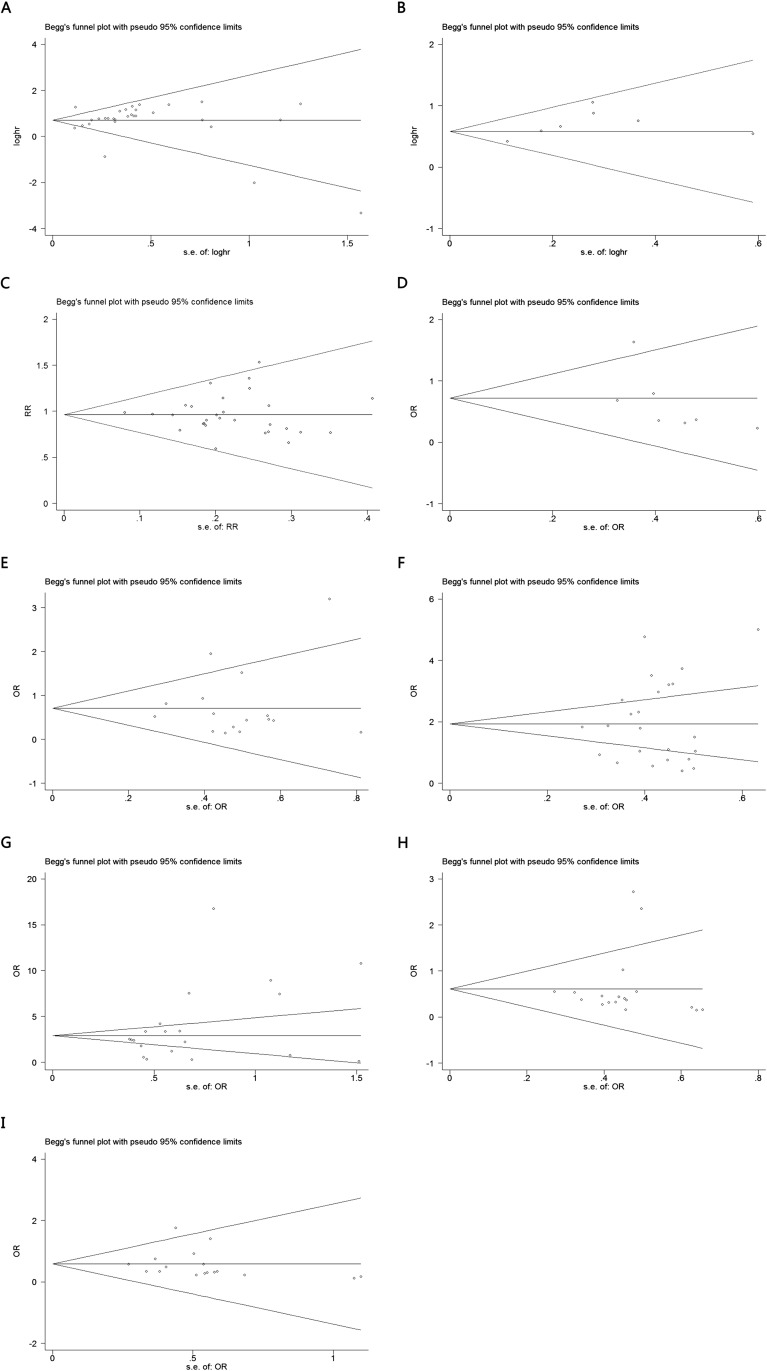
The publication bias of the present meta-analysis was evaluated by Bgger’s test. All groups had no bias, due to all the values of P>0.05(Fig 6). For DSS group, the publication bias was not analyzed because of the small number of studies.
Fig 6. Begg’s test for publication bias.
A. overall survival; B. disease- free survival C. gender; D. tumor size (<5 vs ≥5); E. tumor differentiation; F. lymph node metastasis; G. distant metastasis; H. TNM stage; I. Tumor invasion depth.
Discussion
Nowadays, CRC remains a major health problem and it is the third most common cause of cancer-related death wordwide[53–55]. Molecular Pathologic Epidemiology [56] is a relatively new, evolving field of epidemiology that is designed to clarify how various exposures affect initiation, transformation and progression of neoplasia[57]. It will continue to provide insights into carcinogenic process and help us to optimize prevention and treatment strategies[58]. Recently, a large number of studies demonstrated that the occurrence and progression of CRC was a multi-step process involving in lncRNAs dysregulation of multiple oncogenes and tumor suppressors. It is urgent to identify sensitive and specific biomarkers for early diagnosis and prognosis evaluation[35]. The up-regulation of FTX could serve as an important prognostic factor in CRC patients by promoting growth, migration, invasion and increasing colony formation in colorectal cancer cells[43]. Study from Cao et al. showed that up-regulated expression of SPRY4-IT1 dramatically shortened patients’ survival time by EMT pathway[45]. In the CRC, MEG3 might act as a tumor suppressor gene and contribute to tumorigenesis through inhibiting cancer cell proliferation[30]. In addition, some researchers pointed out that H19 was associated with a poor prognosis in colorectal cancer, which promoted tumor growth by recruiting and binding to eIF4A3[37]. In order to search for a prognostic potential target for CRC, we performed a meta-analysis to examine the relationship between lncRNAs and prognosis of patients with CRC.
Emerging evidence have demonstrated that the association between lncRNAs and CRC. In 2014, Yin et al. suggested that the downregulated GAS5 was related with a poor prognosis and could serve as a candidate prognostic biomarker for OS[29]. Meanwhile, the elevated expression of 91H was regarded as a novel prognosis indicator that contributed to predict tumor metastasis and poor survival of patients with CRC[18]. In our meta-analysis, we explored the prognostic role of lncRNAs in CRC. The results implied that high lncRNA transcription level exhibited a significant risk factor for OS, DFS and DSS. For OS, the test for heterogeneity of included studies was significant (I2 = 70, P<0.001). Although we employed subgroup analysis, meta-regression analysis and sensitivity analysis, all the methods failed to confirm the source of heterogeneity. For DSS, there were only two studies revealed its correlation with LncRNA expression, thus in the further, large-scale and more detailed studies may be needed to be recruited in the meta-analysis.
In our meta-analysis, HOTAIR was investigated in three studies and Urothelial carcinoma-associated 1 (UCA1) was detected in four studies. The increased expression of the two lncRNAs was associated with low survival rate of patients of CRC. UCA1 is an oncofoetal gene involved in embryonic development and carcinogenesis[35]. In 2015, Ni et al. illuminated that UCA1 could significantly enhance migration and invasion of CRC cells. Bian et al. found UCA1 promoted cell proliferation and chemoresistance in colorectal cancer by inhibiting miR-204-5p[48]. Furthermore, HOTAIR regulated expression of multiple genes in cooperation with PRC2 and was a novel molecule involved in the progression of CRC[40]. Therefore, it also was an independent prognostic factor for patients with CRC. Our results showed that the two lncRNAs had a significantly prognostic value in CRC.
In addition, we assessed the association of lncRNA transcription levels with the main clinicopathological features of CRC. Subgroup analysis indicated that lncRNA transcription was related to tumor size, tumor differentiation, lymph node metastasis, distant metastasis, TNM stage and tumor invasion depth. However, there was no correlation between lncRNAs expression and gender.
We have to admit that there are some limitations in our study. Firstly, most of the population in our studies were Chinese, so the conclusion of this study cannot be extended to all populations. Secondly, multiple lncRNAs were used to evaluate the prognosis of CRC, and there was a lack of specific CRC-related lncRNA for clinical evaluation. Thirdly, since there was only one study for each lncRNA in most cases, the prognostic value of each lncRNA may be overestimated. Furthermore, data of HR and 95% CIs were estimated from Kaplan–Meier survival curves in eight studies, which might be less accurate than that acquired directly from published statistics and might increase the potential bias.
Together, this meta-analysis for the first time evaluated the expressions of lncRNAs and clinical values of lncRNAs in CRC. The above analysis showed that lncRNAs were closely related with colorectal cancer, and it can be used as a promising indicator for prognostic of patients with CRC. However, larger-size and higher-quality studies are required to offer better insights into the prognostic value of lncRNAs patients with CRC.
Supporting information
A. sample size; B. analysis type; C. cut-off valus; D. Nos score; E. weight.
(TIF)
A. gender; B. tumor size (<5 vs ≥5); C. tumor differentiation; D. lymph node metastasis; E. distant metastasis; F. TNM stage; G. Tumor invasion depth.
(TIF)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This study was supported by National Natural Science Funds (No. 81472033 and No. 30901308), the National Science Foundation of Hubei Province (No. 2013CFB233 and No. 2013CFB235), the Scientific and technological project of Wuhan City (No. 2014060101010045), Hubei Province health and family planning scientific research project (WJ2015Q021) and Training Program of the science and technology innovation from Zhongnan Hospital of Wuhan University (cxpy20160054). There was no additional external funding received for this study.
References
- 1.Miller KD, Siegel RL, Lin CC, Mariotto AB, Kramer JL, Rowland JH, et al. Cancer treatment and survivorship statistics, 2016. CA Cancer J Clin. 2016;66(4):271–89. doi: 10.3322/caac.21349 . [DOI] [PubMed] [Google Scholar]
- 2.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. International journal of cancer. 2015;136(5):E359–86. doi: 10.1002/ijc.29210 . [DOI] [PubMed] [Google Scholar]
- 3.Dickinson BT, Kisiel J, Ahlquist DA, Grady WM. Molecular markers for colorectal cancer screening. Gut. 2015;64(9):1485–94. doi: 10.1136/gutjnl-2014-308075 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maurel J, Postigo A. Prognostic and Predictive Biomarkers in Colorectal Cancer. From the Preclinical Setting to Clinical Practice. Current cancer drug targets. 2015;15(8):703–15. Epub 2015/10/11. . [DOI] [PubMed] [Google Scholar]
- 5.Zauber AG, Winawer SJ, O'Brien MJ, Lansdorp-Vogelaar I, van Ballegooijen M, Hankey BF, et al. Colonoscopic polypectomy and long-term prevention of colorectal-cancer deaths. N Engl J Med. 2012;366(8):687–96. doi: 10.1056/NEJMoa1100370 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rupnarain C, Dlamini Z, Naicker S, Bhoola K. Colon cancer: genomics and apoptotic events. Biological chemistry. 2004;385(6):449–64. Epub 2004/07/17. doi: 10.1515/BC.2004.053 . [DOI] [PubMed] [Google Scholar]
- 7.Ponjavic J, Ponting CP, Lunter G. Functionality or transcriptional noise? Evidence for selection within long noncoding RNAs. Genome research. 2007;17(5):556–65. doi: 10.1101/gr.6036807 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23(13):1494–504. doi: 10.1101/gad.1800909 ;. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Han Li C, Chen Y. Small and Long Non-Coding RNAs: Novel Targets in Perspective Cancer Therapy. Curr Genomics. 2015;16(5):319–26. doi: 10.2174/1389202916666150707155851 ;. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheetham SW, Gruhl F, Mattick JS, Dinger ME. Long noncoding RNAs and the genetics of cancer. British journal of cancer. 2013;108(12):2419–25. doi: 10.1038/bjc.2013.233 ;. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hansji H, Leung EY, Baguley BC, Finlay GJ, Askarian-Amiri ME. Keeping abreast with long non-coding RNAs in mammary gland development and breast cancer. Frontiers in genetics. 2014;5:379 doi: 10.3389/fgene.2014.00379 ;. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guan Y, Kuo WL, Stilwell JL, Takano H, Lapuk AV, Fridlyand J, et al. Amplification of PVT1 contributes to the pathophysiology of ovarian and breast cancer. Clinical cancer research: an official journal of the American Association for Cancer Research. 2007;13(19):5745–55. doi: 10.1158/1078-0432.CCR-06-2882 . [DOI] [PubMed] [Google Scholar]
- 13.Hashad D, Elbanna A, Ibrahim A, Khedr G. Evaluation of the Role of Circulating Long Non-Coding RNA H19 as a Promising Novel Biomarker in Plasma of Patients with Gastric Cancer. J Clin Lab Anal. 2016;30(6):1100–5. doi: 10.1002/jcla.21987 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang R, Wang X, Zhang W, Zhangyuan G, Jin K, Yu W, et al. Down-Regulation of LncRNA DGCR5 Correlates with Poor Prognosis in Hepatocellular Carcinoma. Cell Physiol Biochem. 2016;40(3–4):707–15. doi: 10.1159/000452582 . [DOI] [PubMed] [Google Scholar]
- 15.Ke L, Xu SB, Wang J, Jiang XL, Xu MQ. High expression of long non-coding RNA ATB indicates a poor prognosis and regulates cell proliferation and metastasis in non-small cell lung cancer. Clin Transl Oncol. 2016. doi: 10.1007/s12094-016-1572-3 . [DOI] [PubMed] [Google Scholar]
- 16.Li C, Zhou L, He J, Fang XQ, Zhu SW, Xiong MM. Increased long noncoding RNA SNHG20 predicts poor prognosis in colorectal cancer. BMC cancer. 2016;16:655 doi: 10.1186/s12885-016-2719-x ;. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun J, Ding C, Yang Z, Liu T, Zhang X, Zhao C, et al. The long non-coding RNA TUG1 indicates a poor prognosis for colorectal cancer and promotes metastasis by affecting epithelial-mesenchymal transition. Journal of translational medicine. 2016;14:42 doi: 10.1186/s12967-016-0786-z ;. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deng Q, He B, Gao T, Pan Y, Sun H, Xu Y, et al. Up-regulation of 91H promotes tumor metastasis and predicts poor prognosis for patients with colorectal cancer. PloS one. 2014;9(7):e103022 doi: 10.1371/journal.pone.0103022 ;. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen N, Guo D, Xu Q, Yang M, Wang D, Peng M, et al. Long non-coding RNA FEZF1-AS1 facilitates cell proliferation and migration in colorectal carcinoma. Oncotarget. 2016;7(10):11271–83. doi: 10.18632/oncotarget.7168 ;. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.He X, Tan X, Wang X, Jin H, Liu L, Ma L, et al. C-Myc-activated long noncoding RNA CCAT1 promotes colon cancer cell proliferation and invasion. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine. 2014;35(12):12181–8. doi: 10.1007/s13277-014-2526-4 . [DOI] [PubMed] [Google Scholar]
- 21.Lu M, Liu Z, Li B, Wang G, Li D, Zhu Y. The high expression of long non-coding RNA PANDAR indicates a poor prognosis for colorectal cancer and promotes metastasis by EMT pathway. Journal of cancer research and clinical oncology. 2017;143(1):71–81. doi: 10.1007/s00432-016-2252-y . [DOI] [PubMed] [Google Scholar]
- 22.Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. European journal of epidemiology. 2010;25(9):603–5. doi: 10.1007/s10654-010-9491-z . [DOI] [PubMed] [Google Scholar]
- 23.Tierney JF, Stewart LA, Ghersi D, Burdett S, Sydes MR. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials. 2007;8:16 Epub 2007/06/09. doi: 10.1186/1745-6215-8-16 ;. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang W, Xing C. Upregulation of long noncoding RNA ZFAS1 predicts poor prognosis and prompts invasion and metastasis in colorectal cancer. Pathology, research and practice. 2016;212(8):690–5. Epub 2016/07/28. doi: 10.1016/j.prp.2016.05.003 . [DOI] [PubMed] [Google Scholar]
- 25.Wang F, Ni H, Sun F, Li M, Chen L. Overexpression of lncRNA AFAP1-AS1 correlates with poor prognosis and promotes tumorigenesis in colorectal cancer. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie. 2016;81:152–9. doi: 10.1016/j.biopha.2016.04.009 . [DOI] [PubMed] [Google Scholar]
- 26.Sun Y, Zheng ZP, Li H, Zhang HQ, Ma FQ. ANRIL is associated with the survival rate of patients with colorectal cancer, and affects cell migration and invasion in vitro. Molecular medicine reports. 2016;14(2):1714–20. Epub 2016/06/18. doi: 10.3892/mmr.2016.5409 . [DOI] [PubMed] [Google Scholar]
- 27.Lu M, Liu Z, Li B, Wang G, Li D, Zhu Y. The high expression of long non-coding RNA PANDAR indicates a poor prognosis for colorectal cancer and promotes metastasis by EMT pathway. Journal of cancer research and clinical oncology. 2016. doi: 10.1007/s00432-016-2252-y . [DOI] [PubMed] [Google Scholar]
- 28.Ge X, Chen Y, Liao X, Liu D, Li F, Ruan H, et al. Overexpression of long noncoding RNA PCAT-1 is a novel biomarker of poor prognosis in patients with colorectal cancer. Medical oncology (Northwood, London, England). 2013;30(2):588 doi: 10.1007/s12032-013-0588-6 . [DOI] [PubMed] [Google Scholar]
- 29.Yin D, He X, Zhang E, Kong R, De W, Zhang Z. Long noncoding RNA GAS5 affects cell proliferation and predicts a poor prognosis in patients with colorectal cancer. Medical oncology. 2014;31(11):253 doi: 10.1007/s12032-014-0253-8 . [DOI] [PubMed] [Google Scholar]
- 30.Yin DD, Liu ZJ, Zhang E, Kong R, Zhang ZH, Guo RH. Decreased expression of long noncoding RNA MEG3 affects cell proliferation and predicts a poor prognosis in patients with colorectal cancer. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine. 2015;36(6):4851–9. doi: 10.1007/s13277-015-3139-2 . [DOI] [PubMed] [Google Scholar]
- 31.Ye LC, Ren L, Qiu JJ, Zhu DX, Chen T, Chang WJ, et al. Aberrant expression of long noncoding RNAs in colorectal cancer with liver metastasis. Tumour Biol. 2015;36(11):8747–54. doi: 10.1007/s13277-015-3627-4 . [DOI] [PubMed] [Google Scholar]
- 32.Takahashi Y, Sawada G, Kurashige J, Uchi R, Matsumura T, Ueo H, et al. Amplification of PVT-1 is involved in poor prognosis via apoptosis inhibition in colorectal cancers. British journal of cancer. 2014;110(1):164–71. doi: 10.1038/bjc.2013.698 ;. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Svoboda M, Slyskova J, Schneiderova M, Makovicky P, Bielik L, Levy M, et al. HOTAIR long non-coding RNA is a negative prognostic factor not only in primary tumors, but also in the blood of colorectal cancer patients. Carcinogenesis. 2014;35(7):1510–5. doi: 10.1093/carcin/bgu055 . [DOI] [PubMed] [Google Scholar]
- 34.Zheng HT, Shi DB, Wang YW, Li XX, Xu Y, Tripathi P, et al. High expression of lncRNA MALAT1 suggests a biomarker of poor prognosis in colorectal cancer. International journal of clinical and experimental pathology. 2014;7(6):3174–81. ;. [PMC free article] [PubMed] [Google Scholar]
- 35.Ni B, Yu X, Guo X, Fan X, Yang Z, Wu P, et al. Increased urothelial cancer associated 1 is associated with tumor proliferation and metastasis and predicts poor prognosis in colorectal cancer. International journal of oncology. 2015;47(4):1329–38. doi: 10.3892/ijo.2015.3109 . [DOI] [PubMed] [Google Scholar]
- 36.Li Y, Li Y, Chen W, He F, Tan Z, Zheng J, et al. NEAT expression is associated with tumor recurrence and unfavorable prognosis in colorectal cancer. Oncotarget. 2015;6(29):27641–50. doi: 10.18632/oncotarget.4737 ;. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Han D, Gao X, Wang M, Qiao Y, Xu Y, Yang J, et al. Long noncoding RNA H19 indicates a poor prognosis of colorectal cancer and promotes tumor growth by recruiting and binding to eIF4A3. Oncotarget. 2016;7(16):22159–73. doi: 10.18632/oncotarget.8063 ;. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu T, Zhang X, Yang YM, Du LT, Wang CX. Increased expression of the long noncoding RNA CRNDE-h indicates a poor prognosis in colorectal cancer, and is positively correlated with IRX5 mRNA expression. OncoTargets and therapy. 2016;9:1437–48. Epub 2016/04/05. doi: 10.2147/OTT.S98268 ;. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu ZH, Wang XL, Tang HM, Jiang T, Chen J, Lu S, et al. Long non-coding RNA HOTAIR is a powerful predictor of metastasis and poor prognosis and is associated with epithelial-mesenchymal transition in colon cancer. Oncology reports. 2014;32(1):395–402. Epub 2014/05/21. doi: 10.3892/or.2014.3186 . [DOI] [PubMed] [Google Scholar]
- 40.Kogo R, Shimamura T, Mimori K, Kawahara K, Imoto S, Sudo T, et al. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer research. 2011;71(20):6320–6. doi: 10.1158/0008-5472.CAN-11-1021 . [DOI] [PubMed] [Google Scholar]
- 41.Han Y, Yang YN, Yuan HH, Zhang TT, Sui H, Wei XL, et al. UCA1, a long non-coding RNA up-regulated in colorectal cancer influences cell proliferation, apoptosis and cell cycle distribution. Pathology. 2014;46(5):396–401. doi: 10.1097/PAT.0000000000000125 . [DOI] [PubMed] [Google Scholar]
- 42.Liu Y, Zhang M, Liang L, Li J, Chen YX. Over-expression of lncRNA DANCR is associated with advanced tumor progression and poor prognosis in patients with colorectal cancer. International journal of clinical and experimental pathology. 2015;8(9):11480–4. Epub 2015/12/01. ;. [PMC free article] [PubMed] [Google Scholar]
- 43.Guo XB, Hua Z, Li C, Peng LP, Wang JS, Wang B, et al. Biological significance of long non-coding RNA FTX expression in human colorectal cancer. International journal of clinical and experimental medicine. 2015;8(9):15591–600. Epub 2015/12/03. ;. [PMC free article] [PubMed] [Google Scholar]
- 44.Ren YK, Xiao Y, Wan XB, Zhao YZ, Li J, Li Y, et al. Association of long non-coding RNA HOTTIP with progression and prognosis in colorectal cancer. International journal of clinical and experimental pathology. 2015;8(9):11458–63. Epub 2015/12/01. ;. [PMC free article] [PubMed] [Google Scholar]
- 45.Cao D, Ding Q, Yu W, Gao M, Wang Y. Long noncoding RNA SPRY4-IT1 promotes malignant development of colorectal cancer by targeting epithelial-mesenchymal transition. OncoTargets and therapy. 2016;9:5417–25. doi: 10.2147/OTT.S111794 ;. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jiang H, Chen Y-T, Fu X-G. Tissue expression level of lncRNA UCA1 is a prognostic biomarker for colorectal cancer. International journal of clinical and experimental pathology. 2016;9(4):4241–6. [Google Scholar]
- 47.Qiu JJ, Yan JB. Long non-coding RNA LINC01296 is a potential prognostic biomarker in patients with colorectal cancer. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine. 2015;36(9):7175–83. Epub 2015/04/22. doi: 10.1007/s13277-015-3448-5 . [DOI] [PubMed] [Google Scholar]
- 48.Bian Z, Jin L, Zhang J, Yin Y, Quan C, Hu Y, et al. LncRNA-UCA1 enhances cell proliferation and 5-fluorouracil resistance in colorectal cancer by inhibiting miR-204-5p. Scientific reports. 2016;6:23892 doi: 10.1038/srep23892 ;. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Iguchi T, Uchi R, Nambara S, Saito T, Komatsu H, Hirata H, et al. A long noncoding RNA, lncRNA-ATB, is involved in the progression and prognosis of colorectal cancer. Anticancer research. 2015;35(3):1385–8. Epub 2015/03/10. . [PubMed] [Google Scholar]
- 50.Qi P, Xu MD, Ni SJ, Huang D, Wei P, Tan C, et al. Low expression of LOC285194 is associated with poor prognosis in colorectal cancer. Journal of translational medicine. 2013;11:122 doi: 10.1186/1479-5876-11-122 ;. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shi D, Zheng H, Zhuo C, Peng J, Li D, Xu Y, et al. Low expression of novel lncRNA RP11-462C24.1 suggests a biomarker of poor prognosis in colorectal cancer. Medical oncology (Northwood, London, England). 2014;31(7):31 doi: 10.1007/s12032-014-0031-7 ;. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhou M, Zhang XY, Yu X. Overexpression of the long non-coding RNA SPRY4-IT1 promotes tumor cell proliferation and invasion by activating EZH2 in hepatocellular carcinoma. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie. 2016. doi: 10.1016/j.biopha.2016.11.035 . [DOI] [PubMed] [Google Scholar]
- 53.Colussi D, Brandi G, Bazzoli F, Ricciardiello L. Molecular pathways involved in colorectal cancer: implications for disease behavior and prevention. International journal of molecular sciences. 2013;14(8):16365–85. doi: 10.3390/ijms140816365 ;. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kocarnik JM, Shiovitz S, Phipps AI. Molecular phenotypes of colorectal cancer and potential clinical applications. Gastroenterology report. 2015;3(4):269–76. doi: 10.1093/gastro/gov046 ;. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kudryavtseva AV, Lipatova AV, Zaretsky AR, Moskalev AA, Fedorova MS, Rasskazova AS, et al. Important molecular genetic markers of colorectal cancer. Oncotarget. 2016;7(33):53959–83. doi: 10.18632/oncotarget.9796 ;. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tampe B, Steinle U, Tampe D, Carstens JL, Korsten P, Zeisberg EM, et al. Low-dose hydralazine prevents fibrosis in a murine model of acute kidney injury-to-chronic kidney disease progression. Kidney Int. 2017;91(1):157–76. doi: 10.1016/j.kint.2016.07.042 . [DOI] [PubMed] [Google Scholar]
- 57.Ogino S, Chan AT, Fuchs CS, Giovannucci E. Molecular pathological epidemiology of colorectal neoplasia: an emerging transdisciplinary and interdisciplinary field. Gut. 2011;60(3):397–411. doi: 10.1136/gut.2010.217182 ;. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ogino S, Nishihara R, VanderWeele TJ, Wang M, Nishi A, Lochhead P, et al. Review Article: The Role of Molecular Pathological Epidemiology in the Study of Neoplastic and Non-neoplastic Diseases in the Era of Precision Medicine. Epidemiology. 2016;27(4):602–11. doi: 10.1097/EDE.0000000000000471 ;. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A. sample size; B. analysis type; C. cut-off valus; D. Nos score; E. weight.
(TIF)
A. gender; B. tumor size (<5 vs ≥5); C. tumor differentiation; D. lymph node metastasis; E. distant metastasis; F. TNM stage; G. Tumor invasion depth.
(TIF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.