Abstract
Background
Neurocysticercosis (NCC), a neglected tropical disease, inflicts substantial health and economic costs on people living in endemic areas such as India. Nevertheless, accurate diagnosis using brain imaging remains poorly accessible and too costly in endemic countries. The goal of this study was to test if blood monocyte gene expression could distinguish patients with NCC-associated epilepsy, from NCC-negative imaging lesion-free patients presenting with idiopathic epilepsy or idiopathic headaches.
Methods/Principal findings
Patients aged 18 to 51 were recruited from the Department of Neurological Sciences, Christian Medical College and Hospital, Vellore, India, between January 2013 and October 2014. mRNA from CD14+ blood monocytes was isolated from 76 patients with NCC, 10 Recovered NCC (RNCC), 29 idiopathic epilepsy and 17 idiopathic headaches patients. A preliminary microarray analysis was performed on six NCC, six idiopathic epilepsy and four idiopathic headaches patients to identify genes differentially expressed in NCC-associated epilepsy compared with other groups. This analysis identified 1411 upregulated and 733 downregulated genes in patients with NCC compared to Idiopathic Epilepsy. Fifteen genes up-regulated in NCC patients compared with other groups were selected based on possible relevance to NCC, and analyzed by qPCR in all patients’ samples. Differential gene expression among patients was assessed using linear regression models. qPCR analysis of 15 selected genes showed generally higher gene expression among NCC patients, followed by RNCC, idiopathic headaches and Idiopathic Epilepsy. Gene expression was also generally higher among NCC patients with single cyst granulomas, followed by mixed lesions and single calcifications.
Conclusions/Significance
Expression of certain genes in blood monocytes can distinguish patients with NCC-related epilepsy from patients with active Idiopathic Epilepsy and idiopathic headaches. These findings are significant because they may lead to the development of new tools to screen for and monitor NCC patients without brain imaging.
Author summary
Taenia solium is a parasite normally transmitted between humans and pigs in areas with poor sanitation. Neurocysticercosis (NCC) occurs when humans are infected with larvae of T. solium that are shed with human feces and the larvae establish in the brain. NCC is often accompanied by neurological symptoms such as epilepsy. In fact, NCC causes approximately one-third of epilepsy cases in areas where T. solium is common. Unfortunately, diagnosis of NCC requires brain computerized tomography or magnetic resonance imaging, tools rarely accessible to people living where NCC is prevalent. This study tested whether genes expressed in blood monocytes, a type of white blood cell, could distinguish between people with epilepsy caused by NCC from those with epilepsy of unknown cause (idiopathic). We compared gene expression in people with NCC and epilepsy, people with idiopathic epilepsy, people cured of NCC and people without NCC or epilepsy but with headaches. We identified 15 genes which were expressed differently in the four different groups indicating that monocyte gene expression patterns in people with NCC and epilepsy are different than people with idiopathic epilepsy. These findings could lead to better understanding how humans respond to NCC and to diagnostic tests which would not require brain imaging.
Introduction
Neurocysticercosis (NCC) is a brain infection by Taenia solium larvae which is a common cause of acquired epilepsy. NCC may consist of single or multiple larvae (cysts) that progress from viable vesicular to colloidal and granuloma states and finally calcify. The time required for cysts to evolve and degenerate varies from a few months to several years. NCC is responsible for nearly half of all acquired epilepsies and about one third of active epilepsies in endemic areas [1–3]. This is particularly important since epilepsy affects from 5.8 to 15.4 people per 1,000 population worldwide with a preponderance in developing countries [4]. NCC causes the largest number of disability adjusted life years among foodborne diseases [5].
Diagnosis of NCC-associated epilepsy relies on brain imaging by computerized tomography (CT) or magnetic resonance imaging (MRI), technology that is unavailable or too expensive for most people in endemic countries [6, 7]. Moreover, current blood antibody and antigen tests are not optimal for NCC diagnosis, especially for cases with low cyst numbers [8]. A reliable diagnostic test for NCC-associated epilepsy not requiring brain imaging would greatly aid its clinical management and could lead to a more comprehensive understanding of the infection.
Inflammatory host responses to degenerating cysts are related to NCC seizures development [9]. Although the relevant pathophysiologic lesions of NCC reside in the central nervous system, salient changes in peripheral blood leukocytes may be detectible. For example, Cardenas et al. detected differential expression of immune-response genes in Taenia-antigen stimulated peripheral blood mononuclear cells from NCC patients who did or did not demonstrate radiological response to anti-helminthic drug treatment [10]. It has also been reported that the network of monocytes and astrocytes play a role in the inflammatory response to intraparenchymal cysts of NCC [11, 12]. Furthermore, other studies show that blood monocytes produce bioactive mediators including CXCL8 (IL-8), CCL2 (MCP-1) and TNF-α, in response to stimulation with T. solium metacestode antigens [11, 13]. Moreover, emerging data show that gene expression in peripheral monocytes can be used as biomarkers to diagnose some psychiatric diseases and monitor response to therapy [14, 15]. The objective of this study was to determine the degree to which gene expression in peripheral blood monocytes could discriminate among patients with NCC-associated epilepsy, recovered NCC patients, patients with idiopathic epilepsy and patients with headaches without brain lesions.
Methods
Study design and sampling strategy
This cross-sectional study sequentially recruited eligible patients aged 18 to 51 years seeking care at the Department of Neurological Sciences, Christian Medical College (CMC) and Hospital, Vellore, India, between January 2013 and October 2014.
Ethics
A research nurse explained the study objectives to eligible patients. Written consent was obtained from each patient agreeing to join the study. The study was approved by the Institutional Review Boards of Christian Medical College, Vellore, India and the University of Oklahoma HSC, Oklahoma, USA.
Study population
Participants were categorized into four groups. Group 1 included new patients with NCC-associated epilepsy with at least one seizure in the 7 months prior to enrollment. This group was sub-divided into patients with: i) solitary cysticercus granuloma (SCG), ii) single calcified cysts (SCC) with or without edema and iii) multiple cysts at various stages of degeneration (MNCC). Group 2 included previously-diagnosed NCC patients who had recovered based on absence of seizures and brain lesions for two years or more (recovered NCC (RNCC)); Group 3 included new patients with idiopathic epilepsy with at least one seizure in the 7 months prior to enrollment, no evidence of NCC or other lesions on brain imaging and seronegative for antigens and antibodies to larval stages of T solium; Group 4 (idiopathic headaches) were seizure-free patients seeking consultation for headaches. These subjects had normal brain imaging, were free of seizures, head trauma, HIV, HBV HCV, and serum cysticercus antigens or antibodies. Healthy, asymptomatic subjects were not recruited as such individuals would not typically undergo serological testing for NCC. Moreover, subjecting healthy cysticercus sero-negative individuals to brain imaging would have raised ethical concerns. Patients in all groups were free of anti-inflammatory drugs (i.e. acetaminophen, ibuprofen etc) at least 7 days prior to enrollment and were not acutely ill at the time of blood sampling. NCC and RNCC patients with extra-parenchymal lesions were excluded.
The maximum age of all patients was 51 years to reduce possible confounding effects of co-existing chronic diseases. Patients were tested for HIV, HCV and HBV as clinically indicated, but not routinely tested as part of this study. Results from tests ordered during routine medical care were used to determine inclusion or exclusion. For Groups 1 and 3, patients with seizures in the past 7 months were included to maximize the potential presence of inflammatory markers circulating in the peripheral blood following a seizure.
Definition of neurocysticercosis
The recommended NCC diagnostic criteria [16] based on brain imaging showing cystic lesions with scolex, lesions highly suggestive of NCC, or lesions compatible with NCC in combination with clinical manifestations suggestive of NCC such as epilepsy/seizures were used. All participants underwent CT or MRI of the brain. All patients in the SCG and MNCC groups and most in the SCC group had contrast enhanced studies. Images were read by one of the authors (VR). NCC lesions were categorized as described above using published recommendations [17, 18]. The MNCC group was subdivided into patients having granulomas only, calcified lesions only, or both lesion types.
Definition of epilepsy
Epilepsy was defined according to the operational definition of the International League Against Epilepsy [19]. Thus, patients with only one epileptic seizure associated with NCC met the definition of epilepsy. All patients with Idiopathic Epilepsy had at least two lifetime seizures thereby meeting the definition.
Measurement of exposure and current infection with T. solium cysticercosis
All patients were tested for serum antibodies to T. solium cysts by a modified enzyme-linked immunoelectrotransfer blot (EITB) [20, 21]. Additionally, all patients were tested for active infection with the AgELISA that identifies excretory/secretory products of T. saginata metacestodes in serum [22, 23].
Blood sample collection
Blood (15–18 mL) was collected by venipuncture in heparinized BD Vacutainer tubes and processed for the isolation of monocytes. In addition, 2mL of blood was collected in clotting tubes for cyst antigen and antibody assays. Serum was separated and stored at -20°C until the assays were performed.
Isolation and purification of monocytes
Peripheral blood mononuclear cells were isolated within one hour of blood collection over Histopaque-1077 by standard methods (Sigma-Aldrich Chemical Co, USA) and were >98% viable as measured by Trypan blue exclusion. CD14+ monocytes were isolated from mononuclear cells by immunomagnetic cell sorting using the MACS system (Miltenyi Biotec) according to the manufacturer’s recommendations. Monocyte purity was >95% as measured by flow cytometry on representative samples.
Isolation of RNA and cDNA synthesis
RNA was extracted from monocytes with TRI Reagent following the manufacturer’s instructions (Sigma-Aldrich Chemical Co, USA) and stored at -20°C until use. RNA (10μgm) was treated with DNase to remove contaminating DNA (Ambion Life Technologies Co method, USA) and then reverse-transcribed to cDNA as described by Invitrogen Life Technologies Co, USA and stored at -80°C until used.
Micro-array analysis
A preliminary micro-array study of a sub-group of 16 patients was performed to identify genes significantly up or down-regulated in Group 1 compared to Groups 3 and 4. cDNA samples from 16 patients (six samples each from Groups 1 and 3 and four samples from Group 4) were hybridized to U95Av2 microarrays (Affymetrix) containing 50,683 gene-probe loci and micro-array assays carried out according to the protocol of Agilent Technologies, USA. The assays were performed at Genotypic Technology Pvt. Ltd., Bangalore, India.
Initial quality check of the hybridized micro-arrays assessed the signal intensity of each gene-probe for each subject. Genes with signal intensity above background were flagged as “detected” and were further analyzed for differential expression among groups.
Genes significantly up- or down-regulated according to the p-value or Naïve Bayes classifier (see statistical analyses section below) in Group 1 compared to Groups 3 or 4 were identified. A subset of 14 genes and one pseudogene, hereafter referred to as the 15 target genes, were identified for further study based on literature review of those with biological relevance to helminthic infection, inflammatory response, and/or neurological disorders.
The genes were categorized (Table 1) based on open source databases including PubMed Gene and PANTHER [24]. Six genes encoded proteins with GTPase, GTPase activator or GTP binding activity were termed “GTP-related” and included CHN2, GBP1, GBP1P1, PLCG2, RAP1A, and TAGAP. Four genes, IL20RB, LRRFIP2, PPP2R2D, and TAX1BP1, were termed “immune-related” [25–28]. Three genes, MZB1, PECAM1, and SLC8A1, were termed “miscellaneous” and displayed protein-protein or cation binding capabilities. Two genes, FEZ2 and TOR3A, were related to “neural processes” based on Slim Molecular Function annotation [24].
Table 1. Peripheral blood monocyte genes initially identified by micro-array analysis selected for further study to differentiate between NCC-associated epilepsy, Idiopathic Epilepsy and idiopathic headaches.
| Gene category | Gene | p-value | Bayes Classifier | FDR | Protein name | Gene Ontology Function |
|---|---|---|---|---|---|---|
| GTP-related | CHN2 | 0.0003 | 0.91 | 0.09 | beta-chimerin | GTPase activator activity |
| GBP1 | 0.0176 | 0.65 | 0.35 | guanylate-binding protein 1 | GTP binding/GTPase activity | |
| GBP1P1 | 0.0175 | 0.72 | 0.28 | guanylate-binding protein 1 pseudogene | GTP binding/GTPase activity pseudogene | |
| PLCG2 | 0.0003 | 0.84 | 0.16 | phospholipase C gamma 2 | PIPLC activity | |
| RAP1A | 0.0002 | 0.93 | 0.07 | ras-related protein Rap-1A | GTP binding/GTPase activity | |
| TAGAP | 0.0060 | 0.74 | 0.26 | T-cell activation Rho GTPase-activating protein | GTPase activator activity | |
| Immune—related | IL20RB | 0.0015 | 0.92 | 0.08 | IL-20 receptor subunit beta | cytokine receptor activity |
| LRRFIP2 | 0.0001 | 0.99 | 0.01 | leucine-rich repeat flightless-interacting protein 2 | LRR domain binding | |
| PPP2R2D | 0.0006 | 0.90 | 0.10 | protein phosphatase 2 regulatory subunit Bdelta | protein phosphatase type 2A regulator activity | |
| TAX1BP1 | 0.0003 | 0.75 | 0.25 | tax1-binding protein 1 | Immune regulator | |
| Miscellaneous | MZB1 | 0.0004 | 0.86 | 0.14 | marginal zone B- and B1-cell-specific protein | protein binding |
| PECAM1a (NP113779) |
0.248 (0.0006) |
0.52 (0.92) |
0.48 (0.08) |
platelet endothelial cell adhesion molecule | protein binding | |
| SLC8A1 | 0.0008 | 0.96 | 0.04 | solute carrier family 8 member A1 | Calcium/sodium antiporter activity | |
| Neural processes | FEZ2a (FEZ2) |
0.3260 (0.0001) |
0.43 (0.91) |
0.57 (0.09) |
fasciculation and elongation protein zeta-2 | protein binding |
| TOR3A | 0.0005 | 0.96 | 0.04 | torsin family 3 member A | ATP binding/ATPase activity |
aDifferent primer sets for these genes showed discordant results, therefore, both are reported. Names in parenthesis is the name of the gene used in micro-array
Quantitative PCR (qPCR)
cDNA samples from all participants were subject to qPCR using the 15 target genes. The primers used for qPCR were based on probes selected from a NCBI Pubmed complete cDNA search of each target gene. qPCR reactions were carried out in a volume of 20 μl containing 1μl of 3X-diluted cDNA using SYBR Green PCR Master Mix (2X dilution) (Applied Biosystems) and consensus primers (synthesized at Sigma-Aldrich Chemicals Pvt Ltd (Bangalore, India)) (S1 Table). The reactions were performed in duplicate and each gene assayed twice. A melting curve was obtained for each PCR product after each run, confirming that the SYBR Green signal corresponded to a unique and specific amplicon.
Standard curves were generated for every qPCR run and were obtained by using serial 3-fold dilutions of a sample containing the sequence of interest. Their plots were used to convert cycle thresholds (Ct) into arbitrary quantities of initial template for a given sample. Gene expression normalized to two housekeeping genes, β2 microglobulin and 18S, was calculated by the 2-ΔΔCt method [29]. Both housekeeping genes returned similar results with respect to direction (up- or down-regulation compared with comparator) and statistical significance among groups compared, but did vary in the absolute magnitude of the fold-change.
Statistical analyses
Descriptive analyses were used to characterize the study groups. Significance among the four groups was determined using chi-square tests and Student’s t-tests.
Micro-array results were analyzed by calculating the mean log2 expression levels of each detected gene. Expression levels in Group 1 were compared with that in Groups 3 (Idiopathic Epilepsy) and 4 (idiopathic headaches) by 2 tailed Student’s t-test. The corresponding p-values were sorted from smallest to largest for all genes. Because of the exploratory nature of this analysis, a multiple correction procedure was not applied. A separate naïve Bayes classifier, similar to that of Anderson and Dubnicka [30] but modified for micro-array data, was used to compute a probability of differential expression and describe the estimated false discovery rate for each gene. This method computes areas from kernel density estimates of the log2 gene expressions for use in quantifying the probability of group affiliation for each subject. Averaging these probabilities across subjects, but within a gene, describes the “typical” probability of differential expression of that gene.
The natural logarithm (ln) of all qPCR fold changes was used to meet assumptions for parametric statistics. Significant differences of ln-transformed fold change of normalized gene expression among groups, including NCC subgroups, were determined by Student’s t-test with a p-value <0.05. Additionally, three dichotomous variables were created to characterize the type of NCC lesions: any calcification vs granuloma only, any granuloma vs calcification only, and presence of edema (yes/no). Separate analyses were conducted using the idiopathic headaches and the Idiopathic Epilepsy groups as the calibrator groups. Since using the Idiopathic Epilepsy group for calibration did not modify the conclusions regarding the comparison among NCC groups, only the results using the idiopathic headaches group are presented here. Simple linear regression models were fitted to estimate the mean difference (95% CI) in ln-transformed fold changes among all groups. Multivariable linear regression models were fitted to estimate the mean difference (95%CI) in fold changes while adjusting for potential confounders such as age, gender, time since last seizure, number of lifetime seizures and the presence of antigens or antibodies to cysticercosis among the NCC group. The linearity, normality and homoscedasticity of the studentized errors were verified visually and using the Shapiro-Wilk tests for normality and the Breush-Pagan test for homoscedasticity. All non-Bayesian analyses were conducted in Stata 14.
Results
Study population
Seventy-six NCC (29 SCG, 20 SCC, 27 MNCC), 10 RNCC, 29 Idiopathic Epilepsy and 17 idiopathic headaches subjects were included in the study.
Characteristics of the study population are shown in Table 2. NCC and RNCC patients showed a higher frequency of living near a pig rearing household (p = 0.01). There was a non-statistically significant larger proportion of females in the idiopathic headaches group. The NCC group had a higher percentage of patients with only one seizure in their lifetime, due to the epilepsy definition used. There was no statistically significant difference in the time since the last seizure and the type of the last seizure. Most subjects in Groups 1 and 3 had had a seizure in the past 3 months, but only two subjects had a seizure in the past 48 hours. Exclusion of these subjects did not modify the results, and hence these subjects were kept in all analyses. All subjects in Group 4 presented with headaches, but detailed data on the type, duration, and intensity of the headaches were not collected.
Table 2. Socio-demographic and clinical characteristics of eligible patients with different NCC lesions, RNCC, Idiopathic Epilepsy and idiopathic headaches recruited from 2013 to 2014 at the Department of Neurological Sciences, Christian Medical College (CMC) and Hospital, Vellore, India.
| Study Group | ||||||||
|---|---|---|---|---|---|---|---|---|
| Variable | Category | SCG | SCC | MNCC | All NCC | RNCC | Idiopathic Epilepsy | Idiopathic Headaches |
| Sample size | 29 | 20 | 27 | 76 | 10 | 29 | 17 | |
| Gender | Male | 24 (83%) | 12 (60%) | 18 (67%) | 54 (71%) | 8 (80%) | 19 (66%) | 7 (41%) |
| Age | Mean (SD) | 30.4 (10.3) | 27.1 (8.0) | 31.1 (8.4) | 29.8 (9.1) | 30.0 (7.7) | 26.9 (7.2) | 34.0 (9.2) |
| Schooling | <Secondary | 12 (41%) | 8 (40%) | 15 (56%) | 35 (46%) | 2 (20%) | 11 (38%) | 5 (29%) |
| ≥ Secondary | 17 (59%) | 12 (60%) | 12 (44%) | 41 (54%) | 8 (80%) | 18 (62%) | 12 (71%) | |
| Pork eating | Yes | 5 (17%) | 3 (15%) | 4 (15%) | 12 (16%) | 1 (10%) | 2 (7%) | 1 (6%) |
| Living near a house that raised pigsa | Yes | 7 (24%) | 5 (25%) | 8 (30%) | 20 (26%) | 5 (50%) | 4 (14%) | 0 (0%) |
| Owning pigs | Yes | 1 (3%) | 0 (0%) | 2 (7%) | 3 (4%) | 0 (0%) | 0 (0%) | 0 (0%) |
| How often do you use the toilet to defecate? | Never | 23 (79%) | 14 (70%) | 18 (67%) | 55 (72%) | 9 (90%) | 22 (76%) | 16 (94%) |
| Cigarette smokingc | Yes | 4 (14%) | 6 (30%) | 7 (26%) | 17 (22%) | 2 (20%) | 4 (14%) | 0 (0%) |
| Paan usec,d | Yes | 4 (14%) | 4 (20%) | 8 (30%) | 16 (21%) | 1 (10%) | 3 (10%) | 0 (0%) |
| Headaches | Yes | 1 (3%) | 0 (0%) | 2 (7%) | 3 (4%) | 0 (0%) | 0 (0%) | 17 (100%) |
| EITBb | Positive | 11 (38%) | 8 (40%) | 16 (59%) | 35 (46%) | 3 (30%) | 0 (0%) | 0 (0%) |
| AgELISAb,e | Positive | 5 (17%) | 5 (25%)§ | 16 (59%) | 26 (35%)§ | 1 (10%) | 0 (0%) | 0 (0%) |
| Type of NCC lesion | Edema | 26 (90%) | 4 (20%) | 18 (67%) | 48 (63%) | NA | NA | NA |
| Only calcification | 0 (0%) | 20 (100%) | 10 (37%) | 30 (39%) | NA | NA | NA | |
| Only granuloma | 29 (100%) | 0 (0%) | 11 (41%) | 40 (53%) | NA | NA | NA | |
| Time since last seizure | ≤ 3 months | 25 (86%) | 17 (85%) | 21 (78%) | 63 (83%) | NA | 21 (72%) | NA |
| 3–7 months | 4 (14%) | 3 (15%) | 6 (22%) | 13 (17%) | NA | 8 (28%) | NA | |
| No of lifetime seizuresa,b | 1 | 9 (31%) | 0 (0%) | 5 (18%) | 14 (18%) | NA | 0 (0%) | NA |
| 2–5 | 16 (55%) | 5 (25%) | 7 (26%) | 28 (37%) | NA | 9 (31%) | NA | |
| 6–10 | 1 (3%) | 1 (5%) | 4 (15%) | 6 (8%) | NA | 2 (7%) | NA | |
| >10 | 3 (11%) | 14 (70%) | 11 (41%) | 28 (37%) | NA | 18 (62%) | NA | |
| Type of last seizure | Generalized | 20 (69%) | 15 (75%) | 19 (70%) | 54 (71%) | 9 (90%) | 24 (83%) | NA |
| Partial | 6 (21%) | 3 (15%) | 3 (11%) | 12 (16%) | 1 (10%) | 1 (3%) | NA | |
| Partial then generalized | 3 (10%) | 2 (10%) | 5 (19%) | 10 (13%) | 0 (0%) | 4 (14%) | NA | |
SCG: single cysticercus granuloma; SCC: single calcified cyst; MNCC: multiple neurocysticercosis, NCC: neurocysticercosis; RNCC: recovered neurocysticercosis;
NA: not applicable.
aStatistically significant difference among Groups 1, 2, 3 and 4 using chi-square at p < .05
bStatistically significant difference among SCG, MNCC and SCG using chi-square at p < .05
cThese questions were only asked to male patients. Percentages are reported among males.
d “Paan” is a betel leaf with areca nut and considered a stimulant with psychoactive properties. It is chewed.
eAgELISA: One AgELISA test result was missing for one patient in the SCC group. Percentages are reported among those with a test result available.
There were no statistical differences among the various NCC groups with regard to age, gender, schooling, pork eating, headaches, the type of seizure and time from last seizure. However, SCG and MNCC patients were more likely to have had only one seizure than the SCC group. There was a statistically significant higher percentage of positivity by EITB and AgELISA in the MNCC patients (48% for both) compared to the SCC (11% for both) and SCG (7% for both) groups
Micro-array analyses
Micro-array analysis found 19,778 uncompromised gene readings for all groups and showed that monocytes from patients with NCC and Idiopathic Epilepsy shared 397 up-regulated and 676 down-regulated genes compared to idiopathic headaches. Comparison of 20,739 uncompromised gene expressions of NCC with Idiopathic Epilepsy showed 1,411 up-regulated and 733 down-regulated genes specific to NCC. The probability of differential expression obtained using the naïve Bayes classifier for the up- or down-regulated genes in the latter comparison ranged from 51.3% to 98.5% and 53.3% to 98.4%, respectively.
Pathway analysis of up-regulated genes in NCC compared to Idiopathic Epilepsy revealed involvement of multiple inflammatory/immune pathways. The 10 pathways with the highest number of differentially upregulated genes included cytokine-cytokine receptor interaction, MAPK signaling, JAK-STAT signaling, Toll-like receptor signaling, and chemokine signaling (S2 Table). A pathway related to toxoplasmosis was also identified, a common protozoal brain infection. Analysis of the differentially expressed down-regulated genes also revealed pathways related to inflammation and immunity. Pathways with the greatest numbers of differentially down-regulated genes included cytokine-cytokine receptor interaction, T-cell receptor signaling, primary immunodeficiency, and MAPK signaling. These data suggest that NCC-related epilepsy elicits a monocyte gene signature that is distinct from Idiopathic Epilepsy and encompass multiple immune and inflammatory pathways.
qPCR analysis of gene expression in NCC-associated epilepsy, Idiopathic Epilepsy and RNCC
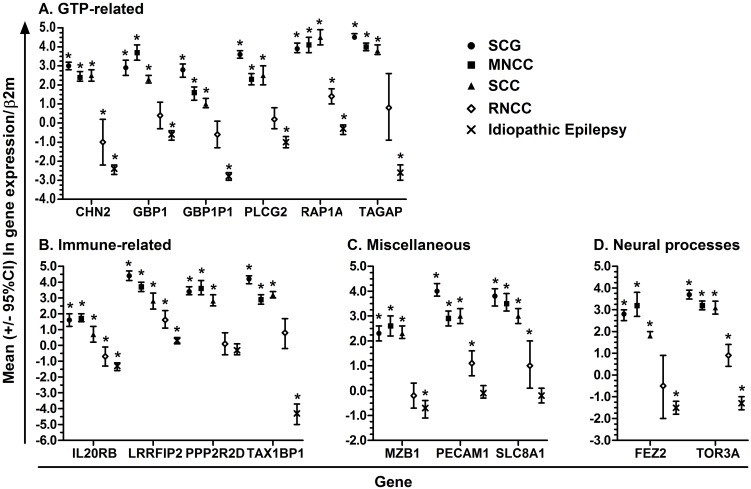
Expression of each of these 15 genes was significantly increased in SCG, SCC, and MNCC compared with idiopathic headaches (Fig 1). The magnitude of expression was typically lower in RNCC patients than in patients with SCG, SCC, and MNCC and expression values of several genes in the RNCC group were not significantly different from idiopathic headaches. Fold-changes in patients with Idiopathic Epilepsy ranged from -4.3 to 0.3, with 11 genes being down-regulated compared with idiopathic headaches (Fig 1).
Fig 1. Mean (95%CI) ln-fold change in gene expression of GTP-related (Panel A), Immune-related (Panel B), Miscellaneous (Panel C) and Neural processes-related (Panel D) genes among subjects with SCG (full circle), MNCC (full square), SCC (full triangle), RNCC (open diamond) and Idiopathic Epilepsy (X) measured by qPCR, normalized to β2 microglobulin and calibrated with idiopathic headaches.
*Indicates significant differences in ln-transformed fold-changes compared with idiopathic headaches using Student’s t-tests with a p-value <0.05.
Comparison of gene expression among patients with NCC and Idiopathic Epilepsy, and according to type of NCC lesion
Linear regression analysis showed all 15 genes were over-expressed in each NCC sub-group as compared with Idiopathic Epilepsy (Table 3). All genes except for IL20RB, PPP2R2D and MZB1 were more expressed in the RNCC group as compared to the Idiopathic Epilepsy group. Adjusting for age, gender, time since last seizures and total number of lifetime seizures, and sero-positivity to cysticercus antigens or antibodies did not alter these conclusions.
Table 3. Linear regression coefficient (95%CI) of the ln-fold change in gene expression comparing NCC and Idiopathic Epilepsy groups.
β2M was used for normalization and the idiopathic headaches for calibration.
| Comparison group | Idiopathic Epilepsy | MNCC | SCC | SCC | |||
|---|---|---|---|---|---|---|---|
| Study group | SCG | SCC | MNCC | RNCC | SCG | SCG | MNCC |
| CHN2 | 5.5 (5.1; 5.8)a | 4.9 (4.5; 5.4)a | 4.9 (4.5; 5.3)a | 1.4 (0.9; 2.0)a | 0.6 (0.2; 1.0)a | 0.5 (0.1; 1.0)a | 0 (-0.5; 0.4) |
| GBP1 | 3.5 (3.0; 4.0)a | 2.9 (2.4; 3.4)a | 4.3 (3.9; 4.8)a | 1.0 (0.4; 1.6)a | -0.8 (-1.3; -0.4)a | 0.6 (0.1; 1,1)a | 1.4 (0.9; 2.0)a |
| GBP1P1 | 5.6 (5.1; 6.0)a | 3.8 (3.4; 4.3)a | 4.3 (3.9; 4.8)a | 2.2 (1.6; 2.8)a | 1.2 (0.8; 1.7)a | 1.7 (1.3; 2.2)a | 0.5 (0.02; 1.0)a |
| PLCG2 | 4.6 (4.2; 5.0)a | 3.5 (3.1; 4.0)a | 3.3 (2.9; 3.7)a | 1.2 (0.7; 1.8)a | 1.3 (0.9; 1.7)a | 1.1 (0.6; 1.5)a | -0.2 (-0.7; 0.2) |
| RAP1A | 4.3 (3.8; 4.7)a | 4.8 (4.3; 5.3)a | 4.4 (4.0; 4.8)a | 1.7 (1.1; 2.3)a | -0.1 (-0.6; 0.3) | -0.6 (-1.0; -0.1)a | -0.4 (-0.9; 0.06) |
| TAGAP | 7.2 (6.7; 7.7)a | 6.5 (5.9; 7.0)a | 6.7 (6.2; 7.2)a | 3.5 (2.8; 4.1)a | 0.5 (0; 1.0)a | 0.7 (0.2; 1.3)a | 0.2 (-0.3; 0.8) |
| IL20RB | 2.9 (2.4; 3.3)a | 2.1 (1.6; 2.6)a | 3.0 (2.6; 3.5)a | 0.6 (-0.03; 1.2) | -0.1 (-0.6; 0.3) | 0.8 (0.3; 1.3) | 1.0 (0.5; 1.5)a |
| LRRFIP2 | 4.1 (3.7; 4.5)a | 2.5 (2.0; 2.9)a | 3.4 (3.0; 3.8)a | 1.3 (0.8; 1.9)a | 0.7 (0.3; 1.1)a | 1.6 (1.2; 2.1)a | 0.9 (0.5; 1.4)a |
| PPP2R2D | 3.7 (3.3; 4.2)a | 3.1 (2.6; 3.6)a | 3.9 (3.4; 4.4)a | 0.4 (-0.2; 1.0) | -0.2 (-0.7; 0.3) | 0.6 (0.1; 1.1)a | 0.8 (0.3; 1.3)a |
| TAX1P1 | 8.5 (7.9; 9.1)a | 7.6 (6.9; 8.2)a | 7.3 (6.7; 7.8)a | 5.1 (4.3; 5.9)a | 1.2 (0.7; 1.8)a | 0.9 (0.3; 1.6)a | -0.3 (-0.9; 0.3) |
| MZB1 | 3.1 (2.6; 3.5)a | 3.1 (2.6; 3.5)a | 3.3 (2.9; 3.8)a | 0.5 (-0.1; 1.1) | -0.3 (-0.7; 0.1) | -0.01 (-0.5; 0.5) | 0.3 (-0.2; 0.8) |
| PECAM1 | 4.1 (3.8; 4.5)a | 3.1 (2.7; 3.5)a | 3.0 (2.6; 3.3)a | 1.2 (0.7; 1.7)a | 1.1 (0.8; 1.5)a | 1.0 (0.6; 1.4)a | -0.1 (-0.5; 0.3) |
| SLC8A1 | 4.0 (3.5; 4.4)a | 3.2 (2.7; 3.7)a | 3.7 (3.3; 4.2)a | 1.2 (0,6; 1.8)a | 0.2 (-0.2; 0.7) | 0.8 (0.3; 1.3)a | 0.6 (0.1; 1.0)a |
| FEZ2 | 4.2 (3.7; 4.8)a | 3.3 (2.8; 3.9) | 4.7 (4.2; 5.3)a | 1.0 (0.2; 1.7)a | -0.5 (-1.0; 0.05) | 0.9 (0.3; 1.5)a | 1.4 (0.8; 2.0)a |
| TOR3A | 5.0 (4.7; 5.3)a | 4.4 (4.0; 4.7)a | 4.4 (4.1; 4.8)a | 2.2 (1.7; 2.6)a | 0.6 (0.2; 0.9)a | 0.6 (0.3; 1.0)a | 0 (-0.3; 0.4) |
aIndicates a significant difference at p < .05
Twelve of the 15 genes decreased significantly with the resolution of solitary cyst infections, i.e., as SCG evolved to SCC (Table 3). One exception was observed with RAP1A which was higher among those with SCC as compared to SCG.
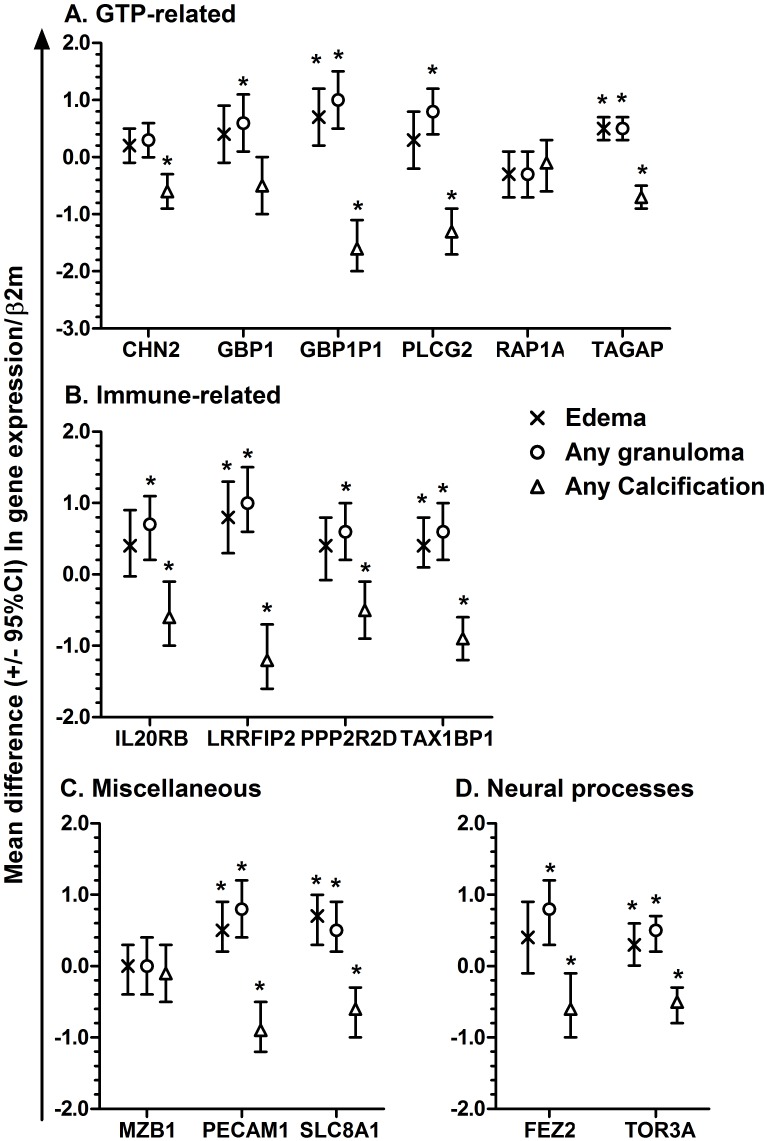
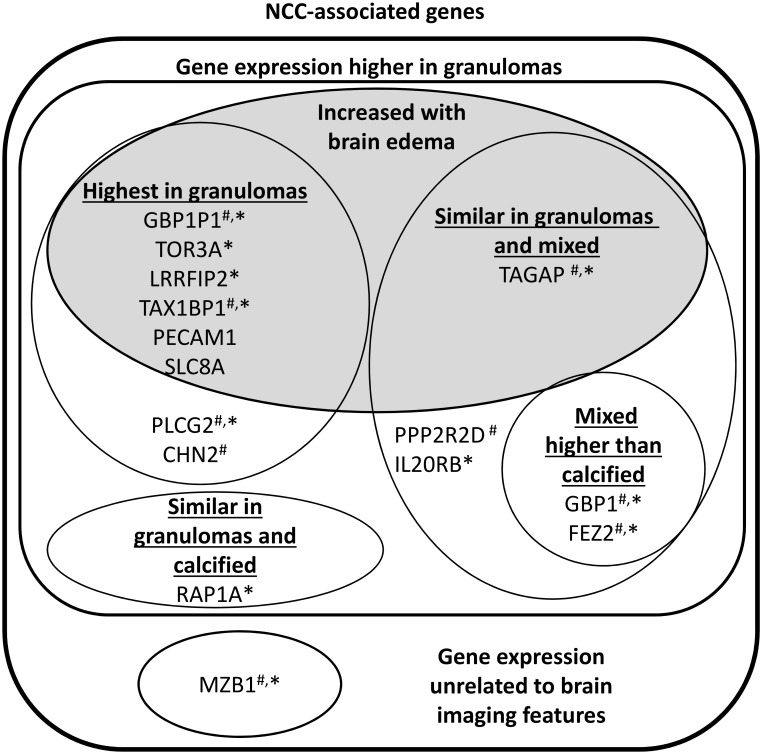
The distribution of lesions in each NCC subgroup is presented in Table 2. Twelve genes were over expressed among patients who only had granulomas as compared to patients with only calcifications (Table 4). Seven genes were over expressed in patients with edema, all of which were also over expressed among patients with any granulomas (Fig 2). RAP1A and MZB1 did not show any difference in expression according to any granuloma, any calcification or edema (Fig 2). Fig 3 summarizes associations of the gene expressions with brain lesions and edema.
Table 4. Linear regression coefficients (95%CI) comparing the granuloma only, calcified only, and combined (granuloma and calcified) type of NCC.
| Comparison group | Calcification only (30 with) | Calcification only (30 with) | Mixed (6) |
| Lesion | Granuloma only (40 with) | Mixed (6) | Granuloma only (40) |
| CHN2 | 0.4 (0.1; 0.6)a | -0.2 (-0.7; 0.4) | 0.5 (0.008; 1.1)a |
| GBP1 | 0.5 (-0.006; 1.0) | 1.0 (0.03; 1.9)a | -0.5 (-1.4; 0.5) |
| GBP1P1 | 1.3 (0.8; 1.7)a | -0.7 (-1.5; 0.1) | 1.9 (1.2; 2.7)a |
| PLCG2 | 1.0 (0.6; 1.4)a | -0.5 (-1.2; 0.2) | 1.5 (0.8; 2.2)a |
| RAP1A | -0.2 (-0.6; 0.2) | -1.0 (-1.8; -0.2)a | 0.8 (0.009; 1.5)a |
| TAGAP | 0.5 (0.3; 0.8)a | 0.2 (-0.3; 0.7) | 0.3 (-0.1; 0.8) |
| IL20RB | 0.7 (0.2; 1.4)a | 0.5 (-0.3; 1.4) | 0.1 (-0.7; 1.0) |
| LRRFIP2 | 1.2 (0.7; 1.6)a | 0.1 (-0.8; 0.9) | 1.1 (0.3; 1.9)a |
| PPP2R2D | 0.6 (0.2; 1.0)a | 0.6 (-0.2; 1.3) | 0 (-0.7; 0.8) |
| TAX1P1 | 0.8 (0.5; 1.1)a | -0.8 (-1.4; -0.2)a | 1.6 (1.0; 2.1)a |
| MZB1 | 0.1 (-0.3; 0.5) | -0.2 (-1.0; 0.5) | 0.3 (-0.4; 1.0) |
| PECAM1 | 0.9 (0.5; 1.3)a | 0.2 (-0.5; 0.9) | 0.7 (0.1; 1.4)a |
| SLC8A1 | 0.6 (0.3; 1.0)a | -0.1 (-0.8; 0.7) | 0.7 (-0.005; 1.4) |
| FEZ2 | 0.8 (0.3; 1.2)a | 1.0 (0.2; 1.9)a | -0.3 (-1.2; 0.6) |
| TOR3A | 0.5 (0.3; 0.8)a | 0 (-0.5; 0.5) | 0.5 (0.03; 1.0)a |
aIndicates a significant difference at p < .05
Fig 2. Mean differences (95%CI) in ln-fold change in gene expression of GTP-related (Panel A), Immune-related (Panel B), Miscellaneous (Panel C) and Neural processes-related (Panel D) genes among subjects with edema (X), any granuloma (open circle) and any calcification (open triangle) measured by qPCR, normalized to β2 microglobulin and calibrated with idiopathic headaches.
*Indicates significant differences in ln-transformed fold-changes compared with subjects not showing that characteristic using Student’s t-tests with a p-value <0.05.
Fig 3. Schematic representation of monocyte genes grouped accordingly to their relationship with brain imaging features.
#Expression of the gene in patients with resolved NCC is comparable to its expression in normal idiopathic headaches. *Expression of the gene in patients with idiopathic epilepsy is lower than found in idiopathic headaches.
Discussion
Analysis of host responses in peripheral blood is one possible avenue for developing new diagnostic tools [31]. Results presented here suggest that gene expression in peripheral blood monocytes can distinguish patients with NCC-related epilepsy from patients with Idiopathic Epilepsy and from subjects negative to standard antigen and antibody assays, with normal brain imaging, no seizures and presenting with headaches. Moreover, we found that expression of 15 selected target genes in carefully selected patients with different NCC-related brain lesions including SCG, SCC, and MNCC showed differences in the magnitude of gene expression between active and recovered infections. Such differences were also observed according to the type of lesion (granuloma, calcification, edema). In general, patients with NCC showed higher magnitude of expression than did patients with recovered NCC. In addition, gene expression among these target genes was reduced as lesions resolved, i.e. as seen by the granuloma to calcification progression. Our data add to the evidence that responses in blood monocytes may be used to investigate peripheral markers that differentiate patients with and without NCC-associated epilepsy [12].
Since adjustment for age, gender, the time since the last seizure, number of lifetime seizures did not alter the conclusions and that the type of seizures, pork eating habits and headaches among the NCC group were similar, the increased gene expression in these patients compared to the idiopathic epilepsy and NCC-free headache groups may indicate expression specific to T. solium infection of the brain per se and not to seizures. However, the expressions may also reflect responses to brain infections in general. Increased expression of genes being due to T. solium infection of the brain and not to seizures would also be supported by their higher expression in RNCC patients compared to those with idiopathic epilepsy. Although headaches are a common symptom of NCC [32], there were large differences in the expression of target genes in patients with NCC-associated epilepsy as compared to idiopathic headaches subjects.
Results presented here agree with studies by Gupta et al [33] showing perilesional inflammation decreased as solitary granulomas calcified. The transition to calcification was accompanied by recovery of a breached blood brain barrier measured by reduced serum matrix metalloproteinase-9 levels. The observation that perilesional inflammation that was higher in symptomatic (seizures) compared to asymptomatic NCC patients with solitary cyst calcifications [34] may in fact be reflected by our finding of low inflammatory gene expression among RNCC patients. Together these may reflect our findings of reduced inflammatory gene expression with resolution of cysts and of low expression among RNCC patients.
There is a clear need for sensitive and accurate diagnostic tests for NCC that do not rely upon brain imaging technology and perform better than current serological tests. Results presented here support the idea that blood-based diagnostics, in particular monocyte gene expression, may be a useful tool for diagnosing and staging NCC, and perhaps also for assessing resolution. Further studies controlling for other brain infections and space occupying lesions, e.g. malignancies, are needed to establish a specific association between up-regulation of these genes and NCC.
Supporting information
(DOC)
(DOC)
Please refer to the readme tab for more details.
(XLSX)
Please refer to the readme tab for more details.
(XLSX)
Please refer to the readme tab for more details.
(XLSX)
Acknowledgments
The authors wish to thank all participants for their time and commitment to this project. We also wish to thank the staff at Genotypic Technology Pvt. Ltd., Bangalore, for the time they spent with our group going over the methods to be used and how the micro-array data was generated.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by the National Institute of Neurological Diseases and Stroke (https://www.ninds.nih.gov/) in the U.S. (R21NS077466) with HC as the PI and by the Department of Biotechnology (www.dbtindia.nic.in) in India (BT/MB/BRCP/06/2011) with VR as the PI under the U.S.-India Bilateral Brain Research Collaborative Partnerships (U.S.–India BRCP). Further support was received from the National Institute of Neurological Diseases and Stroke and the Fogarty International Center (https://www.fic.nih.gov) (R01NS098891) with HC and VR as MPI under the Global Brain and Nervous System Disorders Research Across the Lifespan program. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Rajshekhar V, Raghava MV, Prabhakaran V, Oommen A, Muliyil J. Active epilepsy as an index of burden of neurocysticercosis in Vellore district, India. Neurology. 2006;67(12):2135–9. Epub 2006/12/28. doi: 10.1212/01.wnl.0000249113.11824.64 . [DOI] [PubMed] [Google Scholar]
- 2.Garcia HH, Del Brutto OH, Cysticercosis Working Group in P. Neurocysticercosis: updated concepts about an old disease. Lancet Neurol. 2005;4(10):653–61. doi: 10.1016/S1474-4422(05)70194-0 . [DOI] [PubMed] [Google Scholar]
- 3.Ndimubanzi PC, Carabin H, Budke CM, Nguyen H, Qian YJ, Rainwater E, et al. A systematic review of the frequency of neurocyticercosis with a focus on people with epilepsy. PLoS neglected tropical diseases. 2010;4(11):e870 Epub 2010/11/13. doi: 10.1371/journal.pntd.0000870 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bell GS, Neligan A, Sander JW. An unknown quantity—the worldwide prevalence of epilepsy. Epilepsia. 2014;55(7):958–62. doi: 10.1111/epi.12605 . [DOI] [PubMed] [Google Scholar]
- 5.Torgerson PR, Devleesschauwer B, Praet N, Speybroeck N, Willingham AL, Kasuga F, et al. World Health Organization Estimates of the Global and Regional Disease Burden of 11 Foodborne Parasitic Diseases, 2010: A Data Synthesis. PLoS Med. 2015;12(12):e1001920 doi: 10.1371/journal.pmed.1001920 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Del Brutto OH. Diagnostic criteria for neurocysticercosis, revisited. Pathogens and global health. 2012;106(5):299–304. Epub 2012/12/26. doi: 10.1179/2047773212Y.0000000025 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coyle CM. Neurocysticercosis: an update. Current infectious disease reports. 2014;16(11):437 Epub 2014/10/24. doi: 10.1007/s11908-014-0437-6 . [DOI] [PubMed] [Google Scholar]
- 8.Rodriguez S, Wilkins P, Dorny P. Immunological and molecular diagnosis of cysticercosis. Pathogens and global health. 2012;106(5):286–98. doi: 10.1179/2047773212Y.0000000048 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nash TE, Mahanty S, Loeb JA, Theodore WH, Friedman A, Sander JW, et al. Neurocysticercosis: A natural human model of epileptogenesis. Epilepsia. 2015;56(2):177–83. doi: 10.1111/epi.12849 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Uddin J, Garcia HH, Gilman RH, Gonzalez AE, Friedland JS. Monocyte-astrocyte networks and the regulation of chemokine secretion in neurocysticercosis. Journal of immunology (Baltimore, Md: 1950). 2005;175(5):3273–81. . [DOI] [PubMed] [Google Scholar]
- 11.Uddin J, Gonzalez AE, Gilman RH, Garcia HH, Verastegui M, Moore LJ, et al. Neurocysticercal antigens stimulate chemokine secretion from human monocytes via an NF-kappaB-dependent pathway. Microbes and infection / Institut Pasteur. 2006;8(7):1732–40. Epub 2006/07/04. doi: 10.1016/j.micinf.2006.02.009 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cardenas G, Fragoso G, Rosetti M, Uribe-Figueroa L, Rangel-Escareno C, Saenz B, et al. Neurocysticercosis: the effectiveness of the cysticidal treatment could be influenced by the host immunity. Med Microbiol Immunol. 2014;203(6):373–81. doi: 10.1007/s00430-014-0345-2 . [DOI] [PubMed] [Google Scholar]
- 13.Uddin J, Gonzalez AE, Gilman RH, Thomas LH, Rodriguez S, Evans CA, et al. Mechanisms regulating monocyte CXCL8 secretion in neurocysticercosis and the effect of antiparasitic therapy. Journal of immunology (Baltimore, Md: 1950). 2010;185(7):4478–84. Epub 2010/09/10. doi: 10.4049/jimmunol.0904158 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beumer W, Gibney SM, Drexhage RC, Pont-Lezica L, Doorduin J, Klein HC, et al. The immune theory of psychiatric diseases: a key role for activated microglia and circulating monocytes. Journal of leukocyte biology. 2012;92(5):959–75. Epub 2012/08/10. doi: 10.1189/jlb.0212100 . [DOI] [PubMed] [Google Scholar]
- 15.Bergink V, Gibney SM, Drexhage HA. Autoimmunity, inflammation, and psychosis: a search for peripheral markers. Biological psychiatry. 2014;75(4):324–31. Epub 2013/11/30. doi: 10.1016/j.biopsych.2013.09.037 . [DOI] [PubMed] [Google Scholar]
- 16.Del Brutto OH, Rajshekhar V, White AC Jr, Tsang VC, Nash TE, Takayanagui OM, et al. Proposed diagnostic criteria for neurocysticercosis. Neurology. 2001;57(2):177–83. ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garcia HH, Del Brutto OH. Imaging findings in neurocysticercosis. Acta Trop. 2003;87(1):71–8. . [DOI] [PubMed] [Google Scholar]
- 18.Rajshekhar V, Oommen A. Serological studies using ELISA and EITB in patients with solitary cysticercus granuloma and seizures. Neurological Infections and Epidemiology. 1997;(2):177–0. [Google Scholar]
- 19.Fisher RS, Acevedo C, Arzimanoglou A, Bogacz A, Cross JH, Elger CE, et al. ILAE official report: a practical clinical definition of epilepsy. Epilepsia. 2014;55(4):475–82. doi: 10.1111/epi.12550 . [DOI] [PubMed] [Google Scholar]
- 20.Tsang VC, Brand JA, Boyer AE. An enzyme-linked immunoelectrotransfer blot assay and glycoprotein antigens for diagnosing human cysticercosis (Taenia solium). J Infect Dis. 1989;159(1):50–9. . [DOI] [PubMed] [Google Scholar]
- 21.Prabhakaran V, Rajshekhar V, Murrell KD, Oommen A. Conformation-sensitive immunoassays improve the serodiagnosis of solitary cysticercus granuloma in Indian patients. Transactions of the Royal Society of Tropical Medicine and Hygiene. 2007;101(6):570–7. Epub 2006/12/16. doi: 10.1016/j.trstmh.2006.10.001 . [DOI] [PubMed] [Google Scholar]
- 22.Brand JA, Tsang VC. A rapid immunoblot assay (western blot) to detect specific antibodies for human immunodeficiency virus, Schistosoma mansoni, and Taenia solium (Cysticercosis). J Immunoassay. 1989;10(2–3):237–55. doi: 10.1080/01971528908053239 . [DOI] [PubMed] [Google Scholar]
- 23.Dorny P, Brandt J, Zoli A, Geerts S. Immunodiagnostic tools for human and porcine cysticercosis. Acta Trop. 2003;87(1):79–86. . [DOI] [PubMed] [Google Scholar]
- 24.Mi H, Poudel S, Muruganujan A, Casagrande JT, Thomas PD. PANTHER version 10: expanded protein families and functions, and analysis tools. Nucleic Acids Res. 2016;44(D1):D336–42. doi: 10.1093/nar/gkv1194 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jin J, Yu Q, Han C, Hu X, Xu S, Wang Q, et al. LRRFIP2 negatively regulates NLRP3 inflammasome activation in macrophages by promoting Flightless-I-mediated caspase-1 inhibition. Nat Commun. 2013;4:2075 doi: 10.1038/ncomms3075 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Logsdon NJ, Deshpande A, Harris BD, Rajashankar KR, Walter MR. Structural basis for receptor sharing and activation by interleukin-20 receptor-2 (IL-20R2) binding cytokines. Proc Natl Acad Sci U S A. 2012;109(31):12704–9. doi: 10.1073/pnas.1117551109 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.An in vivo screen implicates PPP2R2D as an inhibitor of T-cell function. Cancer Discov. 2014;4(3):OF13 doi: 10.1158/2159-8290.CD-RW2014-032 . [DOI] [PubMed] [Google Scholar]
- 28.Iha H, Peloponese JM, Verstrepen L, Zapart G, Ikeda F, Smith CD, et al. Inflammatory cardiac valvulitis in TAX1BP1-deficient mice through selective NF-kappaB activation. EMBO J. 2008;27(4):629–41. doi: 10.1038/emboj.2008.5 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods (San Diego, Calif). 2001;25(4):402–8. Epub 2002/02/16. doi: 10.1006/meth.2001.1262 . [DOI] [PubMed] [Google Scholar]
- 30.Anderson MP, Dubnicka SR. A sequential naive Bayes classifier for DNA barcodes. Stat Appl Genet Mol Biol. 2014;13(4):423–34. doi: 10.1515/sagmb-2013-0025 . [DOI] [PubMed] [Google Scholar]
- 31.Holcomb ZE, Tsalik EL, Woods CW, McClain MT. Host-Based Peripheral Blood Gene Expression Analysis for Diagnosis of Infectious Diseases. J Clin Microbiol. 2017;55(2):360–8. doi: 10.1128/JCM.01057-16 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carabin H, Ndimubanzi PC, Budke CM, Nguyen H, Qian Y, Cowan LD, et al. Clinical manifestations associated with neurocysticercosis: a systematic review. PLoS neglected tropical diseases. 2011;5(5):e1152 Epub 2011/06/02. doi: 10.1371/journal.pntd.0001152 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gupta RK, Awasthi R, Garg RK, Kumar N, Gupta PK, Singh AK, et al. T1-weighted dynamic contrast-enhanced MR evaluation of different stages of neurocysticercosis and its relationship with serum MMP-9 expression. AJNR Am J Neuroradiol. 2013;34(5):997–1003. doi: 10.3174/ajnr.A3346 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gupta RK, Awasthi R, Rathore RK, Verma A, Sahoo P, Paliwal VK, et al. Understanding epileptogenesis in calcified neurocysticercosis with perfusion MRI. Neurology. 2012;78(9):618–25. doi: 10.1212/WNL.0b013e318248deae . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOC)
(DOC)
Please refer to the readme tab for more details.
(XLSX)
Please refer to the readme tab for more details.
(XLSX)
Please refer to the readme tab for more details.
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.