Abstract
Background
X-chromosome inactivation (XCI) is the mechanism by which gene dosage uniformity is achieved between female mammals with two X chromosomes and male mammals with a single X chromosome, and is thought to occur randomly. For molecular genetic testing, accessible tissues (eg blood) are commonly studied, but the relationship with inaccessible tissues (eg brain) is poorly understood. For accessible tissues to be informative for genetic analysis, a high degree of concordance of genetic findings among tissue types is required.
Objective
To determine the relationship among multiple tissues within females at different ages (fetus to 82 years).
Methods
XCI patterns were analysed using the polymorphic androgen receptor (AR) gene assay. DNA was isolated from 26 different human females without history of malignancy, using 34 autopsy tissues representing the three embryonic germ layers.
Results
33 of the 280 tissue samples analysed from 13 of the 26 females showed skewed XCI values (>80:20%). Average XCI value was not significantly different among the tissues, but a trend for increasing XCI variability was observed with age in blood and other tissues studied (eg the SD for all tissues studied for the 0–2 years group was 9.9% compared with 14.8% in the >60 years group). We found a significant correlation (rs = 0.51, p = 0.035) between XCI values for blood and/or spleen and brain tissue, and in most other tissues representing the three embryonic germ layers.
Conclusions
In our study, XCI data were comparable among accessible (eg blood) and inaccessible tissues (eg brain) in females at various ages, and may be useful for genetic testing. A trend was seen for greater XCI variability with increasing age, particularly in older women (>60 years).
Each tissue of an adult female is composed of cells that actively transcribe genes from either the maternal or the paternal X chromosome. X-chromosome inactivation (XCI) is the mechanism by which gene dosage equivalence is achieved between female mammals with two X chromosomes and male mammals with a single X chromosome.1 XCI is thought to take place within 7–10 days after fertilisation, when the embryonic cell mass contains no more than a few dozen cells,2,3 and is subsequently stable in daughter cells. XCI is thought to occur at random in the general female population, resulting in an average XCI ratio of 50:50 with normal distribution. Thus, an equal number of cells in any tissue will have a transcriptionally active maternal or paternal X chromosome, and the other X chromosome will be transcriptionally silenced. Given a normal distribution of maternal versus paternal X inactivation, a small percentage of females (8%) will have skewness (>80%:20%), where there is a bias in the inactivation of the X chromosomes.4 Extreme skewness (95%:5%) in peripheral blood cells is rare, occurring in approximately 1% of human females, but is more common in older women (>60 years of age).5,6 However, in the case of an X-chromosome aberration, the ratio is skewed in favour of the normal X chromosome, which may be due to post-inactivation cell selection, and the resulting cell population is not random.
Skewed XCI has been described in many disorders or conditions, including in female carriers of X-linked mental retardation,7 in women with recurrent spontaneous abortions8 and in lymphocytes from females with Rett syndrome, a severely disabling neurological disorder caused by MECP2 gene mutations located at Xq28.9 Clinical severity in patients with Rett syndrome appears to correlate with the direction and degree of skewing of XCI.10 Furthermore, we have perviously reported that non-random X inactivation occurs at a higher frequency in females with autism compared with unaffected females, indicating that X-linked genes may play a role in the reported higher male:female ratio seen in this neurodevelopmental disorder.11
Females with Prader–Willi syndrome (PWS) due to maternal disomy 15 also show more XCI skewness compared with PWS females with the paternal 15q11–q13 deletion or in control females.12 Fragile X tremor/ataxia syndrome (FXTAS) consisting of tremor, ataxia, parkinsonism, and executive dysfunction, results from a premutation in the fragile X gene (FMR1). The severity of clinical symptoms in females carrying a premutation in this gene has been shown to correlate with a molecular pattern of X inactivation favouring higher expression of the premutation FMR1 allele.13 However, X-inactivation skewness is seen occasionally in healthy females, possibly indicating unknown genetic factors involved in the process.14–16 The small population of embryonic cells present when a specific X chromosome is committed to inactivation could lead to skewed patterns seen in healthy individuals. Studies have shown that skewness increases with age.5,6 A finite pool of stem cells may exist that could diminish over time, creating a small sample size of precursor cells, which eventually lead to a higher degree of XCI skewness.17
X-inactivation patterns can be assessed using the human androgen receptor (AR) gene located at Xq11.2. This gene contains a highly polymorphic in-frame CAG triplet, repeated 11–31 times in exon 1. Assessment is possible for females with different alleles (informative) at the CAG repeat using DNA digestion with methyl-sensitive restriction enzymes (eg HpaII), which will not cut the methylated (inactive) alleles, followed by PCR amplification. After capillary electrophoresis, the resulting peak sizes (the signal intensity is directly related to the amount of PCR product made) can be compared to determine the skewness ratio. XCI status (one X chromosome may be more or less active (unmethylated) compared with the second X chromosome in somatic cells) is defined as being highly skewed (>80:20%) or extremely skewed (>90:10%).8,11,12,18,19 In healthy females, XCI is considered to follow a Gaussian or bell-shaped distribution, with highly skewed patterns being uncommon events.20
For molecular genetic testing, accessible tissues (eg blood) are commonly used; however, their relationship with inaccessible tissues (eg brain) is poorly understood. For accessible tissues to be informative for genetic analysis, a high degree of concordance of genetic findings among tissue types would be required. A previous report indicated a general concordance between XCI pattern in three different tissues, but was limited in scope.21 Therefore, we examined the XCI pattern in 26 human females (20 weeks gestation to 82 years old), using multiple autopsy tissues representing the three primordial germ layers, to characterise the relationship of XCI patterns within and among females at all ages. We examined tissues collected at autopsy from 26 females ranging in age from fetus (20 weeks gestation) to 82 years.
METHODS
We analysed 280 frozen autopsy tissues from 26 females without a history of malignancy grouped by age as follows: fetus (n = 4); 0–2 years (n = 5); 5–8 years (n = 3); 15–20 years (n = 4); 21–40 years (n = 4); 41–60 years (n = 3) and >60 years (n = 3). In total, 34 different tissues were collected, representing the three embryonic germ layers, with an average of 11 different tissues from each female (table 1). Most tissue samples were evaluated with a single assay, with the exception of those performed multiple times to test the reliability of the assay (see Results section).
Table 1.
Description of tissues analysed for X-chromosome inactivation
| Group | Age | No. of tissues | Endoderm | Mean (SD)* | Mesoderm | Mean (SD)* | Ectoderm | Mean (SD)* |
|---|---|---|---|---|---|---|---|---|
| Fetus (n = 4) | 20 weeks’ gestation | 16 | 6 | 48.1 (24.1) | 8 | 47.9 (27.9) | 2 | 44.4 (23.3) |
| 22 weeks’ gestation | 16 | 6 | 7 | 3 | ||||
| 27 weeks’ gestation | 11 | 5 | 6 | 0 | ||||
| 34 weeks’ gestation | 11 | 3 | 6 | 2 | ||||
| 0–2 years (n = 5) | 4 days | 13 | 4 | 47.2 (16.8) | 8 | 46.6 (15.4) | 1 | 55.1 (23.4) |
| 7 days | 16 | 7 | 8 | 1 | ||||
| 8 days | 13 | 4 | 7 | 2 | ||||
| 5 weeks | 15 | 6 | 7 | 2 | ||||
| 2 years | 12 | 4 | 6 | 2 | ||||
| 5–8 years (n = 3) | 5 years | 8 | 1 | 59.9 (22.6) | 5 | 52.2 (23.3) | 2 | 53.5 (7.0) |
| 6 years | 15 | 6 | 8 | 1 | ||||
| 8 years | 7 | 3 | 3 | 1 | ||||
| 15–20 years (n = 4) | 15 years | 10 | 2 | 53.3 (16.5) | 5 | 55.4 (16.1) | 3 | 48.3 (17.7) |
| 17 years | 7 | 3 | 3 | 1 | ||||
| 18 years | 14 | 3 | 8 | 3 | ||||
| 19 years | 9 | 3 | 5 | 1 | ||||
| 21–40 years (n = 4) | 25 years | 5 | 1 | 47.4 (5.4) | 3 | 48.5 (11.2) | 1 | 48.5 (10.1) |
| 29 years | 12 | 3 | 6 | 3 | ||||
| 33 years | 8 | 2 | 4 | 2 | ||||
| 36 years | 10 | 2 | 6 | 2 | ||||
| 41–60 years (n = 3) | 42 years | 8 | 3 | 49.6 (5.3) | 4 | 47.8 (10.3) | 1 | 38.0 (14.1) |
| 47 years | 9 | 3 | 6 | 0 | ||||
| 53 years | 8 | 3 | 4 | 1 | ||||
| >60 years (n = 3) | 61 years | 10 | 2 | 54.3 (30.1) | 6 | 53.2 (18.5) | 2 | 41.8 (6.0) |
| 79 years | 5 | 2 | 3 | 0 | ||||
| 82 years | 12 | 4 | 6 | 2 |
Group mean (SD) value of X-chromosome inactivation calculated using the individual mean for each female for the first or smaller allele for tissues from each germ layer; there appeared to be more variability among germ layer tissues for older women—that is, a wider range of means (SD) seen in those aged >60 years.
Androgen receptor assay
Genomic DNA was extracted from tissue samples (about 1 cm3) using a DNA purification kit (Gentra, Madison, Wisconsin, USA) according to the manufacturer’s instructions. The AR gene contains a highly polymorphic region consisting of CAG repeats. Template DNA from each tissue was amplified by PCR using primers flanking the polymorphic region of the AR gene. The size of the PCR product and signal intensity was determined by capillary electrophoresis using an ABI 3100 DNA sequencer (Applied Biosystems, Foster City, California, USA). Subsequently, 200 ng of genomic DNA was digested with the methyl-sensitive restriction enzyme HpaII, as previously described.22 The reverse primer was labelled with the fluorescent marker 6-FAM and the resulting PCR fragments analysed by capillary electrophoresis, using established protocols.11,12 Genomic DNA from a male with a different CAG repeat size was added to the digestion reaction as an internal control to ensure that the restriction was complete. XCI was calculated as the ratio of the height of the shorter peak to the sum of the two peaks, using genotyping software after digestion as previously described.11,12 To account for preferential allele amplification, values for the digested DNA were normalised with those for the undigested DNA for each subject and XCI was calculated using the following formula: (phd1/phu1)/(phd1/phu1)+(phd2/phu2), where phd1 is peak height of first allele (digested DNA), phd2 is peak height of second allele (digested DNA), phu1 is peak height of first allele (undigested DNA) and phu2 is peak height of second allele (undigested DNA), as previously described.23 Spearman’s rank correlation (rs) analysis was performed using SPSS version 12 statistical software package on the XCI data obtained from the 26 females.
RESULTS
We analysed XCI patterns in multiple tissues from 26 human females to determine the relationship of XCI among tissues within individuals at different ages (See supplementary table S1). We repeated the AR assay on 22 randomly selected tissue samples to evaluate precision of the technique, including re-isolating DNA from the same tissue source of an individual. The mean value of the XCI difference between repeated measures was 3.1% (SD 2.3%) (range 0% to 8%).
The variation in XCI pattern was reasonably consistent within individuals. We assessed the variation of X inactivation between females by determining the standard deviation (SD) of the difference between the percentage X inactivation of the two alleles (first and second) and calculated the average of the SD for all individuals. This value relates variation in XCI among tissues within an individual to the entire set of subjects. The average SD was 16.8%. We also examined the variation of the percentage X inactivation of the first (smaller) allele by determining the SD and comparing among individuals. The average SD of the first (smaller) allele among all individuals was 10.7% (range 4.7% to 18.7%).
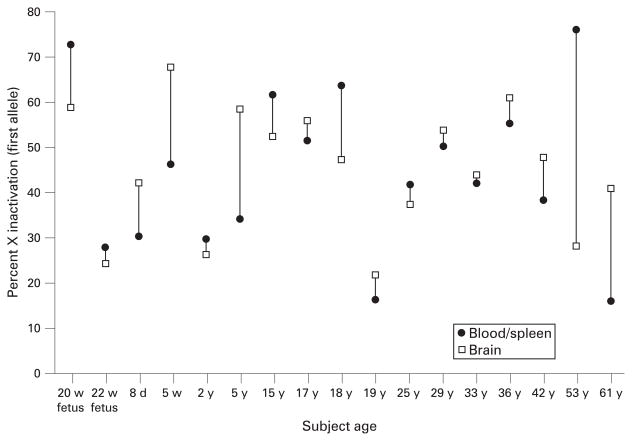
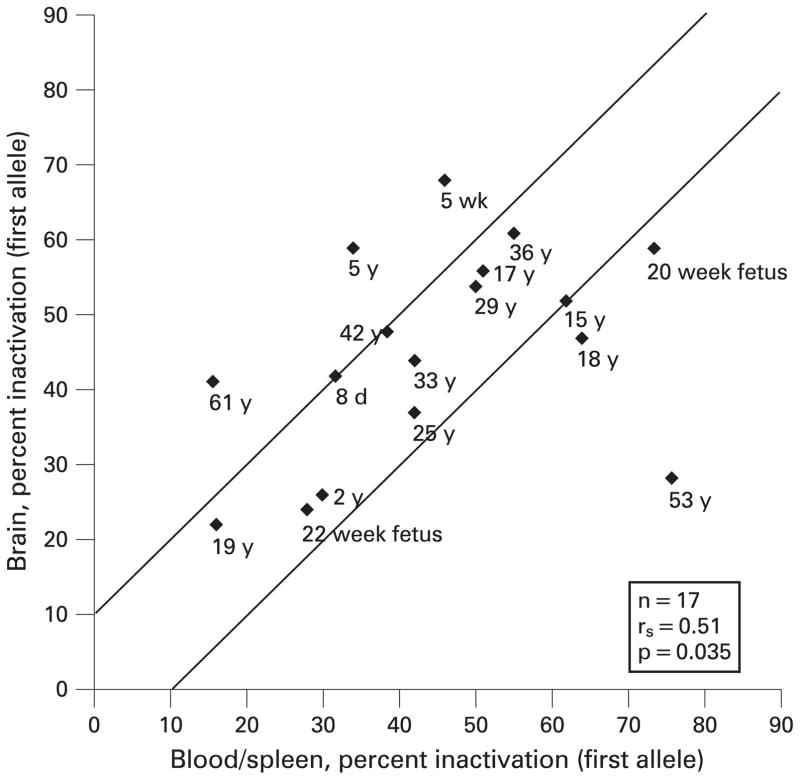
We compared haematopoietic tissues (eg blood and/or spleen) with the central nervous system (CNS; eg brain) to test a paradigm of accessible versus inaccessible tissue. Available blood and/or spleen and brain tissue from 17 of our 26 subjects were compared after determination of a high concordance of XCI between blood and spleen obtained from the same individuals (n = 17, rs = 0.94, p = 0.002). Figure 1 shows a comparison of percentage inactivation of the first (smaller) allele for blood/ spleen with brain, beginning from the youngest (fetus, 20 weeks’ gestation) to one of the oldest (61 years) females. Figure 2 shows a scatter plot of percentage X inactivation in blood/spleen versus brain. The XCI pattern found in the brain was similar to that found in blood/spleen, with 11 of 17 subjects differing by ≤10% (n = 17, rs = 0.51, p = 0.035). The SD was 12% for the difference between percentage X inactivation of the first allele for blood/spleen samples and brain. The two females with the greatest discrepancy between the XCI pattern for blood/spleen and brain were >50 years of age (figs 1,2). These data suggest that most females, particularly younger subjects, have a relatively high level of concordance in XCI pattern between haematopoietic and CNS tissues.
Figure 1.
Scatterplot of X-chromosome inactivation values for blood/spleen and brain (cerebrum) versus age. d, Days; w, weeks; y, years.
Figure 2.
Scatterplot of X-chromosome inactivation values for blood/ spleen versus brain. Lines are +/−10% variation in allele inactivation to allow for technical variation. When both blood and spleen were available from an individual, the average of the X-chromosome inactivation values was taken. w, Weeks; y, years.
We also found significant correlations between samples of blood/spleen and skin (n = 14, rs = 0.82, p<0.001), heart (n = 18, rs = 0.66, p = 0.002), lung (n = 13, rs = 0.77, p = 0.002), muscle (n = 17, rs = 0.66, p = 0.004), oesophagus/ stomach/intestine (n = 14, rs = 0.72, p = 0.004) and kidney (n = 14, rs = 0.54, p = 0.018) (supplementary figs 1–6), suggesting concordance between haematopoietic and other tissues, including those from other embryonic germ layers. Some tissues in individuals, particularly older women, did not demonstrate a significant XCI correlation pattern but showed a trend towards correlation (eg blood/spleen and liver (n = 20, rs = 0.40, p = 0.083); supplementary fig 7). However, small sample sizes may have contributed to the comparisons failing to reach significance.
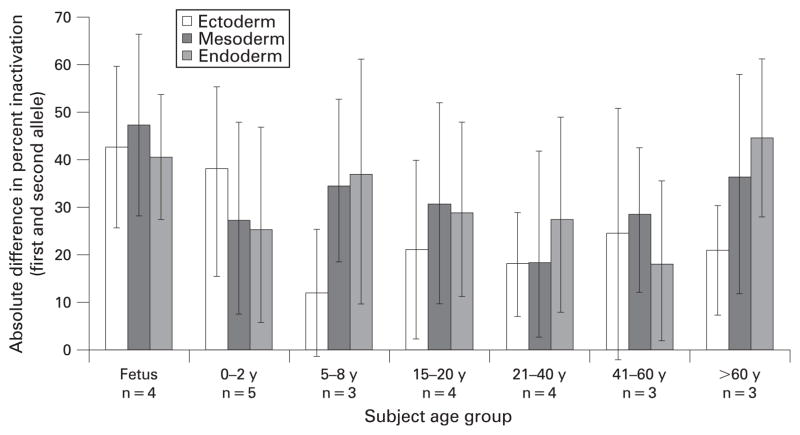
We also examined the variation of XCI in germ layers as a function of age and found no obvious patterns. We compared the difference in the two alleles for tissues within each age group for the separate germ layers (ie absolute value of the difference in percentage inactivation of the first and second alleles, fig 3). We also compared the mean percentage inactivation of tissues within a germ layer across age groups (fig 4). This chart shows the proportion (in percentages) of the total standard deviations within groups, calculated from the variance of X-inactivation values for all tissues from a subject. Variation among subjects within each age group was noted for each germ layer, but the greatest variation was observed in the three oldest subject groups (>21 years). However, the sample sizes were relatively small, which made identifying trends more difficult.
Figure 3.
Mean difference in X-chromosome inactivation for germ layers by age group. Mean absolute value of the difference between X-chromosome inactivation value for the first (smaller) and second alleles was calculated for each individual. Mean (SD) were determined for each subject age group and for tissues representing each of the three germ layers.
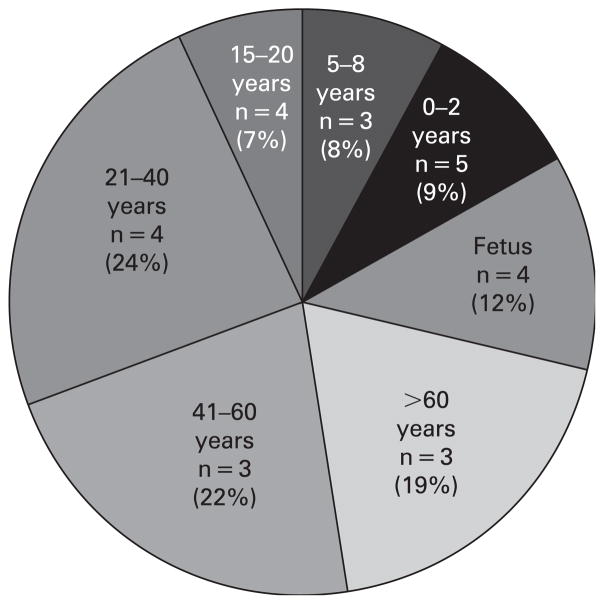
Figure 4.
Pie chart illustrating the proportion (%) of the age-group average SD, calculated from the variance of each individual within a group. Group mean variance in percentage X-chromosome inactivation of the first allele was calculated for all tissues from each individual and grouped by subject age, and the SD calculated from the individual variances.
XCI values for tissues within an individual tended to be clustered around the mean value, but nearly all females studied had one or two tissues that were >1 SD above or below the mean XCI value. These were collected to identify patterns. A total of 81 samples had inactivation values >1 SD from the mean XCI value for that individual. Tissues with XCI values >1 SD from the mean did not appear to originate in any particular germ layer: there were 27 such tissues (8 of which were liver) out of a total of 91 in endoderm; 39/148 tissues (7 of which were kidney) in mesoderm; and 15/41 (7 of which were skin) in ectoderm. Nearly half of these tissues with atypical XCI values in the ectoderm were from skin. A possible explanation may be the regenerating nature of skin, which could allow for selection of a particular cell type, leading to more XCI variation and resulting in a less valuable accessible candidate tissue for study.
We attempted to examine the variation associated with X inactivation related to age and germ layer origin. We determined the variance of X inactivation values from the smaller AR allele for all tissues measured from each individual. We then determined the mean (SD) of the variance for individuals within an age group and plotted the total SD as a pie chart to illustrate the variation within age groups (fig 4). It is clear that there is greater variation (greater SD of the variance) in the oldest three groups than in the younger groups, indicating a trend toward greater variability in XCI values with increasing age.
DISCUSSION
Several studies have reported on the relationship between XCI skewing and developmental disorders in females. These studies often examined XCI patterns in blood because the tissue of interest (eg CNS) was unavailable. Thus, the rationale for our study was to examine concordance of X inactivation patterns in blood compared with other tissues representing the three embryonic germ layers (accessible and inaccessible tissues), and to compare changes in X inactivation patterns with age among the different tissues. Therefore, we examined the XCI pattern in 34 tissues representing all three embryonic germ layers from 26 females at various ages (supplementary table S1).
Although the sample size is relatively small, a general concordance was seen in XCI pattern between haematopoietic (blood and/or spleen) and several other tissues (eg brain, skin, heart, lung, muscle, kidney and gastrointestinal; fig 2, supplementary figs 1–7). Perhaps not surprisingly however, the general concordance of XCI between tissues does not hold for all tissues in every individual examined; a 53-year-old woman in our study had greater variation in her tissues than did other subjects. The concordance of XCI among tissues may weaken with age. For example, in comparing haematopoietic tissues with brain (fig 2), the two oldest women had the greatest difference between inactivation values of the two tissues studied. This might result from a changing prevalence of cell lineage with age, perhaps due to cell selection.
In our study, the germ layer of origin did not seem to affect XCI variability between tissues (figs 3, 4). All tissues had non-significant differences in variation when examined by germ layer versus age. Greater variability in these comparisons may reflect the small sample sizes.
Furthermore, a general concordance of XCI patterns was seen between tissues within the same person, but occasionally one or more tissues had an XCI pattern in one tissue that was different from other tissues in that person; this applied particularly in older females. This has implications for expression of genes on the X chromosome in those tissues showing X-chromosome skewness (ie the expression may be primarily from one X chromosome, resulting in expression that may be detrimental and cannot be predicted by examining the XCI pattern of other tissues). Hopefully, our study will stimulate additional research with larger subject numbers and tissue types to further characterise the relationship (similarities and differences) among different tissues within an individual and the effect of age on XCI data and clinical outcomes.
Supplementary Material
Key points.
X-chromosome inactivation (XCI) patterns were studied using the polymorphic androgen receptor (AR) gene assay with DNA isolated from 26 different human females without history of malignancy, using 34 autopsy tissues representing the three embryonic germ layers.
A significant correlation (rs = 0.51, p = 0.035) was found between XCI values for blood/spleen and brain tissue, and for most other tissues representing the three embryonic germ layers.
A trend was seen for greater XCI variability with increasing age, particularly in older women (>60 years).
XCI data were comparable among accessible (eg blood) and inaccessible tissues (eg brain) in females at various ages and suggests that accessible tissues may be useful for genetic testing.
Acknowledgments
We thank Amon Holt III for helpful analysis of the data.
Funding: This study was partially supported by the Hall Foundation of Kansas City and a Physician Scientist Award (MGB) and the Fraternal Order of Eagles of the State of Kansas (DCB). Human tissues were obtained from the NICHD Brain and Tissue Bank for Developmental Disorders, University of Maryland, Baltimore, MD under contracts NO1-HD-4-3368 and NO1-HD-4-3383.
Footnotes
Supplementary material is available at http://jmg.bmj.com/content/vol45/issue5
Competing interests: None.
References
- 1.Lyon MF. Gene action in the X-chromosome of the mouse (Mus musculus L.) Nature. 1961;190:372–3. doi: 10.1038/190372a0. [DOI] [PubMed] [Google Scholar]
- 2.Avner P, Heard E. X-chromosome inactivation: counting, choice and initiation. Nat Rev Genet. 2001;2:59–67. doi: 10.1038/35047580. [DOI] [PubMed] [Google Scholar]
- 3.Bacher CP, Guggiari M, Brors B, Augui S, Clerc P, Avner P, Eils R, Heard E. Transient colocalization of X-inactivation centres accompanies the initiation of X inactivation. Nat Cell Biol. 2006;8:293–9. doi: 10.1038/ncb1365. [DOI] [PubMed] [Google Scholar]
- 4.Amos-Landgraf JM, Cottle A, Plenge RM, Friez M, Schwartz CE, Longshore J, Willard HF. X chromosome-inactivation patterns of 1,005 phenotypically unaffected females. Am J Hum Genet. 2006;79:493–9. doi: 10.1086/507565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hatakeyama C, Anderson CL, Beever CL, Penaherrera MS, Brown CJ, Robinson WP. The dynamics of X-inactivation skewing as women age. Clin Genet. 2004;6:327–32. doi: 10.1111/j.1399-0004.2004.00310.x. [DOI] [PubMed] [Google Scholar]
- 6.Sharp A, Robinson D, Jacobs P. Age- and tissue-specific variation of X chromosome inactivation ratios in normal women. Hum Genet. 2000;107:343–9. doi: 10.1007/s004390000382. [DOI] [PubMed] [Google Scholar]
- 7.Plenge RM, Stevenson RA, Lubs HA, Schwartz CE, Willard HF. Skewed X-chromosome inactivation is a common feature of X-linked mental retardation disorders. Am J Hum Genet. 2002;71:168–73. doi: 10.1086/341123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sangha KK, Stephenson MD, Brown CJ, Robinson WP. Extremely skewed X-chromosome inactivation is increased in women with recurrent spontaneous abortion. Am J Hum Genet. 1999;65:913–17. doi: 10.1086/302552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krepischi AC, Kok F, Otto PG. X chromosome-inactivation patterns in patients with Rett syndrome. Hum Genet. 1998;102:319–21. doi: 10.1007/s004390050698. [DOI] [PubMed] [Google Scholar]
- 10.Archer H, Evans J, Leonard H, Colvin L, Ravine D, Christodoulou J, Williamson S, Charman T, Bailey ME, Sampson J, de Klerk N, Clarke A. Correlation between clinical severity in patients with Rett syndrome with a p.R168X or p.T158M MECP2 mutation, and the direction and degree of skewing of X-chromosome inactivation. J Med Genet. 2007;44:148–52. doi: 10.1136/jmg.2006.045260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Talebizadeh Z, Bittel DC, Veatch OJ, Kibiryeva N, Butler MG. Brief report: non-random X chromosome inactivation in females with autism. J Autism Dev Disord. 2005;35:675–81. doi: 10.1007/s10803-005-0011-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Butler MG, Theodoro MF, Bittel DC, Kuipers PJ, Driscoll DJ, Talebizadeh Z. X-chromosome inactivation patterns in females with Prader-Willi syndrome. Am J Med Genet A. 2007;143:469–75. doi: 10.1002/ajmg.a.31506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berry-Kravis E, Potanos K, Weinberg D, Zhou L, Goetz CG. Fragile X-associated tremor/ataxia syndrome in sisters related to X-inactivation. Ann Neurol. 2005;57:144–7. doi: 10.1002/ana.20360. [DOI] [PubMed] [Google Scholar]
- 14.Naumova AK, Plenge RM, Bird LM, Leppert M, Morgan K, Willard HF, Sapienza C. Heritability of X chromosome--inactivation phenotype in a large family. Am J Hum Genet. 1996;58:1111–19. [PMC free article] [PubMed] [Google Scholar]
- 15.Plenge RM, Hendrich BD, Schwartz C, Arena JF, Naumova A, Sapienza C, Winter RM, Willard HF. A promoter mutation in the XIST gene in two unrelated families with skewed X-chromosome inactivation. Nat Genet. 1997;17:353–6. doi: 10.1038/ng1197-353. [DOI] [PubMed] [Google Scholar]
- 16.Hendrich BD, Plenge RM, Willard HF. Identification and characterization of the human XIST gene promoter: implications for models of X chromosome inactivation. Nucleic Acids Res. 1997;25:2661–71. doi: 10.1093/nar/25.13.2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sandovici I, Naumova AK, Leppert M, Linares Y, Sapienza C. A longitudinal study of X-inactivation ratio in human females. Hum Genet. 2004;115:387–92. doi: 10.1007/s00439-004-1177-8. [DOI] [PubMed] [Google Scholar]
- 18.Harris A, Collins J, Vetrie D, Cole C, Bobrow M. X inactivation as a mechanism of selection against lethal alleles: further investigation of incontinentia pigmenti and X linked lymphoproliferative disease. J Med Genet. 1992;29:608–14. doi: 10.1136/jmg.29.9.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maier EM, Kammerer S, Muntau AC, Wichers M, Braun A, Roscher AA. Symptoms in carriers of adrenoleukodystrophy relate to skewed X inactivation. Ann Neurol. 2002;52:683–8. doi: 10.1002/ana.10376. [DOI] [PubMed] [Google Scholar]
- 20.Migeon BR. Non-random X chromosome inactivation in mammalian cells. Cytogenet Cell Genet. 1998;80:142–8. doi: 10.1159/000014971. [DOI] [PubMed] [Google Scholar]
- 21.Gale RE, Wheadon H, Boulos P, Linch DC. Tissue specificity of X-chromosome inactivation patterns. Blood. 1994;83:2899–905. [PubMed] [Google Scholar]
- 22.Allen RC, Zoghbi HY, Moseley AB, Rosenblatt HM, Belmont JW. Methylation of HpaII and HhaI sites near the polymorphic CAG repeat in the human androgen-receptor gene correlates with X chromosome inactivation. Am J Hum Genet. 1992;51:1229–39. [PMC free article] [PubMed] [Google Scholar]
- 23.Lau AW, Brown CJ, Penaherrera M, Langlois S, Kalousek DK, Robinson WP. Skewed X-chromosome inactivation is common in fetuses or newborns associated with confined placental mosaicism. Am J Hum Genet. 1997;61:1353–61. doi: 10.1086/301651. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.