Abstract
Brain tumor analysis is moving towards volumetric assessment of magnetic resonance imaging (MRI), providing a more precise description of disease progression to better inform clinical decision-making and treatment planning. While a multitude of segmentation approaches exist, inherent variability in the results of these algorithms may incorrectly indicate changes in tumor volume. In this work, we present a systematic approach to characterize variability in tumor boundaries that utilizes equivalence tests as a means to determine whether a tumor volume has significantly changed over time. To demonstrate these concepts, 32 MRI studies from 8 patients were segmented using four different approaches (statistical classifier, region-based, edge-based, knowledge-based) to generate different regions of interest representing tumor extent. We showed that across all studies, the average Dice coefficient for the superset of the different methods was 0.754 (95% confidence interval 0.701–0.808) when compared to a reference standard. We illustrate how variability obtained by different segmentations can be used to identify significant changes in tumor volume between sequential time points. Our study demonstrates that variability is an inherent part of interpreting tumor segmentation results and should be considered as part of the interpretation process.
I. Introduction
Gliomas are the most frequent primary brain tumors in adults, accounting for 70% of all malignant primary brain tumors [1–2]. The development of computational methods to objectively extract insights out of imaging data is an ongoing challenge [3–4]. For instance, current tumor guidelines to assess tumor progression in the clinical setting are often limited to one- or two- dimensional orthogonal measurements; standards like the Revised Assessment in Neuro-Oncology (RANO) [5] attempt to quantify lesions by assigning different evaluation options depending on these linear measurements (longest diameter measures) to ascertain significant change.
Assessing tumor change during treatment is important to estimate the effectiveness of a therapy or medical procedure, and for comparing results of different treatment options among different populations. More sophisticated quantified analyses, such as volumetrics, require segmentation – that is, identifying a key region of interest (ROI) on multiple imaging sequences. In routine clinical practice, tumor segmentations are drawn manually by an expert (e.g., radiologist), which while providing meaningful results are highly labor intensive and time consuming. Automating such tumor segmentation is thus an active area of research [6–7].
In this paper, we present an approach for characterizing the variability that occurs when segmenting complex tumor shapes with heterogeneous textures, and its impact when comparing volumetric assessments across time points. We first propose a method for generating probabilistic maps of error, characterizing the variability in how edges are segmented by different algorithms. We then utilize equivalence testing to determine whether volumes measured at two neighboring time points have significantly changed (e.g., disease has progressed) given the variability in the ROIs generated by the different segmentation algorithms. We believe the presented approach provides a more realistic assessment of volumetric change in the presence of variability.
II. Methods
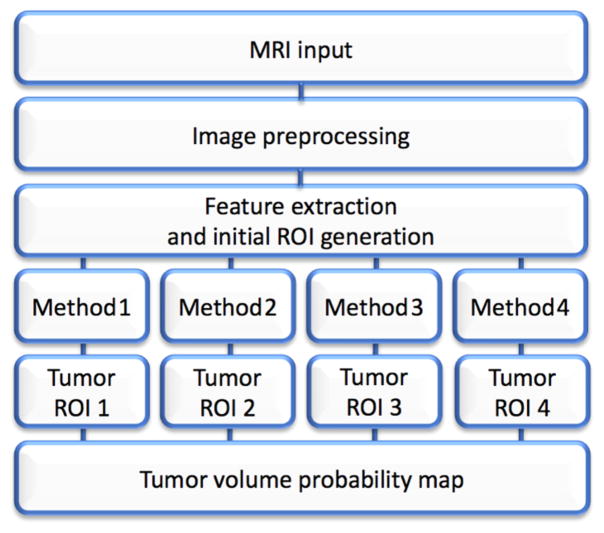
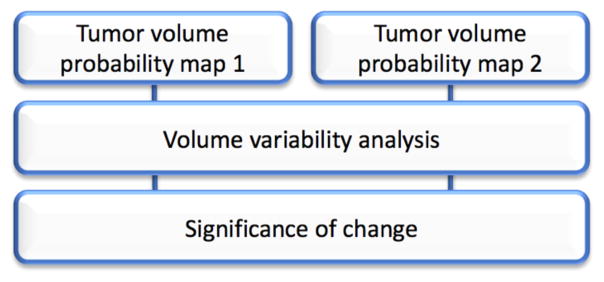
The steps to quantify volumetric variability in terms of a tumor probability map is summarized in Figure 1. Subsequently, the process of taking into account the resultant range of volumes given variability in ROIs in order to determine true change is given in Figure 2.
Figure 1.
Flowchart that exemplifies the general workflow from the MRI input until the generation of tumor probability maps.
Figure 2.
The comparison between different time points to define statistical significance can be achieved by doing a standard analysis of variance on the output tumor volumes
A. Input data
We randomly selected eight patients diagnosed with glioblastoma multiforme with preoperative imaging studies available. In total, 32 studies acquired using a 3.0T Magnetic Resonance Imaging (MRI) system were analyzed. Each study consisted of standard T1, T1 with contrast, T2, and Fluid Attenuated Inversion Recovery (FLAIR) sequences.
A reference standard for total tumor volume was manually created. Total tumor volume was defined as the contrast enhancing portion and the necrotic core found on T1-weighted with contrast enhancement scans. Six manual segmentations were generated for each study by three trained annotators (each study was segmented twice during two independent sessions to assess intra- and inter-rater reliability). The annotators achieved an inter-rater agreement of 91 ± 2% and intra-rater agreement of 93 ±1 %. The reference standard was used to assess the accuracy of the four segmentation methods below.
B. Segmentation methods
Significant progress in brain tumor segmentation has been made in recent years due to the rapid development of machine learning techniques [3], but none of these methods have examined the level of variability that may occur, particularly at the boundary of the tumor. Such tumor segmentation is specifically challenging, as tumors are irregular in shape, inhomogeneous in texture and have discontinuous edges; along with other challenges associated with the acquisition and post-processing processes (e.g., image registration to spatially co-align acquired studies for comparison) [3,8]. As such, determining changes in size over time is challenging to estimate and comes with several caveats (differences in acquisition parameters or segmentation method, intra- and inter-observer variability, movement during the scan, etc.).
To explore variability in tumor segmentation, three different approaches were selected based on the availability of the code and their capacity to provide automatic contours of the tumor boundaries. Additionally, a fourth segmentation method was developed.
Classifier-based segmentation: We applied a support vector machine classifier that was trained on a set of brain tumors to sort each pixel as normal or not normal (tumor) [9]. This generative model outputs a color coded mask that contains the tumor boundaries.
Region-based active contour segmentation: This geometric active contour model is based on the Chan-Vese algorithm [10]. This model evolves an automatically specified initial ROI (e.g., bounding box) that contains the expected intensities that compose the tumor by using a level-set approach.
Edge-based segmentation: This method follows a geodesic segmentation [11] approach by taking an input ROI that surrounds the tumor and then evolve it until a state of lower energy located at the boundary is found.
Knowledge-based segmentation: An in-house developed algorithm that finds the tumor boundary by doing a histogram analysis on the three-dimensional image. This algorithm finds the pixels that have the lowest probability of being normal, according to a prior distribution of the different cerebral tissues. This prior information is obtained in the form of multiple probability masks for each tissue (gray matter, white matter, cerebro-spinal fluid and soft tissue) using SPM12 [12]. Afterwards the ROI is obtained by identifying the regions where the intensities on each slice deviate from the expected value for each tissue (i.e. lower probability). We then select the boundary of the tumor by doing standard histogram analysis.
Prior to the execution of any of the aforementioned segmentation algorithms, different image preprocessing steps were employed, including image normalization, bias field image correction, rigid registration to a reference atlas using FSL [13] and skull stripping [14].
C. Probability map
Once the segmentation was completed on all input images, the individual results (masks) were overlaid and aggregated into a probability map. In this work, as we only examined four different methods (i.e., four sources of variation), the map consists of five discrete states: 0% (regions without any ROI), 25% (regions with 1 ROI), 50% (2 ROIs), 75% (3 ROIs), and 100% (full overlap). In summary, higher probabilities correspond to higher agreement (overlap) between the methods.
D. Temporal progression
A principal task when evaluating tumor progression over time is to define if an important difference between two observed volumes exists. To provide an objective way to determine if a tumor has significantly changed over time, an equivalence test can be used to evaluate if the volume estimates for both time points are sufficiently similar to be considered equivalent or not. This hypothesis can be tested by doing a one-way analysis of variance (ANOVA) by taking the different segmentation results that compose the probability map for the different follow-ups and computing the actual tumor change over time relative to the significance of the statistical test result.
III. Results
The previously described segmentation methods were used to segment the total tumor volume found on each of the available sequences for all subjects. A pair of representative examples of the tumor outline achieved by each method as well as the probability map can be observed on Figures 3 (FLAIR) and 4 (T1 with contrast enhancement).
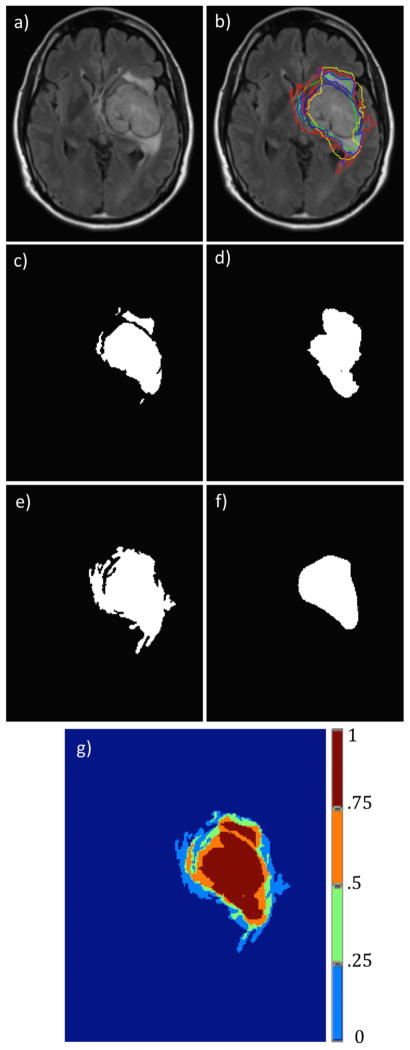
Figure 3.
Segmentation results for the different segmentation approaches on an example T2-weighted FLAIR image. Figure 3a is the original input image, Figure 3b is the image with the different segmentation results overlapped, Figure 3c–f show the binary images associated with the different segmentation methods (knowledge-based, region-based, classifier and edge-based). Figure 3g shows the output probability map.
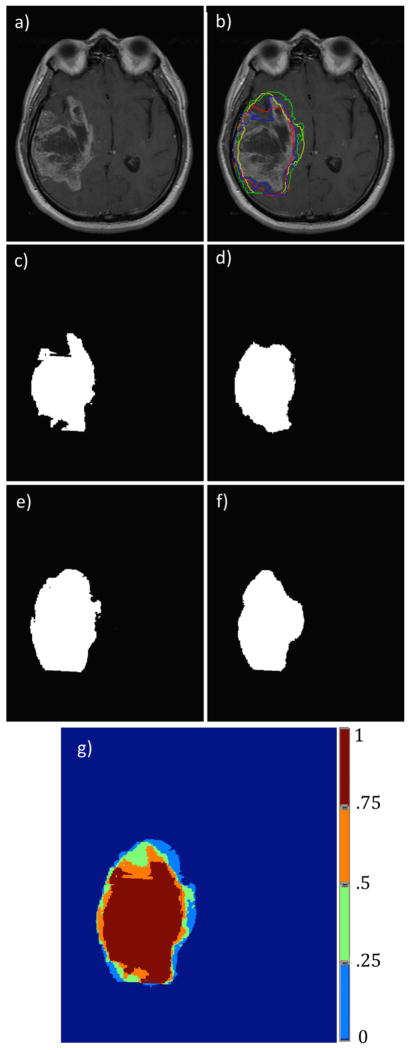
Figure 4.
Segmentation results for the different segmentation approaches on an example T1-weighted image with contrast enhancement. Figure 4a is the original input image, Figure 4b is the image with the different segmentation results overlapped, Figure 4c–f show the binary images associated with the different segmentation methods (knowledge-based, region-based, classifier and edge-based). Figure 4g shows the output probability map.
A comparison against the reference standard was performed to assess the validity of each of the individual tumor outlines. Additionally, a tumor mask was produced by taking the superset of the overlapped results for each method. The Dice coefficient found when comparing the gold standard and the total abnormality found on T1-weighted contrast enhancement sequences for each subject as well as for each method is presented on Table 1.
TABLE 1.
Comparison With Gold Standard
The Dice coefficient obtained when comparing the gold standard and the abnormality found on contrast enhancement sequence is shown for the different segmentation approaches (classifier, region-based, edge-based, knowledge-based) as well as the superset of them. It can be noted that for the majority of the cases the mask obtained by the combination of the individual methods is more similar to the reference (higher Dice coefficient).
| Subject | Method1 | Method2 | Method3 | Method4 | Superset |
|---|---|---|---|---|---|
| 1 | .673 | .583 | .705 | .770 | .747 |
| 2 | .634 | .603 | .564 | .706 | .736 |
| 3 | .812 | .681 | .707 | .766 | .852 |
| 4 | .713 | .547 | .539 | .788 | .798 |
| 5 | .763 | .608 | .685 | .710 | .718 |
| 6 | .793 | .647 | .756 | .774 | .785 |
| 7 | .669 | .668 | .595 | .630 | .709 |
| 8 | .649 | .691 | .655 | .694 | .694 |
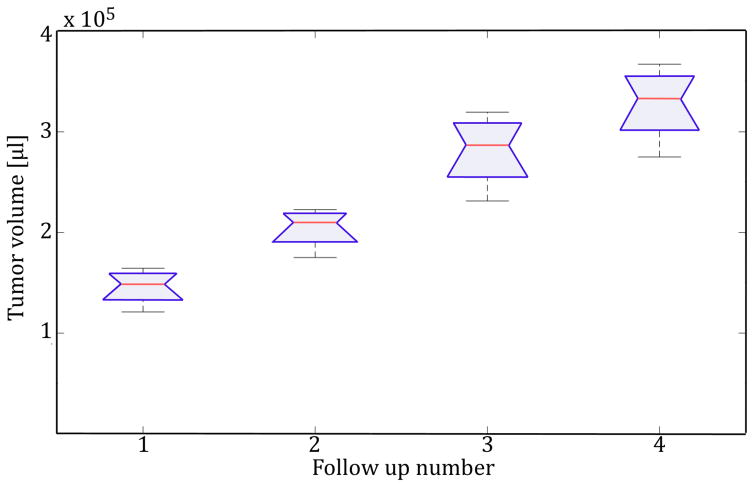
Figure 5 and Table 2 show the tumor volume progression over time on a single subject with multiple follow-ups, as well as the result of the ANOVA test to determine statistical significance of volume change for each pair of sequential tumor volume measures.
Figure 5.
Boxplot that indicates tumor volume progression over time as well as the variability observed at each time point.
TABLE 2.
Volume Comparison Results
The results from the analysis of variance show statistically significant difference between the different measures. Intergroup refers to the variance found between segmentation methods and intragroup to the variance between different time points. The F-statistic is computed using the ratio of the intergroup and intragroup mean square error (MSE) for each pair of time points.
| Scan # | MSE | F-statistic | P value | |
|---|---|---|---|---|
| 1 vs. 2 | Intergroup | .0085 | 25.68 | 0.001 |
| Intragroup | .0033 | |||
| 2 vs. 3 | Intergroup | .0148 | 18.7 | 0.0025 |
| Intragroup | .0007 | |||
| 3 vs. 4 | Intergroup | .0079 | 6.27 | 0.0367 |
| Intragroup | .0013 |
IV. Discussion
On this work we explored the idea of characterizing variability by aggregating the results of several tumor segmentation methods and defining an objective way to define if the overall volume mass is significantly changing, which plays an important role in decision making.
It was observed that while in general each algorithm shows some error while segmenting the tumor, the probability map was better at describing the distribution of the abnormality and was informative in determining which regions of the image were more likely to be part of the tumor. This finding leads to the conclusion that repeated measures of tumor volume tends to do better than any single measurement and is more robust against errors.
While many have argued that three-dimensional evaluation of tumor size provides a more accurate assessment of disease progression/regression over traditional two-dimensional measurements, the methods for generating these volumes should not be taken for granted: errors that may occur during the segmentation process, whether manual or automated, should be considered as part of the interpretation process. Accurately characterizing change in time is necessary to understand the aggressiveness of the disease and to evaluate treatment response.
Future work includes to explore the variability found on the various tumor sub-components of the tumor such as necrosis, edema and enhancing tumor to progressively move to a more quantifiable way to understand the disease and ultimately improve treatment effectiveness.
Footnotes
Research reported in this paper was supported by the National Cancer Institute of the National Institutes of Health under award number R01CA1575533. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Contributor Information
Edgar A. Rios Piedra, Department of Radiological Sciences at the University of California, Los Angeles, CA. Department of Bioengineering at the University of California, Los Angeles, CA. Medical Imaging Informatics (MII) at the University of California, Los Angeles, CA.
Ricky K. Taira, Department of Radiological Sciences at the University of California, Los Angeles, CA. Department of Bioengineering at the University of California, Los Angeles, CA. Medical Imaging Informatics (MII) at the University of California, Los Angeles, CA.
Suzie El-Saden, Department of Radiological Sciences at the University of California, Los Angeles, CA. Department of Bioengineering at the University of California, Los Angeles, CA. Medical Imaging Informatics (MII) at the University of California, Los Angeles, CA. Department of Radiology, Veterans Administration Greater Los Angeles Healthcare, Los Angeles, CA.
Benjamin M. Ellingson, Department of Radiological Sciences at the University of California, Los Angeles, CA. Department of Bioengineering at the University of California, Los Angeles, CA. Department of Biomedical Physics and Neurology at the University of California, Los Angeles, CA.
Alex A. T. Bui, Department of Radiological Sciences at the University of California, Los Angeles, CA. Department of Bioengineering at the University of California, Los Angeles, CA. Medical Imaging Informatics (MII) at the University of California, Los Angeles, CA.
William Hsu, Department of Radiological Sciences at the University of California, Los Angeles, CA. Department of Bioengineering at the University of California, Los Angeles, CA. Medical Imaging Informatics (MII) at the University of California, Los Angeles, CA.
References
- 1.Lima Flavia RS, et al. Glioblastoma: therapeutic challenges, what lies ahead. Biochimica et Biophysica Acta (BBA)-Reviews on Cancer. 2012;1826(2):338–349. doi: 10.1016/j.bbcan.2012.05.004. [DOI] [PubMed] [Google Scholar]
- 2.Veliz Ignacio, et al. Advances and challenges in the molecular biology and treatment of glioblastoma—is there any hope for the future? Annals of translational medicine. 2015;3(1) doi: 10.3978/j.issn.2305-5839.2014.10.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bauer Stefan, et al. A survey of MRI-based medical image analysis for brain tumor studies. Physics in medicine and biology. 2013;58(13):R97–129. doi: 10.1088/0031-9155/58/13/R97. [DOI] [PubMed] [Google Scholar]
- 4.Hevia-Montiel Nidiyare, et al. Neuromorphometry of primary brain tumors by magnetic resonance imaging. Journal of Medical Imaging. 2015;2(2):024503. doi: 10.1117/1.JMI.2.2.024503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wen Patrick Y, et al. Updated response assessment criteria for high-grade gliomas: response assessment in neuro-oncology working group. Journal of Clinical Oncology. 2010;28(11):1963–1972. doi: 10.1200/JCO.2009.26.3541. [DOI] [PubMed] [Google Scholar]
- 6.Lin Lin, et al. Significant predictors of patients’ uncertainty in primary brain tumors. Journal of neuro-oncology. 2015;122(3):507–15. doi: 10.1007/s11060-015-1756-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao Liang, et al. Semi-automatic brain tumor segmentation by constrained MRFS using structural trajectories. Medical Image Computing and Computer-Assisted Intervention–MICCAI 2013; Berlin Heidelberg: Springer; 2013. pp. 567–575. [DOI] [PubMed] [Google Scholar]
- 8.Menze Bjoern, Reyes Mauricio, Van Leemput Koen. The Multimodal Brain TumorImage Segmentation Benchmark (BRATS) IEEE Trans Med Imaging. 2015;34(10):1993–2024. doi: 10.1109/TMI.2014.2377694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Porz Nicole, et al. Multi-modal glioblastoma segmentation: man versus machine. PloS one. 2014;9(5):e96873. doi: 10.1371/journal.pone.0096873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chan Tony F, Vese Luminita. Active contours without edges. Image processing, IEEE transactions on. 2001;10(2):266–277. doi: 10.1109/83.902291. [DOI] [PubMed] [Google Scholar]
- 11.Caselles Vicent, Kimmel Ron, Sapiro Guillermo. Geodesic active contours. International journal of computer vision. 1997;22(1):61–79. [Google Scholar]
- 12.Ashburner John, Friston Karl J. Unified segmentation. Neuroimage. 2005;26(3):839–851. doi: 10.1016/j.neuroimage.2005.02.018. [DOI] [PubMed] [Google Scholar]
- 13.Jenkinson M, Beckmann CF, Behrens TE, Woolrich MW, Smith SM, FSL NeuroImage. 2012;62:782–90. doi: 10.1016/j.neuroimage.2011.09.015. [DOI] [PubMed] [Google Scholar]
- 14.The AFNI program (Analysis of Functional NeuroImages) National Institute of Health; USA: [Google Scholar]