Abstract
Exosomes offer new insight into cancer biology with both diagnostic and therapeutic implications. Because of their cell-to-cell communication, exosomes influence tumor progression, metastasis, and therapeutic efficacy. They can be isolated from blood and other bodily fluids to reveal disease processes occurring within the body, including cancerous growth. In addition to being a reservoir of cancer biomarkers, they can be re-engineered to reinstate tumor immunity. Tumor exosomes interact with various cells of the microenvironment to confer tumor-advantageous changes that are responsible for stromal activation, induction of the angiogenic switch, increased vascular permeability, and immune escape. Exosomes also contribute to metastasis by aiding in the epithelial-to-mesenchymal transition and formation of the pre-metastatic niche. Furthermore, exosomes protect tumor cells from the cytotoxic effects of chemotherapy drugs and transfer chemoresistance properties to nearby cells. Thus, exosomes are essential to many lethal elements of cancer and it is important to understand their biogenesis and role in cancer.
Keywords: Signaling, Immunosurveillance, Fibroblast, Targeted therapy, Multivesicular endosome, Vaccine, ESCRT, Biogenesis
Introduction
Despite promising advances, cancer remains the second leading cause of death in the US and new insights into cancer biology are necessary to drive therapeutic innovation [1, 2]. Once viewed as a passive, insignificant appendage to cancer, the microenvironment has moved into the limelight as an integral contributor to cancer suppression [3–5], cancer promotion [6, 7], and drug resistance [8, 9]. It is now recognized that a tumor is really a system, more organ-like than homogenous, with complex interactions between the microenvironment and tumor cells [10]. The tumor microenvironment, or stroma, is composed of fibroblasts and the extracellular matrix they deposit, immune cells, and vascular cells [10]. Not only does the microenvironment provide prognostic information since changes in the microenvironment correspond to a better or worse patient outcome, but it also reflects treatment efficacy and has therapeutic potential, since it presents a wealth of new targets [11]. Reviews covering multiple aspects of the tumor microenvironment have been published, including bipolar effects [12], therapeutic targeting [13], immunology [14], and chemoprevention [15]. In this review, we focus instead on an emerging aspect of the tumor microenvironment that enables the crucial communication between the tumor and microenvironment and is implicated in tumor progression, metastasis, and chemoresistance: exosomes (see Box 1 for historical perspective). Tumor progression results from active partnership between the tumor and microenvironment that would be impossible without efficient modes of exchanging information: direct cell-to-cell contact, secretion of signaling molecules, and release of vesicles like exosomes into the extracellular space [16]. In this review, we present the biogenesis of exosomes via multivesicular endosomes (MVEs), the unique contribution they make to cancer progression through interaction with multiple microenvironmental cell types, the role they play in metastasis and chemoresistance, and discuss their application to diagnostics and therapy [17].
Exosome biogenesis in cancer
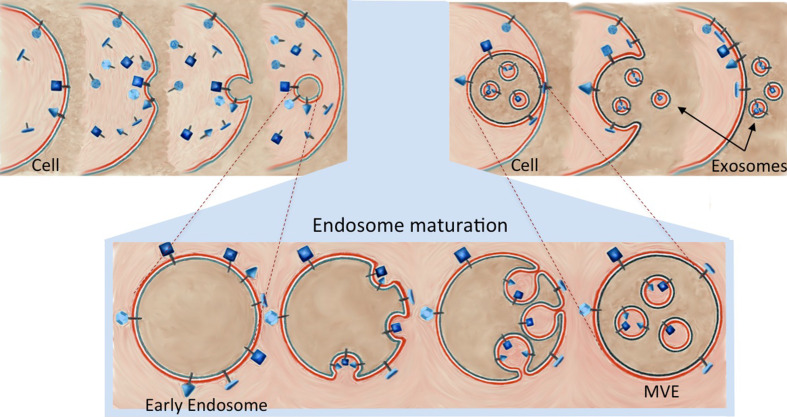
Although there has been some confusion with regards to exosome identification, exosomes are defined by certain shared characteristics, including size (50–100 nm), density, morphology, and general protein composition (Fig. 1, see Box 2 for use of “exosome” in scientific literature) [18, 19]. Unlike other extracellular vesicles such as microvesicles, ectosomes, and membrane particles, exosomes do not originate by the direct budding or shedding of the plasma membrane (shedding microvesicles reviewed in [20]) [18]. Instead, exosomes are derived from MVEs, commonly referred to as multivesicular bodies (MVBs). Exosome formation occurs when the membrane of the MVE bulges inward and pinches off to create small membranous vesicles within the MVE, packed with cytoplasmic contents, including proteins, RNAs, and recently genomic DNA was also found [21] (refer to [22] for a free database of more than 14,000 biomolecules identified in exosomes). Subsequent exosome secretion occurs when MVEs fuse with the plasma membrane and release their membrane-bound contents (exosomes) into the extracellular environment (Fig. 2). It should be noted that not all MVEs go on to release exosomes; an alternative fate of MVE cargo is degradation by fusion with the lysosome. It has been shown that the presence of high concentrations of the ceramide lipid family appear to help MVE contents escape lysosomal digestion in favor of release as exosomes [23].
Fig. 1.
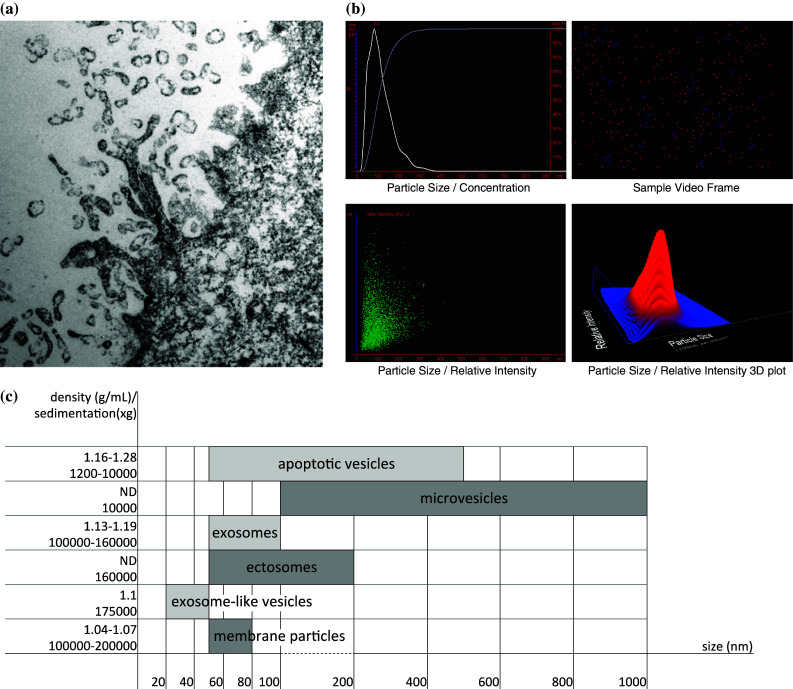
Exosomes. a TEM of plectin-positive pancreatic ductal adenocarcinoma (PDAC) cells with enhanced exosome production. Plectin is necessary for exosome production in PDAC and is aberrantly expressed on the cell surface through an exosome-mediated process (see [39] for details about PDAC exosomes). b Nanosight analysis of exosomes isolated from mouse serum using ExoQuick-TC isolation reagent shows the exosome size/concentration, a sample video frame of the exosomes, as well as the exosome size/relative intensity in both a scatter and 3D plot (unpublished image). c Exosomes can be differentiated from other secreted vesicles by their size (nm), density in sucrose (g/mL), and sedimentation speed (×g) (numbers from [18])
Fig. 2.
Exosome Biogenesis. The plasma membrane buds inward, forming a membrane-bound vacuole. This endosome goes through several changes as it matures from an early endosome to a late endosome. Most notably, the endosomal membrane buds inward and pinches off to make membrane-bound vesicles inside the endosome and the endosome is now titled a multivesicular endosome (MVE). The MVE may travel to the lysosome and degrade its contents or it may travel to and fuse with the plasma membrane, releasing its contents, which, once existing outside the cell, are called exosomes (see [26] for more details about MVEs)
The consequence of the mechanism of exosome biogenesis is an extracellular particle whose membrane reflects the composition of the MVE, but with the same orientation as the parental cell plasma membrane (Fig. 2). The exosomal membrane is enriched in endosome-related membrane transport and fusion proteins (flotillin, Annexins, GTPases), certain lipids (sphingomyelin, cholesterol, ceramide) and, if from antigen presenting cells, MHC-II. Exosomes also contain endosome-specific tetraspanins (CD9, CD63, CD81, CD82), and MVE biogenesis-related proteins (Alix, TSG101) on their membrane surfaces [24–26]. Beyond their characteristic collection of membrane markers, the molecular content of exosomes can vary significantly based on the physiological conditions and the original cell type [27]. Importantly, the composition of an exosome is not a mere reflection of the donor cell, and it has been shown that the profiles of exosomal cargo can be substantially different from the originating cell, which indicates the existence of a highly controlled sorting process [28]. The exact mechanisms involved in exosome packaging have not been fully delineated, but appear to be similar to how lysosomal-bound MVEs are packaged since endosomal-sorting complexes required for transport (ESCRT) proteins are found in exosomes. [29].
The proteins of the ESCRT pathway divide into four complexes that identify and aggregate ubiquinated proteins in the endosomal membrane (ESCRT-0), cause membrane budding (ESCRT-I, -II), and instigate separation from the membrane (ESCRT-III). However, there is also evidence for ESCRT-independent packaging: MVEs are not obliterated by simultaneous depletion of the subunits belonging to the four ESCRT proteins [29]. These pathways may involve lipids such as sphingosine-1-phosphate (S1P) [30] and the tetraspanin-enriched microdomains [31]. Colombo et al. [29] confirmed that while some exosome production may be ESCRT-independent, at least some exosomes are ESCRT-dependent; they showed that silencing genes of ESCRT-0 (HRS, STAM1), ESCRT-I (TSG101), or VPS4B altered the amount of exosomes produced and their cargo. Insight into the selective sorting of miRNAs into exosomes has recently been reported, involving sumoylated heterogeneous nuclear ribonucleoproteins, mainly hnRNPA2B1, which bind to a subset of miRNAs and control their loading into exosomes [32].
In addition to exosomal packaging, each step of exosome biogenesis—trafficking to the plasma membrane, docking, fusion, and release—appears to be highly organized and regulated and, therefore, is potentially altered in cancer. Notably, exosome release tends to be exacerbated in tumor cells as compared to other proliferating cell types, which can be a result of stimulation in response to stressful conditions due to excess growth and cell damage caused by chemotherapeutics. In many cancers, aberrant p53 stimulation results in over-expression of tumor suppressor-activated pathway 6 (TSAP6), which, in turn, increases exosome production [33, 34]. Additionally, heparanase, which is an enzyme up-regulated in many tumor cell lines, has been implicated in exosome secretion [35]. However, tumor cell exosome release is inhibited by previously secreted tumor exosomes that are still present in the microenvironment, creating a balancing negative feedback control loop [36]. The GTPases Rab27a and Rab27b control secretory pathways, including exosome release, probably by regulating the trafficking of secretory vesicles to the plasma membrane, tethering the vesicles to the plasma membrane, and/or helping fuse the vesicles to the plasma membrane [37]. In the absence of Rab27a, other Rab proteins, especially Rab27b, appear to be able to act as compensatory pathways for exosome secretion; Rab2b, Rab5a, Rab9a, Rab27a, and Rab27b have all been shown to decrease exosome secretion in cervical cancer cells [38]. Similarly, knockdown of plectin in three types of pancreatic cancer cells decreased exosome production about fivefold [39]. Another Rab protein, Rab11, has been shown to regulate the exosome pathway of a leukemia cell line by influencing calcium-dependent fusion of MVEs with the plasma membrane [40, 41]. Recently, invadopodia have emerged as a docking site for exosomes, promoting cancer invasion. In fact, exosome secretion and invadopodia formation appear to be interdependent; inhibition of exosome biogenesis affects invadopodia formation and stability and, conversely, inhibition of invadopodia formation greatly decreases exosome secretion [42].
Exosomes and the tumor microenvironment
Tumor progression, metastasis, and chemoresistance all depend on the ability of the tumor and its microenvironment to communicate. Exosomes are a unique form of transferring information both locally and to a distant site. Cells of the microenvironment release exosomes to affect each other and the tumor cells. Meanwhile, tumor cells release exosomes to reprogram their surroundings to be tumor permissive and even tumor promoting. Here we discuss the evidence of how exosomes influence each major type of stromal cell—cancer-associated fibroblasts, vascular endothelial cells, and immune cells—as well as how exosomes accelerate metastasis and chemoresistance.
Cancer-associated fibroblasts
Fibroblasts become “activated” as they progress through changes during the neoplastic process, including changes in morphology and protein expression, with disorganized and uncontrolled growth as well as increased production of collagen and stimulation of hyaluronate synthesis [43]. These activated fibroblasts can both hinder and promote tumor growth and progression, depending on the molecular state of the tumor epithelial cells, and are certainly capable of accelerating growth and promoting tumor cell invasion [6, 44–48]. Cancer exosomes have been shown to trigger such fibroblast transformation through the TGFβ/Smad pathway and elicit effects unique from soluble TGFβ [49–51]. It has even been proposed that the stroma may go beyond promoting pre-existing tumors to causing tumorigenicity in adjacent cells through oncogenic signals [43]. Co-cultures of tumor fibroblasts with non-transformed epithelial cells caused immature, pleomorphic epithelial cells with enlarged nuclei and aberrant mitosis. Cells also increased in their rate of proliferation, lost polarity, and had altered cell cycle protein expression; p53, PCNA, Ki67and cytokeratin expression were increased, whereas p21 nuclear expression and Bcl2 were decreased [52].
Vascular endothelial cells
Tumors must have access to nutrients, oxygen, and waste removal to grow beyond a few cubic millimeters and to accomplish this, the tumor cells must access the host vasculature and divert blood to the tumor [53, 54]. Creation of a tumor blood supply requires induction of the angiogenic switch, in other words sufficient increase of pro-angiogenic factors to overcome anti-angiogenic factors [54]. Many soluble factors such as VEGF contribute to flipping this angiogenic switch [55]. In addition, exosomes have also been shown to play a role in increasing angiogenesis [55]. As hypoxia is sensed throughout the tumor, carcinoma cells secrete exosomes into the microenvironment to initiate signaling and flip the angiogenic switch to ensure adequate oxygenation [55, 56]. Hypoxia-induced proteins secreted in the tumor exosomes are taken up by the normal host endothelial cells, where the exosomal cargo stimulates new tubule formation, eventually creating a network of blood vessels to supply that area [57]. Endothelial cells that have taken up hypoxic tumor exosomes start releasing growth factors and cytokines that stimulate pericytes via the PI3 K/AKT pathway [58]. Moreover, exosomes transfer miR-105 to endothelial cells, increasing vascular permeability by damaging tight junctions [59].
Immune cells
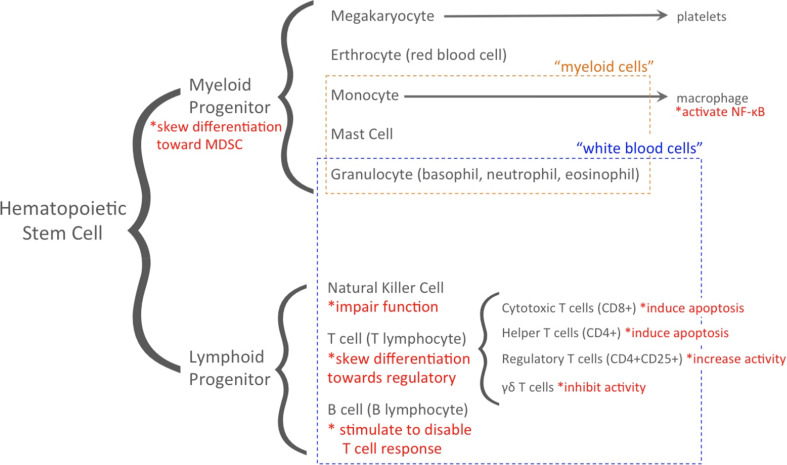
The immune system responds to cancer through both innate and adaptive immunity, the latter of which involves antigen-presentation. Hematopoietic stem cells from the bone marrow give rise to blood cells, including immune cells (Fig. 3). Dendritic cells are a separate class of immune cells that claim multiple origins and facilitate communication between the innate and adaptive components [60, 61]. Exosomes produced by immune cells and those produced by tumor cells have opposite effects. Thus, in the initial stages of cancer, the host immune system may stifle tumor progression; however, growing tumors activate suppressive pathways and eventually manage to evade immune surveillance and exosomes are involved in both responses [60]. Exosomes produced by immune cells ignite an anti-tumor immune response. Mast cell exosomes induce dendritic cell differentiation and activate T and B cells and dendritic cell exosomes sensitize other immune cells to tumor antigens [62, 63]. In contrast, tumor-derived exosomes participate in immune escape by (1) increasing the influencing myeloid progenitor cell differentiation, (2) decreasing T cell proliferation and effector functions, and (3) cancelling the natural cytotoxic responses mediated by natural killer cells (Fig. 3) [64, 65].
Fig. 3.
Role of exosomes in immune escape. This immune cell lineage groups cell types in dashed boxes that are commonly classified as myeloid cells or white blood cells. Highlighted in red with asterisks are the immune escape mechanisms conferred to each immune cell type by tumor-derived exosomes (functions from [60–81])
The first part of exosome participation in immune escape involves myeloid cell precursor differentiation. Tumor exosome uptake by myeloid progenitor cells inhibits differentiation into dendritic cells and instead channels them into myeloid-derived suppressor cell (MDSC) differentiation, causing MDSCs to accumulate in tumor tissue, primary and secondary lymphoid organs, and blood [66, 67]. This creates a dual pro-tumor effect: dendritic cells are no longer present to display antigens to simulate the host immune system to act against the tumor and MDSCs supply the tumor with growth- and angiogenesis-stimulating molecules and factors [65–67]. Such exosome-induced accumulation of MDSCs is MyD88 dependent and requires tumor exosomal expression of TGFβ and prostaglandin E2 (PGE2) [65, 68, 69]. While toll-like receptor 2/Stat3 has also been shown to be essential for tumor exosomes’ effect on MDSCs, additional studies revealed that this might be a cell culture artifact [70]. Additionally, MDSCs create a powerful immunosuppressive effect because they deplete arginine and produce nitric oxide to decrease T cell functionality [71].
Second, exosomes dysregulate T cells, by impeding proliferation, decreasing differentiation into helper T cells, increasing differentiation into regulatory T cells, and varying the levels of cytokines in stimulated T cells [72]. In particular, tumor exosomes impair T cell response to IL-2, which inhibits differentiation of naïve T cells into Th1 and Th17 helper T cells. Th1 helper T cells affect macrophages, cytotoxic T cells, and helper T cells; whereas Th17 helper T cells affect neutrophils, B cells, and helper T cells [73]. Tumor exosomes also deplete cytotoxic T cell populations by inducing apoptosis via FasL and deplete mature Th1 populations by inducing apoptosis via galectin-9 [74, 75]. Moreover, tumor exosomes stimulate B cells to disable helper T cells so that the immune system is less responsive to tumor antigen presentation [76, 77].
The third arm of immunosurveillance concerns the cancelling of cytotoxic responses mediated by natural killer cells. Negating natural killer cells’ killing capacity represents a mode of immunosuppression independent of the effect of regulatory T cells [73]. Unlike T and B cells, natural killer cells do not depend on MHC to respond to antigens and instead scan a cell and activate if the cell is missing self-identifiers or engages activating receptors, such as the NKG2D immunoreceptor, which recognizes self-proteins that are rarely expressed on healthy cells, but frequently are expressed by cells stressed by infection, tumorigenesis, or damage [78]. To escape this mode of destruction, cancer cells release NKG2D-ligand-expressing exosomes, essentially packaging destruction tags into exosomes to remove them from the cancer cell membrane [78]. These NKG2D-ligand-containing exosomes also reduce the amount of NKG2D in natural killer cells’ membranes and lessen natural killer cell cytotoxicity in an NKG2D-independent way [79]. Exosome immunomodulation has been exclusively covered in recent reviews [80, 81].
Other roles of exosomes in cancer: metastasis and chemoresistance
Metastasis
The stage at diagnosis dictates patient prognosis with the presence of distal metastatic disease being almost universally fatal. For many cancers, the process of metastasis is initiated by the tumor cells undergoing an epithelial-to-mesenchymal transition (EMT) making them capable of migrating and gaining access to the vascular or lymphatic channels, where they can establish a metastasis or circulate to a different organ and produce a metastasis there [82]. Exosome composition changes with gain of metastatic capacity, becoming enriched in EMT proteins and other proteins that help coordinate metastatic efforts between the primary tumor and microenvironment [83]. Additionally, exosomes influence surrounding microenvironment cells to aid in metastasis. For example, when macrophages engulf tumor exosomes, NF-κB is activated, causing secretion of pro-inflammatory cytokines that have been linked to metastasis development [84]. Furthermore, a hypoxic tumor microenvironment induces expression of factors (HIFs) that are linked to metastasis and poor prognosis in patients [85]. HIFs have been implicated in the hypoxia-induced increase in exosome production mentioned in relation to angiogenesis. In breast cancer, exosomes secreted by hypoxic tumor cells stimulated focal adhesion formation, invasion into the extracellular matrix, and metastasis to the lungs [86]. While migratory capability is essential to metastasis, reaching a new organ is not sufficient for establishment of a metastasis; rather, the tumor cell must reach a niche that has the correct conditions for growth and where a new metastatic cell-niche microenvironment crosstalk is created [87]. Both HIFs and tumor exosomes have also been implicated in formation of a premetastatic niche [88, 89]. Upon exposure to tumor exosomes, adipose stem cells (ASC) have been shown to be recruited to the metastatic effort [90]. In a study by Abd Elmageed, et al. [90], ASC from prostate cancer patients were primed with exosomes isolated from prostate cancer cells and then transplanted into mice. These ASC became genetically unstable after exposure to the tumor exosomes, underwent reverse EMT and oncogenic transformation, then developed into tumors; thus, it is hypothesized that ASC in cancer patients may circulate to a new location and follow the same pattern to establish metastases [90].
Chemoresistance
Often, even after surgery, radiation, and chemotherapy, a small number of drug-resistant cancer cells will remain and cause recurrence. In breast cancer, for example, recurrent cancer strikes the vast majority of patients [91]. Since tumor cells are a heterogeneous population, the level of chemoresistance varies from tumor cell to tumor cell. As one mechanism of drug resistance, residual tumor cells will transmit resistance properties via exosomes to sensitive cells, creating a larger population of cells that are unaffected by cancer treatments [92]. Chen et al. [92] compared adriamycin- and docetaxel-resistant breast cancer cell lines to sensitive cell lines, establishing certain miRNAs (miR-100, miR-222, miR-30a, miR-17) that appear to be transferred intracellularly to confer resistance. Once transferred to sensitive cells, these miRNAs alter the cell cycle distribution and affect apoptosis pathways to decrease drug susceptibility. Docetaxel resistance has also been studied in prostate cancer, where the conferred resistance was found to be partly attributable to transfer of MDR-1/P-gp, a P-glycoprotein transporter protein that is over-expressed in drug resistant tumors [93]. High levels of other transporter proteins, including MRP2, ATP7A, and ATP7B, have been found in exosomes from cisplatin-resistant ovarian cancer cells [94]. In addition to these transporter proteins, multiple genes (PI3 kinase, rho GTPases, annexins, XIAP) whose products function in lysosome formation and the lysosome protein LAMP1 have also been implicated in cisplatin sensitivity [94]. Other studies have revealed that modulating the chemosusceptibility of other cells is not the only means by which exosomes increase chemoresistance. In fact, tumor cells can also package chemotherapy drugs into exosomes to protect the tumor cell from cytotoxic effects [94, 95]. Therefore, disabling exosome-mediated elimination could increase drug efficacy. With these exciting advances, it should be noted that drug resistance is a multi-faceted problem of which exosomes likely play only a contributing part. Still, the discovery of the ability of exosomes to affect chemoresistance has led researchers to suggest the need for more comprehensive profiling of the molecular contents of exosomes to deduce the key contributors to the spreading of drug resistance [92, 93].
Applications of exosomes to cancer diagnostics and therapy
Biomarkers
The inherent properties of exosomes make them ideal candidates for facilitating cancer diagnosis and prognosis, as well as the prediction and monitoring of therapeutic response. Because exosomes are readily accessible in nearly all bodily fluids (blood, saliva [24], urine [96], breast milk [97], ascites [98], amniotic fluid [99], bronchoalveolar fluid [100], cerebrospinal fluid [101], seminal fluid [102], synovial fluid [103]), they offer the key advantage of non-invasive testing opportunities [104]. Notably, analysis of circulating exosomes may be a powerful tool to facilitate early cancer detection, since a solid tumor is not required for testing. The mere fact that exosome production is increased in cancer allows for exosome analysis to be useful for cancer detection and assessment of disease progression. Indeed, studies have demonstrated elevated levels of exosomes in the plasma of cancer patients as compared to control groups and even noted a positive correlation between the abundance of tumor exosomes and cancer stage in the case of ovarian cancer [105, 106]. Perhaps more importantly, however, is the fact that exosomes are packed full of biologically active molecules that reflect the pathological state of the host cell, making exosomes a reservoir of potential diagnostic and prognostic markers. Further, exosomes offer an enriched source of biomarkers (a result of the concentration of highly selected molecules during exosomal packaging), which would otherwise constitute a mere fraction of the total proteome/transcriptome of bodily fluids, and even of the tumor cells [107].
A number of studies have identified tumor-specific markers in circulating exosomes in a plethora of cancers, including elevated levels of claudin-3 in ascites-derived exosomes of colorectal cancer patients [108], the known prostate cancer mRNA biomarkers PCA-3 and TMPRSS2:ERG in urine exosomes [109], as well as the tumor-specific gene variant EGFRvIII in serum exosomes of glioblastoma patients [57]. In addition to proteins and mRNAs, circulating microRNAs (miRNAs) have gained substantial attention with regard to being highly promising biomarker candidates. In a report by Taylor et al. [106] it was demonstrated that malignant ovarian cancers could be reliably distinguished from benign disease based on the levels of eight specific exosomal miRNAs. Other exosomal miRNAs associated with specific cancers include urine-derived exosomal miR-107 and miR-574-3p in prostate cancer [110], plasma-derived exosomal miR-141 and miR-195 in breast cancer [111], and serum-derived miR-21 in glioblastoma [57] as well as esophageal squamous cell carcinoma [112]. All of the aforementioned exosomal miRNAs are present at elevated levels in cancer patients as compared to normal control groups. It should be noted that many studies investigating circulating miRNAs as cancer biomarkers are not necessarily exosomal in origin, although it is believed that the encapsulation of miRNAs into exosomes is a primary mechanism by which miRNAs exist stably in the extracellular environment.
Vaccines
Exosomes are uniquely suited for drug or gene delivery because they are naturally biocompatible, stable while circulating in the blood, and capable of crossing the blood brain barrier [113]. Dendritic cell-derived exosomes (dexosomes) have been successfully engineered to target helper T cells to stimulate cytotoxic T cell proliferation, influence T cell differentiation, and create an anti-tumor environment [114]. Dexosomes have entered clinical trials for colorectal cancer, metastatic melanoma, and non-small cell lung cancer. In a phase I clinical trial, dexosomes isolated from ascites combined with granulocyte–macrophage colony-stimulating factor (GM–CSF), which increases immunity induction, were nontoxic and able to incite an antitumor cytotoxic T cell response; without GM–CSF, dexosomes were insufficient to produce a response [115]. In a separate phase I clinical trial, dexosomes purified from cell culture and pulsed with tumor peptides displayed low toxicity and some tumor regression was observed, supporting continuation to a phase II clinical trial [116]. Likewise, dexosomes generated only grade 1–2 adverse effects and were able to stabilize disease during a phase I clinical trial for non-small cell lung cancer patients [117]. Reviews have been recently published for exosomes as cancer vaccines [113] and dexosome immunotherapy [118].
Conclusion
With all of the important functions of exosomes to tumors, the study of exosomes is of vital importance for understanding cancer mechanisms. Although many studies have been published, we have just begun to understand the biogenesis from MVEs, especially in the context of cancer when exosome production is increased. Exosomes appear to have both an ESCRT-dependent and -independent means of sorting that enriches exosomes secreted from tumors with proteins that affect the three major stromal cell types; tumor exosomes can activate fibroblasts to increase cancer aggression, have altered cargo content under hypoxic conditions to stimulate pro-angiogenic signals in endothelial cells, and promote immune escape through MDSCs, T-cells, and natural killer cells [29, 30]. Because exosomes affect metastasis through many different routes, including EMT proteins, pro-inflammatory cytokines, and HIFs, therapeutic intervention of exosomes signifies a new approach to controlling tumor growth and spread. Furthermore, by disabling tumor-enabled communication, it may be possible to abrogate spread of chemoresistant properties.
The potential of exosomes to contain biomarkers of carcinogenesis, therapeutic response, and disease progression, has led to profiling the cargo of various types of exosomes and careful isolation/purification is necessary to yield unadulterated results [119]. Exosomes would be ideal biomarkers because they can be collected in a non-invasive procedure, and reflect the current state of the tumor. Exosomes derived from immune cells and engineered to stimulate an anti-tumor immune response are naturally advantageous for drug or gene delivery [113]. Since exosomes are involved in so many of the processes that make cancer dangerous, it will be important to consider how they impact cancer biology in developing new therapies.
Given the fact that exosomes from discrete cancers could be quite unique in terms of biological activity, a thorough interrogation of isolated and purified exosomes from molecularly to genomically well-characterized cancers is of paramount importance. Combinatorial data sets of proteomic, transcriptomic, metabolomic and lipidomic data are needed to define cancer exosomes as a function of metastasis and virulence. Mass spectrometry-based metabolomics and lipidomic analyses are beginning to expand our understanding of exosomal biochemistry and biophysics. Even though there are multiple studies defining the lipid content of exosomes (reviewed in [120]) that note elevated levels of disaturated phospholipids, sphingolipids and cholesterol, very few lipidomic studies on defined cancer cell populations [121] or immunological cells from the tumor microenvironment [122] are available. Beyond defining new biomarker or signaling elements with MS-based lipidomic and metabolomic studies that utilize well-characterized patient cells, these types of studies will begin to redefine the biophysical properties that underlie exosomal generation, release, and fusion in the tumor microenvironment. Critical to these types of studies are the validation of new bioengineering techniques to improve the yield of isolating circulating cancer-derived exosomes from exosomes released from non-transformed cells. New microfluidic platforms that utilize immunoaffinity-, magnetic- or electrical field-based separations offer the potential to isolate these discrete exosome populations [123–125].
Abbreviations
- MVE
Multivesicular endosome
- ESCRT
Endosomal-sorting complexes required for transport
- MDSC
Myeloid-derived suppressor cells
- EMT
Epithelial-to-mesenchymal transition
- HIF
Hypoxia-inducible factor
- GM-CSF
Granulocyte–macrophage colony-stimulating factor
- ASC
Adipose stem cell
Box 1: historical perspective on exosome formation
In the 1970s, plasma membrane fragments were being isolated from human bodily fluids as well as cultured cells [27, 126, 127]. However, it was not until 1983 that two teams of researchers from the laboratories of Stahl and Johnstone independently discovered what we now term exosomes [128, 129]. The actual title of “exosome” was applied by the Johnstone group to these vesicles in 1987 [130]. Although “exosome” had previously been used by Trams et al. to describe small enzyme-containing vesicles that they observed being released from both normal and cancerous cells in culture that they postulated to have a role in communication, these vesicles do not fit the current definition of exosome [25, 131]. It was later discovered that the intracellular origin of exosomes is multivesicular endosomes (MVEs) that fuse with the plasma membrane and release their contents into the extracellular space; the connection between exosomes and MVEs lead scientists to believe that exosomes were an additional means of waste removal from cells [132, 133]. Indeed, immunological cell waste, including Major Histocompatability Complex (MHC), was found to be discarded via exosomes, but Raposo et al. [134] re-popularized the idea that exosomes were more than garbage receptacles and further postulated that exosomes could function in antigen presentation and stimulate a T cell response. Presently, exosomes are even categorized as a novel mechanism of cell-to-cell communication, particularly between tumor and stromal cells [135, 136]. For a more in-depth look at the history and nomenclature of exosomes, see [131, 137].
Box 2: use of “exosome” in scientific literature
It is important to recognize many different systems of classification of small, secreted vesicles have been used and, thus, terminology cannot necessarily be relied upon. Typically, it is agreed that exosomes are about 50–100 nm; sediment at 100,000–160,000×g or float on a sucrose gradient of 1.13–1.19 g/mL; look like a flattened sphere (transmission electron microscope, TEM), round trilobed structure with a central depression (2nN amplitude modulated—atomic force microscopy, AM–AFM), or round bulging sphere (field emission scanning electron microscope, FESEM). To ensure that research articles are indeed referring to exosomes as now defined, a few precautions should be taken:
careful search of the parameters used by the authors to classify the described vesicles
analysis of purification methods–since some are more robust than others, many false positive identifications of exosome proteins have arisen from analysis of contaminated samples [119]
many papers describe exosomes but incorrectly refer to them as some other type of vesicle, so it may be easy to overlook valuable exosome research
while some noted the presence of DNA as a difference between apoptotic vesicles and exosomes, others later concluded that exosomes also contain DNA [22, 138]
sometimes LAMP1 or LAMP2 is used to differentiate exosomes from other secretory vesicles; however, because of their lysosomal escape, some exosomes lack these lysosomal proteins [139–141].
For a more detailed analysis, refer to [137], which reviews the inconsistencies in nomenclature for exosomes.
References
- 1.Hoyert DL, Xu J. Deaths: preliminary data for 2011. Natl Vital Stat Rep. 2012;61:1–52. [PubMed] [Google Scholar]
- 2.Heron M. Deaths: leading causes for 2009. Natl Vital Stat Rep. 2012;61:1–94. [PubMed] [Google Scholar]
- 3.DeNardo DG, Coussens LM. Inflammation and breast cancer. Balancing immune response: crosstalk between adaptive and innate immune cells during breast cancer progression. Breast Cancer Res. 2007;9:212. doi: 10.1186/bcr1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ishigami S, Natsugoe S, Tokuda K, Nakajo A, Che X, Iwashige H, Aridome K, Hokita S, Aikou T. Prognostic value of intratumoral natural killer cells in gastric carcinoma. Cancer. 2000;88:577–583. [PubMed] [Google Scholar]
- 5.Bissell MJ, Hines WC. Why don’t we get more cancer? A proposed role of the microenvironment in restraining cancer progression. Nat Med. 2011;17:320–329. doi: 10.1038/nm.2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brentnall TA. Arousal of cancer-associated stromal fibroblasts: palladin-activated fibroblasts promote tumor invasion. Cell Adh Migr. 2012;6:488–494. doi: 10.4161/cam.21453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li H, Fan X, Houghton J. Tumor microenvironment: the role of the tumor stroma in cancer. J Cell Biohem. 2007;101:805–815. doi: 10.1002/jcb.21159. [DOI] [PubMed] [Google Scholar]
- 8.Johnson LM, Price DK, Figg WD. Treatment-induced secretion of WNT16B promoted tumor growth and acquired resistance to chemotherapy: implications for potential use of inhibitors in cancer treatment. Cancer Biol Ther. 2013;14:90–91. doi: 10.4161/cbt.22636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hanahan D, Coussens LM. Accessories to the crime: functions of cells recruited to the tumor microenvironment. Cancer Cell. 2012;21:309–322. doi: 10.1016/j.ccr.2012.02.022. [DOI] [PubMed] [Google Scholar]
- 10.Sund M, Kalluri R. Tumor stroma derived biomarkers in cancer. Cancer Metastasis Rev. 2009;28:177–183. doi: 10.1007/s10555-008-9175-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Allen M, Louis Jones J. Jekyll and Hyde: the role of the microenvironment on the progression of cancer. J Pathol. 2011;223:162–176. doi: 10.1002/path.2803. [DOI] [PubMed] [Google Scholar]
- 12.Mueller MM, Fusenig NE. Friends or foes: bipolar effects of the tumour stroma in cancer. Nat Rev Cancer. 2004;4:839–849. doi: 10.1038/nrc1477. [DOI] [PubMed] [Google Scholar]
- 13.Joyce JA. Therapeutic targeting of the tumor microenvironment. Cancer Cell. 2005;7:513–520. doi: 10.1016/j.ccr.2005.05.024. [DOI] [PubMed] [Google Scholar]
- 14.Rosenberg SA. Progress in human tumour immunology and immunotherapy. Nature. 2001;411:380–384. doi: 10.1038/35077246. [DOI] [PubMed] [Google Scholar]
- 15.Albini A, Sporn MB. The tumour microenvironment as a target for chemoprevention. Nat Rev Cancer. 2007;7:139–147. doi: 10.1038/nrc2067. [DOI] [PubMed] [Google Scholar]
- 16.Kucharzewska P, Belting M. Emerging roles of extracellular vesicles in the adaptive response of tumour cells to microenvironmental stress. J Extracell Vesicles. 2013 doi: 10.3402/jev.v2i0.20304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khan S, Jutzy JM, Aspe JR, McGregor DW, Neidigh JW, Wall NR. Survivin is released from cancer cells via exosomes. Apoptosis. 2011;16:1–12. doi: 10.1007/s10495-010-0534-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Théry C, Ostrowski M, Segura E. Membrane vesicles as conveyors of immune responses. Nat Rev Immunol. 2009;9:581–593. doi: 10.1038/nri2567. [DOI] [PubMed] [Google Scholar]
- 19.Sharma S, Rasool HI, Palanisamy V, Mathisen C, Schmidt M, Wong DT, Gimzewski JK. Structural-mechanical characterization of nanoparticle exosomes in human saliva, using correlative AFM, FESEM, and force spectroscopy. ACS Nano. 2010;4:1921–1926. doi: 10.1021/nn901824n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cocucci E, Racchetti G, Meldolesi J. Shedding microvesicles: artefacts no more. Trends Cell Biol. 2009;19:43–51. doi: 10.1016/j.tcb.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 21.Kahlert C, Melo SA, Protopopov A, Tang J, Seth S, Koch M, Zhang J, Weitz J, Chin L, Futreal A, Kalluri R. Identification of double-stranded genomic DNA spanning all chromosomes with mutated KRAS and p53 DNA in the serum exosomes of patients with pancreatic cancer. J Biol Chem. 2014;289:3869–3875. doi: 10.1074/jbc.C113.532267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mathivanan S, Fahner CJ, Reid GE, Simpson RJ. ExoCarta 2012: database of exosomal proteins, RNA and lipids. Nucl Acids Res. 2012;40:D1241–D1244. doi: 10.1093/nar/gkr828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trajkovic K, Hsu C, Chiantia S, Rajendran L, Wenzel D, Wieland F, Schwille P, Brügger B, Simons M. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science. 2008;319:1244–1247. doi: 10.1126/science.1153124. [DOI] [PubMed] [Google Scholar]
- 24.Michael A, Bajracharya SD, Yuen PS, Zhou H, Star RA, Illei GG, Alevizos I. Exosomes from human saliva as a source of microRNA biomarkers. Oral Dis. 2010;16:34–38. doi: 10.1111/j.1601-0825.2009.01604.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trams EG, Lauter CJ, Salem N, Jr, Heine U. Exfoliation of membrane ecto-enzymes in the form of micro-vesicles. Biochim Biophys Acta. 1981;645:63–70. doi: 10.1016/0005-2736(81)90512-5. [DOI] [PubMed] [Google Scholar]
- 26.Théry C, Zitvogel L, Amigorena S. Exosomes: composition, biogenesis and function. Nat Rev Immunol. 2002;2:569–579. doi: 10.1038/nri855. [DOI] [PubMed] [Google Scholar]
- 27.De Broe ME, Wieme RJ, Logghe GN, Roels F. Spontaneous shedding of plasma membrane fragments by human cells in vivo and in vitro. Clin Chim Acta. 1977;81:237–245. doi: 10.1016/0009-8981(77)90054-7. [DOI] [PubMed] [Google Scholar]
- 28.Villarroya-Beltri C, Baixauli F, Gutiérrez-Vázquez C, Sánchez-Madrid F, Mittelbrunn M. Sorting it out: regulation of exosome loading. Semin Cancer Biol. 2014;28C:3–13. doi: 10.1016/j.semcancer.2014.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Colombo M, Moita C, van Niel G, Kowal J, Vigneron J, Benaroch P, Manel N, Moita LF, Théry C, Raposo G. Analysis of ESCRT functions in exosome biogenesis, composition and secretion highlights the heterogeneity of extracellular vesicles. J Cell Sci. 2013;126:5553–5565. doi: 10.1242/jcs.128868. [DOI] [PubMed] [Google Scholar]
- 30.Kajimoto T, Okada T, Miya S, Zhang L, Nakamura S. Ongoing activation of sphingosine 1-phosphate receptors mediates maturation of exosomal multivesicular endosomes. Nat Commun. 2013;4:2712. doi: 10.1038/ncomms3712. [DOI] [PubMed] [Google Scholar]
- 31.Perez-Hernandez D, Gutiérrez-Vázquez C, Jorge I, López-Martín S, Ursa A, Sánchez-Madrid F, Vázquez J, Yáñez-Mó M. The intercellular interactome of tetraspanin-enriched microdomains reveals their function as sorting machineries toward exosomes. J Biol Chem. 2013;288:11649–11661. doi: 10.1074/jbc.M112.445304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Villarroya-Beltri C, Gutiérrez-Vázquez C, Sánchez-Cabo F, Pérez-Hernández D, Vázquez J, Martin-Cofreces N, Martinez-Herrera DJ, Pascual-Montano A, Mittelbrunn M, Sánchez-Madrid F. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat Commun. 2013;4:2980. doi: 10.1038/ncomms3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu X, Harris SL, Levine AJ. The regulation of exosome secretion: a novel function of the p53 protein. Cancer Res. 2006;66:4795–4801. doi: 10.1158/0008-5472.CAN-05-4579. [DOI] [PubMed] [Google Scholar]
- 34.Lespagnol A, Duflaut D, Beekman C, Blanc L, Fiucci G, Marine JC, Vidal M, Amson R, Telerman A. Exosome secretion, including DNA damage-induced p53-dependent secretory pathway, is severly compromised in TSAP6/Steap3-null mice. Cell Death Differ. 2008;15:1723–1733. doi: 10.1038/cdd.2008.104. [DOI] [PubMed] [Google Scholar]
- 35.Thompson CA, Purushothaman A, Ramani VC, Vlodavsky I, Sanderson RD. Heparanase regulates secretion, composition, and function of tumor cell-derived exosomes. J Biol Chem. 2013;288(14):10093–10099. doi: 10.1074/jbc.C112.444562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Riches A, Campbell E, Borger E, Powis S. Regulation of exosome release from mammary epithelial and breast cancer cells: a new regulatory pathway. Eur J Cancer. 2014;50:1025–1034. doi: 10.1016/j.ejca.2013.12.019. [DOI] [PubMed] [Google Scholar]
- 37.Tolmachova T, Anders R, Stinchcombe J, Bossi G, Griffiths GM, Huxley C, Seabra MC. A general role for Rab27a in secretory cells. Mol Biol Cell. 2004;15:332–344. doi: 10.1091/mbc.E03-07-0452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barral DC, Ramalho JS, Anders R, Hume AN, Knapton HJ, Tolmachova T, Collinson LM, Goulding D, Authi KS, Seabra MC. Functional redundancy of Rab27 proteins and the pathogenesis of Griscelli syndrome. J Clin Invest. 2002;110:247–257. doi: 10.1172/JCI15058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shin SJ, Smith JA, Rezniczek GA, Pan S, Chen R, Brentnall TA, Wiche G, Kelly KA. Unexpected gain of function for the scaffolding protein plectin due to mislocalization in pancreatic cancer. Proc Natl Acad Sci USA. 2013;110:19414–19419. doi: 10.1073/pnas.1309720110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Savina A, Vidal M, Colombo MI. The exosome pathway in K562 cells is regulated by Rab11. J Cell Sci. 2002;115:2505–2515. doi: 10.1242/jcs.115.12.2505. [DOI] [PubMed] [Google Scholar]
- 41.Savina A, Fader CM, Damiani MT, Colombo MI. Rab11 promotes docking and fusion of multivesicular bodies in a calcium-dependent manner. Traffic. 2005;6:131–143. doi: 10.1111/j.1600-0854.2004.00257.x. [DOI] [PubMed] [Google Scholar]
- 42.Hoshino D, Kirkbride KC, Costello K, Clark ES, Sinha S, Grega-Larson N, Tyska MJ, Weaver AM. Exosome secretion is enhanced by invadopodia and drives invasive behavior. Cell Rep. 2013;5:1159–1168. doi: 10.1016/j.celrep.2013.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tlsty TD, Hein PW. Know thy neighbor: stromal cells can contribute oncogenic signals. Curr Opin Genet Dev. 2001;11:54–59. doi: 10.1016/s0959-437x(00)00156-8. [DOI] [PubMed] [Google Scholar]
- 44.Orimo A, Gupta PB, Sgroi DC, Arenzana-Seisdedos F, Delaunay T, Naeem R, Carey VJ, Richardson AL, Weinberg RA. Stromal fibroblasts present in invasive human breast carcinomas promote tumor growth and angiogenesis through elevated SDF-1/CXCL12 secretion. Cell. 2005;121:335–348. doi: 10.1016/j.cell.2005.02.034. [DOI] [PubMed] [Google Scholar]
- 45.Camps JL, Chang SM, Hsu TC, Freeman MR, Hong SJ, Zhau HE, von Eschenbach AC, Chung LW. Fibroblast-mediated acceleration of human epithelial tumor growth in vivo. Proc Natl Acad Sci USA. 1990;87:75–79. doi: 10.1073/pnas.87.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gleave M, Hsieh JT, Gao CA, von Eschenbach AC, Chung LW. Acceleration of human prostate cancer growth in vivo by factors produced by prostate and bone fibroblasts. Cancer Res. 1991;51:3753–3761. [PubMed] [Google Scholar]
- 47.Kanekura T, Chen X, Kanzaki T. Basigin (CD147) is expressed on melanoma cells and induces tumor cell invasion by stimulating production of matrix metalloproteinases by fibroblasts. Int J Cancer. 2002;99:520–528. doi: 10.1002/ijc.10390. [DOI] [PubMed] [Google Scholar]
- 48.Sameshima T, Nabeshima K, Toole BP, Yokogami K, Okada Y, Goya T, Koono M, Wakisaka S. Glioma cell extracellular matrix metalloproteinase inducer (EMMPRIN) (CD147) stimulates production of membrane-type matrix metalloproteinases and activated gelatinase A in co-cultures with brain-derived fibroblasts. Cancer Lett. 2000;157:177–184. doi: 10.1016/s0304-3835(00)00485-7. [DOI] [PubMed] [Google Scholar]
- 49.Gu J, Qian H, Shen L, Zhang X, Zhu W, Huang L, Yan Y, Mao F, Zhao C, Shi Y, Xu W. Gastric cancer exosomes trigger differentiation of umbilical cord derived mesenchymal stem cells to carcinoma-associated fibroblasts through TGF-β/Smad pathway. PLoS One. 2012 doi: 10.1371/journal.pone.0052465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Webber J, Steadman R, Mason MD, Tabi Z, Clayton A. Cancer exosomes trigger fibroblast to myofibroblast differentiation. Cancer Res. 2010;70:9621–9630. doi: 10.1158/0008-5472.CAN-10-1722. [DOI] [PubMed] [Google Scholar]
- 51.Webber JP, Spary LK, Sanders AJ, Chowdhury R, Jiang WG, Steadman R, Wymant J, Jones AT, Kynsaston H, Mason MD, Tabi Z, Clayton A. Differentiation of tumou-promoting stromal myofibroblasts by cancer exosomes. Oncogene. 2014 doi: 10.1038/onc.2013.560. [DOI] [PubMed] [Google Scholar]
- 52.Atula S, Grenman R, Syrjänen S. Fibroblasts can modulate the phenotype of malignant epithelial cells in vitro. Exp Cell Res. 1997;235(1):180–187. doi: 10.1006/excr.1997.3676. [DOI] [PubMed] [Google Scholar]
- 53.Gesierich S, Berezovskiy I, Ryschich E, Zöller M. Systemic induction of the angiogenesis switch by the tetraspanin D6.1A/CO-029. Cancer Res. 2006;66:7083–7094. doi: 10.1158/0008-5472.CAN-06-0391. [DOI] [PubMed] [Google Scholar]
- 54.Bergers G, Benjamin LE. Tumorigenesis and the angiogenic switch. Nat Rev Cancer. 2003;3:401–410. doi: 10.1038/nrc1093. [DOI] [PubMed] [Google Scholar]
- 55.Giordano FJ, Johnson RS. Angiogenesis: the role of the microenvironment in flipping the switch. Curr Opin Genet Dev. 2001;11:35–40. doi: 10.1016/s0959-437x(00)00153-2. [DOI] [PubMed] [Google Scholar]
- 56.Park JE, Tan HS, Datta A, Lai RC, Zhang H, Meng W, Lim SK, Sze SK. Hypoxic tumor cell modulates its microenvironment to enhance angiogenic and metastatic potential by secretion of proteins and exosomes. Mol Cell Proteomics. 2010;9:1085–1099. doi: 10.1074/mcp.M900381-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Skog J, Würdinger T, van Rijn S, Meijer DH, Gainche L, Curry WT, Jr, Carter BS, Krichevsky AM, Breakefield XO. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10:1470–1476. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kucharzewska P, Christianson HC, Welch JE, Svensson KJ, Fredlund E, Ringnér M, Mörgelin M, Bourseau-Guilmain E, Bengzon J, Belting M. Exosomes reflect the hypoxic status of glioma cells and mediate hypoxia-dependent activation of vascular cells during tumor development. Proc Natl Acad Sci USA. 2013;110:7312–7317. doi: 10.1073/pnas.1220998110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhou W, Fong MY, Min Y, Somlo G, Liu L, Palomares MR, Yu Y, Chow A, O’Connor ST, Chin AR, Yen Y, Wang Y, Marcusson EG, Chu P, Wu J, Wu X, Li AX, Li Z, Gao H, Ren X, Boldin MP, Lin PC, Wang SE. Cancer-secreted miR-105 destroys vascular endothelial barriers to promote metastasis. Cancer Cell. 2014;25:501–515. doi: 10.1016/j.ccr.2014.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lakshmi Narendra B, Eshvendar Reddy K, Shantikumar S, Ramakrishna S. Immune system: a double-edged sword in cancer. Inflamm Res. 2013;62:823–834. doi: 10.1007/s00011-013-0645-9. [DOI] [PubMed] [Google Scholar]
- 61.Kawamoto H, Minato N. Myeloid cells. Int J Biochem Cell Biol. 2004;36:1374–1379. doi: 10.1016/j.biocel.2004.01.020. [DOI] [PubMed] [Google Scholar]
- 62.Skokos D, Botros HG, Demeure C, Morin J, Peronet R, Birkenmeier G, Boudaly S, Mécheri S. Mast cell-derived exosomes induce phenotypic and functional maturation of dendritic cells and elicit specific immune responses in vivo. J Immunol. 2003;170:3037–3045. doi: 10.4049/jimmunol.170.6.3037. [DOI] [PubMed] [Google Scholar]
- 63.Théry C, Regnault A, Garin J, Wolfers J, Zitvogel L, Ricciardi-Castagnoli P, Raposo G, Amigorena S. Molecular characterization of dendritic cell-derived exosomes. Selective accumulation of the heat shock protein hsc73. J Cell Biol. 1999;147:599–610. doi: 10.1083/jcb.147.3.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Valenti R, Huber V, Iero M, Filipazzi P, Parmiani G, Rivoltini L. Tumor-released microvesicles as vehicles of immunosuppression. Cancer Res. 2007;67:2912–2915. doi: 10.1158/0008-5472.CAN-07-0520. [DOI] [PubMed] [Google Scholar]
- 65.Xiang X, Poliakov A, Liu C, Liu Y, Deng ZB, Wang J, Cheng Z, Shah SV, Wang GJ, Zhang L, Grizzie WE, Mobley J, Zhang HG. Induction of myeloid-derived suppressor cells by tumor exosomes. Int J Cancer. 2009;124:2621–2633. doi: 10.1002/ijc.24249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yu S, Liu C, Su K, Wang J, Liu Y, Zhang L, Li C, Cong Y, Kimberly R, Grizzle WE, Falkson C, Zhang HG. Tumor exosomes inhibit differentiation of bone marrow dendritic cells. J Immunol. 2007;178:6867–6875. doi: 10.4049/jimmunol.178.11.6867. [DOI] [PubMed] [Google Scholar]
- 67.Serafini P, Borrello I, Bronte V. Myeloid suppressor cells in cancer: recruitment, phenotype, properties, and mechanisms of immune suppression. Semin Cancer Biol. 2006;16:53–65. doi: 10.1016/j.semcancer.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 68.Liu Y, Xiang X, Zhuang X, Zhang S, Liu C, Cheng Z, Michalek S, Grizzle W, Zhang HG. Contribution of MyD88 to the tumor exosome-mediated induction of myeloid derived suppressor cells. Am J Pathol. 2010;176:2490–2499. doi: 10.2353/ajpath.2010.090777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hong EH, Chang SY, Lee BR, Kim YS, Lee JM, Kang CY, Kweon MN, Ko HJ. Blockade of Myd88 signaling induces antitumor effects by skewing the immunosuppressive function of myeloid-derived suppressor cells. Int J Cancer. 2013;132:2839–2848. doi: 10.1002/ijc.27974. [DOI] [PubMed] [Google Scholar]
- 70.Xiang X, Liu Y, Zhuang X, Zhang S, Michalek S, Taylor DD, Grizzle W, Zhang HG. TLR2-mediated expansion of MDSCs is dependent on the source of tumor exosomes. Am J Pathol. 2010;177:1606–1610. doi: 10.2353/ajpath.2010.100245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Luczyński W, Krawczuk-Rybak M, Stasiak-Barmuta A. Myeloid-derived suppressor cells: the new mechanism of immunosuppression in cancer. Postepy Hig Med Dosw. 2008;62:18–22. [PubMed] [Google Scholar]
- 72.Ye SB, Li ZL, Luo DH, Huang BJ, Chen YS, Zhang XS, Cui J, Zeng YX, Li J (2014) Tumor-derived exosomes promote tumor progression and T-cell dysfunction through the regulation of enriched exosomal microRNAs in human nasopharyngeal carcinoma. Oncotarget [Epub ahead of print] [DOI] [PMC free article] [PubMed]
- 73.Clayton A, Mitchell JP, Court J, Mason MD, Tabi Z. Human tumor-derived exosomes selectively impair lymphocyte responses to interleukin-2. Cancer Res. 2007;67:7458–7466. doi: 10.1158/0008-5472.CAN-06-3456. [DOI] [PubMed] [Google Scholar]
- 74.Abusamra AJ, Zhong A, Zheng X, Li M, Ichim TE, Chin JL, Min WP. Tumor exosomes expressing Fas ligand mediate CD8 + T-cell apoptosis. Blood Cells Mol Dis. 2005;35:169–173. doi: 10.1016/j.bcmd.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 75.Klibi J, Niki T, Riedel A, Pioche-Durieu C, Souquere S, Rubinstein E, Le Moulec S, Guigay J, Hirashima M, Guemira F, Adhikary D, Mautner J, Busson P. Blood diffusion and Th1-suppressive effects of galectin-9-containing exosomes released by Epstein-Barr virus-infected nasopharyngeal carcinoma cels. Blood. 2009;113:1957–1966. doi: 10.1182/blood-2008-02-142596. [DOI] [PubMed] [Google Scholar]
- 76.Yang C, Chalasani G, Ng YH, Robbins PD. Exosomes released from Mycoplasma infected tumor cells activate inhibitory B cells. PLoS One. 2012 doi: 10.1371/journal.pone.0036138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Qin Z, Richter G, Schüler T, Ibe S, Cao X, Blankenstein T. B cells inhibit induction of T cell-dependent tumor immunity. Nat Med. 1998;4:627–630. doi: 10.1038/nm0598-627. [DOI] [PubMed] [Google Scholar]
- 78.Mincheva-Nilsson L, Baranov V. Cancer exosomes and NKG2D receptor-ligand interactions: impairing NKG2D-mediated cytotoxicity and anti-tumour immune surveillance. Semin Cancer Biol. 2014 doi: 10.1016/j.semcancer.2014.02.010. [DOI] [PubMed] [Google Scholar]
- 79.Ashiru O, Boutet P, Fernández-Messina L, Agüera-González S, Skepper JN, Valés-Gómez M, Reyburn HT. Natural killer cell cytotoxicity is suppressed by exposure to the human NKG2D ligand MICA*008 that is shed by tumor cells in exosomes. Cancer Res. 2010;70:481–489. doi: 10.1158/0008-5472.CAN-09-1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gehrmann U, Näslund TI, Hiltbrunner S, Larssen P, Gabrielsson S. Harnessing the exosome-induced immune response for cancer immunotherapy. Semin Cancer Biol. 2014 doi: 10.1016/j.semcancer.2014.05.003. [DOI] [PubMed] [Google Scholar]
- 81.Altevogt P, Bretz NP, Ridinger J, Utikal J, Umansky V. Novel insights into exosome-induced, tumor-associated inflammation and immunomodulation. Semin Cancer Biol. 2014 doi: 10.1016/j.semcancer.2014.04.008. [DOI] [PubMed] [Google Scholar]
- 82.Fidler IJ. The organ microenvironment and cancer metastasis. Differentiation. 2002;70:498–505. doi: 10.1046/j.1432-0436.2002.700904.x. [DOI] [PubMed] [Google Scholar]
- 83.Jeppesen DK, Nawrocki A, Jensen SG, Thorsen K, Whitehead B, Howard KA, Dyrskjøt L, Ørntoft TF, Larsen MR, Ostenfeld MS. Quantitative proteomics of fractionated membrane and lumen exosome proteins from isogenic metastatic and nonmetastatic bladder cancer cells reveal differential expression of EMT factors. Proteomics. 2014;14:699–712. doi: 10.1002/pmic.201300452. [DOI] [PubMed] [Google Scholar]
- 84.Chow A, Zhou W, Liu L, Fong MY, Champer J, Van Haute D, Chin AR, Ren X, Gugiu BG, Meng Z, Huang W, Ngo V, Kortylewski M, Wang SE. Macrophage immunomodulation by breast cancer-derived exosomes requires Toll-like receptor 2-mediated activation of NF-κB. Sci Rep. 2014;4:5750. doi: 10.1038/srep05750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kimbro KS, Simons JW. Hypoxia-inducible factor-1 in human breast and prostate cancer. Endocr Relat Cancer. 2006;13:739–749. doi: 10.1677/erc.1.00728. [DOI] [PubMed] [Google Scholar]
- 86.Wang T, Gilkes DM, Takano N, Xiang L, Luo W, Bishop CJ, Chaturvedi P, Green JJ, Semenza FL (2014) Hypoxia-inducible factors and RAB22 mediate formation of microvesicles that stimulate breast cancer invasion and metastasis. Proc Natl Acad Sci USA [Epub ahead of print] [DOI] [PMC free article] [PubMed]
- 87.Irmisch A, Huelsken J. Metastasis: new insights into organ-specific extravasation and metastatic niches. Exp Cell Res. 2013 doi: 10.1016/j.yexcr.2013.02.012. [DOI] [PubMed] [Google Scholar]
- 88.Wong CC, Gilkes DM, Zhang H, Chen J, Wei H, Chaturvedi P, Fraley SI, Wong CM, Khoo US, Ng IO, Wirtz D, Semenza GL. Hypoxia-inducible factor 1 is a master regulator of breast cancer metastatic niche formation. Proc Natl Acad Sci USA. 2011;108:16369–16374. doi: 10.1073/pnas.1113483108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Thuma F, Zöller M. Outsmart tumor exosomes to steal the cancer initiating cell its niche. Semin Cancer Biol. 2014 doi: 10.1016/j.semcancer.2014.02.011. [DOI] [PubMed] [Google Scholar]
- 90.Abd Elmageed ZY, Yang Y, Thomas R, Ranjan M, Mondal D, Moroz K, Fang Z, Rezk BM, Moparty K, Sikka SC, Sartor O, Abdel-Mageed AB. Neoplastic reprogramming of patient-derived adipose stem cells by prostate cancer cell-associated exosomes. Stem Cells. 2014;32:983–997. doi: 10.1002/stem.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wang T, Diaz AJ, Yen Y. The role of peroxiredoxin II in chemoresistance of breast cancer cells. Breast Cancer. 2014;6:73–80. doi: 10.2147/BCTT.S61281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chen WX, Liu XM, Lv MM, Chen L, Zhao JH, Zhong SL, Ji MH, Hu Q, Luo Z, Wu JZ, Tang JH. Exosomes from drug-resistance breast cancer cells transmit chemoresistance by a horizontal transfer of microRNAs. PLoS One. 2014 doi: 10.1371/journal.pone.0095240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Corcoran C, Rani S, O’Brien K, O’Neill A, Prencipe M, Sheikh R, Webb G, McDermott R, Watson W, Crown J, O’Driscoll L. Docetaxel-resistance in prostate cancer: evaluating associated phenotypic changes and potential for resistance transfer via exosomes. PLoS One. 2012;7(12):e50999. doi: 10.1371/journal.pone.0050999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Safaei R, Larson BJ, Cheng TC, Gibson MA, Otani S, Naerdemann W, Howell SB. Abnormal lysosomal trafficking and enhanced exosomal export of cisplatin in drug-resistant human ovarian carcinoma cells. Mol Cancer Ther. 2005;4(10):1595–1604. doi: 10.1158/1535-7163.MCT-05-0102. [DOI] [PubMed] [Google Scholar]
- 95.Federici C, Petrucci F, Caimi S, Cesolini A, Logozzi M, Borghi M, D’Ilio S, Lugini L, Violante N, Azzarito T, Majorani C, Brambilla D, Fais S. Exosome release and low pH belong to a framework of resistance of human melanoma cells to cisplatin. PLoS One. 2014 doi: 10.1371/journal.pone.0088193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gonzales PA, Zhou H, Pisitkun T, Wang NS, Star RA, Knepper MA, Yuen PS. Isolation and purification of exosomes in urine. Methods Mol Biol. 2010;641:89–99. doi: 10.1007/978-1-60761-711-2_6. [DOI] [PubMed] [Google Scholar]
- 97.Zhou Q, Li M, Wang X, Li Q, Wang T, Zhu Q, Zhou X, Wang X, Gao X, Li X. Immune-related microRNAs are abundant in breast milk exosomes. Int J Bio Sci. 2012;8:118–123. doi: 10.7150/ijbs.8.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Peng P, Yan Y, Keng S. Exosomes in the ascites of ovarian cancer patients: origin and effects on anti-tumor immunity. Oncol Rep. 2011;25:749–762. doi: 10.3892/or.2010.1119. [DOI] [PubMed] [Google Scholar]
- 99.Asea A, Jean-Pierre C, Kaur P, Rao P, Linhares IM, Skupski D, Witkin SS. Heat shock protein-containing exosomes in mid-trimester amniotic fluids. J Reprod Immunol. 2008;79:12–17. doi: 10.1016/j.jri.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 100.Rodriguez M, Silva J, López-Alfonso A, López-Muñiz MB, Peña C, Domínguez G, García JM, López-Gónzalez A, Méndez M, Provencio M, García V, Bonilla F. Different exosome cargo from plasma/bronchoalveolar lavage in non-small-cell lung cancer. Genes Chromosome Cancer. 2014;53:713–724. doi: 10.1002/gcc.22181. [DOI] [PubMed] [Google Scholar]
- 101.Teplyuk NM, Mollenhauer B, Gabriely G, Giese A, Kim E, Smolsky M, Kim RY, Saria MG, Pastorino S, Kesari S, Krichevsky AM. MicroRNAs in cerebrospinal fluid identify glioblastoma and metastatic brain cancers and reflect disease activity. Neuro Oncol. 2012;14:689–700. doi: 10.1093/neuonc/nos074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Vojtech L, Woo S, Hughes S, Levy C, Ballweber L, Sauteraud RP, Strobi J, Westerberg K, Gottardo R, Tewari M, Hladik F. Exosomes in human semen carry a distinctive repertoire of small non-coding RNAs with potential regulatory functions. Nucleic Acids Res. 2014;42:7290–7304. doi: 10.1093/nar/gku347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Skriner K, Adolph K, Jungblut PR, Burmester GR. Association of citrullinated proteins with synovial exosomes. Arthritis Rheum. 2006;54:3809–3814. doi: 10.1002/art.22276. [DOI] [PubMed] [Google Scholar]
- 104.Properzi F, Logozzi M, Fais S. Exosomes: the future of biomarkers in medicine. Biomark Med. 2013;7:769–778. doi: 10.2217/bmm.13.63. [DOI] [PubMed] [Google Scholar]
- 105.Rabinowits G, Gerçel-Taylor C, Day JM, Taylor DD, Kloecker GH. Exosomal microRNA: a diagnostic marker for lung cancer. Clin Lung Cancer. 2009;10(1):42–46. doi: 10.3816/CLC.2009.n.006. [DOI] [PubMed] [Google Scholar]
- 106.Taylor DD, Gercel-Taylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol Oncol. 2008;110(1):13–21. doi: 10.1016/j.ygyno.2008.04.033. [DOI] [PubMed] [Google Scholar]
- 107.Properzi F, Logozzi M, Fais S. Exosomes: the future of biomarkers in medicine. Biomark Med. 2013;7(5):769–778. doi: 10.2217/bmm.13.63. [DOI] [PubMed] [Google Scholar]
- 108.Choi DS, Park JO, Jang SC, Yoon YJ, Jung JW, Choi DY, Kim JW, Kang JS, Park J, Hwang D, Lee KH, Park SH, Kim YK, Desiderio DM, Kim KP, Gho YS. Proteomic analysis of microvesicles derived from human colorectal cancer ascites. Proteomics. 2011;11(13):2745–2751. doi: 10.1002/pmic.201100022. [DOI] [PubMed] [Google Scholar]
- 109.Nilsson J, Skog J, Nordstrand A, Baranov V, Mincheva-Nilsson L, Breakefield XO, Widmark A. Prostate cancer-derived urine exosomes: a novel approach to biomarkers for prostate cancer. Br J Cancer. 2009;100(10):1603–1607. doi: 10.1038/sj.bjc.6605058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Bryant RJ, Pawlowski T, Catto JW, Marsden G, Vessella RL, Rhees B, Kuslich C, Visakorpi T, Hamdy FC. Changes in circulating microRNA levels associated with prostate cancer. Br J Cancer. 2012;106(4):768–774. doi: 10.1038/bjc.2011.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Corcoran C, Friel AM, Duffy MJ, Crown J, O’Driscoll L. Intracellular and extracellular microRNAs in breast cancer. Clin Chem. 2011;57(1):18–32. doi: 10.1373/clinchem.2010.150730. [DOI] [PubMed] [Google Scholar]
- 112.Kosaka N, Iguchi H, Ochiya T. Circulating microRNA in body fluid: a new potential biomarker for cancer diagnosis and prognosis. Cancer Sci. 2010;101(10):2087–2092. doi: 10.1111/j.1349-7006.2010.01650.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Natasha G, Gundogan B, Tan A, Farhatnia Y, Wu W, Rajadas J, Seifalian AM. Exosomes as immunotheranostic nanoparticles. Clin Ther. 2014;36:820–829. doi: 10.1016/j.clinthera.2014.04.019. [DOI] [PubMed] [Google Scholar]
- 114.Hao S, Liu Y, Yuan J, Zhang X, He T, Wu X, Wei Y, Sun D, Xiang J. Novel exosome-targeted CD4 + T cell vaccine counteracting CD4 + 25 + regulatory T cell-mediated immune suppression and stimulating efficient central memory CD8 + CTL response. J Immunol. 2007;179:2731–2740. doi: 10.4049/jimmunol.179.5.2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Dai S, Wei D, Wu Z, Zhou X, Wei X, Huang H, Li G. Phase I clinical trial of autologous ascites-derived exosomes combined with GM-CSF for colorectal cancer. Mol Ther. 2008;16:782–790. doi: 10.1038/mt.2008.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Escudier B, Dorval T, Chaput N, André F, Caby MP, Novault S, Flament C, Leboulaire C, Borg C, Amigorena S, Boccaccio C, Bonnerot C, Dhellin O, Movassagh M, Piperno S, Robert C, Serra V, Valente N, Le Pecq JB, Spatz A, Lantz O, Tursz T, Angevin E, Zitvogel L. Vaccination of metastatic melanoma patients with autologous dendritic cell (DC) derived-exosomes: results of the first phase I clinical trial. J Transl Med. 2005;3:10. doi: 10.1186/1479-5876-3-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Morse MA, Garst J, Osada T, Khan S, Hobeika A, Clay TM, Valente N, Shreeniwas R, Sutton MA, Delcayre A, Hsu DH, Le Pecq JB, Lyerly HK. A phase I study of dexosome immunotherapy in patients with advanced non-small cell lung cancer. J Transl Med. 2005;3:9. doi: 10.1186/1479-5876-3-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Pitt JM, Charrier M, Viaud S, André F, Besse B, Chaput N, Zitvogel L. Dendritic cell-derived exosomes as immunotherapies in the fight against cancer. J Immunol. 2014;193:1006–1011. doi: 10.4049/jimmunol.1400703. [DOI] [PubMed] [Google Scholar]
- 119.Welton JL, Khanna S, Giles PJ, Brennan P, Brewis IA, Staffurth J, Mason MD, Clayton A. Proteomics analysis of bladder cancer exosomes. Mol Cell Proteomics. 2010;9:1324–1338. doi: 10.1074/mcp.M000063-MCP201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Choi DS, Kim DK, Kim YK, Gho YS. Proteomics, transcriptomics and lipidomics of exosomes and ectosomes. Proteomics. 2013;13(10–11):1554–1571. doi: 10.1002/pmic.201200329. [DOI] [PubMed] [Google Scholar]
- 121.Llorente A, Skotland T, Sylvänne T, Kauhanen D, Róg T, Orlowski A, Vattulainen I, Ekroos K. Sandvig K (2013) Molecular lipidomics of exosomes released by PC-3 prostate cancer cells. Biochim Biophys Acta. 1831;7:1302–1309. doi: 10.1016/j.bbalip.2013.04.011. [DOI] [PubMed] [Google Scholar]
- 122.Laulagnier K, Motta C, Hamdi S, Roy S, Fauvelle F, Pageaux JF, Kobayashi T, Salles JP, Perret B, Bonnerot C, Record M. Mast cell- and dendritic cell-derived exosomes display a specific lipid composition and an unusual membrane organization. Biochem J. 2004;380(Pt 1):161–171. doi: 10.1042/BJ20031594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.He M, Crow J, Roth M, Zeng Y, Godwin AK. Integrated immunoisolation and protein analysis of circulating exosomes using microfluidic technology. Lab Chip. 2014;14(19):3773–3780. doi: 10.1039/c4lc00662c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Petersen KE, Manangon E, Hood JL, Wickline SA, Fernandez DP, Johnson WP, Gale BK (2014) A review of exosome separation techniques and characterization of B16-F10 mouse melanoma exosomes with AF4-UV-MALS-DLS-TEM. Anal Bioanal Chem [Epub ahead of print] [DOI] [PMC free article] [PubMed]
- 125.Mizutani K, Terazawa R, Kameyama K, Kato T, Horie K, Tsuchiya T, Seike K, Ehara H, Fujita Y, Kawakami K, Ito M, Deguchi T. Isolation of prostate cancer-related exosomes. Anticancer Res. 2014;34(7):3419–3423. [PubMed] [Google Scholar]
- 126.De Broe ME, Borgers M, Wieme RJ. The separation and characterization of liver plasma membrane fragments circulating in the blood of patients with cholestasis. Clin Chim Acta. 1975;59(3):369–372. doi: 10.1016/0009-8981(75)90013-3. [DOI] [PubMed] [Google Scholar]
- 127.Brocklehurst D, Wilde CE, Doar JW. The incidence and likely origins of serum particulate alkaline phosphatase and lipoprotein-X in liver disease. Clin Chim Acta. 1978;88(3):509–515. doi: 10.1016/0009-8981(78)90285-1. [DOI] [PubMed] [Google Scholar]
- 128.Harding C, Stahl P. Transferrin recycling in reticulocytes: pH and iron are important determinants of ligand binding and processing. Biochem Biophys Res Commun. 1983;113(2):650–658. doi: 10.1016/0006-291x(83)91776-x. [DOI] [PubMed] [Google Scholar]
- 129.Pan BT, Johnstone RM. Fate of the transferrin receptor during maturation of sheep reticulocyte in vitro: selective externalization of the receptor. Cell. 1983;33(3):967–978. doi: 10.1016/0092-8674(83)90040-5. [DOI] [PubMed] [Google Scholar]
- 130.Johnstone RM, Adam M, Hammond JR, Orr L, Turbide C. Vesicle formation during reticulocyte maturation. Association of plasma membrane activities with released vesicles (exosomes) J Biol Chem. 1987;262(19):9412–9420. [PubMed] [Google Scholar]
- 131.Harding CV, Heuser JE, Stahl PD. Exosomes: looking back three decades and into the future. J Cell Biol. 2013;200(4):367–371. doi: 10.1083/jcb.201212113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Johnstone RM, Matthew A, Mason AB, Teng K. Exosome formation during maturation of mammalian and avian reticulocytes: evidence that exosome release is a major route for externalization of obsolete membrane proteins. J Cell Physiol. 1991;147:27–36. doi: 10.1002/jcp.1041470105. [DOI] [PubMed] [Google Scholar]
- 133.Gruenberg J. The endocytic pathway: a mosaic of domains. Nat Rev Mol Cell Biol. 2001;2:721–730. doi: 10.1038/35096054. [DOI] [PubMed] [Google Scholar]
- 134.Raposo G, Nijman HW, Stoorvogel W, Liejendekker R, Harding CV, Melief CJ, Geuze HJ. B lymophocytes secrete antigen-presenting vesicles. J Exp Med. 1996;183:1161–1172. doi: 10.1084/jem.183.3.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Ratajczak J, Wysoczynski M, Hayek F, Janowska-Wieczorek A, Ratajczak MZ. Membrane-derived microvesicles: important and underappreciated mediators of cell-to-cell communication. Leukemia. 2006;20:1487–1495. doi: 10.1038/sj.leu.2404296. [DOI] [PubMed] [Google Scholar]
- 136.Théry C. Exosomes: secreted vesicles and intercellular communications. F1000 Biol Rep. 2011;3:15. doi: 10.3410/B3-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Gould SJ, Raposo G. As we wait: coping with an imperfect nomenclature for extracellular vesicles. J Extracell Vesicles. 2013 doi: 10.3402/jev.v2i0.20389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Orozco AF, Lewis DE. Flow cytometric analysis of circulating microparticles in plasma. Cytometry. 2010;77:502–514. doi: 10.1002/cyto.a.20886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Morelli AE, Larregina AT, Shufesky WJ, Sullivan ML, Stolz DB, Papworth GD, Zahorchak AF, Logar AJ, Wang Z, Watkins SC, Falo LD, Jr, Thomson AW. Endocytosis, intracellular sorting, and processing of exosomes by dendritic cells. Blood. 2004;104:3257–3266. doi: 10.1182/blood-2004-03-0824. [DOI] [PubMed] [Google Scholar]
- 140.Hawari FI, Rouhani FN, Cui X, Yu ZX, Buckley C, Kaler M, Levine SJ. Release of full-length 55-kDa TNF receptor 1 in exosome-like vesicles: a mechanism for generation of soluble cytokine receptors. Proc Natl Acad Sci USA. 2004;101:1297–1302. doi: 10.1073/pnas.0307981100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Escola JM, Kleijmeer MJ, Stoorvogel W, Griffith JM, Yoshie O, Geuze HJ. Selective enrichment of tetraspan proteins on the internal vesicles of multivesicular endosomes and on exosomes secreted by human B-lymphocytes. J Biol Chem. 1998;273:20121–20127. doi: 10.1074/jbc.273.32.20121. [DOI] [PubMed] [Google Scholar]