Abstract
Objective
REDD1 is an endogenous inhibitor of mTOR that regulates cellular stress responses. REDD1 expression is decreased in aged and osteoarthritis (OA) cartilage and it regulates mTOR signaling and autophagy in articular chondrocytes in vitro. The present study investigated the effects of REDD1 deletion in vivo using a mouse model of experimental OA.
Methods
Severity of OA was histologically assessed in 4-month-old wild-type and in Redd1−/− mice subjected to surgical destabilization of the medial meniscus (DMM). Chondrocyte autophagy, apoptosis, mitochondrial content, and expression of mitochondrial biogenesis makers were determined in cartilage and cultured chondrocytes from Redd1+/+ and Redd1−/− mice.
Results
REDD1 deficiency increased severity of changes in cartilage, menisci subchondral bone and synovium in the DMM model of OA. Chondrocyte death was increased in the cartilage of Redd1−/− mice and in cultured Redd1−/− chondrocytes under oxidative stress conditions. Expression of key autophagy markers (LC3 and ATG5) was markedly reduced in cartilage from Redd1−/− mice and in cultured human and mouse chondrocytes with REDD1 depletion. Mitochondrial content, ATP levels, and expression of the mitochondrial biogenesis markers PGC1α and TFAM were also decreased in REDD1 deficient chondrocytes. REDD1 was required for AMPK-induced PGC1α transcriptional activation in chondrocytes.
Conclusion
Our observations suggest that REDD1 is a key mediator of cartilage homeostasis through regulation of autophagy and mitochondrial biogenesis and that REDD1 deficiency exacerbates the severity of injury-induced OA.
Keywords: REDD1, cartilage, osteoarthritis, autophagy, mitochondria
INTRODUCTION
Osteoarthritis (OA) is the most prevalent joint disease and causes pain and disability (1). Although OA is considered a disease of the whole synovial joint, degradation of articular cartilage is a critical event in disease initiation and progression. Cartilage damage is a result of an imbalance between anabolic and catabolic processes that compromise the function of articular chondrocytes, the cells that maintain tissue homeostasis (2). OA is a multifactorial disease and aging is its main risk factor (2). Despite the fact that not all persons develop OA with age, there are significant age-related changes in articular cartilage that may serve as a basis for OA development (3). These changes include reduced thickness and cell density, abnormal secretory activity, cell senescence, and defective cellular defense mechanisms (3). Understanding the molecular mechanisms that drive cartilage aging has become a main focus of OA research.
Macroautophagy (hereafter autophagy) is a conserved process that serves to recycle defective cellular organelles and macromolecules, and that is also activated during hypoxic and energy stress to provide energy for the cell (4). Our group has shown that autophagy is constitutively active in articular chondrocytes and that there is a reduction in the expression of key autophagic markers during aging and OA (5). A major regulator of autophagy is the serine/threonine kinase mTOR (6). Under nutrient replete conditions, mTOR suppresses autophagy through phosphorylation of ULK1 and Atg13 (6). As part of the normal cellular response to stress, endogenous regulators such as FOXO, AMPK or REDD1 inhibit mTOR to arrest cell growth and proliferation while enhancing stress resistance and cell survival (6). However, these regulatory mechanisms seem to be defective in articular cartilage during aging and OA. We and others have reported a reduction of FOXO, AMPK and REDD1 expression and activity in cartilage during aging and OA (7–10), and there is evidence of increased mTOR expression in OA cartilage (11). Moreover, genetic or pharmacological inhibition of mTOR signaling results in autophagy activation and protection against experimental OA in mice (11, 12). These findings suggest that increased mTOR signaling may compromise autophagy in chondrocytes and contribute to the development of OA.
REDD1 (Regulated in development and DNA damage 1, encoded by DDIT4) is an evolutionary conserved protein that is ubiquitously expressed in adult tissues and regulates cellular stress responses. REDD1 expression is induced by hypoxia and other stresses (13, 14) and acts primarily as a canonical mTOR inhibitor (15–17). Interestingly, REDD1 is able to regulate autophagy through an mTOR independent mechanism that involves a direct interaction with TXNIP to induce ROS production and activation of ATG4B, a key component of the autophagic machinery (18). We have previously reported that REDD1 is abundantly expressed in human and mouse knee articular cartilage and that its expression is reduced during aging and OA (10). Moreover, REDD1 positively regulates autophagy in human and mouse articular chondrocytes, also in an mTOR independent manner (10) suggesting that reduced REDD1 expression in cartilage may contribute to increased mTOR activity and autophagy deficiency that are characteristic of OA pathophysiology.
The present study was designed to evaluate the role of REDD1 in OA development in vivo using a mouse model of experimental OA. Our results show that genetic disruption of REDD1 impairs autophagy in cartilage and exacerbates the severity of experimental OA. Furthermore, we identified a novel function of REDD1 as a regulator of mitochondrial biogenesis in articular chondrocytes.
METHODS
Mouse knees
All animal experiments were performed in compliance with protocols approved by the Institutional Animal Care and Use Committee at The Scripps Research Institute. Global REDD1 knockout mice (Redd1−/−) on a C57BL/6J background (19) were a kind gift from Dr. Rubin Tuder (University of Colorado Boulder) and Dr. Irina Budunova (Northwestern University). For studies on joint and skeletal development, growth and maturation, mice were sacrificed after 7 days (P7), or at 1, 2 and 6 months of age. For aging studies, wild-type mice were sacrificed at 6 or 27 months of age. For the surgical OA model, 4-month-old male mice (n=12 per genotype) were anesthetized and transection of the medial meniscotibial ligament (DMM) and the medial collateral ligament was performed in the right knee as described (12, 20). To serve as control, sham surgery was performed on the left knee and consisted in a small incision in the medial side and opening of the joint capsule and closing with surgical suture. Animals were euthanized 10 weeks after surgery and knee joints were embedded in a standardized fixed angle of the femur relative to the tibia so that the sections that were scored represent the center of the weight bearing areas of the tibial plateau and femoral condyle (9). Five-μm thick sagittal sections of the medial compartment of the knee were stained with Safranin-O and fast green (9) and OA related changes were scored in one section per animal as previously described for articular cartilage (summed OARSI score for femur and tibia) (21), synovium (22), anterior and posterior meniscal horns (23), and subchondral bone (24). Samples were graded by two different individuals blinded to the genotype.
Mouse cell culture
Immature mouse articular chondrocytes were isolated from knees and hips of 6-days-old wild-type and Redd1−/− mice (19) following the protocol described by Gosset et al (25). Synovial membranes were collected from knee joints of 4-month-old Redd1+/+ and Redd1−/− mice (n=4 per genotype) and synoviocytes were isolated after 4 hour digestion with collagenase (2mg/ml, Sigma) at 37ºC. Cells were maintained in media containing 10% calf serum (CS) and second passage cells were plated at a density of 105 cells/mL, incubated overnight, and treated for 24 hours with tBHP (tert-Butyl hydroperoxide, Sigma), 1nM IL-1β (interleukin 1β, Preprotech), chloroquine (25μM, Sigma), or 1mM AICAR (5-Aminoimidazole-4-carboxamide ribonucleotide, Sigma) as indicated.
Human chondrocyte culture
Human primary chondrocytes were isolated as described previously (8) and maintained in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% CS at 37ºC in 5% CO2. First passage chondrocytes were used in all experiments. For REDD1 knockdown, chondrocytes were grown to confluence and transfected with small interfering RNA (siRNA) for DDIT4/REDD1 (Life Technologies) using lipofectamine RNAiMAX (Life Technologies) in media containing 1% CS for 48 hours.
Immunohistochemistry
Paraffin-embedded knees from Redd1+/+ mice were processed as described (9) and sections were incubated overnight with antibodies against REDD1 (1:100, Proteintech), LC3 (1:50, MBL) or ATG5 (1:100, ABCAM). After washing with phosphate buffered saline (PBS), sections were incubated for 30 minutes with ImmPRESS-AP anti-rabbit IgG polymer detection reagent (Vector Laboratories), dehydrated and mounted. Positive cells were quantified in the articular cartilage and both meniscal horns of the proximal tibial plateau in two different pictures per section taken at 20x magnification. Results are reported as percentage of immunopositive cells.
TUNEL staining
Five-μm thick sections were digested with pepsin (Dako) for 8 minutes at 37º and endogenous peroxidase was blocked using Vector’s Bloxall solution at room temperature for 10 minutes. TUNEL (Terminal deoxynucleotidyl transferase (TdT) dUTP Nick-End Labeling) staining was completed using an in situ detection kit with fluorescein (Roche). Cartilage cellularity and TUNEL positive cells were measured in two different fields per section and results are expressed as cells per mm2 and percentage of fluorescent cells, respectively.
Cell viability
Mouse articular chondrocytes from Redd1+/+ and Redd1−/− mice were cultured in 96-well plates and treated with the oxidant tBHP at 0, 50, 100 or 250 μM for 24 hours. Cell viability was assessed using RealTime-Glo™ MT Cell Viability Assay (Promega). Four independent experiments were performed in triplicate. Results are expressed as relative cell viability with respect to untreated chondrocytes from Redd1+/+ mice.
Intracellular ATP measurement
Mouse articular chondrocytes were cultured in 96-well plates in media containing 25mM glucose or galactose for 24 hours and intracellular ATP levels were measured using CellTiter-Glo® Assay (Promega).
DNA isolation and mitochondrial DNA quantification
Knee cartilage, spleen and brain were collected from 6-month-old mice, flash frozen in liquid nitrogen, and DNA was isolated using a Blood and Tissue DNA isolation kit (Qiagen). To quantify mitochondrial DNA content, quantitative polymerase chain reaction (qPCR) was performed for mtDNA-encoded cytochrome c oxidase subunit 2 (COX2) and nucleus-encoded 18S ribosomal DNA as previously described (26). The COX2/18S ratio represents the relative mitochondrial DNA copy number (26).
RNA isolation and gene expression analysis
Mouse and human cartilage were homogenized in Qiazol Lysis Reagent (Qiagen) and RNA was isolated using the RNeasy Mini kit (Qiagen). In cultured cells, RNA was collected using Direct-Zol RNA miniprep kit (Zymo Research).
Gene expression was measured by qPCR using pre-designed TaqMan gene expression assays for MAP1LC3, ATG5, PPARGC1A and TFAM. GAPDH was measured as a reference gene.
Protein isolation and western blotting
Cultured cells were washed twice in PBS and lysed in ice-cold RIPA buffer (Pierce) supplemented with protease and phosphatase inhibitor cocktail (Thermo Scientific). Twenty micrograms of protein were resolved in 4–12% acrylamide gels, transferred to nitrocellulose membranes, and blotted overnight with antibodies against REDD1 (1:500, Proteintech), cleaved caspase 3 (1:500, Cell Signaling), LC3 (1:1000, Cell Signaling), p62 (1:1000, Cell Signaling), OXPHOS cocktail (1:1000, Abcam), PGC1α (1:1000, BioVision), TFAM (1:500, Cell Signaling), and GAPDH (1:5000, Abcam). After incubating with secondary antibodies for 1 hour at room temperature, blots were visualized using the Odyssey Infrared Imaging System (LI-COR). Intensity values were analyzed with the ImageStudioLite software and normalized to those of GAPDH.
Statistical analysis
Data are reported as the mean ± SD. Differences between groups were assessed by one-way analysis of variance (ANOVA) followed by a post-hoc Tukey test. Comparisons between two groups were assessed by an unpaired, two-tailed T-test after testing for equal variance using an F-test. All statistical analyses were performed using Prism 6 software (GraphPad Software). P-values less than 0.05 were considered significant.
RESULTS
Normal embryonic skeletal development and postnatal growth and maturation in REDD1 deficient mice
Mice with global deletion of REDD1 developed normally, and had similar weight and length as their wild type (WT) littermates (Supplemental Figure 1). Histological analysis of the knees showed no differences in articular cartilage morphology between Redd1+/+ and Redd1−/− mice up to 6 months of age (Supplemental Figure 1).
Increased severity of experimental OA in REDD1 deficient mice
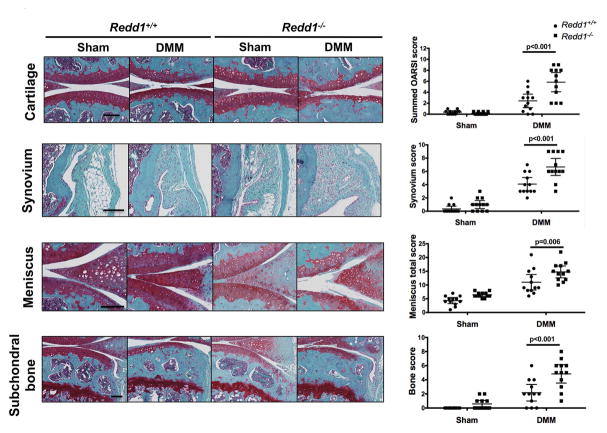
To evaluate the role of REDD1 in cartilage homeostasis during OA pathogenesis, we experimentally induced OA by destabilization of the medial meniscus (DMM). Redd1−/− mice showed increased articular cartilage degradation 10 weeks after DMM surgery and significantly higher histopathological scores (p<0.001) when compared with WT mice (Figure 1). In addition, Redd1−/− mice had significantly higher synovitis scores (p<0.001) with increased synovial hyperplasia and abundant cell infiltration, increased meniscus degeneration (p=0.006) with loss of meniscus cells, safranin-O staining and partial erosion of tissue; and more severe bone pathology (p<0.001) with thickening of subchondral bone, reduction of medullary cavities and increased osteophyte formation (Figure 1).
Figure 1.
Increased severity of experimental OA in Redd1−/− mice. A) Representative images of knees showing histological changes in the medial articular cartilage, synovium, meniscus and subchondral bone from 6.5-month-old Redd1+/+ and Redd1−/− mice (n=12 per genotype) subjected to DMM or sham procedure. Right panels show quantification of the histological scores for each joint tissue. All values are mean ± SD. Scale bar = 100 μm.
Reduced REDD1 expression in mouse joint tissues with aging
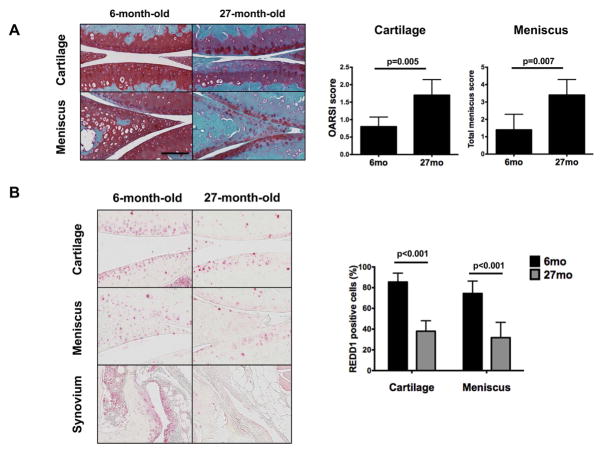
REDD1 deficiency resulted in widespread pathological changes in the joint during experimental OA. Our previous work focused only on articular cartilage, and we showed decreased expression of REDD1 with aging (10). Here, we sought to evaluate the changes in REDD1 expression during aging in different tissues of the knee joint from 6 and 27-month-old mice. Histological analysis of knees from old mice revealed OA like changes with aging that resulted in significantly increased histopathological scores when compared with young controls (Figure 2A). REDD1 immunohistochemical staining was abundant in cartilage, meniscus and synovium at 6 months of age. However, REDD1 expression was significantly reduced in articular cartilage and medial meniscus of 27-month-old mice when compared with 6-month-old controls (Figure 2B) and a similar reduction in REDD1 staining was observed in synovium.
Figure 2.
Reduced expression of REDD1 in join tissues during mouse aging. A) Representative histological images of knee medial articular cartilage and meniscus from 6 and 27-month-old mice (n=5 per group). Panel on the right shows quantification of the histological scores for cartilage and meniscus. B) Representative images of REDD1 immunohistochemical staining in knee joint tissues from 6 and 27-month-old mice (n=5 per group). Panel on the right shows quantification of REDD1 positive cells in cartilage and meniscus. All values are mean ± SD. Scale bar = 100 μm.
REDD1 deficiency and increased chondrocyte death
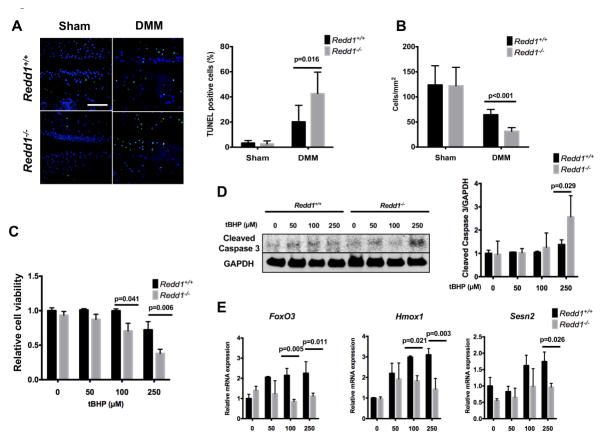
Chondrocyte death following injury is a pivotal event that is closely linked with cartilage degradation (27). We next evaluated whether REDD1 deficiency altered chondrocyte viability. TUNEL staining revealed an increased rate of chondrocyte apoptosis (p<0.001) in the knee articular cartilage of Redd1−/− mice when compared with controls and there was a concomitant reduction in cartilage cellularity (Figure 3A–B). To test whether articular chondrocytes with REDD1 depletion are more susceptible to undergo apoptotic cell death, cultured primary chondrocytes from Redd1−/− and WT mice were treated with increasing concentrations of tBHP (Figure 3C) to induce oxidative stress. Cells from mutant mice showed significantly higher rates of cell death than WT cells and a significant increase of cleaved caspase 3, and indicator of apoptotic cell death (Figure 3D). In addition, REDD1 deficient chondrocytes exhibited a significant reduction in the expression of key antioxidant genes (FoxO3, Hmox1, and Sesn2) under tBHP treatment when compared with WT cells (Figure 3E). On the other hand, REDD1 deficiency did not alter IL-1β induced expression of inflammatory mediators or apoptosis induction by tBHP (data not shown). Taken together, these results indicate that articular chondrocytes lacking REDD1 are more susceptible to apoptotic cell death under oxidative stress conditions.
Figure 3.
Increased chondrocyte death during experimental OA in Redd1−/− mice. A) Representative images of chondrocyte apoptosis in knee articular cartilage from 6.5-month-old Redd1+/+ and Redd1−/− mice (n=6 per genotype) subjected to DMM or sham procedure and assessed by TUNEL staining. Right panel shows quantification of TUNEL positive cells. Magnification bar = 100 μm. B) Quantification of articular cartilage cellularity in knees from 6.5-month-old Redd1+/+ and Redd1−/− mice (n=6 per genotype) subjected to DMM or sham procedure. C) Cell viability in cultured mouse articular chondrocytes treated with 0, 50, 100 or 250 mM tBHP for 24 hours. Data are expressed as relative cell viability with respect to untreated chondrocytes from Redd1+/+ mice. Four independent experiments were performed in triplicate. D) Western blot analysis of cleaved caspase 3 in cultured chondrocytes isolated from Redd1+/+ and Redd1−/− mice treated with different doses of tBHP to induce oxidative stress. Right panel shows quantification. Values are mean ± SD from three different experiments. E) qPCR analysis of FoxO3, Hmox1 and Sesn2 expression in cultured chondrocytes isolated from Redd1+/+ and Redd1−/− mice treated with different doses of tBHP. Values are mean ± SD from three different experiments performed in duplicate.
Redd1 deficiency reduces autophagy protein expression
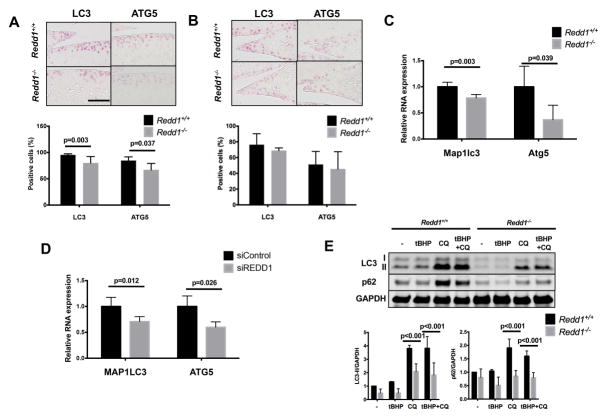
The expression levels of two key autophagic proteins, LC3 and ATG5, was significantly lower in the cartilage of 6-month-old Redd1−/− mice as compared to WT mice (Figure 4A). No differences in LC3 or ATG5 expression were seen in medial meniscus (Figure 4B) or synovium (Supplemental Figure 2A). In cultured primary chondrocytes, LC3 and Atg5 mRNA levels were significantly lower in cells from Redd1−/− mice when compared with WT cells (Figure 4C), and this effect was mimicked by siRNA-mediated REDD1 knockdown in human articular chondrocytes (Figure 4D) but it was not present in primary mouse synoviocytes (Supplemental Figure 2B). In addition, autophagy flux analysis by western blot revealed a significant reduction in LC3 processing and p62 protein levels in REDD1 deficient chondrocytes when compared with WT cells under normal and stress conditions (Figure 4E). These results suggest that REDD1 is a key regulator of articular cartilage autophagy in vivo and in vitro.
Figure 4.
Reduced expression of autophagy markers in REDD1 deficient articular chondrocytes. A) Representative images of LC3 and ATG5 immunohistochemical staining in knee articular cartilage from 6-month-old Redd1+/+ and Redd1−/− mice (n=6 per genotype). Right panel shows quantification of positive cells. Values are mean ± SD. Magnification bar = 100 μm. B) Representative images of LC3 and ATG5 immunohistochemical staining in medial meniscus from 6-month-old Redd1+/+ and Redd1−/− mice (n=6 per genotype). Right panel shows quantification of positive cells. Values are mean ± SD. Magnification bar = 100 μm. C) qPCR analysis of Map1lc3 and Atg5 expression in cultured chondrocytes isolated from Redd1+/+ and Redd1−/− mice. Values are mean ± SD from three different experiments. D) qPCR analysis of LC3 and ATG5 expression in human cultured chondrocytes transfected with siRNA control or siRNA against REDD1. Values are mean ± SD from four different donors. E) Western blot analysis of p62 and LC3 in cultured chondrocytes isolated from Redd1+/+ and Redd1−/− mice and treated with tBHP or chloroquine. Bottom panel shows quantification. Values are mean ± SD from three different experiments.
REDD1 controls mitochondrial biogenesis through PGC1α signaling
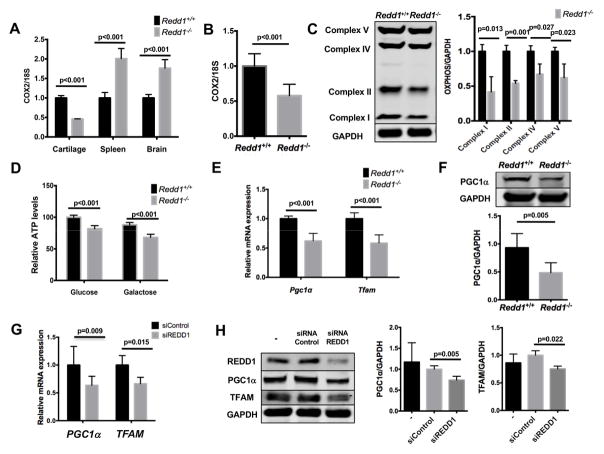
It has been recently reported that REDD1 deficiency results in accumulation of dysfunctional mitochondria (18). We measured the levels of mitochondrial (mtDNA) and nuclear (nDNA) DNA in the cartilage of Redd1−/− and WT mice to estimate the number of mitochondria. Surprisingly, whereas there was a significant increase in the mtDNA/nDNA ratio in the spleen and brain of mutant mice consistent with previous reports (18), we found a marked reduction in mtDNA in the cartilage of Redd1−/− mice (Figure 5A) suggesting that REDD1 controls mitochondrial number in a tissue specific manner. Moreover, Redd1−/− primary chondrocytes had significantly lower mtDNA content (Figure 5B), mitochondrial complex proteins (Figure 5C), and ATP levels (Figure 5D) than WT cells.
Figure 5.
Reduced mitochondrial biogenesis in REDD1 deficient articular chondrocytes. A) Mitochondrial DNA copy number in cartilage, spleen and brain from 6-month-old Redd1+/+ and Redd1−/− mice represented as the COX2/18S DNA ratio measured by qPCR. Values are mean ± SD from four different mice. B) Mitochondrial DNA copy number in cultured articular chondrocytes isolated from Redd1+/+ and Redd1−/− mice represented as the COX2/18S DNA ratio measured by qPCR. Values are mean ± SD from three different experiments. C) Western blot analysis of mitochondrial complex proteins expression in cultured chondrocytes isolated from Redd1+/+ and Redd1−/− mice. Right panel shows quantification. Values are mean ± SD from three different experiments. D) Intracellular ATP levels in cultured chondrocytes isolated from Redd1+/+ and Redd1−/− mice in the presence of 25mM glucose or galactose. Values are mean ± SD from four different experiments. E) qPCR analysis of Pgc1α and Tfam expression in cultured chondrocytes isolated from Redd1+/+ and Redd1−/− mice. Values are mean ± SD from three different experiments. F) Western blot analysis of PGC1α expression in cultured chondrocytes isolated from Redd1+/+ and Redd1−/− mice. Bottom panel shows quantification. Values are mean ± SD from three different experiments. G) qPCR analysis of PGC1α and TFAM expression in human cultured chondrocytes transfected with siRNA control or specific against REDD1. Values are mean ± SD from four different donors. H) Western blot analysis of PGC1α and TFAM expression in human cultured chondrocytes transfected with siRNA control or specific against REDD1. Values are mean ± SD from four different donors.
These findings prompted us to investigate whether mitochondrial biogenesis was impaired in the absence of REDD1. Redd1−/− chondrocytes, but not synoviocytes, had significantly lower mRNA levels of Pgc1a and Tfam, two key regulators of mitochondrial biogenesis (Figure 5E, Supplemental Figure 2C). PGC1α protein levels were also reduced in chondrocytes form mutant mice (Figure 5E), but not in synoviocytes (Supplemental Figure 2D). In a similar fashion, REDD1 knockdown in cultured human articular chondrocytes resulted in a significant decrease of PGC1α and TFAM mRNA and protein levels (Figures 5F–G). These data indicate that REDD1 controls mitochondrial biogenesis in articular chondrocytes through transcriptional regulation of PGC1α and TFAM.
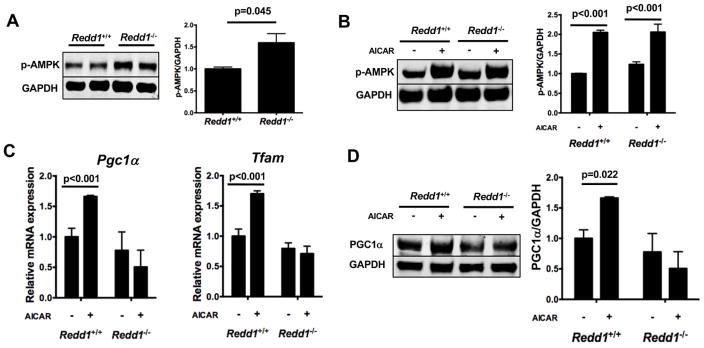
AMPK signaling is known to act upstream of PGC1α to promote mitochondrial biogenesis and regulate energy metabolism (28). To elucidate whether AMPK signaling is involved in REDD1-induced PGC1α expression, levels of phosphorylated AMPK (p-AMPK) were measured in cultured chondrocytes from Redd1+/+ and Redd1−/− mice by western blot. Chondrocytes from mutant mice showed higher levels of p-AMPK than control cells (Figure 6A). Upon treatment with AICAR, p-AMPK levels were similar in Redd1+/+ and Redd1−/− chondrocytes (Figure 6B) indicating that REDD1 is not required for AMPK activation by AICAR. However, AMPK activation induced expression of PGC1α and TFAM mRNA (Figure 6C) and protein levels (Figure 6D) in normal chondrocytes but not in REDD1 deficient cells. These findings suggest that REDD1 acts downstream of AMPK and is required for AMPK mediated transcriptional control of PGC1α.
Figure 6.
REDD1 is required for AMPK dependent PGC1α expression. A) Western blot analysis of AMPK phosphorylation in cultured chondrocytes isolated from Redd1+/+ and Redd1−/− mice. Right panel shows quantification. Values are mean ± SD from three different experiments. B) Western blot analysis of AMPK phosphorylation in cultured chondrocytes isolated from Redd1+/+ and Redd1−/− mice treated with AICAR or vehicle for 24 hours. Right panel shows quantification. Values are mean ± SD from three different experiments. C) qPCR analysis of Pgc1α and Tfam expression in cultured chondrocytes isolated from Redd1+/+ and Redd1−/− mice and treated with AICAR or vehicle for 24 hours. Values are mean ± SD from three different experiments. Right panel shows quantification. Values are mean ± SD from three different experiments. D) Western blot analysis of PGC1α expression in cultured chondrocytes isolated from Redd1+/+ and Redd1−/− mice and treated with AICAR or vehicle for 24 hours. Right panel shows quantification. Values are mean ± SD from three different experiments.
DISCUSSION
The main finding of the present study is that REDD1 deficiency increases the severity of experimental OA in mice. Articular cartilage damage was more severe in Redd1−/− mice as compared to WT and similar differences were observed in synovium, meniscus and in subchondral bone. These findings are of particular relevance in the context of joint aging, since REDD1 expression is reduced in human and mouse cartilage (10), and in mouse meniscus and synovium with age.
Another important finding of this study is that cartilage cellularity in Redd1−/− mice was significantly reduced. Extensive death of chondrocytes is a hallmark of post-traumatic OA pathophysiology and linked to tissue damage (29–31). After the initial injury that leads to disruption of the cartilage matrix and death of chondrocytes in the impacted area, multiple factors are generated in response to the trauma that can contribute to the activation of apoptotic pathways, further extending the zone of chondrocyte death. These factors include inflammatory cytokines, reactive oxygen species, nitric oxide and various damage-associated molecular pattern molecules (DAMPs) (32, 33). Our data showed that REDD1 preserves chondrocyte viability in vivo and in vitro and supports the notion that this function of REDD1 partly mediates the effects of REDD1 in posttraumatic OA. While the precise molecular mechanisms underlying this function of REDD1 in cartilage remains to be entirely elucidated, our data support the role of REDD1 as an important survival factor against oxidative stress in chondrocytes by activating the expression of important antioxidant genes and protecting cells from apoptotic cell death. In keeping with this, it has been reported that a fraction of REDD1 is located in the mitochondria, and that mitochondrial localization is required for REDD1 to suppress mitochondrial reactive oxygen species (ROS) production (34). REDD1 deficient murine embryonic fibroblasts exhibit increased levels of ROS and ectopic expression of REDD1 is sufficient to normalize cellular ROS production (34). Furthermore, treatment with antioxidant prevents hydrogen peroxide-induced apoptosis of human umbilical vein endothelial cells in a REDD1-dependent manner (35). Taken together, these findings highlight the role of REDD1 as a pivotal factor in the cellular defense mechanisms against oxidative stress.
Autophagy is an essential cellular process that maintains organismal homeostasis by degradation and turnover of the defective organelles within the cell. Mounting evidence supports that autophagy is an important regulator of articular cartilage homeostasis and may play a key role on OA pathogenesis (5, 12). REDD1 regulates autophagy in an mTOR independent fashion involving redox-dependent regulation of ATG4B, a key enzyme in autophagosome assembly and maturation (18). In the absence of REDD1, ATG4B activity is high and prevents the correct processing of LC3, thereby decreasing autophagic flux. In addition to our previous findings that REDD1 is essential for autophagy in articular chondrocytes (10), here we show that MAP1LC3 and ATG5 expression is reduced in cartilage from Redd1−/− mice and in human chondrocytes with REDD1 knockdown, indicating that REDD1 also regulates autophagy at the transcriptional level. This finding sheds additional light on the mechanisms that regulate autophagy in cartilage and underscores the importance of REDD1 as a regulator of autophagy at multiple levels.
Mitochondrial dysfunction has been well characterized in OA chondrocytes (26, 36–38) and alterations in mitochondrial biogenesis have been recently reported in OA (26). Emerging evidence suggests that REDD1 is a key regulator of mitochondrial integrity and oxidative capacity (39–41). Redd1−/− murine embryonic fibroblasts exhibited decreased basal oxygen consumption, oxidative ATP generation, and maximal respiratory capacity (18). Modulation of mitochondrial biogenesis can also be tissue specific and might vary depending on the cell-specific metabolic state (42). We found large differences in the relative content of mtDNA in spleen, brain and chondrocytes of Redd1−/− mice when compared with WT controls suggesting that REDD1 functions in mitochondrial biogenesis may be tissue dependent. Since cartilage is an avascular tissue with low oxygen tension (43), the role of hypoxia in controlling mitochondrial biogenesis might be particularly relevant to cartilage biology. The chief mediator of hypoxic response is the transcription factor HIF1α (hypoxia-inducible factor 1 α) (44). Although its role in mitochondrial biogenesis has not yet been studied in cartilage, HIF1α has been reported to regulate mitochondrial biogenesis in adipose tissue (45), skeletal muscle (46), and neurons (47). Since REDD1 is induced by hypoxia in an HIF1α-dependent manner (48) and basal REDD1 expression is high in articular cartilage under normal conditions, it could be hypothesized that the HIF1α-REDD1 pathway is a regulator of mitochondrial biogenesis in cartilage. Based on this rationale, it is thus conceivable that the impact of REDD1 deficiency is more severe in articular cartilage than in other tissues.
Here we have shown that articular chondrocytes from Redd1−/− mice have decreased mtDNA content, mitochondrial complex proteins, and ATP levels and when compared with wild type littermates. Expression of PGC1α and TFAM, two regulators of mitochondrial biogenesis, was significantly reduced in the absence of REDD1 in human and mouse chondrocytes, but not in mouse synoviocytes. Importantly, activation of the AMPK-PGC1α pathway by AICAR required REDD1 and this indicates that REDD1 acts downstream of AMPK to transcriptionally regulate PGC1α expression (Supplementary Figure 3). Thus, the present study provides a novel mechanism whereby REDD1 controls mitochondrial biogenesis in articular chondrocytes.
In summary, our study identifies REDD1 as a novel regulator or autophagy and mitochondrial biogenesis in articular chondrocytes and demonstrates that REDD1 deficiency increases the severity of experimental OA. The present results thus support the notion that the reduced REDD1 expression in human aging and OA-affected joints is a factor that contributes to OA progression and severity. These findings suggest that REDD1 is a central mediator of articular cartilage homeostasis and a potential target for the development of novel therapeutic strategies for OA.
Supplementary Material
Normal skeletal development and maturation in Redd1−/− mice. A) Representative images of 6-month-old Redd1+/+ and Redd1−/− mice. Quantification of nose-to-tail-tip length (B) and weight (C) of 6-month-old Redd1+/+ and Redd1−/− mice (n=6 per genotype). D) Representative images of the knee joints of P7, 1, 2 and 6-month-old Redd1+/+ and Redd1−/− mice. Scale bar = 100 μm.
Expression of autophagy and mitochondrial biogenesis markers in synoviocytes from Redd1+/+ and Redd1−/− mice. A) Representative images of LC3 and ATG5 immunohistochemical staining in knee synovium from 6-month-old Redd1+/+ and Redd1−/− mice (n=6 per genotype). Magnification bar = 100μM. B) qPCR analysis of Map1lc3 and Atg5 expression in cultured synoviocytes isolated from Redd1+/+ and Redd1−/− mice. Values are mean ± SD from three different experiments. C) qPCR analysis of Pgc1α and Tfam expression in cultured synoviocytes isolated from Redd1+/+ and Redd1−/− mice. Values are mean ± SD from three different experiments. D) Western blot analysis of PGC1α expression in cultured synoviocytes isolated from Redd1+/+ and Redd1−/− mice. Right panel shows quantification. Values are mean ± SD from three different experiments.
Novel mechanisms whereby REDD1 regulates articular cartilage homeostasis.
Acknowledgments
This study was supported by NIH grants AG007996 and AG049617.
We thank Josan Chung, Stuart Duffy, and Lilo Creighton for their technical assistance. We also thank Drs. Irina Budunova and Rubin Tuder for kindly providing the Redd1−/− mice. This study was supported by NIH grants AG007996 and AG049617.
Footnotes
The authors have no conflicts of interest.
AUTHOR CONTRIBUTIONS
All authors were involved in drafting the article or revising it critically for important intellectual content, and all authors approved the final version to be published. Dr. Lotz had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Study conception and design: Alvarez-Garcia, Kelly, Lotz.
Acquisition of data: Alvarez-Garcia, Matsuzaki, Olmer, Plate.
Analysis and interpretation of data: Alvarez-Garcia, Matsuzaki, Lotz.
References
- 1.Hunter DJ, Schofield D, Callander E. The individual and socioeconomic impact of osteoarthritis. Nat Rev Rheumatol. 2014;10(7):437–41. doi: 10.1038/nrrheum.2014.44. [DOI] [PubMed] [Google Scholar]
- 2.Loeser RF, Goldring SR, Scanzello CR, Goldring MB. Osteoarthritis: a disease of the joint as an organ. Arthritis Rheum. 2012;64(6):1697–707. doi: 10.1002/art.34453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lotz M, Loeser RF. Effects of aging on articular cartilage homeostasis. Bone. 2012;51(2):241–8. doi: 10.1016/j.bone.2012.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Choi AM, Ryter SW, Levine B. Autophagy in human health and disease. N Engl J Med. 2013;368(19):1845–6. doi: 10.1056/NEJMc1303158. [DOI] [PubMed] [Google Scholar]
- 5.Carames B, Taniguchi N, Otsuki S, Blanco FJ, Lotz M. Autophagy is a protective mechanism in normal cartilage, and its aging-related loss is linked with cell death and osteoarthritis. Arthritis Rheum. 2010;62(3):791–801. doi: 10.1002/art.27305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149(2):274–93. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Terkeltaub R, Yang B, Lotz M, Liu-Bryan R. Chondrocyte AMP-activated protein kinase activity suppresses matrix degradation responses to proinflammatory cytokines interleukin-1beta and tumor necrosis factor alpha. Arthritis Rheum. 2011;63(7):1928–37. doi: 10.1002/art.30333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Akasaki Y, Alvarez-Garcia O, Saito M, Carames B, Iwamoto Y, Lotz MK. FoxO transcription factors support oxidative stress resistance in human chondrocytes. Arthritis Rheumatol. 2014;66(12):3349–58. doi: 10.1002/art.38868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Akasaki Y, Hasegawa A, Saito M, Asahara H, Iwamoto Y, Lotz MK. Dysregulated FOXO transcription factors in articular cartilage in aging and osteoarthritis. Osteoarthritis Cartilage. 2014;22(1):162–70. doi: 10.1016/j.joca.2013.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alvarez-Garcia O, Olmer M, Akagi R, Akasaki Y, Fisch KM, Shen T, et al. Suppression of REDD1 in osteoarthritis cartilage, a novel mechanism for dysregulated mTOR signaling and defective autophagy. Osteoarthritis Cartilage. 2016;24(9):1639–47. doi: 10.1016/j.joca.2016.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Y, Vasheghani F, Li YH, Blati M, Simeone K, Fahmi H, et al. Cartilage-specific deletion of mTOR upregulates autophagy and protects mice from osteoarthritis. Ann Rheum Dis. 2014 doi: 10.1136/annrheumdis-2013-204599. [DOI] [PubMed] [Google Scholar]
- 12.Carames B, Hasegawa A, Taniguchi N, Miyaki S, Blanco FJ, Lotz M. Autophagy activation by rapamycin reduces severity of experimental osteoarthritis. Ann Rheum Dis. 2012;71(4):575–81. doi: 10.1136/annrheumdis-2011-200557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ellisen LW, Ramsayer KD, Johannessen CM, Yang A, Beppu H, Minda K, et al. REDD1, a developmentally regulated transcriptional target of p63 and p53, links p63 to regulation of reactive oxygen species. Molecular cell. 2002;10(5):995–1005. doi: 10.1016/s1097-2765(02)00706-2. [DOI] [PubMed] [Google Scholar]
- 14.Shoshani T, Faerman A, Mett I, Zelin E, Tenne T, Gorodin S, et al. Identification of a novel hypoxia-inducible factor 1-responsive gene, RTP801, involved in apoptosis. Molecular and cellular biology. 2002;22(7):2283–93. doi: 10.1128/MCB.22.7.2283-2293.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brugarolas J, Lei K, Hurley RL, Manning BD, Reiling JH, Hafen E, et al. Regulation of mTOR function in response to hypoxia by REDD1 and the TSC1/TSC2 tumor suppressor complex. Genes & development. 2004;18(23):2893–904. doi: 10.1101/gad.1256804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Corradetti MN, Inoki K, Guan KL. The stress-inducted proteins RTP801 and RTP801L are negative regulators of the mammalian target of rapamycin pathway. The Journal of biological chemistry. 2005;280(11):9769–72. doi: 10.1074/jbc.C400557200. [DOI] [PubMed] [Google Scholar]
- 17.DeYoung MP, Horak P, Sofer A, Sgroi D, Ellisen LW. Hypoxia regulates TSC1/2-mTOR signaling and tumor suppression through REDD1-mediated 14-3-3 shuttling. Genes & development. 2008;22(2):239–51. doi: 10.1101/gad.1617608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qiao S, Dennis M, Song X, Vadysirisack DD, Salunke D, Nash Z, et al. A REDD1/TXNIP pro-oxidant complex regulates ATG4B activity to control stress-induced autophagy and sustain exercise capacity. Nat Commun. 2015;6:7014. doi: 10.1038/ncomms8014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brafman A, Mett I, Shafir M, Gottlieb H, Damari G, Gozlan-Kelner S, et al. Inhibition of oxygen-induced retinopathy in RTP801-deficient mice. Investigative ophthalmology & visual science. 2004;45(10):3796–805. doi: 10.1167/iovs.04-0052. [DOI] [PubMed] [Google Scholar]
- 20.Glasson SS, Blanchet TJ, Morris EA. The surgical destabilization of the medial meniscus (DMM) model of osteoarthritis in the 129/SvEv mouse. Osteoarthritis Cartilage. 2007;15(9):1061–9. doi: 10.1016/j.joca.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 21.Glasson SS, Chambers MG, Van Den Berg WB, Little CB. The OARSI histopathology initiative - recommendations for histological assessments of osteoarthritis in the mouse. Osteoarthritis Cartilage. 2010;18(Suppl 3):S17–23. doi: 10.1016/j.joca.2010.05.025. [DOI] [PubMed] [Google Scholar]
- 22.Krenn V, Morawietz L, Haupl T, Neidel J, Petersen I, Konig A. Grading of chronic synovitis--a histopathological grading system for molecular and diagnostic pathology. Pathol Res Pract. 2002;198(5):317–25. doi: 10.1078/0344-0338-5710261. [DOI] [PubMed] [Google Scholar]
- 23.Kwok J, Onuma H, Olmer M, Lotz MK, Grogan SP, D’Lima DD. Histopathological analyses of murine menisci: implications for joint aging and osteoarthritis. Osteoarthritis Cartilage. 2016;24(4):709–18. doi: 10.1016/j.joca.2015.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carames B, Olmer M, Kiosses WB, Lotz MK. The relationship of autophagy defects to cartilage damage during joint aging in a mouse model. Arthritis Rheumatol. 2015;67(6):1568–76. doi: 10.1002/art.39073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gosset M, Berenbaum F, Thirion S, Jacques C. Primary culture and phenotyping of murine chondrocytes. Nat Protoc. 2008;3(8):1253–60. doi: 10.1038/nprot.2008.95. [DOI] [PubMed] [Google Scholar]
- 26.Wang Y, Zhao X, Lotz M, Terkeltaub R, Liu-Bryan R. Mitochondrial biogenesis is impaired in osteoarthritis chondrocytes but reversible via peroxisome proliferator-activated receptor gamma coactivator 1alpha. Arthritis Rheumatol. 2015;67(8):2141–53. doi: 10.1002/art.39182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Olson SA, Horne P, Furman B, Huebner J, Al-Rashid M, Kraus VB, et al. The role of cytokines in posttraumatic arthritis. J Am Acad Orthop Surg. 2014;22(1):29–37. doi: 10.5435/JAAOS-22-01-29. [DOI] [PubMed] [Google Scholar]
- 28.Liang H, Ward WF. PGC-1alpha: a key regulator of energy metabolism. Adv Physiol Educ. 2006;30(4):145–51. doi: 10.1152/advan.00052.2006. [DOI] [PubMed] [Google Scholar]
- 29.Borrelli J., Jr Chondrocyte apoptosis and posttraumatic arthrosis. J Orthop Trauma. 2006;20(10):726–31. doi: 10.1097/01.bot.0000249882.77629.5c. [DOI] [PubMed] [Google Scholar]
- 30.Buckwalter JA, Brown TD. Joint injury, repair, and remodeling: roles in post-traumatic osteoarthritis. Clin Orthop Relat Res. 2004;(423):7–16. [PubMed] [Google Scholar]
- 31.Furman BD, Olson SA, Guilak F. The development of posttraumatic arthritis after articular fracture. J Orthop Trauma. 2006;20(10):719–25. doi: 10.1097/01.bot.0000211160.05864.14. [DOI] [PubMed] [Google Scholar]
- 32.Lieberthal J, Sambamurthy N, Scanzello CR. Inflammation in joint injury and post-traumatic osteoarthritis. Osteoarthritis Cartilage. 2015;23(11):1825–34. doi: 10.1016/j.joca.2015.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martin JA, Buckwalter JA. Post-traumatic osteoarthritis: the role of stress induced chondrocyte damage. Biorheology. 2006;43(3–4):517–21. [PubMed] [Google Scholar]
- 34.Horak P, Crawford AR, Vadysirisack DD, Nash ZM, DeYoung MP, Sgroi D, et al. Negative feedback control of HIF-1 through REDD1-regulated ROS suppresses tumorigenesis. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(10):4675–80. doi: 10.1073/pnas.0907705107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu MC, Shi HM, Wang H, Gao XF. Salidroside protects against hydrogen peroxide-induced injury in HUVECs via the regulation of REDD1 and mTOR activation. Molecular medicine reports. 2013;8(1):147–53. doi: 10.3892/mmr.2013.1468. [DOI] [PubMed] [Google Scholar]
- 36.Blanco FJ, Rego I, Ruiz-Romero C. The role of mitochondria in osteoarthritis. Nat Rev Rheumatol. 2011;7(3):161–9. doi: 10.1038/nrrheum.2010.213. [DOI] [PubMed] [Google Scholar]
- 37.Coleman MC, Ramakrishnan PS, Brouillette MJ, Martin JA. Injurious Loading of Articular Cartilage Compromises Chondrocyte Respiratory Function. Arthritis Rheumatol. 2016;68(3):662–71. doi: 10.1002/art.39460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lane RS, Fu Y, Matsuzaki S, Kinter M, Humphries KM, Griffin TM. Mitochondrial respiration and redox coupling in articular chondrocytes. Arthritis Res Ther. 2015;17:54. doi: 10.1186/s13075-015-0566-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lipina C, Hundal HS. Is REDD1 a Metabolic Eminence Grise? Trends Endocrinol Metab. 2016 doi: 10.1016/j.tem.2016.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Regazzetti C, Dumas K, Le Marchand-Brustel Y, Peraldi P, Tanti JF, Giorgetti-Peraldi S. Regulated in development and DNA damage responses -1 (REDD1) protein contributes to insulin signaling pathway in adipocytes. PloS one. 2012;7(12):e52154. doi: 10.1371/journal.pone.0052154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dungan CM, Wright DC, Williamson DL. Lack of REDD1 reduces whole body glucose and insulin tolerance, and impairs skeletal muscle insulin signaling. Biochemical and biophysical research communications. 2014;453(4):778–83. doi: 10.1016/j.bbrc.2014.10.032. [DOI] [PubMed] [Google Scholar]
- 42.Wenz T. Regulation of mitochondrial biogenesis and PGC-1alpha under cellular stress. Mitochondrion. 2013;13(2):134–42. doi: 10.1016/j.mito.2013.01.006. [DOI] [PubMed] [Google Scholar]
- 43.Schipani E, Ryan HE, Didrickson S, Kobayashi T, Knight M, Johnson RS. Hypoxia in cartilage: HIF-1alpha is essential for chondrocyte growth arrest and survival. Genes & development. 2001;15(21):2865–76. doi: 10.1101/gad.934301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Semenza GL, Wang GL. A nuclear factor induced by hypoxia via de novo protein synthesis binds to the human erythropoietin gene enhancer at a site required for transcriptional activation. Molecular and cellular biology. 1992;12(12):5447–54. doi: 10.1128/mcb.12.12.5447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang X, Lam KS, Ye H, Chung SK, Zhou M, Wang Y, et al. Adipose tissue-specific inhibition of hypoxia-inducible factor 1{alpha} induces obesity and glucose intolerance by impeding energy expenditure in mice. The Journal of biological chemistry. 2010;285(43):32869–77. doi: 10.1074/jbc.M110.135509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.O’Hagan KA, Cocchiglia S, Zhdanov AV, Tambuwala MM, Cummins EP, Monfared M, et al. PGC-1alpha is coupled to HIF-1alpha-dependent gene expression by increasing mitochondrial oxygen consumption in skeletal muscle cells. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(7):2188–93. doi: 10.1073/pnas.0808801106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhao J, Li L, Pei Z, Li C, Wei H, Zhang B, et al. Peroxisome proliferator activated receptor (PPAR)-gamma co-activator 1-alpha and hypoxia induced factor-1alpha mediate neuro- and vascular protection by hypoxic preconditioning in vitro. Brain Res. 2012;1447:1–8. doi: 10.1016/j.brainres.2012.01.059. [DOI] [PubMed] [Google Scholar]
- 48.Schwarzer R, Tondera D, Arnold W, Giese K, Klippel A, Kaufmann J. REDD1 integrates hypoxia-mediated survival signaling downstream of phosphatidylinositol 3-kinase. Oncogene. 2005;24(7):1138–49. doi: 10.1038/sj.onc.1208236. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Normal skeletal development and maturation in Redd1−/− mice. A) Representative images of 6-month-old Redd1+/+ and Redd1−/− mice. Quantification of nose-to-tail-tip length (B) and weight (C) of 6-month-old Redd1+/+ and Redd1−/− mice (n=6 per genotype). D) Representative images of the knee joints of P7, 1, 2 and 6-month-old Redd1+/+ and Redd1−/− mice. Scale bar = 100 μm.
Expression of autophagy and mitochondrial biogenesis markers in synoviocytes from Redd1+/+ and Redd1−/− mice. A) Representative images of LC3 and ATG5 immunohistochemical staining in knee synovium from 6-month-old Redd1+/+ and Redd1−/− mice (n=6 per genotype). Magnification bar = 100μM. B) qPCR analysis of Map1lc3 and Atg5 expression in cultured synoviocytes isolated from Redd1+/+ and Redd1−/− mice. Values are mean ± SD from three different experiments. C) qPCR analysis of Pgc1α and Tfam expression in cultured synoviocytes isolated from Redd1+/+ and Redd1−/− mice. Values are mean ± SD from three different experiments. D) Western blot analysis of PGC1α expression in cultured synoviocytes isolated from Redd1+/+ and Redd1−/− mice. Right panel shows quantification. Values are mean ± SD from three different experiments.
Novel mechanisms whereby REDD1 regulates articular cartilage homeostasis.