Abstract
The practice of pre-emptive individualized medicine is predicated on the discovery, development and application of biomarkers in specific clinical settings. Mycosis fungoides and Sézary syndrome are the two most common type of cutaneous T-cell lymphoma, yet diagnosis, prognosis and disease monitoring remain a challenge. In this review, we discuss the current state of biomarker discovery in mycosis fungoides and Sézary syndrome, highlighting the most promising molecules in different compartments. Further, we emphasize the need for continued multicentre efforts to validate available and new biomarkers and to develop prospective combinatorial panels of already discovered molecules.
Keywords: cell surface marker, cutaneous T-cell lymphoma, gene expression, screening, secretory protein
1 INTRODUCTION
In the era of personalized medicine, the study of malignancy-related biomarkers has led to advances in early detection of various cancers, improved prognostication and monitoring of disease burden.
Cutaneous T-cell lymphomas (CTCL) are a heterogeneous group of extranodal lymphomas with mycosis fungoides (MF) the most common variant, and Sézary syndrome (SzS) an aggressive, leukaemic variant.[1] Differential diagnosis of early-stage MF is a clinically challenging task as many patients present with non-specific eczematous patches. With histological examination, a 40% false-negative rate and a 44% false-positive rate have been estimated in the diagnosis of early disease.[2,3] Furthermore, biopsy interpretation of early MF is frequently subjective, with studies demonstrating poor reproducibility of diagnoses rendered by different pathologists or even the same pathologist.[4–6] Even molecular techniques have may have low sensitivity and specificity in early stages. One such technique, clonal rearrangements of the T-cell receptor (TCR) gamma gene by polymerase chain reaction (PCR) are positive only in 74% of early MF biopsies.[7] Further, sensitivity of a given marker has been shown to vary with technique used and the density of tumor cells in a sample.[8] Early-stage MF is generally indolent with little effect on overall life expectancy; however, rapid progression and extracutaneous spread of malignant T cells occur in a subset of patients.[1,9] Prediction of which MF/SzS patients will progress is difficult, although recent collaborative efforts have been undertaken to better prognosticate outcomes.[10]
Taken together, the challenges of diagnosis and prognosis underscore the need for reliable markers for MF/SzS. In this review, we provide an overview of diagnostic and prognostic biomarkers in MF/SzS and discuss their potential clinical applicability. Current ongoing collaborative efforts will enable us to overcome the above-mentioned limits and determine combinations of biomarkers with the most clinical relevance.
2 FUNCTIONS OF BIOMARKERS IN MF/SZS
Ideal characteristics of molecular biomarkers vary depending on their intended clinical application.[11] We propose four functions biomarkers can perform in MF/SzS:
Biomarkers used for screening purposes must be inexpensive, easily and robustly measurable using minimally invasive routine laboratory techniques, and should discriminate between malignant and benign processes. Early-stage MF may be confused with benign inflammatory conditions such as psoriasis, eczema or other dermatoses (e.g. lymphomatoid drug eruptions, pityriasis rubra pilaris, prodromal bullous dermatoses).[12] Considering the low prevalence of MF and the high prevalence of these benign dermatoses, a screening strategy must achieve high specificity and sensitivity of more than 75% to avoid an unacceptable level of false-positive results. Such markers should also be used prior to potentially harmful therapy. Misdiagnosis of MF can lead to a choice of treatment harmful to patients; for example, cyclosporine and TNF-α inhibitors cause rapid lymphoma progression in patients who are treated for presumed psoriasis, but in fact have undiagnosed MF.[12]
Diagnostic biomarkers should accurately identify malignant T cells in peripheral blood and in tissue including skin, lymph nodes and bone marrow. They should assess tumor burden and differentiate MF/SzS from other diseases. Diagnostic markers should also allow for subtyping of cutaneous lymphoma, as some variants need to be precisely diagnosed in order to provide appropriate therapeutic guidance. Specific diagnostic markers may require more elaborate, time-consuming and complex methods compared with screening markers and are only applied to cases with a high level of suspicion for MF/SzS. Such biomarkers should have very high levels of sensitivity and specificity of more than 95%.
Prognostic Biomarkers, which predict biological behaviour, should accurately differentiate aggressive from indolent disease. Current staging of MF/SzS uses TNMB classification with disease presentation in the skin (T), lymph nodes (N), viscera (M) and blood (B) stratifying patients into early stage (IA to IIA) and advanced stage (IIB to IVB).[13] Advanced stage at the time of presentation correlates with poor prognosis, although outcomes within a particular TNMB stage fall into a wide range. For example, stage IVA/B disease which signifies either blood, nodal or visceral involvement has an overall 5-year survival rate of 0%–40%.[14] An improvement of prognostication within a given TNMB stages may be achieved with a combination of biomarkers.
Biomarkers of disease activity must reflect changing tumor dynamics, meaning the marker should quantitatively or qualitatively change before clinically significant progression or remission.[11] These markers are used for detecting occult recurrences in patients who are in clinical remission and for monitoring response to therapy.
Some existing and emerging markers fall into more than one of these four categories. To more closely examine specific putative markers and biomarker panels, we will review serum-based, cell surface, genetic and epigenetic markers (Figure 1).
FIGURE 1.
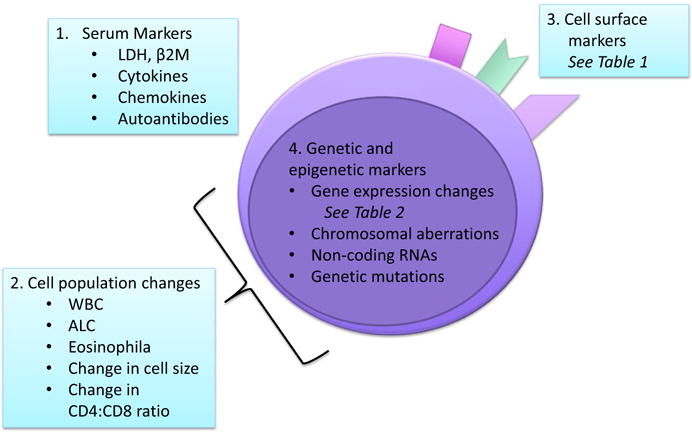
Biomarker Classes in MF/SzS—Putative markers include[1] molecules measured in serum samples,[2] leucocyte cell population changes measured by whole blood or peripheral mononuclear cell analysis,[3] cell surface markers measured by flow cytometry or immunohistochemistry and[4] genetic and epigenetic markers measured by a variety of molecular techniques. Biomarkers may belong to several categories at the same time. LDH, lactate dehydrogenase; β2M, beta-2 microglobulin; WBC, white blood cell count; ALC, absolute lymphocyte count
3 SERUM MARKERS AND SECRETORY PROTEINS
A number of serum markers have been studied in MF/SzS, and while these molecules are non-specific, many are already routinely tested via simple, cheap serum studies. Elevated serum lactate dehydrogenase (LDH) has been associated with advanced stage[15] and aggressive disease in MF/SzS patients.[16–19] Elevated beta-2 microglobulin (β2M)[18] and specific IgE levels towards environmental or food allergens have each been associated with poor prognosis in SzS patients; although unlike LDH, data are limited and they are not widely used.[20] Due to their low specificity, these markers are unlikely to have screening or diagnostic applications, but may be useful in prognostic panels or measures of disease activity.
Secretory proteins including interleukins, growth factors, interferons and necrosis factors are measured in a blood draw with an enzyme-linked immunosorbant assay (ELISA) or in a tissue section using immunohistochemistry (IHC). Elevated soluble IL-2 receptor was upregulated in the serum of SzS patients compared with cutaneous B-cell lymphoma and T-cell inflammatory diseases but did not correlate with disease outcome in CTCL patients.[21,22] Elevated IL-13 was identified in the skin of MF/SzS patients and AD patients but not normal skin or psoriasis, and levels of IL-13 increased with increasing MF/SzS stage.[23] Elevated IL-31 has been identified in MF/SzS with conflicting data on whether higher levels are associated with pruritus.[24–26] Thymus and activation-regulated chemokine (TARC/CCL17) is a chemoattractant elevated in MF compared with psoriasis and healthy controls, with higher levels in tumor-stage disease compared with early stage.[27] Additionally, CCR4, one of the receptors for TARC, is expressed on epidermotropic cells in patch, plaque and tumor-stage MF.[27] Technological advances in cytokine measurement have produced addressable bead or chip-based assays for quantitation of up 100 serum proteins in a single sample. A panel of upregulated TNFR1, TNFR2 and IL-12 tested using a Luminex multiplex assessment of serum proteins in MF/SzS reliably distinguished cases from normal controls with 88% sensitivity and 98% specificity.[22] Application of these technologies could allow for testing of a multiple-marker panel with a single blood draw to screen patients with eczematous and psoriasiform dermatoses as well as to diagnose and prognosticate MF/ SzS patients.
Multiplex immunoassays have another potential application in a promising biomarker class: cancer-specific autoantibodies.[28] Prior to clinical signs and symptoms of cancer, the immune system senses tumor cells and produces autoantibodies to tumor-associated antigens. In solid organ cancers, these autoantibodies have been detected years before clinical manifestations of disease, making them excellent early screening tools.[29,30] Interestingly, several chaperons proteins (HSP60, HSP71 and HSPA5) were identified as antigens for cutaneous lymphoma by proteome serology.[31] Further exploration of the role these autoantibodies may play in detecting MF/SzS is warranted.
4 LEUCOCYTE POPULATION CHANGES
A complete blood count is a non-specific test with useful prognostic implications. Elevated white blood cell (WBC) count, absolute lymphocyte count and absolute eosinophilia have been associated with increased disease progression and disease-specific death in MF/SzS patients.[10,32] Severe cytopenias can also be a sign of marrow involvement and an indication for bone marrow biopsy. More specific than total WBCs, changes in the T-cell population can correlate with disease activity. Low numbers of CD8+ T cells (<600/mL) have been associated with a worse prognosis in MF/SzS patients.[33–35] T-cell size can also be prognostic as large-cell transformation (LCT) to ≥4 times a normal lymphocyte portends a worse prognosis.[36,37] Atypical lymphocytes are larger with irregular nuclear contour, identified by high scatter on flow cytometry.[38] LCT of disease in MF/SzS patients is associated with overall reduction in life expectancy to 37 months compared to 163 months in untransformed disease.[36] As LCT is temporally intertwined with disease progression, it may be more clinically relevant to consider predictive markers of pending LCT to identify patients at risk before their disease progresses. Elevated β2M, LDH and CD25 have been identified as possible predictors of transformation.[36,39] Additionally, cells that demonstrate large-cell transformation may be CD30+ or CD30−. In a study of patients with large-cell transformation of mycosis fungoides, CD30 positivity was a predictor of improved survival.[40]
5 CELL SURFACE MARKERS
Significant efforts have been made to identify changes in cell surface markers specific to circulating Sézary cells or infiltrating cells in MF, summarized in Table 1. Flow cytometry on peripheral blood mononuclear cells (PBMCs) and IHC on formalin-fixed paraffin-embedded skin biopsies are used to characterize surface marker expression. Importantly, multicolour flow cytometry allows for the simultaneous measurement of eleven or more markers on a single cell.[41] Double immunoenzyme staining and multispectral immunofluorescence imaging allow for more than one marker to be visualized at a time in histopathological analysis.[42] Thus, as new markers continue to be identified, additional parameters can easily be added to these existing approaches.
TABLE 1.
Measurement of cell surface markers in SzS or MF patient samples
| Candidate biomarkers | Sample size and typea | Methodb | Diagnostic value/outcomec | Ref |
|---|---|---|---|---|
| CD26 | 52 SzS, 151 MF (14 stage B1), 88 IE, 72 HD | Flow | CD4+CD26−>30% of PBLs in 93% of B1-MF cases and >40% in all SzS; CD4+CD26−<30% PBLs in 100% of HD, IE and B0-MF | [79] |
| 107 SzS | Flow | 89.7% of SzS patients demonstrate loss of CD26 | [50] | |
|
| ||||
| CD3dim | 17 SzS, 11 HD | Flow | 76% of SzS patients with >1000 CD4+CD7−CD3dim cells μL−1, 0% of HD | [44] |
| 107 SzS | Flow | 76.6% of SzS patients demonstrate dim CD3 | [50] | |
|
| ||||
| CD27 | 40 SzS, 137 IE, 63 HD | Flow, IHC | 40% of PBLs from IE or HD were CD4+CD27−compared with 80.5% of PBLs from SzS | [80] |
|
| ||||
| CTLA-4 | 9 SzS, 9 MF | IHC | 89% staining in MF, 100% staining in SzS | [81] |
|
| ||||
| CD45R0 vs CD45RA | 29 SzS | IHC | 75% of SzS with 75%–100% of infiltrate CD45RO+ | [18] |
| 215 MF | IHC | 8.7% of MF are CD45RA+ with higher # of other pathologic abnormalities | [82] | |
| 6 SzS, 30 MF, 18 HD | Flow | Increase in % and # of CD4+CD45R0+ CD45RA−in late stage MF/SzS compared to early disease or HD | [35] | |
|
| ||||
| Vimentin | 14 MF, 3 SzS, 2 other CTCL | SEREX, qRT-PCR | qRT-PCR found vimentin in 73% of CTCL samples, also expressed in 80% of normal tissues | [83] |
| 87 CTCL | 2D WB + MS | Vimentin identified at multiple sites on 2D blot suggesting splicing variants or variable isoforms | [31] | |
|
| ||||
| CD158k/KIR3DL2 | 17 SzS, 11 HD | Flow | 65% of SzS with CD158k+ cells, 0% of HD | [44] |
| 33 SzS | Flow | 96.9% of SzS with CD158k+ cells, these CD158k+ restricted to phenotypically abnormal T cells | [84] | |
| 34 SzS, 6 IE, 10 HD | Flow | Positive correlation between %CD158k+ and %atypical circulating cells in SzS, malignant clone expresses CD158k | [85] | |
| 25 SzS | Flow | CD158k+ associated with clinical flare | [86] | |
|
| ||||
| NKp46/CD335 | 17 SzS, 5 MF, 10 IE, 4 HD | Flow, immunoblotting | CD4+NKp46+ tumoral cells identified in all SzS patients, no significant CD4+NKp46+ in other groups | [87] |
|
| ||||
| Ganglioside GD3/CD60 | 6 SzS, 30 MF, 18 HD | Flow | Significant increase in % and # of CD4+CD60+ in late stage MF/SzS compared to early disease or HD | [35] |
| 62 SzS, 180 MF, 6 BCL, 19 AD | Flow | Higher # of CD60+ circulating CD4+ T cells at SzS presentation associated with lower probability of survival | [88] | |
|
| ||||
| Syndecan 4 (SD-4) | 6 SzS, 3 MF, 4 HD, 3 AD, 3 psoriasis | Flow | All SzS patients had high expression of SD-4 on CD4+ cells, significantly higher than all other samples | [89] |
|
| ||||
| Sialomucin (CD164) | 59 SzS, 10 MF, 6 AD, 14 HD | Flow | % CD4+CD164+ T cells significantly increased in SzS compared to MF, AD, HD; CD164+ cells disappear with disease remission | [90] |
|
| ||||
| PD-1/CD279 | 7 SzS, 4 MF, 5 HD | Flow | Significant increase in CD4+PD-1+ cells in SzS group compared with MF and HD | [91] |
| 27 SzS, 60 MF | IHC | 89% of SzS cases had >50% neoplastic cells PD-1+, 13% of MF cases had >50% neoplastic cells PD-1+ | [92] | |
| 25 SzS, 30 IE | IHC | >50% of infiltrating T cells PD-1+ in 92% of SzS cases (where PD-1+ cells were CD4+) and of 13% IE cases (where PD-1+ cells were CD8+) | [93] | |
|
| ||||
| CD52 | 16 CTCL | Flow | 87.5% of CTCL samples were CD52+ | [94] |
PBLs, peripheral blood lymphocytes.
Sample type abbreviations include Sézary syndrome (SzS), mycosis fungoides (MF), inflammatory erythroderma (IE), healthy donors (HD), cutaneous T-cell lymphoma (CTCL), B-cell lymphomas (BCL) and atopic dermatitis (AD).
Candidate biomarkers have been studied primarily using peripheral blood samples or skin biopsies with application of flow cytometry (flow) or immunohistochemistry (IHC). Other methods include serological identification of antigens by recombinant expression cloning (SEREX), Western blotting (WB) and mass spectrometry (MS).
Peripheral blood leucocytes (PBLs).
New technology has recently been utilized for TCR analysis. TCR-Vβ analysis either by flow cytometry or IHC in frozen tissue has historically been used to identify clonal populations in circulating T cells or lymphocytic infiltrates, allowing for differentiation between MF/SzS and inflammatory conditions.[13,43,44] Further, patients with a single gene rearrangement clone detected in multiple concurrent biopsy samples at the time of diagnosis were more likely to have progressive disease than those with multiple TCR clones.[45] TCR-Vβ analysis using these methods does not include all TCR-Vβ families, so a clone may be suspected but not identified. TCR gene rearrangements have also been identified with TCR PCR analysis. Recent introduction of high-throughput TCR sequencing of CDR3 has improved the identification of malignant T cells. Next-generation sequencing is on its way to offer full spectrum clonal analysis of α-, β-, γ-and δ-submits. This methodology provides a full spectrum of clones in the sample making it easy to follow a particular clone for identification of minimal residual disease, including malignant cells in lymph nodes missed by traditional histopathological analysis.[46,47] The utility of TCR analysis as a stand-alone biomarker of MF/SzS is limited as other T-cell lymphoproliferative conditions and cutaneous lymphoid dyscrasias also demonstrate clonal rearrangement of TCR genes;[48,49] however, TCR analysis has been successfully combined with other markers to improve identification of SzS.[44]
The prognostic value of a cell surface marker may vary with treatment. Novelli et al.,[50] found SzS patients who had a presence of a variable proportion of CD26+ atypical cells at diagnosis showed a statistically significant higher overall survival. However, in their retrospective cohort study of eleven MF/SzS patients who had undergone treatment, Vandersee et al.[51] calculated a low positive predictive value for changes in CD26 expression and clinically meaningful events. CD26-cell number has also been shown to vary with chemotherapeutic and immunomodulating treatments irrespective of concurrent clinical response.[51,52] Thus, CD26 status may confer prognostic information at diagnosis but may not be indicative of treatment response or disease progression. Chemotherapeutic treatment regimens increase cell turnover, and immunomodulating ones can alter the presence of surface markers related to immune function. As a result, treatment conditions must be carefully considered when trending a surface marker for prognosis.
6 GENETIC AND EPIGENETIC MARKERS
Gene transcription in MF/SzS has been studied extensively using blood samples or skin biopsies with microarray platforms, transcriptome sequencing and quantitative reverse transcriptase PCR (qRT-PCR).[53] A review of this work by Wong highlighted a number of key genetic markers with altered expression in MF/SzS patients.[54] We build on that work and present a list of genes identified in multiple expression studies in Table 2.
TABLE 2.
Measurement of gene expression in SzS or MF patient samples
| Candidate genes | Sample size and typea | Methodb | Diagnostic value/outcome | Ref |
|---|---|---|---|---|
| T-plastin (PLS3) | PBMCs: 18 SzS, 9 Th2-skewed controls | Microarray, qRT-PCR | Increased 14-fold in SzS compared with controls based on array and 479-fold based on qRT-PCR | [95] |
| PBMCs: 49 SzS, 69 HD, 3 IE | qRT-PCR | Increased 520-fold in SzS compared with controls | [56] | |
| PBMCs: 10 SzS, 10 HD | Microarray, qRT-PCR | 0.89 AUC for discrimination between SzS and controls with sensitivity 0.82 and specificity 0.91 | [96] | |
| CD4+ T cells: 81 SzS, 12 HD | qRT-PCR | Increased 145-fold in SzS compared with HD samples and positive in 87% of SzS patients | [57] | |
| CD4+ T cells: 9 SzS, 4 HD; skin biopsies: 2 SzS, 4 MF, 9 psoriasis | qRT-PCR, IHC | Increased 270-fold in SzS compared with controls, PLS3 detected in infiltrate on MF/SzS biopsy but not psoriasis | [97] | |
|
| ||||
| JUNB | Peripheral blood ± skin biopsies: 26 SzS, 13 MF | Real-time PCR, IHC | 26% of SzS/MF had increased copy number of JUNB, 91% of SzS/MF had strong nuclear staining of JUNB | [71] |
| PBMCs: 18 SzS, 9 Th2-skewed controls | Microarray, qRT-PCR | Increased fivefold in SzS compared with controls based on array and 10-fold based on qRT-PCR | [95] | |
| PBMCs: 49 SzS, 69 HD, 3 IE | qRT-PCR | Increased 4.3-fold in SzS compared with controls | [56] | |
|
| ||||
| GATA3 | PBMCs: 49 SzS, 69 HD, 3 IE | qRT-PCR | Increased 6.4-fold in SzS compared with controls | [56] |
| PBMCs: 18 SzS, 9 Th2-skewed controls | Microarray, qRT-PCR | Increased 2.5-fold in SzS compared with controls based on array and sevenfold based on qRT-PCR | [95] | |
|
| ||||
| SATB1 | CD4+CD7− T cells: 9 SzS; CD4+ T cells: 9 HD | Microarray, qRT-PCR, WB | Decreased 4.2-fold in SzS samples compared with controls based on array, confirmed by qRT-PCR and WB | [98] |
| PBMCs: 10 SzS; CD4+ T cells: 5 IE, 3 HD | Microarray, qRT-PCR | Decreased 4.6-fold in SzS compared with controls based on array, confirmed by qRT-PCR | [99] | |
| Skin biopsies: 90 MF, 19 benign lesional skin | IHC | Higher expression of SATB1 in MF compared with controls, and SATB1 higher in patients with lymph node involvement | [100] | |
|
| ||||
| STAT4 | PBMCs: 49 SzS, 69 HD, 3 IE | qRT-PCR | Decreased 4.7-fold in SzS compared with controls | [56] |
| PBMCs: 18 SzS, 9 Th2-skewed controls | Microarray, qRT-PCR | Decreased 3.7-fold in SzS compared with controls based on array and 4.5-fold based on qRT-PCR | [95] | |
| Skin biopsies: 60 MF/SzS, 19 benign lesional skin, 6 HD, 19 benign lesional skin | qRT-PCR, WB | STAT4 was expressed in MF/SzS lesional skin but not normal skin or benign lesions, loss of STAT4 was associated with progressive disease | [101] | |
|
| ||||
| Twist1 | PBMCs: 10 SzS; CD4+ T cells: 5 IE, 3 HD; skin biopsies: 9 MF, 24 benign lesions | Microarray, qRT-PCR | Increased 19.8-fold in SzS compared with controls based on array, Overexpressed in all SzS and 4/9 MF samples by qRT-PCR but not control lesional skin | [99] |
| CD4+ T cells: 81 SzS, 12 HD | qRT-PCR | Increased 150-fold in SzS compared with HD samples and positive in 91% of SzS patients | [57] | |
| Skin biopsies: 68 MF/SzS, 3 HD, 3 psoriasis, 3 SCC; CD4+ T cells: 5 SzS | IHC, qRT-PCR | Twist was found in 12.5% of T1, 33.3% of T2, 50.0% of T3, 84.6% of T4 biopsies by IHC. All SzS and no HD had expression of Twist by qRT-PCR in CD4+ T cells | [102] | |
|
| ||||
| Fas | CD4+ T cells: 16 MF, 4 SzS, 25 benign lesions, 15 HD | Flow cytometry | Fas expression was lower in MF/SzS compared with controls, Fas increased after response to treatment | [103] |
| Skin biopsies: 23 MF, 10 LyP, 10 CD30+ LTCL, 9 CD30-LTCL | IHC | Fas expression present in 100% of plaque-stage MF, LyP and CD30+ LTCL, but only 33% of tumor-stage MF. Decrease in Fas was observed with progression. | [104] | |
|
| ||||
| TOX | CD4+ cells: 9 SzS, 4 HD; skin biopsies: 2 SzS, 4 MF, 9 psoriasis | qRT-PCR, IHC | Increased sevenfold in SzS compared with HD on qRT-PCR, Strong nuclear staining of TOX on IHC in MF/SzS but not psoriasis | [97] |
| Skin biopsies: 21 MF, 15 benign lesional skin, 21 HD | Microarray, qRT-PCR, IF, IHC | Increased 10.3-fold in MF compared with HD, confirmed with qRT-PCR, highly specific staining of TOX in MF biopsies on IHC and IF | [105] | |
Sample type abbreviations include Sézary syndrome (SzS), mycosis fungoides (MF), inflammatory erythroderma (IE), healthy donors (HD), peripheral blood mononuclear cells (PBMCs), lymphomatoid papulosis (LyP), large T-cell lymphoma (LTCL).
Methodology abbreviations include quantitative reverse transcription polymerase chain reaction (qRT-PCR), immunohistochemistry (IHC), Western blotting (WB), immunofluorescence (IF).
Clinically, upregulated or downregulated genes can be identified with qRT-PCR, and genes translated to proteins can be identified with Western blotting, ELISA, IHC or flow cytometry. In the case of biomarkers, the best candidate genes are those which can identified with little additional processing of the patient sample, that is whole blood rather than CD4+CD7−T cells.[53] In addition to the genes presented in Table 2, a number of the cell surface markers in Table 1 including KIR3DL2 and CD26 are also altered at the gene expression level; however, cell surface markers can be up-or downregulated at the transcription, translation or localization level, so gene expression does not always correlate with presence on the cell surface.[55]
A panel of genes can easily be tested simultaneously on a single qRT-PCR plate. Two such panels have been proposed. Nebozhyn et al.[56] identified a five gene SzS panel including STAT4, GATA3, PLS3, CD1D and TRAIL which displayed an average accuracy of 90% over 1000 resamplings of PBMCs from 49 SzS patients and 65 healthy controls. Michel et al.[57] demonstrated qRT-PCR analysis of PLS3, Twist1, CD158k/KIR3DL2 and NKp46 accurately classified 100% of 81 SzS patients; however, qRT-PCR detection of CD158k/KIR3DL2 and NKp46 in PBMCs may be complicated by expression of these two markers on NK cells of healthy patients. Instead, the authors propose flow cytometry analysis could be used on these two markers in combination with qRT-PCR analysis. Both of these expression panels were tested in single-centre efforts, and future study of these panels or others would benefit from multicentre, international trials. Prognostic panels based on gene expression have also been proposed; Litvinov et al. identified a group of 17 genes including IL2RA, CCR4, STAT5A and TOX that could identify patients at risk of progression and distinguish MF from SzS (Litvinov et al. 2015).
Non-coding RNAs represent another set of possible biomarkers in MF/SzS, and differential expression of microRNAs (miRNAs) and long non-coding RNAs (lnRNAs) has been studied. Ballabio et al.[58] found an increased level of miR-223 in CD4+ T cells was 90% accurate with 91% specificity and 90% sensitivity in correctly predicting diagnosis of SzS compared with MF or healthy controls. Ralfkiaer et al.[59] reported a three-miRNA panel (miR-155, miR-203 and miR-205) that distinguished CTCL from benign skin diseases with a classification accuracy of 95%. Narducci et al.[60] identified miR-214 and miR-486 overexpression in the majority of SzS patients as well as a signature of 14 miRNAs including miR-21, a miRNA upregulated in a number of cancers, which grouped SzS patients into favourable vs unfavourable outcomes. Benner et al.[61] identified differential expression of five miRNAs, including miR-155, between tumor-stage MF and primary cutaneous anaplastic large-cell lymphoma (cALCL). Conversely, Sandoval et al.[62] identified upregulated miR-155 in both tumor-stage MF and cALCL as well as upregulation of miR-42-5p and miR146a in MF. MiRNA expression has also been correlated with transformation of MF[63] and disease progression to advanced stage.[64] Lee et al.[65] identified 12 long non-coding RNAs in 3 SzS patient samples with transcriptome sequencing and confirmed the presence of long non-coding RNAs in MF/SzS tumors. While the number of studies of miRNAs and lnRNAs in MF/SzS is still relatively small, the results are promising.
Potential biomarkers also exist at the chromosomal level where translocations, duplications or deletions can be detected clinically using fluorescent in situ hybridization (FISH). To date, detected chromosomal imbalances in SzS have included gains of 17p11.2–q25.3 and 8q24.1–8q24.3 and losses of 17p13.2–p11.2, 10p12.1–q26.3 which each occurred in >40% of SzS cases.[66–68] Chromosomal alterations have been more commonly identified in SzS than MF, although duplication of 17q11.2 approximately q12 was identified in both.[66] Individual chromosomal aberrations have not been correlated with SzS prognosis, but an increasing number of chromosomal gains or losses does correlate with decreased survival.[67] Interestingly, TCR loci chromosomal translocations have been described in several T-cell malignancies but were not identified by FISH in MF/SzS.[69] Gains of TCRB and TCRG genes were, however, observed in 23% of SzS and 50% of tumor-stage MF.[69] Gains or losses of additional individual gene loci have also been demonstrated, and FISH panels to screen for genetic changes have been proposed.[70] Analysis of SzS cases with FISH using an IgH/BCL2 probe revealed loss of at least one copy of BCL2 in 83% of samples,[71] and digital droplet PCR showed gains the TNFRSF1B (TNFR2) locus in 10 of 73 SzS/MF patients.[72] In a comprehensive study of chromosomal alterations in SzS, gain of cMYC and loss of cMYC antagonists (MXI1 and MNT) was observed.[68]
In the last year, several reports of exome and whole-genome sequencing in MF and SzS have emerged. Frequent somatic mutations (single nucleotide variants or small insertion and deletions) were reported in TCR/NFκB signalling (NFKB2 truncations, TNFAIP3, PLCG1, PRKCQ and TNFAIP3 mutations),[73,74] Th2 differentiation (ZEB1),[75] cell survival and fate (PDGFR, ERK, JAK/STAT, MAPK),[74–76] epigenetic regulation (DNMT3A, ASLX3, TET1-3),[77] homologous recombination (RAD51C, BRCA2, POLD1)[74] and cell-cycle control (TP53).[75,77] These pathways represent new targets for treatment, and mutated genes represent potential biomarkers particularly for prognostication or monitoring of disease activity in a patient already diagnosed with MF/SzS.
7 WHERE DO WE GO FROM HERE?
To take the discovery of possible biomarkers through the process of validation, collaboration amongst groups will be key. We suggest a group of molecules for future studies based on the various functions of biomarkers in MF/SzS (Table 3). Head-to-head comparisons of various biomarkers in multicentre large-cohort studies will be necessary to select those markers with the highest clinically meaningful applications. Fortunately, collaborative efforts have already begun. The European Organization for Research and Treatment of Cancer evaluated 57 SzS and 40 erythrodermic inflammatory dermatoses cases to determine which histopathological features were the best indicators. CD7 loss, increased small cerebriform lymphocytes, decreased CD8+ lymphocytes and increased proliferation (Ki-67+ lymphocytes) were the best features for differentiation.[78] The Cutaneous Lymphoma International Consortium collected data at 29 specialist centres on 1275 patients diagnosed with advanced-stage MF/SzS examining and identified four independent prognostic markers for a worse survival: stage IV, age>60 years, large-cell transformation and increased LDH.[10] Moving forward, continued work of this type that incorporates new available technological platforms or possibly combinations of platforms along with rigorous biostatistical analysis will allow for additional integration of the bench discovery work summarized in this review with the clinical care of MF/SzS patients.
TABLE 3.
Selected promising biomarkers for future validation studies
| Diagnostic biomarkers | Biomarkers of advanced disease | Biomarkers of aggressive disease | Biomarkers of active disease |
|---|---|---|---|
| TOX (MF) T-plastin (SzS) Twist1 (SzS) CD158k/KIR3DL2 (SzS) | LDH CD4+CD45R0+CD45RA- SATB1 ganglioside GD3/CD60 STAT4 (loss) IL-13 TARC/CCL17 | LDH ganglioside GD3/CD60+ Fas (decrease) β2M IgE | CD158k/ KIR3DL2 sialomucin (CD164) TCR clone |
Acknowledgments
BD and OA conceived the review. BD, LG, JG and OA selected biomarkers for inclusion. BD and OA wrote the paper. LG and JG provided critical feedback and edited the paper.
Footnotes
CONFLICT OF INTERESTS
The authors have declared no conflicting interests.
References
- 1.Willemze R, Jaffe ES, Burg G, Cerroni L, Berti E, Swerdlow SH, Ralfkiaer E, Chimenti S, Diaz-Perez JL, Duncan LM, Grange F, Harris NL, Kempf W, Kerl H, Kurrer M, Knobler R, Pimpinelli N, Sander C, Santucci M, Sterry W, Vermeer MH, Wechsler J, Whittaker S, Meijer CJ. Blood. 2005;105:3768. doi: 10.1182/blood-2004-09-3502. [DOI] [PubMed] [Google Scholar]
- 2.Herrmann JJ, Kuzel TM, Rosen ST, Roenigk HH., Jr J Am Acad Dermatol. 1994;31:819. doi: 10.1016/s0190-9622(09)80056-x. [DOI] [PubMed] [Google Scholar]
- 3.Foss FM, Sausville EA. Hematol Oncol Clin North Am. 1995;9:1011. [PubMed] [Google Scholar]
- 4.Lefeber WP, Robinson JK, Clendenning WE, Dunn JL, Colton T. Arch Dermatol. 1981;117:408. [PubMed] [Google Scholar]
- 5.Olerud JE, Kulin PA, Chew DE, Carlsen RA, Hammar SP, Weir TW, Patterson SD, Bolen JW, Kadin ME, Barker E. Arch Dermatol. 1992;128:501. doi: 10.1001/archderm.128.4.501. [DOI] [PubMed] [Google Scholar]
- 6.Santucci M, Biggeri A, Feller AC, Burg G. Arch Dermatol. 2000;136:497. doi: 10.1001/archderm.136.4.497. [DOI] [PubMed] [Google Scholar]
- 7.Tok J, Szabolcs MJ, Silvers DN, Zhong J, Matsushima AY. J Am Acad Dermatol. 1998;38:453. doi: 10.1016/s0190-9622(98)70505-5. [DOI] [PubMed] [Google Scholar]
- 8.Ponti R, Fierro MT, Quaglino P, Lisa B, Paola F, Michela O, Paolo F, Comessatti A, Novelli M, Bernengo MG. J Invest Dermatol. 2008;128:1030. doi: 10.1038/sj.jid.5701109. [DOI] [PubMed] [Google Scholar]
- 9.Kim YH, Liu HL, Mraz-Gernhard S, Hoppe RT. Arch Dermatol. 2003;139:857. doi: 10.1001/archderm.139.7.857. [DOI] [PubMed] [Google Scholar]
- 10.Scarisbrick JJ, Prince HM, Vermeer MH, Quaglino P, Horwitz S, Porcu P, Stadler R, Wood GS, Beylot-Barry M, Pham-Ledard A, Foss F, Girardi M, Bagot M, Michel L, Battistella M, Guitart J, Kuzel TM, Martinez-Escala ME, Estrach T, Papadavid E, Antoniou C, Rigopoulos D, Nikolaou V, Sugaya M, Miyagaki T, Gniadecki R, Sanches JA, Cury-Martins J, Miyashiro D, Servitje O, Muniesa C, Berti E, Onida F, Corti L, Hodak E, Amitay-Laish I, Ortiz-Romero PL, Rodriguez-Peralto JL, Knobler R, Porkert S, Bauer W, Pimpinelli N, Grandi V, Cowan R, Rook A, Kim E, Pileri A, Patrizi A, Pujol RM, Wong H, Tyler K, Stranzenbach R, Querfeld C, Fava P, Maule M, Willemze R, Evison F, Morris S, Twigger R, Talpur R, Kim J, Ognibene G, Li S, Tavallaee M, Hoppe RT, Duvic M, Whittaker SJ, Kim YH. J Clin Oncol. 2015;33:3766. doi: 10.1200/JCO.2015.61.7142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kalnina Z, Meistere I, Kikuste I, Tolmanis I, Zayakin P, Line A. World J Gastroenterol. 2015;21:11636. doi: 10.3748/wjg.v21.i41.11636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zattra E, Belloni Fortina A, Peserico A, Alaibac M. Eur J Dermatol. 2012;22:167. doi: 10.1684/ejd.2011.1569. [DOI] [PubMed] [Google Scholar]
- 13.Olsen E, Vonderheid E, Pimpinelli N, Willemze R, Kim Y, Knobler R, Zackheim H, Duvic M, Estrach T, Lamberg S, Wood G, Dummer R, Ranki A, Burg G, Heald P, Pittelkow M, Bernengo MG, Sterry W, Laroche L, Trautinger F, Whittaker S. Blood. 2007;110:1713. doi: 10.1182/blood-2007-03-055749. [DOI] [PubMed] [Google Scholar]
- 14.Scarisbrick JJ, Kim YH, Whittaker SJ, Wood GS, Vermeer MH, Prince HM, Quaglino P. Br J Dermatol. 2014;170:1226. doi: 10.1111/bjd.12909. [DOI] [PubMed] [Google Scholar]
- 15.Scarisbrick JJ, Whittaker S, Evans AV, Fraser-Andrews EA, Child FJ, Dean A, Russell-Jones R. Blood. 2001;97:624. doi: 10.1182/blood.v97.3.624. [DOI] [PubMed] [Google Scholar]
- 16.Shipp MA. Blood. 1994;83:1165. [PubMed] [Google Scholar]
- 17.Diamandidou E, Colome M, Fayad L, Duvic M, Kurzrock R. J Am Acad Dermatol. 1999;40:914. doi: 10.1016/s0190-9622(99)70079-4. [DOI] [PubMed] [Google Scholar]
- 18.Marti RM, Pujol RM, Servitje O, Palou J, Romagosa V, Bordes R, Gonzalez-Castro J, Miralles J, Gallardo F, Curco N, Gomez X, Domingo A, Estrach T. Leukemia Lymphoma. 2003;44:59. doi: 10.1080/1042819021000054652. [DOI] [PubMed] [Google Scholar]
- 19.Vidulich KA, Talpur R, Bassett RL, Duvic M. Int J Dermatol. 2009;48:243. doi: 10.1111/j.1365-4632.2009.03771.x. [DOI] [PubMed] [Google Scholar]
- 20.Scala E, Abeni D, Palazzo P, Liso M, Pomponi D, Lombardo G, Picchio MC, Narducci MG, Russo G, Mari A. Int Arch Allergy Immunol. 2012;157:159. doi: 10.1159/000327553. [DOI] [PubMed] [Google Scholar]
- 21.Hassel JC, Meier R, Joller-Jemelka H, Burg G, Dummer R. Dermatology. 2004;209:296. doi: 10.1159/000080852. [DOI] [PubMed] [Google Scholar]
- 22.Geskin LJ, Akilov OE, Lin Y, Lokshin AE. Exp Dermatol. 2014;23:598. doi: 10.1111/exd.12455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Geskin LJ, Viragova S, Stolz DB, Fuschiotti P. Blood. 2015;125:2798. doi: 10.1182/blood-2014-07-590398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cedeno-Laurent F, Singer EM, Wysocka M, Benoit BM, Vittorio CC, Kim EJ, Yosipovitch G, Rook AH. Clin Immunol. 2015;158:1. doi: 10.1016/j.clim.2015.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Malek M, Glen J, Rebala K, Kowalczyk A, Sobjanek M, Nowicki R, Ruckemann-Dziurdzinska K, Sokolowska-Wojdylo M. Acta Derm Venereol. 2015;95:283. doi: 10.2340/00015555-1958. [DOI] [PubMed] [Google Scholar]
- 26.Ohmatsu H, Sugaya M, Suga H, Morimura S, Miyagaki T, Kai H, Kagami S, Fujita H, Asano Y, Tada Y, Kadono T, Sato S. Acta Derm Venereol. 2012;92:282. doi: 10.2340/00015555-1345. [DOI] [PubMed] [Google Scholar]
- 27.Kakinuma T, Sugaya M, Nakamura K, Kaneko F, Wakugawa M, Matsushima K, Tamaki K. J Am Acad Dermatol. 2003;48:23. doi: 10.1067/mjd.2003.132. [DOI] [PubMed] [Google Scholar]
- 28.Chapman CJ, Healey GF, Murray A, Boyle P, Robertson C, Peek LJ, Allen J, Thorpe AJ, Hamilton-Fairley G, Parsy-Kowalska CB, MacDonald IK, Jewell W, Maddison P, Robertson JF. Tumour Biol. 2012;33:1319. doi: 10.1007/s13277-012-0379-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chapman C, Murray A, Chakrabarti J, Thorpe A, Woolston C, Sahin U, Barnes A, Robertson J. Ann Oncol. 2007;18:868. doi: 10.1093/annonc/mdm007. [DOI] [PubMed] [Google Scholar]
- 30.Zhong L, Coe SP, Stromberg AJ, Khattar NH, Jett JR, Hirschowitz EA. J Thorac Oncol. 2006;1:513. [PubMed] [Google Scholar]
- 31.Forgber M, Gellrich S, Sharav T, Sterry W, Walden P. PLoS ONE. 2009;4:e8376. doi: 10.1371/journal.pone.0008376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tancrede-Bohin E, Ionescu MA, de La Salmoniere P, Dupuy A, Rivet J, Rybojad M, Dubertret L, Bachelez H, Lebbe C, Morel P. Arch Dermatol. 2004;140:1057. doi: 10.1001/archderm.140.9.1057. [DOI] [PubMed] [Google Scholar]
- 33.Abeni D, Frontani M, Sampogna F, Sera F, Bolli S, Corona R, Baliva G, Russo G. Br J Dermatol. 2005;153:324. doi: 10.1111/j.1365-2133.2005.06755.x. [DOI] [PubMed] [Google Scholar]
- 34.Vermeer MH, van Doorn R, Dukers D, Bekkenk MW, Meijer CJ, Willemze R. J Clin Oncol. 2001;19:4322. doi: 10.1200/JCO.2001.19.23.4322. [DOI] [PubMed] [Google Scholar]
- 35.Scala E, Russo G, Cadoni S, Narducci MG, Girardelli CR, De Pita O, Puddu P. J Invest Dermatol. 1999;113:622. doi: 10.1046/j.1523-1747.1999.00718.x. [DOI] [PubMed] [Google Scholar]
- 36.Diamandidou E, Colome-Grimmer M, Fayad L, Duvic M, Kurzrock R. Blood. 1998;92:1150. [PubMed] [Google Scholar]
- 37.Vonderheid EC, Pena J, Nowell P. Leukemia Lymphoma. 2006;47:1841. doi: 10.1080/10428190600709655. [DOI] [PubMed] [Google Scholar]
- 38.Clark RA, Shackelton JB, Watanabe R, Calarese A, Yamanaka K, Campbell JJ, Teague JE, Kuo HP, Hijnen D, Kupper TS. Blood. 2011;117:1966. doi: 10.1182/blood-2010-05-287664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stefanato CM, Tallini G, Crotty PL. Am J Dermatopathol. 1998;20:1. doi: 10.1097/00000372-199802000-00001. [DOI] [PubMed] [Google Scholar]
- 40.Benner MF, Jansen PM, Vermeer MH, Willemze R. Blood. 2012;119:1643. doi: 10.1182/blood-2011-08-376319. [DOI] [PubMed] [Google Scholar]
- 41.Baumgarth N, Roederer M. J Immunol Methods. 2000;243:77. doi: 10.1016/s0022-1759(00)00229-5. [DOI] [PubMed] [Google Scholar]
- 42.Chen X, Cho DB, Yang PC. N Am J Med Sci. 2010;2:241. doi: 10.4297/najms.2010.2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Finn DT, Jaworsky C, Chooback L, Jensen PJ, Lessin SR. J Cutan Pathol. 1996;23:306. doi: 10.1111/j.1600-0560.1996.tb01302.x. [DOI] [PubMed] [Google Scholar]
- 44.Klemke CD, Brade J, Weckesser S, Sachse MM, Booken N, Neumaier M, Goerdt S, Nebe TC. Br J Dermatol. 2008;159:871. doi: 10.1111/j.1365-2133.2008.08739.x. [DOI] [PubMed] [Google Scholar]
- 45.Vega F, Luthra R, Medeiros LJ, Dunmire V, Lee SJ, Duvic M, Jones D. Blood. 2002;100:3369. doi: 10.1182/blood.V100.9.3369. [DOI] [PubMed] [Google Scholar]
- 46.Kirsch IR, Watanabe R, O’Malley JT, Williamson DW, Scott LL, Elco CP, Teague JE, Gehad A, Lowry EL, LeBoeuf NR, Krueger JG, Robins HS, Kupper TS, Clark RA. Sci Transl Med. 2015;7:308ra158. doi: 10.1126/scitranslmed.aaa9122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Assaf C, Hummel M, Steinhoff M, Geilen CC, Orawa H, Stein H, Orfanos CE. Blood. 2005;105:503. doi: 10.1182/blood-2004-06-2220. [DOI] [PubMed] [Google Scholar]
- 48.Greisser J, Palmedo G, Sander C, Kutzner H, Kazakov DV, Roos M, Burg G, Kempf W. J Cutan Pathol. 2006;33:711. doi: 10.1111/j.1600-0560.2006.00560.x. [DOI] [PubMed] [Google Scholar]
- 49.Stowman AM, Hsia LL, Kanner WA, Mahadevan MS, Bullock GC, Patterson JW. Int J Dermatol. 2016;55:e62. doi: 10.1111/ijd.12847. [DOI] [PubMed] [Google Scholar]
- 50.Novelli M, Fava P, Sarda C, Ponti R, Osella-Abate S, Savoia P, Bergallo M, Lisa F, Fierro MT, Quaglino P. Am J Clin Pathol. 2015;143:57. doi: 10.1309/AJCP1NA3YCHCDEIG. [DOI] [PubMed] [Google Scholar]
- 51.Vandersee S, Humme D, Terhorst D, Almohamad A, Mobs M, Beyer M. J Dtsch Dermatol Ges. 2015;13:30. doi: 10.1111/ddg.12549. [DOI] [PubMed] [Google Scholar]
- 52.Sokolowska-Wojdylo M, Wenzel J, Gaffal E, Steitz J, Roszkiewicz J, Bieber T, Tuting T. Clin Exp Dermatol. 2005;30:702. doi: 10.1111/j.1365-2230.2005.01904.x. [DOI] [PubMed] [Google Scholar]
- 53.Dulmage BO, Geskin LJ. Br J Dermatol. 2013;169:1188. doi: 10.1111/bjd.12578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wong HK. Discov Med. 2013;16:71. [PubMed] [Google Scholar]
- 55.Lotem J, Sachs L. EMBO J. 1986;5:2163. doi: 10.1002/j.1460-2075.1986.tb04480.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nebozhyn M, Loboda A, Kari L, Rook AH, Vonderheid EC, Lessin S, Berger C, Edelson R, Nichols C, Yousef M, Gudipati L, Shang M, Showe MK, Showe LC. Blood. 2006;107:3189. doi: 10.1182/blood-2005-07-2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Michel L, Jean-Louis F, Begue E, Bensussan A, Bagot M. Blood. 2013;121:1477. doi: 10.1182/blood-2012-10-460535. [DOI] [PubMed] [Google Scholar]
- 58.Ballabio E, Mitchell T, van Kester MS, Taylor S, Dunlop HM, Chi J, Tosi I, Vermeer MH, Tramonti D, Saunders NJ, Boultwood J, Wainscoat JS, Pezzella F, Whittaker SJ, Tensen CP, Hatton CS, Lawrie CH. Blood. 2010;116:1105. doi: 10.1182/blood-2009-12-256719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ralfkiaer U, Hagedorn PH, Bangsgaard N, Lovendorf MB, Ahler CB, Svensson L, Kopp KL, Vennegaard MT, Lauenborg B, Zibert JR, Krejsgaard T, Bonefeld CM, Sokilde R, Gjerdrum LM, Labuda T, Mathiesen AM, Gronbaek K, Wasik MA, Sokolowska-Wojdylo M, Queille-Roussel C, Gniadecki R, Ralfkiaer E, Geisler C, Litman T, Woetmann A, Glue C, Ropke MA, Skov L, Odum N. Blood. 2011;118:5891. doi: 10.1182/blood-2011-06-358382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Narducci MG, Arcelli D, Picchio MC, Lazzeri C, Pagani E, Sampogna F, Scala E, Fadda P, Cristofoletti C, Facchiano A, Frontani M, Monopoli A, Ferracin M, Negrini M, Lombardo GA, Caprini E, Russo G. Cell Death Dis. 2011;2:e151. doi: 10.1038/cddis.2011.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Benner MF, Ballabio E, van Kester MS, Saunders NJ, Vermeer MH, Willemze R, Lawrie CH, Tensen CP. Exp Dermatol. 2012;21:632. doi: 10.1111/j.1600-0625.2012.01548.x. [DOI] [PubMed] [Google Scholar]
- 62.Sandoval J, Diaz-Lagares A, Salgado R, Servitje O, Climent F, Ortiz-Romero PL, Perez-Ferriols A, Garcia-Muret MP, Estrach T, Garcia M, Nonell L, Esteller M, Pujol RM, Espinet B, Gallardo F. J Invest Dermatol. 2015;135:1128. doi: 10.1038/jid.2014.487. [DOI] [PubMed] [Google Scholar]
- 63.Marosvari D, Teglasi V, Csala I, Marschalko M, Bodor C, Timar B, Csomor J, Harsing J, Reiniger L. Pathol Oncol Res. 2015;21:821. doi: 10.1007/s12253-015-9897-8. [DOI] [PubMed] [Google Scholar]
- 64.Ralfkiaer U, Lindahl LM, Litman T, Gjerdrum LM, Ahler CB, Gniadecki R, Marstrand T, Fredholm S, Iversen L, Wasik MA, Bonefeld CM, Geisler C, Krejsgaard T, Glue C, Ropke MA, Woetmann A, Skov L, Gronbaek K, Odum N. Anticancer Res. 2014;34:7207. [PubMed] [Google Scholar]
- 65.Lee CS, Ungewickell A, Bhaduri A, Qu K, Webster DE, Armstrong R, Weng WK, Aros CJ, Mah A, Chen RO, Lin M, Sundram U, Chang HY, Kretz M, Kim YH, Khavari PA. Blood. 2012;120:3288. doi: 10.1182/blood-2012-04-423061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Barba G, Matteucci C, Girolomoni G, Brandimarte L, Varasano E, Martelli MF, Mecucci C. Cancer Genet Cytogenet. 2008;184:48. doi: 10.1016/j.cancergencyto.2008.03.007. [DOI] [PubMed] [Google Scholar]
- 67.Caprini E, Cristofoletti C, Arcelli D, Fadda P, Citterich MH, Sampogna F, Magrelli A, Censi F, Torreri P, Frontani M, Scala E, Picchio MC, Temperani P, Monopoli A, Lombardo GA, Taruscio D, Narducci MG, Russo G. Cancer Res. 2009;69:8438. doi: 10.1158/0008-5472.CAN-09-2367. [DOI] [PubMed] [Google Scholar]
- 68.Vermeer MH, van Doorn R, Dijkman R, Mao X, Whittaker S, van Voorst Vader PC, Gerritsen MJ, Geerts ML, Gellrich S, Soderberg O, Leuchowius KJ, Landegren U, Out-Luiting JJ, Knijnenburg J, Ijszenga M, Szuhai K, Willemze R, Tensen CP. Cancer Res. 2008;68:2689. doi: 10.1158/0008-5472.CAN-07-6398. [DOI] [PubMed] [Google Scholar]
- 69.Salgado R, Gallardo F, Servitje O, Estrach T, Garcia-Muret MP, Romagosa V, Florensa L, Serrano S, Salido M, Sole F, Pujol RM, Espinet B. Cancer Genet. 2011;204:405. doi: 10.1016/j.cancergen.2011.05.004. [DOI] [PubMed] [Google Scholar]
- 70.Weed J, Gibson J, Lewis J, Carlson K, Foss F, Choi J, Li P, Girardi M. J Invest Dermatol. 2016;136:S13. doi: 10.1016/j.jid.2016.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mao X, Orchard G, Lillington DM, Child FJ, Vonderheid EC, Nowell PC, Bagot M, Bensussan A, Russell-Jones R, Young BD, Whittaker SJ. Br J Dermatol. 2004;151:546. doi: 10.1111/j.1365-2133.2004.06106.x. [DOI] [PubMed] [Google Scholar]
- 72.Ungewickell A, Bhaduri A, Rios E, Reuter J, Lee CS, Mah A, Zehnder A, Ohgami R, Kulkarni S. Nat Genet. 2015;47:1056. doi: 10.1038/ng.3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Choi J, Goh G, Walradt T, Hong BS, Bunick CG, Chen K, Bjornson RD, Maman Y, Wang T, Tordoff J, Carlson K, Overton JD, Liu KJ, Lewis JM, Devine L, Barbarotta L, Foss FM, Subtil A, Vonderheid EC, Edelson RL, Schatz DG, Boggon TJ, Girardi M, Lifton RP. Nat Genet. 2015;47:1011. doi: 10.1038/ng.3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Woollard WJ, Pullabhatla V, Lorenc A, Patel VM, Butler RM, Bayega A, Begum N, Bakr F, Dedhia K, Fisher J, Aguilar-Duran S, Flanagan C, Ghasemi AA, Hoffmann RM, Castillo-Mosquera N, Nuttall EA, Paul A, Roberts CA, Solomonidis EG, Tarrant R, Yoxall A, Beyers CZ, Ferreira S, Tosi I, Simpson MA, de Rinaldis E, Mitchell TJ, Whittaker SJ. Blood. 2016;127:3387. doi: 10.1182/blood-2016-02-699843. [DOI] [PubMed] [Google Scholar]
- 75.McGirt LY, Jia P, Baerenwald DA, Duszynski RJ, Dahlman KB, Zic JA, Zwerner JP, Hucks D, Dave U, Zhao Z, Eischen CM. Blood. 2015;126:508. doi: 10.1182/blood-2014-11-611194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kiel MJ, Sahasrabuddhe AA, Rolland DC, Velusamy T, Chung F, Schaller M, Bailey NG, Betz BL, Miranda RN, Porcu P, Byrd JC, Jeffrey Medeiros L, Kunkel SL, Bahler DW, Lim MS, Elenitoba-Johnson KS. Nat Commun. 2015;6:8470. doi: 10.1038/ncomms9470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.da Silva Almeida AC, Abate F, Khiabanian H, Martinez-Escala E, Guitart J, Tensen CP, Vermeer MH, Rabadan R, Ferrando A, Palomero T. Nat Genet. 2015;47:1465. doi: 10.1038/ng.3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Klemke CD, Booken N, Weiss C, Nicolay JP, Goerdt S, Felcht M, Geraud C, Kempf W, Assaf C, Ortonne N, Battistella M, Bagot M, Knobler R, Quaglino P, Arheiliger B, Santucci M, Jansen P, Vermeer MH, Willemze R. Br J Dermatol. 2015;173:93. doi: 10.1111/bjd.13832. [DOI] [PubMed] [Google Scholar]
- 79.Bernengo MG, Novelli M, Quaglino P, Lisa F, De Matteis A, Savoia P, Cappello N, Fierro MT. Br J Dermatol. 2001;144:125. doi: 10.1046/j.1365-2133.2001.04014.x. [DOI] [PubMed] [Google Scholar]
- 80.Fierro MT, Novelli M, Quaglino P, Comessatti A, Fava P, Ortoncelli M, Ponti R, Bernengo MG. Dermatology. 2008;216:213. doi: 10.1159/000112928. [DOI] [PubMed] [Google Scholar]
- 81.Kamarashev J, Burg G, Kempf W, Hess Schmid M, Dummer R. J Cutan Pathol. 1998;25:407. doi: 10.1111/j.1600-0560.1998.tb01766.x. [DOI] [PubMed] [Google Scholar]
- 82.Fierro MT, Novelli M, Savoia P, Cambieri I, Quaglino P, Osella-Abate S, Bernengo MG. J Cutan Pathol. 2001;28:356. doi: 10.1034/j.1600-0560.2001.280704.x. [DOI] [PubMed] [Google Scholar]
- 83.Hartmann TB, Thiel D, Dummer R, Schadendorf D, Eichmuller S. Br J Dermatol. 2004;150:252. doi: 10.1111/j.1365-2133.2004.05651.x. [DOI] [PubMed] [Google Scholar]
- 84.Bahler DW, Hartung L, Hill S, Bowen GM, Vonderheid EC. Cytometry B Clin Cytom. 2008;74:156. doi: 10.1002/cyto.b.20395. [DOI] [PubMed] [Google Scholar]
- 85.Poszepczynska-Guigne E, Schiavon V, D’Incan M, Echchakir H, Musette P, Ortonne N, Boumsell L, Moretta A, Bensussan A, Bagot M. J Invest Dermatol. 2004;122:820. doi: 10.1111/j.0022-202X.2004.22326.x. [DOI] [PubMed] [Google Scholar]
- 86.Bouaziz JD, Remtoula N, Bensussan A, Marie-Cardine A, Bagot M. Br J Dermatol. 2010;162:123. doi: 10.1111/j.1365-2133.2009.09364.x. [DOI] [PubMed] [Google Scholar]
- 87.Bensussan A, Remtoula N, Sivori S, Bagot M, Moretta A, Marie-Cardine A. J Invest Dermatol. 2011;131:969. doi: 10.1038/jid.2010.404. [DOI] [PubMed] [Google Scholar]
- 88.Scala E, Abeni D, Pomponi D, Narducci MG, Lombardo GA, Mari A, Frontani M, Picchio MC, Pilla MA, Caprini E, Russo G. Haematologica. 2010;95:1905. doi: 10.3324/haematol.2010.026260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chung JS, Shiue LH, Duvic M, Pandya A, Cruz PD, Jr, Ariizumi K. Blood. 2011;117:3382. doi: 10.1182/blood-2010-08-302034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wysocka M, Kossenkov AV, Benoit BM, Troxel AB, Singer E, Schaffer A, Kim B, Dentchev T, Nagata S, Ise T, Showe LC, Rook AH. J Invest Dermatol. 2014;134:229. doi: 10.1038/jid.2013.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Samimi S, Benoit B, Evans K, Wherry EJ, Showe L, Wysocka M, Rook AH. Arch Dermatol. 2010;146:1382. doi: 10.1001/archdermatol.2010.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cetinozman F, Jansen PM, Vermeer MH, Willemze R. Arch Dermatol. 2012;148:1379. doi: 10.1001/archdermatol.2012.2089. [DOI] [PubMed] [Google Scholar]
- 93.Cetinozman F, Jansen PM, Willemze R. Br J Dermatol. 2014;171:499. doi: 10.1111/bjd.12934. [DOI] [PubMed] [Google Scholar]
- 94.Jiang L, Yuan CM, Hubacheck J, Janik JE, Wilson W, Morris JC, Jasper GA, Stetler-Stevenson M. Br J Haematol. 2009;145:173. doi: 10.1111/j.1365-2141.2009.07606.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kari L, Loboda A, Nebozhyn M, Rook AH, Vonderheid EC, Nichols C, Virok D, Chang C, Horng WH, Johnston J, Wysocka M, Showe MK, Showe LC. J Exp Med. 2003;197:1477. doi: 10.1084/jem.20021726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Booken N, Gratchev A, Utikal J, Weiss C, Yu X, Qadoumi M, Schmuth M, Sepp N, Nashan D, Rass K, Tuting T, Assaf C, Dippel E, Stadler R, Klemke CD, Goerdt S. Leukemia. 2008;22:393. doi: 10.1038/sj.leu.2405044. [DOI] [PubMed] [Google Scholar]
- 97.Dulmage BO, Akilov O, Vu JR, Falo LD, Geskin LJ. Oncotarget. 2015 doi: 10.18632/oncotarget.5742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wang Y, Su M, Zhou LL, Tu P, Zhang X, Jiang X, Zhou Y. Blood. 2011;117:3826. doi: 10.1182/blood-2010-07-294819. [DOI] [PubMed] [Google Scholar]
- 99.van Doorn R, Dijkman R, Vermeer MH, Out-Luiting JJ, van der Raaij-Helmer EM, Willemze R, Tensen CP. Cancer Res. 2004;64:5578. doi: 10.1158/0008-5472.CAN-04-1253. [DOI] [PubMed] [Google Scholar]
- 100.Jankowska-Konsur A, Kobierzycki C, Reich A, Grzegrzolka J, Bieniek A, Dziegiel P. Anticancer Res. 2016;36:189. [PubMed] [Google Scholar]
- 101.Litvinov IV, Cordeiro B, Fredholm S, Odum N, Zargham H, Huang Y, Zhou Y, Pehr K, Kupper TS, Woetmann A, Sasseville D. Cell Cycle. 2014;13:2975. doi: 10.4161/15384101.2014.947759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Goswami M, Duvic M, Dougherty A, Ni X. J Cutan Pathol. 2012;39:500. doi: 10.1111/j.1600-0560.2012.01883.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Dereure O, Portales P, Clot J, Guilhou JJ. Br J Dermatol. 2000;143:1205. doi: 10.1046/j.1365-2133.2000.03889.x. [DOI] [PubMed] [Google Scholar]
- 104.Zoi-Toli O, Vermeer MH, De Vries E, Van Beek P, Meijer CJ, Willemze R. Br J Dermatol. 2000;143:313. doi: 10.1046/j.1365-2133.2000.03656.x. [DOI] [PubMed] [Google Scholar]
- 105.Zhang Y, Wang Y, Yu R, Huang Y, Su M, Xiao C, Martinka M, Dutz JP, Zhang X, Zheng Z, Zhou Y. J Invest Dermatol. 2012;132:1698. doi: 10.1038/jid.2012.13. [DOI] [PubMed] [Google Scholar]


