Abstract
Recipient responses to primary graft dysfunction (PGD) after lung transplantation may have important implications to the fate of the allograft. We therefore evaluated longitudinal differences in peripheral blood gene expression in subjects with PGD. RNA expression was measured throughout the first transplant year in 106 subjects enrolled in the Clinical Trials in Organ Transplantation-03 study using a panel of 100 hypothesis-driven genes. PGD was defined as grade 3 in the first 72 posttransplant hours. Eighteen genes were differentially expressed over the first year based on PGD development, with significant representation from innate and adaptive immunity genes, with most differences identified very early after transplant. Sixteen genes were overexpressed in the blood of patients with PGD compared to those without PGD within 7 days of allograft reperfusion, with most transcripts encoding innate immune/inflammasome-related proteins, including genes previously associated with PGD. Thirteen genes were underexpressed in patients with PGD compared to those without PGD within 7 days of transplant, highlighted by T cell and adaptive immune regulation genes. Differences in gene expression present within 2 hours of reperfusion and persist for days after transplant. Future investigation will focus on the long-term implications of these gene expression differences on the outcome of the allograft.
Introduction
Primary graft dysfunction (PGD) remains the most common cause of early posT transplant morbidity and mortality for lung transplant recipients1. PGD is felt to be predominantly a result of severe ischemia-reperfusion injury, which clinically manifests following the time of allograft reperfusion. Gene expression profiling has been used to identify important cellular pathways in disease states related to ischemia reperfusion injury, including the acute respiratory distress syndrome and delayed graft failure after kidney transplantation. Peripheral blood gene expression profiles differ significantly when comparing sepsis patients with and without ARDS, with an overrepresentation of genes involved in known respiratory and infection pathways2. Likewise, blood gene expression profiles differ significantly among patients with and without delayed graft function, a complication of renal transplantation closely associated with ischemia reperfusion injury3–5.
Gene expression profiling of lung donors has also been used to evaluate the risk for PGD in the resulting lung recipient. Lung biopsies taken prior to cold-flushing revealed differential gene expression based on the development of grade 3 PGD within 6 hours of allograft reperfusion6. We previously utilized gene set enrichment analysis to compare changes in donor lung gene expression in bronchoalveolar lavage (BAL) fluid before transplant with those in BAL fluid after reperfusion, highlighting the importance of inflammasome–mediated and innate immune signaling pathways7. However, the association of differential gene expression has been thus far focused on lung donors and the immediate peri-operative transplant period. The recipient systemic response to the injured lung may differ. For example, recipient neutrophil responses to sterile inflammation in the lung are key to the development of lung injury in the setting of ischemia reperfusion injury in mouse models of lung transplantation8,9. We therefore aimed to evaluate the association of PGD with differences in peripheral blood gene expression longitudinally after lung transplantation using a multicenter prospective cohort study under the NIAID Clinical Trials in Organ Transplant -03 (CTOT 03) study, (ClinicalTrials.gov identifier: NCT00531921). The primary goal was to improve our mechanistic understanding of PGD by: 1) identifying early gene expression markers of injury associated with PGD, and 2) defining differences in longer term gene expression that may help identify potential mediators of the association between early PGD and later chronic lung allograft dysfunction (CLAD).
Materials and Methods
Study Design and Subject Selection
The CTOT 03 study is a United States National Institutes of Health sponsored multicenter, prospective cohort study designed to assess the relationship between donor and recipient gene expression and early organ dysfunction after heart, lung, liver, or kidney transplantation7. All patients presented in this manuscript received lung transplantation. Patients were eligible for enrollment in the lung study if they were between 16 and 70 years old and provided signed consent. Patients were excluded if they had previously received a solid organ transplant, required a multi-organ transplant, had human immunodeficiency virus (HIV) or hepatitis C infection, or received a living donor transplant. Subjects were enrolled at three U.S. transplant centers: the University of Pennsylvania, Columbia University, and the University of Wisconsin, between 2008 and 2010. Clinical parameters were collected prospectively. The institutional review boards at each center approved this study. Sample collection protocols were identical across the three sites.
Outcome Definition
PGD was defined as any grade 3 PGD developing within 72 hours of allograft reperfusion, defined as the presence of diffuse alveolar infiltrates with a PaO2/FiO2 ratio < 200 and the exclusion of secondary causes according to International Society for Heart and Lung Transplantation guidelines10. This is an accepted and well validated outcome construct for PGD that we have utilized extensively in the past11–13. Chest radiographs from immediately after transplant and from 24, 48 and 72 hours after transplant were assessed for the presence of multifocal infiltrates by two independent readers, with adjudication, similar to previous studies7,12,14.
Sample collection
Blood samples were drawn from all 106 study participants at posT reperfusion (within 2 hours) and at 1 week, 6 weeks, 3 months, 6 months, and 1 year posT transplantation. Blood samples were collected in PAXgene Blood RNA tubes for transportation and storage. The RNA was then purified from blood using PAXgene Blood RNA Kit (Qiagen GmbH, Hilden, Germany. The number of samples per time point is shown in Table S1.
Gene expression profiling, Quality Control, and data pre-processing
The Panomics Quantigene Plex assays (Affymetrix, Santa Clara, CA) were employed to measure expression levels of 108 inflammatory and immune related genes in peripheral blood samples collected longitudinally. Transcripts were chosen at the time of assay study design in 2009 based on hypothesized roles in graft function, injury and recovery across four organs (lung, heart, liver, and kidney). Assays were performed by overnight hybridization of the RNA with target specific probe sets. Samples were assayed at a concentration of 250 ng/well and Universal Human Total RNA (500 ng/well) was used as a positive control. Signal amplification was achieved using branch DNA (bDNA) technology, according to the manufacturer’s instructions, and the signals were read and recorded using a Luminex instrument. Background signal subtraction and signal intensity normalization were performed according to manufacturer’s recommendation.
Statistical analysis
Patient characteristics and clinical data were displayed as mean ± standard deviation (SD), unless described otherwise. Categorical clinical data were compared using Fisher’s exact test. Continuous clinical data were compared using the Mann-Whitney test. A p-value <0.05 was considered significant.
Normalized gene expression values were analyzed using Array Studio software (http://www.omicsoft.com/). Outliers implicated by Principal Components Analysis (PCA) clustering and mean absolute deviation (MAD) scores were excluded. In total, 401 peripheral blood samples passed quality control and were included in later statistical analyses. The gene expression levels of twenty-seven out of 108 genes measured were under limit of quantification in all samples and were considered not reliably quantifiable and therefore excluded. In total, eighty-one genes remained for later statistical analyses.
Multivariable linear mixed models were utilized to assess differences in gene expression over the first year, with gene expression as the dependent variable and PGD status, age, and pre-transplant pulmonary diagnosis included as fixed effects. A p-value<0.05 for the interaction between PGD and time after transplant was considered significant. The differences in gene expression for individual time points (within 2 hours of reperfusion and at 1 week after transplant) were assessed using multivariable general linear models, adjusting for age and pre-transplant diagnosis. Significance was defined as a>|1.5| fold difference in expression with false discovery rate adjusted p<0.0515,16.
Results
Of the 106 patients enrolled in the study, 24 (23%) met criteria for PGD. Of these 24 patients, 21 (88%) required mechanical ventilation for longer than 24 hours after transplant, significantly more than those without PGD (n=48, 59%) (p=0.009). There were no significant differences identified in gender, race, pre-transplant pulmonary diagnosis, use of cardiopulmonary bypass, transplant type, or pulmonary arterial pressures, measured via pre-operative right heart catheterization, according to recipient PGD status, likely due to small patient numbers (Table 1).
Table 1.
Patient demographics
| Recipient Characteristics | PGD (n=24) |
Non-PGD (n=82) |
p-value |
|---|---|---|---|
| Male Gender, n, (%) | 19 (79) | 56 (68) | 0.3 |
| Age, years, mean | 57 | 53 | 0.2 |
| Race, n (%) | 0.5 | ||
| Caucasian | 21 (88) | 73 (89) | |
| Black | 3 (13) | 6 (7) | |
| Other | 0 (0) | 3 (4) | |
| Pulmonary Diagnosis, n (%) | 0.5 | ||
| Chronic Obstructive Pulmonary Disease | 5 (21) | 18(22) | |
| Pulmonary Fibrosis | 12 (50) | 36 (44) | |
| Cystic Fibrosis/Bronchiectasis | 4 (17) | 17 (21) | |
| Pulmonary Arterial Hypertension | 1 (4) | 0 | |
| Other | 2 (8) | 11 (13) | |
| Cardiopulmonary Bypass, yes | 15 (63) | 39 (48) | 0.2 |
| Transplant Type, bilateral | 17 (71) | 49 (60) | 0.3 |
| Pulmonary Arterial Systolic Pressure, mean (mmHg) | 39 | 32 | 0.2 |
| Prolonged Mechanical Ventilation, yes | 21 (88) | 48 (59) | 0.009 |
Prolonged Mechanical Ventilation: defined as requiring mechanical ventilation more than 24 hours after transplant
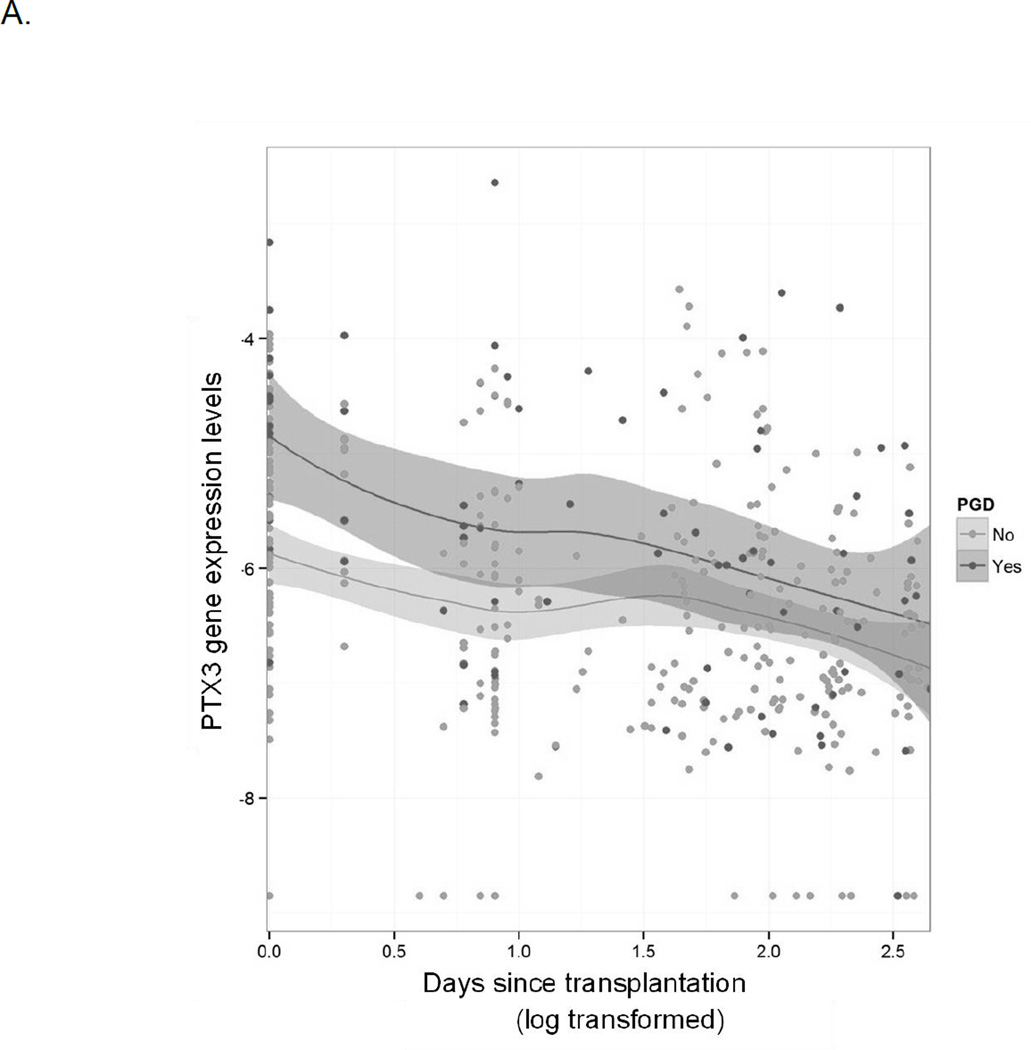
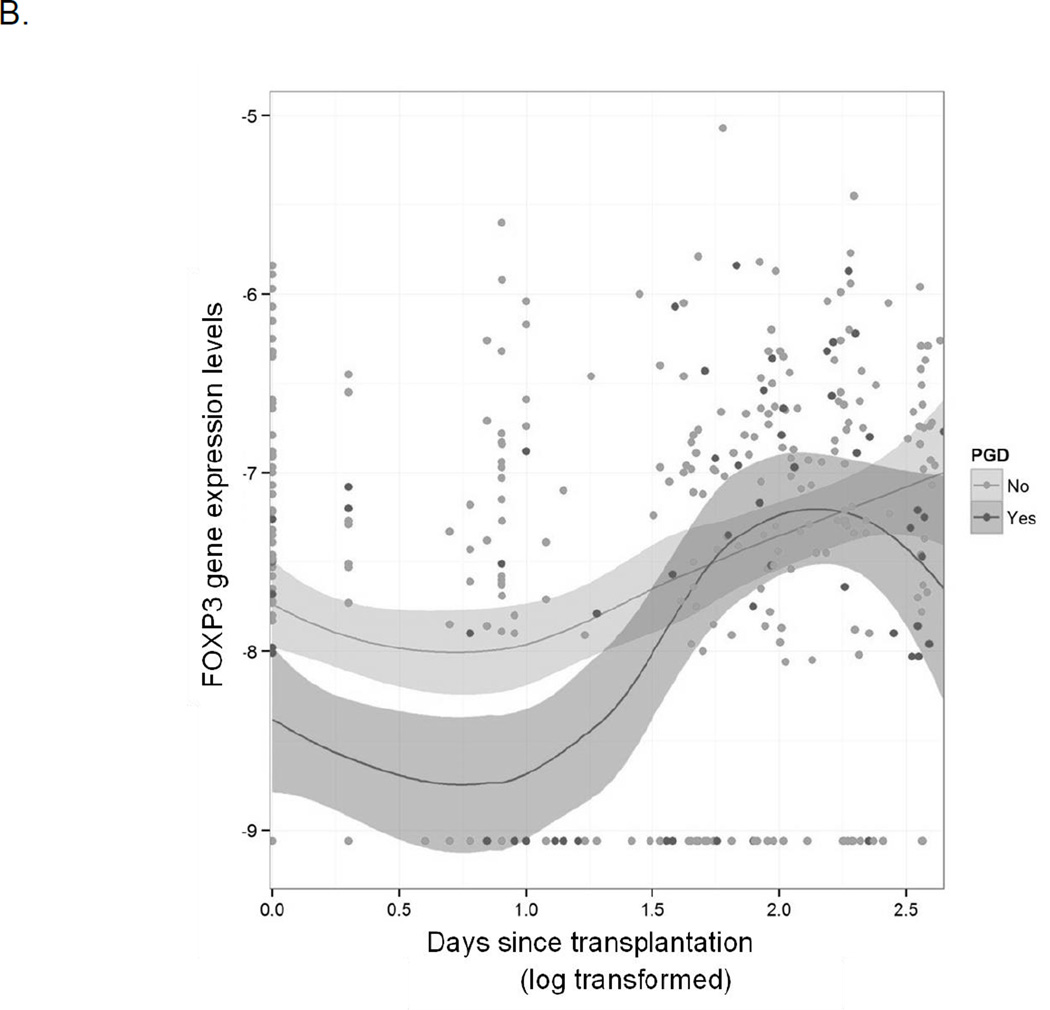
We first evaluated for differences in longitudinal gene expression over the entire first year after lung transplantation. Eighteen genes were differentially expressed over the first year based on the development of PGD after transplant with significant representation from genes involved in both innate and adaptive immunity (Table 2). Much of the differential expression was a result of early differences in gene expression, with later time points showing no significant differences in gene expression based on early PGD, as seen in changes in gene expression patterns for long pentraxin 3 (PTX3) and forkhead box P3 (FOXP3) (Figure 1). Additionally, nearly half of the blood samples available for analysis were collected within the first week after transplant, limiting the power of our longitudinal statistical evaluation.
Table 2.
Longitudinal gene expression differences over the first year after lung transplantation
| Gene | Gene activity | p-value |
|---|---|---|
| CD27 | Required for generation and long-term maintenance of T cell immunity | 0.0007 |
| CD40LG | Regulates B-cell function | 0.001 |
| HSP90AA1 | Inducible form involved in protein stabilization and refolding after stress | 0.001 |
| IL10 | Immunoregulation and inflammation | 0.002 |
| CD80 | T-cell proliferation and cytokine production | 0.006 |
| CD3E | Component of the T-cell receptor complex | 0.007 |
| FASLG | Apoptosis trigger essential for immune system regulation | 0.02 |
| PTX3 | Secreted innate immune mediator | 0.02 |
| FOXP3 | Regulator in development and function of regulatory T-cells | 0.02 |
| NLRP3 | Inflammasome component in macrophages | 0.03 |
| PTGER4 | Central role in immunomodulation and control of inflammation mediated by prostaglandin E2 | 0.03 |
| NLRC4 | Inflammasome complex protein important for inflammatory responses | 0.03 |
| CD28 | Receptor for CD80, T cell activation and survival | 0.03 |
| STAT3 | Acute phase response transcription factor | 0.03 |
| IL18R1 | Receptor for pro-inflammatory cytokine, activating NK cells | 0.04 |
| LDLR | Low density lipoprotein receptor | 0.04 |
| NFE2L2 | Leucine zipper transcription factor required for expression of anti-oxidant enzyme NQO1 | 0.048 |
Genes significantly differentially expressed between patients with PGD and without PGD throughout the first year after lung transplantation. P-value presented is from linear mixed models.
Figure 1.
Longitudinal differences in gene expression over time throughout the first year after transplant. Dark gray indicates patients with PGD and light gray indicates patients without PGD. The x-axis is log transformed days since transplant and the y-axis is normalized gene expression levels.
A: gene expression differences in long pentraxin 3 (PTX3) indicating higher levels of expression in patients with PGD compared to without PGD early after transplant with no differences in expression later in the first year.
B: gene expression differences in forkhead box P3 (FOXP3) indicating lower levels of expression in patients with PGD compared to without PGD early after transplant with no differences in expression later in the first year.
We therefore next focused our evaluation on the early posT transplant period, evaluating gene expression differences within the first week after allograft reperfusion. Of the sixteen genes upregulated early after transplant, eleven genes were overexpressed in patients with PGD compared to those without PGD within 2 hours after reperfusion (Table 3). Genes encoding innate immune/inflammasome-related proteins were highly represented among these genes, including genes and gene products that have been previously associated with PGD risk in lung and blood samples (PTX3, p=0.01 and nod-like receptor family, pyrin domain-containing 3 (NLRP3), p=0.02). Nine genes were overexpressed 7 days after transplant, again with significant representation from innate immune and inflammasome genes, including toll-like receptor-4 (TLR4, p=0.02), TLR2 (p=0.04), Nod-like receptor family, caspase recruitment domain-containing 4 (NLRC4, p=0.01), and NLRP3 (p=0.01), further highlighting the representation of early innate immune activity.
Table 3.
Overexpressed genes within the first seven days after lung transplantation
| Gene | Gene activity | Fold Change - 2 hours |
FDR p- value – 2 hours |
Fold Change - 7 days |
FDR p- value – 7 days |
|---|---|---|---|---|---|
| FOSB | Adaptive neuronal responses |
2.34 | 0.03 | −1.01 | 0.9 |
| PTX3 | Secreted innate immune mediator |
2.07 | 0.01 | 1.56 | 0.08 |
| IL10 | Immunoregulation and inflammation |
1.98 | 0.01 | 1.01 | 0.9 |
| SOCS3 | Cytokine-inducible negative regulators of cytokine signaling |
1.97 | 0.03 | 1.02 | 0.9 |
| HBEGF | Mitogenic and chemotactic glycoprotein produced by monocytes and macrophages |
1.87 | 0.02 | −1.05 | 0.5 |
| LDLR | LDL receptor | 1.74 | 0.03 | 1.46 | 0.07 |
| HMOX1 | Inducible enzyme in heme catabolism |
1.72 | 0.03 | 1.19 | 0.2 |
| IL18R1 | Receptor for pro- inflammatory cytokine, activating NK cells |
2.07 | 0.03 | 3.22 | 0.01 |
| NLRP3 | Inflammasome component in macrophages |
1.73 | 0.02 | 1.62 | 0.01 |
| HGF | Regulation of cell growth, cell motility and morphogenesis |
1.72 | 0.03 | 1.75 | 0.02 |
| NLRC4 | Caspase recruitment protein with essential innate immune role |
1.59 | 0.02 | 1.65 | 0.01 |
| C1QA | Major constituent of complement system |
−1.07 | 0.9 | 1.75 | 0.02 |
| CEBPD | Regulation of immune and inflammatory response genes |
1.34 | 0.03 | 1.72 | 0.02 |
| NOD2 | Innate pattern recognition receptor |
1.33 | 0.047 | 1.62 | 0.01 |
| TLR4 | Innate immune receptor | 1.38 | 0.04 | 1.59 | 0.02 |
| TLR2 | Innate immune receptor | 1.4 | 0.07 | 1.53 | 0.04 |
Of the thirteen genes underexpressed early after transplant, eleven genes were underexpressed in patients with PGD compared to those without PGD as early as 2 hours after reperfusion (Table 4), highlighted by genes related to T cell regulation. Ten genes were underexpressed 7 days after reperfusion, highlighted by T cell regulatory molecules, including FOXP3, a master regulator for regulatory T cell development and function (p=0.011). Eight genes, predominantly involved in T cell and adaptive immune responses, including Fas ligand (FASLG), an apoptosis trigger essential for adaptive immune regulation (p=0.005 at 2 hours and p=0.03 at 7 days after transplant), were underexpressed within 2 hours of reperfusion and remained differentially expressed at 7 days, indicating very early differences that remained persistently decreased in the immediate posT transplant period.
Table 4.
Under expressed genes within the first seven days after lung transplantation
| Gene | Gene activity | Fold Change - 2 hours |
FDR p- value – 2 hours |
Fold Change - 7 days |
FDR p- value – 7 days |
|---|---|---|---|---|---|
| CD80 | T-cell proliferation and cytokine production |
−1.51 | 0.03 | −1.48 | 0.08 |
| IFNG | Activator of macrophages ad MHC expression |
−1.69 | 0.03 | −1.6 | 0.08 |
| GZMB | Serine protease expressed by cytotoxic T-cells |
−1.71 | 0.02 | −1.39 | 0.2 |
| FOXP3 | Regulator in development and function of regulatory T- cells |
−1.52 | 0.03 | −1.8 | 0.01 |
| CD28 | Receptor for CD80, T cell activation and survival |
−1.58 | 0.02 | −2.13 | 0.01 |
| CXCR3 | Regulates leukocyte trafficking |
−1.6 | 0.04 | −1.98 | 0.01 |
| CD3E | Component of the T-cell receptor complex |
−1.67 | 0.02 | −1.97 | 0.02 |
| CD40LG | Regulates B-cell function | −1.75 | 0.02 | −2.12 | 0.01 |
| PRF1 | Key effector molecule for T-cell and NK cell mediated cytolysis |
−1.78 | 0.02 | −1.9 | 0.02 |
| CD27 | Required for generation and long-term maintenance of T cell immunity |
−1.79 | 0.009 | −1.72 | 0.02 |
| FASLG | Apoptosis trigger essential for immune system regulation |
−2.1 | 0.005 | −1.88 | 0.03 |
| CTLA4 | Inhibiting T-cell activity | −1.33 | 0.05 | −1.54 | 0.04 |
| ICOS | Cell-cell signaling, immune responses, and regulation of cell proliferation |
−1.12 | 0.4 | −1.97 | 0.01 |
Discussion
In this multicentered cohort study, we have identified in the recipient very early elevated expression of genes associated with inflammasome activation and innate immunity in PGD subjects within two hours of lung transplantation. Additionally, we identified that genes associated with T cell regulation were significantly down-regulated early after reperfusion in patients with PGD. Importantly, differential gene expression for many of these genes persisted for at least 7 days after the initial allograft insult while others, in similar pathways, had a delayed pattern of differential expression in circulating cells. Taken together, our findings indicate a recipient immune cell response to the injured allograft occurring within hours of reperfusion and lasting over a week, which is characterized by innate immune and inflammasome activation, and suppression of T cell regulatory responses.
The rapid time course of recipient circulating innate immune and inflammasome responses identified in our study is notable, and mirrors what we have previously published in the lung allograft7. Likewise, other organs have shown a similar response. Biopsy samples of liver allografts 90 minutes after reperfusion demonstrate significant upregulation of genes in the allograft involved in activation and function of innate immune cells17. Similarly, elevated peripheral blood gene expression of the innate immune receptors toll-like receptor 2 and 4 (TLR2 and TLR4) measured 3 to 6 days after reperfusion is associated with delayed kidney graft function18. We identified significant differential expression of innate immune and inflammasome pathways in circulating cells within 2 hours of lung reperfusion, indicating that early damage to the allograft in the setting of ischemia reperfusion injury results in an extremely rapid recipient systemic response, likely in response to Damage Associated Molecular Patterns (DAMPs) released by the lung allograft. New knowledge of the timing of recipient responses may affect the type and timing of administration for future potential PGD therapeutics, suggesting efforts aimed at prevention may be most effective.
Innate immune activation and inflammasome pathways are frequently identified to be strongly associated with the development of PGD after lung transplantation. In the current study, the inflammasome component gene NLRP3 was persistently upregulated in the peripheral blood of patients with PGD at least 7 days after transplant. We have previously demonstrated that recipient genetic variants in the innate immune mediator PTX3 and protein levels of PTX3 are associated with differential risk of PGD19,20. In this study, PTX3 was one of the most upregulated genes identified early after lung reperfusion. Likewise, mirroring our blood findings, Nod-like receptor (NLR), toll-like receptor (TLR), and myeloid differentiation primary response gene 88 (MyD88) pathways were among the most highly ranked gene sets in patients with PGD in an analysis of BAL fluid gene expression in a nested population from this CTOT 03 study7. Recipient genetic variation in toll-interacting protein (TOLLIP), a regulator of innate inflammatory TLR signaling cascades, is also significantly associated with risk for PGD after lung transplantation21. Taken together, the pathways identified in our current and previous studies indicate that inflammasome and innate immune activity are upregulated both locally in the allograft and systemically in the recipient in patients with PGD, thus providing further evidence that innate immune and inflammasome signaling pathways are novel targets for future PGD preventative and therapeutic interventions.
We also found that the primary down-regulated genes associated with PGD were clustered in pathways involved in T cell regulation. FOXP3 is notably systemically significantly down-regulated in patients with PGD throughout the first year after lung transplantation, with the most pronounced differences seen in the immediate posT transplant period. T regulatory cells are integral to suppressing immune responses. We have previously demonstrated that genetic variation in PTGER4 at the rs4434423 locus was associated with altered risk for PGD and differential suppressive function of regulatory T cells14. Prostagladin E2 induces FOXP3 gene and protein expression and enhances the inhibitory function of human T regulatory cells22. We also identified decreased cytotoxic T lymphocyte antigen-4 (CTLA4) gene expression in patients with PGD immediately after allograft reperfusion. CTLA4 is a central negative regulator of T cell responses and nonsense mutations in this gene are associated with complex immune dysregulation syndromes with autoimmune features. Decreased expression of CTLA4 and FOXP3 in lung transplant recipients with PGD highlights the potential importance of regulatory T cells in the pathogenesis of PGD. Future study should focus on the therapeutic implications of the association between prostaglandin E2 and T regulatory cell function.
Concordant with our findings, dysregulation of T cell specific pathways have previously been associated with ischemia reperfusion injury in animal models as well as PGD after human lung transplantation. In a mouse model of lung ischemia reperfusion injury, sphingosine-1-phosphate receptor agonists decreased natural killer T cell cytokine production but not migration leading to increased pulmonary compliance and decreased pulmonary artery pressure23. The same investigator group also identified CD4+ invariant natural killer cells to be central in the development of ischemia reperfusion injury after hilar ligation, highlighting the importance of T cells in the development and regulation of lung injury24. In a recent study by Strüber et al., higher levels of CD25+CD4+CD152+ and CD25+CD4+Foxp3+ T cells within 3 weeks of lung transplant were correlated with higher forced expiratory volume in 1 second (FEV1)25. Furthermore, CD25+CD4+ T regulatory cells have been shown to decrease the development of obliterative airways disease lesion in a mouse model of bronchiolitis obliterans syndrome (BOS), a leading cause of chronic lung allograft dysfunction after lung transplantation26. There is a strong association of PGD with the development of later BOS, and a shared mechanism of dysregulated T cell function is a potential pathologic link between the two syndromes. Histone deacetylase (HDAC) inhibitors have been shown to improve T regulatory cell function and also protect against renal ischemia reperfusion injury in a murine renal transplant model27,28. Based on our findings, HDAC inhibitors are therefore attractive targets for preventing and as a potential treatment for PGD.
There are several limitations to our study. First, the total number of patients enrolled in the study was 106, limiting the number of comparisons that could be made. We combated the small numbers by 1) enrolling patients in three sites, adding to the generalizability of the findings; 2) limiting our gene expression analysis to a panel of genes with previous linkage to PGD based on previous gene association, protein biomarker, or animal models studies, early allograft dysfunction in other solid organs, or on known or hypothesized associations with ischemia reperfusion injury; and 3) using a false discovery rate adjustment in our significance testing. Longitudinal evaluation of the effect of PGD on later gene expression changes was limited by the smaller number of samples available for evaluation at the later time points, with 184 of the 401 samples evaluated (46%) clustered in the first 7 days after transplant (Table S1). The drop off in numbers is due to predominantly to loss of follow-up, missed opportunities for sample collection (timing, lack of personnel, etc.) and differences in bronchoscopy protocols across centers. As this was an observational cohort study, we did not protocolize bronchoscopy timing and over time, based on clinical need, not all patients underwent bronchoscopy at the set time points. The longer-term impact on gene and protein expression related to PGD should be the focus of future studies, utilizing longitudinal sample collection and well phenotyped study subjects. As the focus of the CTOT 03 study was early graft dysfunction, we are unable to evaluate the impact of the early gene expression differences identified in the first year with later CLAD development because of the lack of imaging and spirometric measurements necessary for CLAD phenotyping. Additionally, as the gene expression changes identified were concurrent with the development of clinical PGD, we are unable to assign causality to the pathways identified. The overall validity of our findings is buttressed, though, by the significant evidence for the role of innate immunity, inflammasomes, and T cell regulation identified previously in prior studies. While we cannot evaluate the impact of different absolute numbers of NK cells or regulatory T cells, the differences in gene expression are likely due to more than simple numerical differences in number of leukocytes or cellular subsets. Gene expression differences may indicate differences in activation states of circulating immune cells or differences in subpopulations of effector cells. Flow cytometric analysis of changes in circulating immune cell populations in the setting of PGD should be an area of future research. Finally, we are unable to directly compare our findings of blood gene expression differences with our previous findings of differential gene expression in BAL due to small numbers of patients, differences in collection times, and non-overlapping genotyping platforms.
In summary, we found that genes associated with innate immunity and inflammasome function were upregulated in the systemic circulation of patients with PGD within two hours of lung transplantation while genes associated with T cell regulation were systemically down-regulated. This study further highlights the importance of these pathways in the development of ischemia reperfusion induced lung injury after transplantation. Future studies should be focused on identifying potential therapeutics aimed at preventing dysregulation of these pathways to potentially mitigate the development and progression of PGD after lung transplantation.
Supplementary Material
Acknowledgments
This study was supported by NIH grants U01 AI063589, R01 HL087115, R01 HL081619, K24 HL115354, K23 HL121406, and K23 HL116656.
Glossary
- bDNA
branch DNA
- BOS
bronchiolitis obliterans syndrome
- BAL
bronchoalveolar lavage
- CLAD
chronic lung allograft dysfunction
- CTOT
Clinical Trials in Organ Transplantation
- CYLA4
cytotoxic T lymphocyte antigen-4
- DAMP
Damage Associated Molecular Pattern
- FASLG
Fas ligand
- FEV1
forced expiratory volume in 1 second
- FOXP3
forkhead box P3
- HDAC
Histone deacetylase
- HIV
human immunodeficiency virus
- MAD
mean absolute deviation
- MyD88
myeloid differentiation primary response gene 88
- NLR
nod-like receptor
- NLRP3
nod-like receptor family, pyrin domain-containing
- PTX3
pentraxin 3
- PGD
primary graft dysfunction (PGD
- PCA
Principal Components Analysis
- SD
standard deviation
- TOLLIP
toll-interacting protein
- TLR
toll-like receptor
- TLR4
toll-like receptor-4
Footnotes
Disclosures:
The authors of this manuscript have no conflicts of interest to disclose as described by the American Journal of Transplantation.
Supplemental Information:
Supplemental Table 1: Number of samples analyzed at each designated time point.
References
- 1.Porteous MK, Diamond JM, Christie JD. Primary graft dysfunction: lessons learned about the first 72 h after lung transplantation. Current opinion in organ transplantation. 2015 Oct;20(5):506–514. doi: 10.1097/MOT.0000000000000232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kangelaris KN, Prakash A, Liu KD, et al. Increased expression of neutrophil-related genes in patients with early sepsis-induced ARDS. American journal of physiology. Lung cellular and molecular physiology. 2015 Jun 1;308(11):L1102–L1113. doi: 10.1152/ajplung.00380.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andrade-Oliveira V, Campos EF, Goncalves-Primo A, et al. TLR4 mRNA levels as tools to estimate risk for early posttransplantation kidney graft dysfunction. Transplantation. 2012 Sep 27;94(6):589–595. doi: 10.1097/TP.0b013e31825db680. [DOI] [PubMed] [Google Scholar]
- 4.Guerrieri D, Re L, Petroni J, et al. Gene expression profile in delay graft function: inflammatory markers are associated with recipient and donor risk factors. Mediators of inflammation. 2014;2014:167361. doi: 10.1155/2014/167361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Salvadori M, Rosso G, Bertoni E. Update on ischemia-reperfusion injury in kidney transplantation: Pathogenesis and treatment. World journal of transplantation. 2015 Jun 24;5(2):52–67. doi: 10.5500/wjt.v5.i2.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ray M, Dharmarajan S, Freudenberg J, Zhang W, Patterson GA. Expression profiling of human donor lungs to understand primary graft dysfunction after lung transplantation. American journal of transplantation : official journal of the American Society of Transplantation and the American Society of Transplant Surgeons. 2007 Oct;7(10):2396–2405. doi: 10.1111/j.1600-6143.2007.01918.x. [DOI] [PubMed] [Google Scholar]
- 7.Cantu E, Lederer DJ, Meyer K, et al. Gene set enrichment analysis identifies key innate immune pathways in primary graft dysfunction after lung transplantation. American journal of transplantation : official journal of the American Society of Transplantation and the American Society of Transplant Surgeons. 2013 Jul;13(7):1898–1904. doi: 10.1111/ajt.12283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kreisel D, Sugimoto S, Tietjens J, et al. Bcl3 prevents acute inflammatory lung injury in mice by restraining emergency granulopoiesis. The Journal of clinical investigation. 2011 Jan;121(1):265–276. doi: 10.1172/JCI42596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Spahn JH, Li W, Bribriesco AC, et al. DAP12 expression in lung macrophages mediates ischemia/reperfusion injury by promoting neutrophil extravasation. Journal of immunology. 2015 Apr 15;194(8):4039–4048. doi: 10.4049/jimmunol.1401415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Christie JD, Carby M, Bag R, et al. Report of the ISHLT Working Group on Primary Lung Graft Dysfunction part II: definition. A consensus statement of the International Society for Heart and Lung Transplantation. The Journal of heart and lung transplantation : the official publication of the International Society for Heart Transplantation. 2005 Oct;24(10):1454–1459. doi: 10.1016/j.healun.2004.11.049. [DOI] [PubMed] [Google Scholar]
- 11.Christie JD, Bellamy S, Ware LB, et al. Construct validity of the definition of primary graft dysfunction after lung transplantation. The Journal of heart and lung transplantation : the official publication of the International Society for Heart Transplantation. 2010 Nov;29(11):1231–1239. doi: 10.1016/j.healun.2010.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Diamond JM, Lee JC, Kawut SM, et al. Clinical risk factors for primary graft dysfunction after lung transplantation. American journal of respiratory and critical care medicine. 2013 Mar 1;187(5):527–534. doi: 10.1164/rccm.201210-1865OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shah RJ, Diamond JM, Cantu E, et al. Latent class analysis identifies distinct phenotypes of primary graft dysfunction after lung transplantation. Chest. 2013 Aug;144(2):616–622. doi: 10.1378/chest.12-1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Diamond JM, Akimova T, Kazi A, et al. Genetic variation in the prostaglandin E2 pathway is associated with primary graft dysfunction. American journal of respiratory and critical care medicine. 2014 Mar 1;189(5):567–575. doi: 10.1164/rccm.201307-1283OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Butte AJ, Ye J, Haring HU, Stumvoll M, White MF, Kohane IS. Determining significant fold differences in gene expression analysis. Pacific Symposium on Biocomputing. Pacific Symposium on Biocomputing. 2001:6–17. doi: 10.1142/9789814447362_0002. [DOI] [PubMed] [Google Scholar]
- 16.Geiss GK, Bumgarner RE, An MC, et al. Large-scale monitoring of host cell gene expression during HIV-1 infection using cDNA microarrays. Virology. 2000 Jan 5;266(1):8–16. doi: 10.1006/viro.1999.0044. [DOI] [PubMed] [Google Scholar]
- 17.Gehrau RC, Mas VR, Dumur CI, et al. Donor Hepatic Steatosis Induce Exacerbated Ischemia-Reperfusion Injury Through Activation of Innate Immune Response Molecular Pathways. Transplantation. 2015 Dec;99(12):2523–2533. doi: 10.1097/TP.0000000000000857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McDaniel DO, Rigney DA, McDaniel KY, et al. Early expression profile of inflammatory markers and kidney allograft status. Transplantation proceedings. 2013 May;45(4):1520–1523. doi: 10.1016/j.transproceed.2012.08.027. [DOI] [PubMed] [Google Scholar]
- 19.Diamond JM, Lederer DJ, Kawut SM, et al. Elevated plasma long pentraxin-3 levels and primary graft dysfunction after lung transplantation for idiopathic pulmonary fibrosis. American journal of transplantation : official journal of the American Society of Transplantation and the American Society of Transplant Surgeons. 2011 Nov;11(11):2517–2522. doi: 10.1111/j.1600-6143.2011.03702.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Diamond JM, Meyer NJ, Feng R, et al. Variation in PTX3 is associated with primary graft dysfunction after lung transplantation. American journal of respiratory and critical care medicine. 2012 Sep 15;186(6):546–552. doi: 10.1164/rccm.201204-0692OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cantu E, Suzuki Y, Diamond JM, et al. Protein Quantitative Trait Loci Analysis Identifies Genetic Variation in the Innate Immune Regulator TOLLIP in PosT Lung Transplant Primary Graft Dysfunction Risk. American journal of transplantation : official journal of the American Society of Transplantation and the American Society of Transplant Surgeons. 2015 Dec 10; doi: 10.1111/ajt.13525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baratelli F, Lin Y, Zhu L, et al. Prostaglandin E2 induces FOXP3 gene expression and T regulatory cell function in human CD4+ T cells. Journal of immunology. 2005 Aug 1;175(3):1483–1490. doi: 10.4049/jimmunol.175.3.1483. [DOI] [PubMed] [Google Scholar]
- 23.Stone ML, Sharma AK, Zhao Y, et al. Sphingosine-1-phosphate receptor 1 agonism attenuates lung ischemia-reperfusion injury. American journal of physiology. Lung cellular and molecular physiology. 2015 Jun 15;308(12):L1245–11252. doi: 10.1152/ajplung.00302.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sharma AK, LaPar DJ, Zhao Y, et al. Natural killer T cell-derived IL-17 mediates lung ischemia-reperfusion injury. American journal of respiratory and critical care medicine. 2011 Jun 1;183(11):1539–1549. doi: 10.1164/rccm.201007-1173OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakagiri T, Warnecke G, Avsar M, et al. Lung function early after lung transplantation is correlated with the frequency of regulatory T cells. Surgery today. 2012 Feb;42(3):250–258. doi: 10.1007/s00595-011-0087-3. [DOI] [PubMed] [Google Scholar]
- 26.Tiriveedhi V, Takenaka M, Ramachandran S, et al. T regulatory cells play a significant role in modulating MHC class I antibody-induced obliterative airway disease. American journal of transplantation : official journal of the American Society of Transplantation and the American Society of Transplant Surgeons. 2012 Oct;12(10):2663–2674. doi: 10.1111/j.1600-6143.2012.04191.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Beier UH, Wang L, Hancock WW. Combination of isoform-selective histone/protein deacetylase inhibitors improves Foxp3+ T regulatory cell function. Cell cycle. 2012 Sep 15;11(18):3351–3352. doi: 10.4161/cc.21876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Levine MH, Wang Z, Bhatti TR, et al. Class-specific histone/protein deacetylase inhibition protects against renal ischemia reperfusion injury and fibrosis formation. American journal of transplantation : official journal of the American Society of Transplantation and the American Society of Transplant Surgeons. 2015 Apr;15(4):965–973. doi: 10.1111/ajt.13106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.