Abstract
Metastatic urachal carcinoma is a rare, understudied, and aggressive malignancy with limited treatment options. Histologically, urachal carcinomas resemble enteric adenocarcinomas and anecdotally respond to systemic therapies utilized in colorectal cancer. Targeted exome sequencing of archival primary tumor tissue from a patient with metastatic urachal cancer revealed EGFR amplification and wild-type KRAS. The patient was treated with cetuximab, a monoclonal antibody directed against EGFR, as a single agent, and achieved a response lasting more than 8 mo. Subsequent whole-exome sequencing revealed no additional alterations likely to be associated with cetuximab sensitivity. Formalin-fixed, paraffin-embedded tumor specimens from nine additional urachal cancers were subjected to targeted exome sequencing. Mitogen-activated protein kinase (MAPK) pathway mutations were found in four of the nine samples, but no EGFR amplification was detected. Importantly, APC mutations were detected in two of the nine patients. To our knowledge, this is the first report of a response to single-agent cetuximab in a patient with metastatic urachal cancer and of molecular analysis to probe the basis for sensitivity. On the basis of these findings and the histologic, and now genomic, similarities with colorectal cancer, monoclonal antibodies directed at EGFR could be used in the treatment of metastatic urachal cancer.
Patient summary
Urachal cancers are morphologically and genomically similar to colon adenocarcinomas and may respond to drugs targeting the epidermal growth factor receptor.
Keywords: Urachal carcinoma, Genomic alterations, Epidermal growth-factor, Inhibition
Urachal cancer is an aggressive bladder malignancy arising from a vestigial remnant. Patients with metastatic urachal cancer have poor prognosis, with median survival of ~1.3 yr [1]. Given the rarity of urachal cancer, prospective trials to guide the treatment of patients with metastatic disease are lacking, there are no standard chemotherapeutic regimens, and management strategies are empiric and highly institution-dependent. Histologically, urachal cancer resembles enteric adenocarcinoma and anecdotally may respond to chemotherapy used to treat colorectal cancer [1]. Comprehensive molecular analyses of urachal cancer are lacking; however, an analysis of KRAS and BRAF mutations in urachal cancer previously identified KRAS mutations in four out of seven specimens [2].
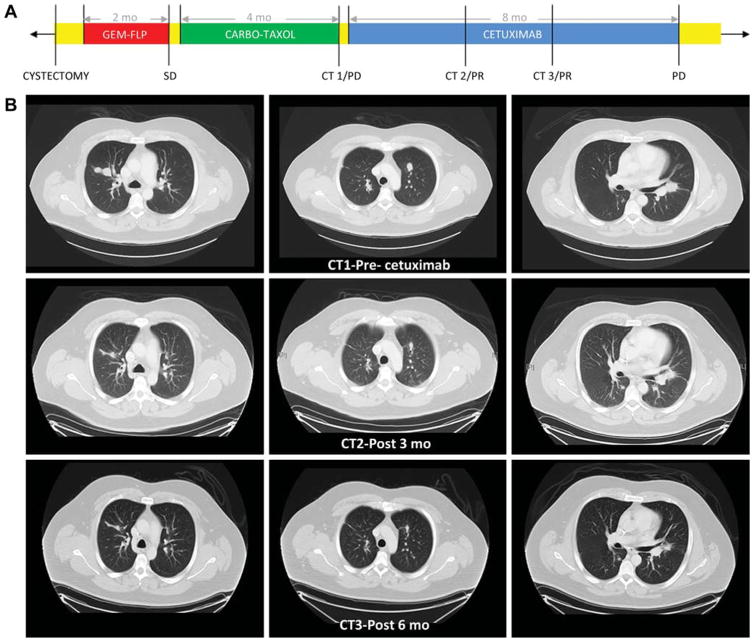
A 35-yr-old male presented with metastatic urachal cancer to the lungs. A partial cystectomy was performed as palliative treatment for severe hematuria. Pathological analysis confirmed the diagnosis of mucinous urachal carcinoma, and the possibility of colorectal adenocarcinoma invading the bladder was excluded. He was subsequently treated with two cycles of gemcitabine-FLP (5-fluorouracil, leucovorin, cisplatin) that resulted in transient disease stabilization. However, treatment was discontinued because of severe treatment-related toxicities including fatigue, nausea, vomiting, and diarrhea. Paclitaxel plus carboplatin was then administered but was discontinued owing to disease progression (Fig. 1A). Given the lack of treatment options, targeted exome sequencing was performed on archival tissue from the primary tumor (Foundation One; Foundation Medicine, Cambridge, MA, USA) and revealed EGFR amplification and wild-type KRAS (Table 1). On the basis of these findings, the patient was started on treatment with the anti-EGFR monoclonal antibody cetuximab, and experienced a 25% decrease in tumor burden that lasted for more than 8 mo (Fig. 1B). Treatment was generally well tolerated with the exception of an acneiform rash that was treated with doxycycline and topical steroids.
Fig. 1.
Clinical information for the index patient. (A) Timeline demonstrating treatment course. CT = computed tomography scan; SD = stable disease; PR = partial response; PD = progression of disease. Yellow represents periods without chemotherapy treatment. (B) CT images of lung metastases before and after 3 and 6 mo of cetuximab treatment.
Table 1.
Histology and genomic alterations in APC, TP53, and MAPK and PI3K pathway genes as determined by next-generation sequencing in a cohort of patients with urachal cancer and by whole-exome sequencing in the index patient with urachal cancer
| Gene | Frequency (%) | Patients | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Index | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | ||
| TP53 | 80 | W47del a | R213* b | L257Rfs*6 b | H179del a | R248Q c | G266R c | R248W c | R175H c | ||
| APC | 20 | DEL | R1450* | R554* b Q1406Efs*2 b |
|||||||
| KRAS | 20 | G13D c | G12V c | ||||||||
| NRAS | 10 | Q61K c | |||||||||
| RASAL1 | 10 | R342H c | |||||||||
| EGFR | 10 | AMP | |||||||||
| MAP2K1 | 10 | C121S c | |||||||||
| MAP3K19 | 10 | R139Q c | |||||||||
| GNAS | 10 | R201C c | |||||||||
| PIK3CA | 10 | E545K c | |||||||||
| MET | 10 | AMP | |||||||||
| ERBB3 | 10 | P583S c | |||||||||
| ERBB4 | 10 | X497_Splice b | |||||||||
| ERBB2 | 0 | ||||||||||
| HRAS | 0 | ||||||||||
| BRAF | 0 | ||||||||||
| RAF1 | 0 | ||||||||||
| NF1 | 0 | ||||||||||
| Histology | Mucinous | Mucinous | SRC | Mucinous | Mucinous | Enteric | SRC | ADC | Mucinous | Mucinous | |
DEL = deletion; AMP = amplification; SRC = signet ring cell; ADC = adenocarcinoma not otherwise specified.
In-frame mutation.
Truncating mutation.
Missense mutation.
Anti-EGFR antibodies have been approved by regulatory agencies for use in metastatic colorectal cancer since 2004 [3]. However, in 2009, on the basis of emerging evidence demonstrating that activating KRAS mutations (resulting in constitutively activated MAPK signaling downstream of EGFR) conferred resistance to treatment, the US Food and Drug Administration restricted use of anti-EGFR antibodies to patients with wild-type KRAS tumors [4,5]. The National Comprehensive Cancer Network guidelines extended this restriction to patients with wild-type KRAS and NRAS tumors. Activating mutations in several other genes downstream of EGFR, such as BRAF and PIK3CA, have also been associated with EGFR inhibitor resistance in some, but not all, studies and therefore have not yet been fully integrated into standard clinical decision-making [6]. Predictors of sensitivity have proven even more elusive; analyses using model systems and human specimens have identified putative treatment response biomarkers such as EGFR amplification, alterations in the tyrosine kinase adaptor gene IRS2, and increased EGFR ligand expression [7–9].
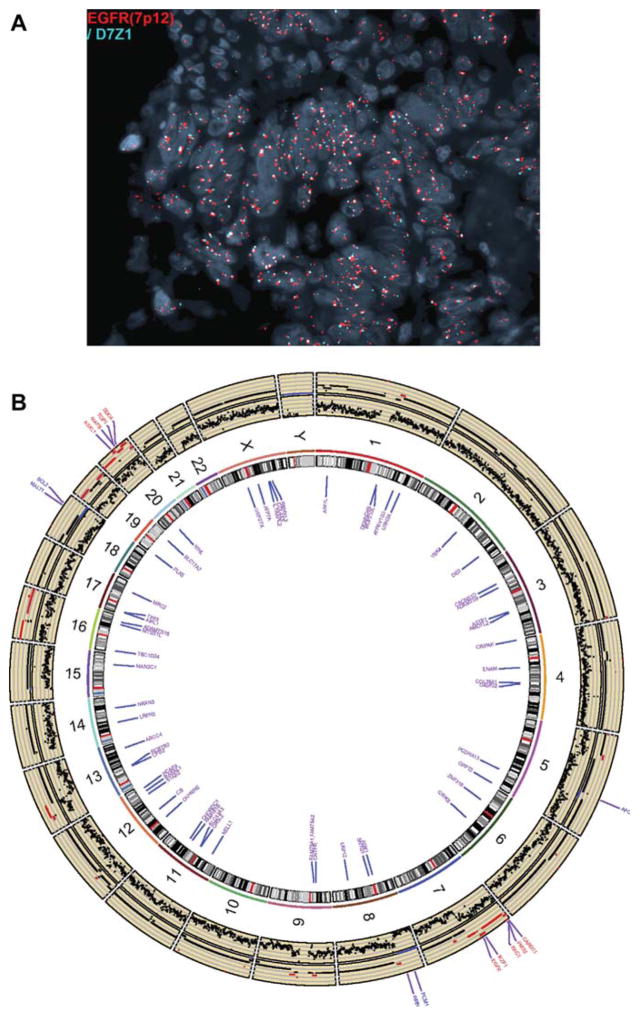
To further explore the mechanistic underpinnings of cetuximab sensitivity in our patient with wild-type KRAS urachal cancer, we first confirmed the EGFR amplification by fluorescent in situ hybridization (FISH; Fig. 2A). Wholeexome sequencing (WES) subsequently performed on the primary tumor to define the pattern of co-altered genes revealed 54 nonsynonymous single-nucleotide variants (SNVs), 106 segmental amplifications (28 segments with estimated copy number ≥6) and 36 segmental deletions (12 segments with homozygous deletions) (Fig. 2B). No additional alterations known to be associated with sensitivity or resistance to anti-EGFR antibodies were identified. Notably, a homozygous APC deletion was identified (Supplementary methods).
Fig. 2.
Molecular alterations observed for the index patient. (A) Fluorescent in situ hybridization confirming EGFR amplification (≥6 copies of EGFR [orange] in the absence of multiple copies of centromere 7 [green]). (B) Circos plot of copy number variant (CNV) and single nucleotide variant (SNV) data. The raw CNV data (log ratio of read number) are plotted in the first track outside the ideogram. The estimated absolute copy number (CN) is plotted in the second track (blue represents CN = 0 and red represents CN ≥ 4). Known cancer genes [9] with CN = 0 (blue) or CN ≥ 6 (red) are labeled in the next track. Genes with nonsynonymous SNVs are labeled in the track inside the ideogram in purple color. Supplementary Tables 1–4 provide complete gene lists for CNV and SNV data.
We next sought to explore the prevalence of mutations downstream of EGFR, determine if EGFR amplification is a common genomic event, and identify additional novel putative therapeutic targets in urachal cancer. We identified archival formalin-fixed, paraffin-embedded tumor specimens from nine additional patients treated at our institution during 2000–2014. Slides were reviewed by an expert genitourinary pathologist (M.C-M.) and targeted exome sequencing was performed using the MSK-IMPACT platform [10] (Table 1 and Supplementary Fig. 1). In addition, FISH was performed to evaluate EGFR amplification. There were no EGFR amplifications detected by next-generation sequencing analysis or FISH. Mutations in the MAPK pathway were present in four of the nine tumors: two KRAS mutation (G13D, G12 V), one NRAS mutation (Q61K), and one MAP2K1 (C121S) kinase domain alteration (Table 1). Importantly, two of the nine tumors had APC truncating mutations (R1450*, R554*) that are frequently found in colorectal adenocarcinoma. Seven of nine tumors had TP53 mutations (2 truncating mutations, 1 in-frame mutation, and 4 missense mutations). Inactivation of tumor suppressor genes such as TP53 is a prototypic event in the cancer genome heretofore thought to be undruggable, although multiple efforts to target TP53 mutant tumors are in preclinical and early-phase clinical development [11].
Several case reports for patients with urachal cancer have described responses to chemotherapy regimens that are effective in patients with colorectal cancer [1,12]. However, to the best of our knowledge this is the first report describing a response to single-agent cetuximab and probing of the molecular basis for sensitivity. Level I evidence, developed in the context of prospective randomized controlled trials, is the gold standard to guide the care of patients with cancer given the often narrow therapeutic index, modest benefits, and high cost of many systemic anticancer therapies. However, there are a multitude of pragmatic challenges associated with the generation of high-level evidence for the care of patients with rare cancers, including inadequate understanding of disease pathogenesis, lack of model systems, insufficient interest/funding from key stakeholders in drug development, and the absence of an international infrastructure to ensure adequate trial accrual. Alternative approaches to evidence development, such as umbrella and n-of-1 trials, may ultimately prove beneficial [13]. In the meantime, use of all the tools currently available is critical for optimizing the care of patients in need of better therapies. Whether the presence of EGFR amplification or the lack of a KRAS mutation, or both, conferred sensitivity to cetuximab in our patient remains unknown. However, given the histologic, and now genomic, similarities between urachal and colonic adenocarcinoma (eg, presence of APC mutations) and the standard role of KRAS mutations in predicting lack of benefit for EGFR monoclonal antibodies in colorectal cancer, EGFR monoclonal antibodies with or without chemotherapy should be considered a potential treatment strategy for metastatic wild-type KRAS urachal cancers. Multicenter and multinational collaborations are needed to validate these findings and advance the care of patients with this rare and understudied malignancy.
Supplementary Material
Acknowledgments
Funding/Support and role of the sponsor: This work was supported by the Spanish Society of Medical Oncology and the National Institutes of Health (grant P01-CA-087497). The sponsors played no direct role in the study.
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.eururo.2016.04.037.
Footnotes
Author contributions: Matthew D. Galsky had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Study concept and design: Galsky, Castillo-Martin, Sfakianos, Collazo-Lorduy.
Acquisition of data: Collazo-Lorduy, Castillo-Martin, Wang, Patel, Leonard, Iyer, Jordan, Al-Ahmadie.
Analysis and interpretation of data: Collazo-Lorduy, Wang, Zhu, Galsky, Castillo-Martin, Al-Ahmadie.
Drafting of the manuscript: Collazo-Lorduy, Castillo-Martin, McBride, Sfakianos, Solit, Wang, Galsky.
Critical revision of the manuscript for important intellectual content: Galsky, Solit, Sfakianos, Castillo-Martin, Cordon-Cardo, Zhu, Oh.
Statistical analysis: Wang.
Obtaining funding: Collazo-Lorduy, Castillo-Martin, Cordon-Cardo, Sfakianos.
Administrative, technical, or material support: Castillo Martin.
Supervision: Galsky.
Other: None.
Financial disclosures: Matthew D. Galsky certifies that all conflicts of interest, including specific financial interests and relationships and affiliations relevant to the subject matter or materials discussed in the manuscript (eg, employment/affiliation, grants or funding, consultancies, honoraria, stock ownership or options, expert testimony, royalties, or patents filed, received, or pending), are the following: None.
References
- 1.Yanagihara Y, Tanji N, Miura N, et al. Modified FOLFOX6 chemotherapy in patients with metastatic urachal cancer. Chemotherapy. 2013;59:402–6. doi: 10.1159/000362400. [DOI] [PubMed] [Google Scholar]
- 2.Sirintrapun SJ, Ward M, Woo J, Cimic A. High-stage urachal adenocarcinoma can be associated with microsatellite instability and KRAS mutations. Hum Pathol. 2014;45:327–30. doi: 10.1016/j.humpath.2013.09.008. [DOI] [PubMed] [Google Scholar]
- 3.Cunningham D, Humblet Y, Siena S, et al. Cetuximab monotherapy and cetuximab plus irinotecan in irinotecan-refractory metastatic colorectal cancer. N Engl J Med. 2004;351:337–45. doi: 10.1056/NEJMoa033025. [DOI] [PubMed] [Google Scholar]
- 4.Karapetis CS, Khambata-Ford S, Jonker DJ, et al. K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N Engl J Med. 2008;359:1757–65. doi: 10.1056/NEJMoa0804385. [DOI] [PubMed] [Google Scholar]
- 5.Van Cutsem E, Kohne CH, Hitre E, et al. Cetuximab and chemotherapy as initial treatment for metastatic colorectal cancer. N Engl J Med. 2009;360:1408–17. doi: 10.1056/NEJMoa0805019. [DOI] [PubMed] [Google Scholar]
- 6.Hsu HC, Thiam TK, Lu YJ, et al. Mutations of KRAS/NRAS/BRAF predict cetuximab resistance in metastatic colorectal cancer patients. Oncotarget. doi: 10.18632/oncotarget.8076. In press. http://dx.doi.org/10.18632/oncotarget.8076. [DOI] [PMC free article] [PubMed]
- 7.Algars A, Lintunen M, Carpen O, Ristamaki R, Sundstrom J. EGFR gene copy number assessment from areas with highest EGFR expression predicts response to anti-EGFR therapy in colorectal cancer. Br J Cancer. 2011;105:255–62. doi: 10.1038/bjc.2011.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bertotti A, Papp E, Jones S, et al. The genomic landscape of response to EGFR blockade in colorectal cancer. Nature. 2015;526:263–7. doi: 10.1038/nature14969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seligmann JF, Elliott F, Richman SD, et al. Combined epiregulin and amphiregulin expression levels as a predictive biomarker for panitumumab therapy benefit or lack of benefit in patients with RAS wild-type advanced colorectal cancer. JAMA Oncol. 2016;2:633–42. doi: 10.1001/jamaoncol.2015.6065. [DOI] [PubMed] [Google Scholar]
- 10.Cheng DT, Mitchell TN, Zehir A, et al. Memorial Sloan Kettering-integrated mutation profiling of actionable cancer targets (MSK-IMPACT): a hybridization capture-based next-generation sequencing clinical assay for solid tumor molecular oncology. J Mol Diagn. 2015;17:251–64. doi: 10.1016/j.jmoldx.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brown CJ, Lain S, Verma CS, Fersht AR, Lane DP. Awakening guardian angels: drugging the p53 pathway. Nat Rev Cancer. 2009;9:862–73. doi: 10.1038/nrc2763. [DOI] [PubMed] [Google Scholar]
- 12.Paner GP, Lopez-Beltran A, Sirohi D, Amin MB. Updates in the pathologic diagnosis and classification of epithelial neoplasms of urachal origin. Adv Anat Pathol. 2016;23:71–83. doi: 10.1097/PAP.0000000000000110. [DOI] [PubMed] [Google Scholar]
- 13.Billingham L, Malottki K, Steven N. Research methods to change clinical practice for patients with rare cancers. Lancet Oncol. 2016;17:e70–80. doi: 10.1016/S1470-2045(15)00396-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.