Abstract
Dicer is the enzyme that cleaves double-stranded RNA (dsRNA) into 21–25-nt-long species responsible for sequence-specific RNA-induced gene silencing at the transcriptional, post-transcriptional, or translational level. We disrupted the dicer-1 (dcr-1) gene in mouse embryonic stem (ES) cells by conditional gene targeting and generated Dicer-null ES cells. These cells were viable, despite being completely defective in RNA interference (RNAi) and the generation of microRNAs (miRNAs). However, the mutant ES cells displayed severe defects in differentiation both in vitro and in vivo. Epigenetic silencing of centromeric repeat sequences and the expression of homologous small dsRNAs were markedly reduced. Re-expression of Dicer in the knockout cells rescued these phenotypes. Our data suggest that Dicer participates in multiple, fundamental biological processes in a mammalian organism, ranging from stem cell differentiation to the maintenance of centromeric heterochromatin structure and centromeric silencing.
Keywords: RNA interference, microRNA, heterochromatin silencing, DNA methylation
Central to the RNA interference (RNAi) machinery is the RNase III-containing enzyme, Dicer (Bernstein et al. 2001). Dicer is required for processing of long double-stranded RNA (dsRNA) or microRNA (miRNA) precursors into mature effector RNA molecules (Carmell and Hannon 2004). The biological function of Dicer proteins has been studied in several organisms. These studies have revealed that Dicer proteins are involved in a variety of gene-silencing phenomena, at the transcriptional (TGS), post-transcriptional (PTGS), or translational level, depending on the organism. Interestingly, there is a remarkable cross-species diversity in the function and number of Dicer-like molecules. Mutation of the single dicer gene in Caenorhabditis elegans showed that Dicer is, indeed, required for RNAi and development (Grishok et al. 2001; Ketting et al. 2001; Knight and Bass 2001; Lee and Ambros 2001). The latter phenotype was primarily attributed to the lack of processing of lin-4 and let-7 pre-miRNAs (Lee et al. 1993; Wightman et al. 1993; Moss et al. 1997; Reinhart et al. 2000; Slack et al. 2000). In mammals, where a single dicer gene likely exists, RNAi-mediated knock-down of Dicer in a human cell line led to defects in both miRNA production and short hairpin RNA (shRNA)-mediated RNAi (Hutvagner et al. 2001; Paddison et al. 2002a). In addition, inactivation of dicer-1 (dcr-1) in the mouse germ line resulted in early embryonic lethality. In Schizosaccharomyces pombe, where no miRNAs have been identified, Dicer and the RNAi pathway have been implicated in the generation of heterochromatic small interfering RNAs (siRNAs) that mediate TGS of centromeric repeats (Hall et al. 2002; Reinhart and Bartel 2002; Volpe et al. 2002, 2003; Verdel et al. 2004). Arabidopsis encodes four Dicer-like (Dcl) proteins. The first one characterized was CAF1/Dicer-like1 (Dcl-1), a nuclear Dicer homolog, which is dispensable for both TGS and PTGS (Finnegan et al. 2003; Papp et al. 2003). Dcl-1 is, however, required for miRNA production and plant development (Park et al. 2002; Reinhart et al. 2002; Xie et al. 2004). Dcl-2 may be important for viral siRNA biogenesis and antiviral defense (Xie et al. 2004), while Dcl-3 is required for generating endogenous siRNAs, which are involved in heterochromatin formation and DNA methylation at homologous loci (Chan et al. 2004; Xie et al. 2004). There is, in addition, a fourth Dicer protein (Dcl-4) whose function has not yet been determined (Matzke et al. 2004).
In Danio rerio, Dicer inactivation abolishes miRNA production and results in a block in development and an overall cellular growth arrest in fish larvae (Wienholds et al. 2003). Drosophila contains two genes of the dcr family. Drosophila Dcr-1 is required for pre-miRNA processing as well as RNAi (Lee et al. 2004), while Dcr-2 is involved in siRNA generation (Lee et al. 2004; Pham et al. 2004). In addition to initiating RNAi, Drosophila Dcr-2 is required for siRNA-induced mRNA degradation and is physically associated with the RNA-induced silencing complex (RISC) in concert with Dcr-1 and R2D2. Dcr-2 homozygous null flies are viable and thus, in vivo, Dcr-2 is dispensable for miRNA generation and development.
Neurospora also possesses two dicer genes, both of which are required for PTGS (Catalanotto et al. 2004). However, neither Dicer-dependent transcriptional nor translational repression has been described in this organism. DNA methylation in Neurospora does not depend on the RNAi pathway (Freitag et al. 2004). Most recently, Dicer ablation in a chicken B-cell line (DT40) was reported to lead to defects in heterochromatin and chromosome segregation (Fukagawa et al. 2004).
RNA-guided DNA methylation was initially described in plants by Wassenegger et al. (1994). It was later shown that small dsRNAs could direct DNA methylation to homologous promoter regions (Jones et al. 1999; Mette et al. 2000; Sijen et al. 2001; Melquist and Bender 2003). In addition, small dsRNAs derived from intergenic regions have been cloned from plants and are homologous to repetitive DNA elements such as endogenous retroelements and transposons (Hamilton et al. 2002; Llave et al. 2002). These endogenous siRNAs are processed by Dcl-3 (Chan et al. 2004) and have been linked to Ago4, an argonaute protein, histone H3 Lys 9 (H3K9) methylation, and CpXpG DNA methylation (Lindroth et al. 2001; Jackson et al. 2002; Zilberman et al. 2003).
Although RNA-dependent TGS is likely to occur in other organisms, it has been most extensively characterized in S. pombe and plants. In Drosophila, a direct link between the Dicer proteins or the small dsRNAs generated by Dicer cleavage and DNA methylation or other epigenetic modifications has not been established. Intriguingly, however, Drosophila expresses repeat-associated siRNAs (rasiRNAs), which have been suggested to participate in defining the chromatin structure of repetitive DNA (Aravin et al. 2001, 2003). Moreover, components of the RNAi pathway in flies, like piwi, aubergine, and spindle-E, have been implicated in heterochromatin-mediated gene silencing (Pal-Bhadra et al. 1999, 2002, 2004; Kogan et al. 2003).
In Tetrahymena, small dsRNAs and a piwi-like gene are required for programmed DNA deletion during somatic macronucleus formation, a process that involves heterochromatin formation (Mochizuki et al. 2002; Taverna et al. 2002).
Recently, TGS of endogenous promoters by synthetic siRNAs or shRNAs was reported in mammalian cells (Kawasaki and Taira 2004; Morris et al. 2004). This is similar to what is seen in S. pombe, where a shRNA is sufficient to silence a homologous locus by recruiting Swi6 and cohesin (Schramke and Allshire 2003). However, in mammalian cells there are also data suggesting that RNAi does not result in DNA methylation of the homologous endogenous sequence (Park et al. 2004; Svoboda et al. 2004).
The most comprehensive and informative series of studies on RNAi-mediated TGS, however, comes from fission yeast, where endogenous siRNAs corresponding to the outer centromeric repeats (otr) have been isolated. These centromeric siRNAs associate with a RNA-induced initiation of transcriptional gene silencing (RITS) complex to direct methylation of H3K9, followed by recruitment of swi6 (an HP1 homolog) and cohesin to centromeres and mating-type loci (Hall et al. 2002; Reinhart and Bartel 2002; Volpe et al. 2002, 2003; Verdel et al. 2004). Mutations within this pathway affect centromere function and manifest themselves as defects in sister-chromatid cohesion, chromosome segregation, and, ultimately, aneuploidy (Volpe et al. 2003).
Similar to S. pombe centromeres, mouse centromeres are rich in heterochromatin-associated histone modifications, which seem to depend largely on the histone methyltransferases Suv39h1 and Suv39h2 (Peters et al. 2001). Suv39h double-deficiency leads to deregulation of DNA methylation and histone modifications at centromeric repeats (Peters et al. 2001; Lehnertz et al. 2003). There is no evidence to date that the heterochromatic structure of mammalian centromeres depends on the RNAi machinery. To investigate this possibility, we have generated Dicer-deficient mouse embryonic stem (ES) cells and analyzed the effect of Dicer deficiency mainly on silencing of centromeric chromosomal regions.
Results
Generation of Dicer-null ES cells
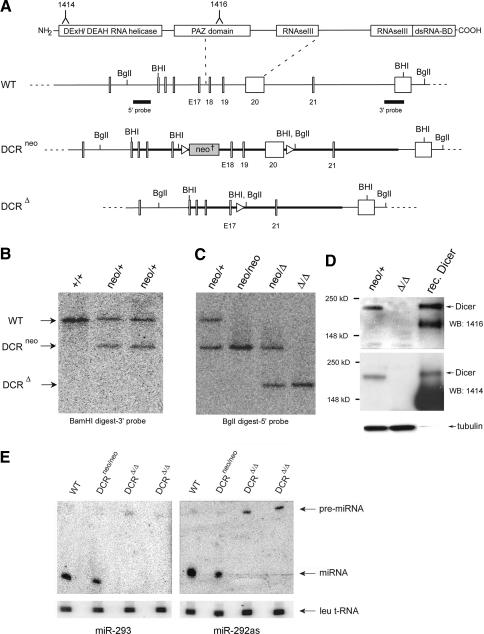
In order to generate cells that do not express a functional Dicer protein, one of the dcr-1 alleles of C57/BL6-derived Bruce-4 ES cells was targeted by homologous recombination. Specifically, loxP sites were inserted flanking exons 18–20 of dcr-1 (Fig. 1A), which encode part of the PAZ and the first RNase III domain. Deletion of this region by Cre-loxP-mediated recombination is predicted to lead to out-of-frame splicing of all downstream exons, including the second RNase III and the dsRNA-binding domains, and should result in complete ablation of Dicer enzymatic activity. Proper targeting of dcr-1 was verified by Southern blot analysis using both a 3′- and a 5′-specific probe (Fig. 1B,C).
Figure 1.
Conditional gene targeting of mouse dcr-1. (A) Depicted at the top is the structure of the Dicer protein composed of an N-terminal RNA helicase domain, PAZ domain, followed by two catalytic ribonuclease III domains and a double-stranded RNA-binding domain. Epitopes used for generation of anti-Dicer antibodies are indicated. Below it, part of the wild-type (WT) dcr-1 gene is depicted. Exons 18–20, encoding half of the PAZ domain and the first RNase III domain, are flanked by loxP sites (triangles). The locations of BamHI (BHI) and BglI restriction sites and 5′ and 3′ probes used for screening of homologous recombinants are indicated. Homologous recombination of our targeting vector (bold line) resulted in the allele depicted as DCRneo. An FRT-flanked PGK-neomycin cassette (neo†) is inserted into intron 17. Upon Cre-mediated recombination of the introduced loxP sites, a Dcr-null allele is generated (depicted as DCRΔ). (B) Southern blot of BamHI-digested genomic DNA from a wild-type (+/+) ES cell clone, and two homologous recombinants (neo/+) were hybridized with the 3′ probe (see A). (C) Southern blot of BglI-digested genomic DNA from a heterozygous (neo/+) ES cell clone, a homozygous (neo/neo) clone, a heterozygous deleted clone (neo/Δ), and a homozygous (Δ/Δ) Dcr-null clone was hybridized with the 5′ probe (see A). (D) Western blot of extracts derived from a heterozygous (neo/+) ES cell clone, a homozygous (Δ/Δ) clone, and recombinant Dicer (rec. Dicer) protein. The Western blot was probed for Dicer protein using antisera raised against two different peptides homologous to mouse Dicer (the corresponding epitopes, 1414 and 1416, are indicated on the schematic representation of Dicer protein, A), and then stripped and reprobed for tubulin (bottom). (E) Northern analysis of miRNA expression in mouse ES cells. Total RNA from a control wild-type and Dicer-proficient clone (neo/neo) and two Dcr-null clones (Δ/Δ) was resolved in a denaturing polyacrylamide gel and transferred onto a nylon membrane. Shown are PhosphorImages of two blots hybridized either to a radiolabeled oligonucleotide complementary to miR-293 (left panel) or miR-292as (right panel). To control for equal RNA loading, the Northern blots were also reprobed for leucine transfer RNA (Leu tRNA). The arrows indicate Leu tRNA, pre-miRNA, and processed mature miRNA.
In order to mutate the second dcr-1 allele, heterozygous mutant cells (DCRneo/+) were cultured in increasing concentrations of the neomycin analog G418. This selection procedure can promote a gene conversion-type modification of the wild-type allele (Mortensen et al. 1992). Indeed, we were able to obtain homozygous DCRneo/neo cells that had properly converted the wild-type allele, as verified by Southern blot analysis (Fig. 1C). DCRneo/neo ES cells were indistinguishable from parental DCRneo/+ cells with respect to proliferation rate and morphology (data not shown). Infection of these cells with a Cre-expressing adenovirus resulted in deletion of exons 18–20 and allowed us to isolate homozygous DCRΔ/Δ clones. Deletion was confirmed by Southern blot analysis (Fig. 1C). Dicer protein ablation was verified by Western blot analysis, using two different rabbit antisera raised against peptides homologous to distinct regions of murine Dicer (Fig. 1A). No Dicer protein was detected in extracts of DCRΔ/Δ cells using either antibody (Fig. 1D).
To determine whether we had generated a functional Dicer knockout, DCRΔ/Δ cells were analyzed for their ability to produce mature miRNAs. Northern blot analyses using probes specific for two ES cell-expressed miRNAs, miR-293 and miR-292as (Houbaviy et al. 2003), demonstrated the absence of mature miRNAs in two, independent DCRΔ/Δ clones. Both of these miRNAs were readily detected in wild-type and DCRneo/neo cells (Fig. 1E). Consistent with the lack of the mature miRNA species, pre-miRNA precursors accumulated in the absence of Dicer (Fig. 1E).
To further determine whether there was any remaining Dicer-like activity in the DCRΔ/Δ cells, we performed an RNAi assay. A green fluorescent protein (GFP) expression vector was stably transfected into DCRΔ/Δ and control ES cells. GFP-expressing cells were sorted by fluorescence-activated cell sorting (FACS) and transduced with a lentivirus encoding a puromycin-resistance gene and a short hairpin RNA (shRNA) homologous to gfp. Following selection with puromycin, cells that had integrated the lentivirus, and thus, should express the shRNA, were assayed for GFP expression by flow cytometry (Supplementary Fig. S1). Lentivirally encoded shRNA should be processed by Dicer into siRNA that can mediate PTGS of gfp. As expected, DCRΔ/Δ cells were incapable of shRNA-mediated RNAi, in contrast to wild-type cells. However, DCRΔ/Δ cells were able to perform RNAi when synthetic siRNAs were supplied exogenously, thus circumventing the requirement for Dicer-mediated processing of the shRNA (Supplementary Fig. S1). Consistent with previous studies (Martinez et al. 2002), Dicer seems dispensable for RISC assembly and siRNA-mediated cleavage of mRNA transcripts.
Dicer-deficient ES cells express ES cell-specific markers but fail to differentiate
Disruption of dcr-1 in the mouse germ line results in early embryonic lethality (Bernstein et al. 2003). dcr-1–/– embryos failed to express the stem cell marker, oct-4, and the primitive streak marker, brachyury. Surprisingly, the DCRΔ/Δ ES cells that we generated were viable and displayed typical ES cell morphology when grown on a mouse embryonic fibroblast (MEF) monolayer, forming oval-shaped compact colonies, indistinguishable from those formed by DCRneo/+ heterozygous or wild-type cells (Fig. 2A). They did, however, proliferate more slowly compared to DCRneo/+ and DCRneo/neo controls (data not shown). Analysis of the knockout cells for expression of ES cell-specific markers, such as the short α6-integrin isoform and oct-4, showed that these markers were expressed at comparable levels in DCRneo/+, DCRneo/neo, and DCRΔ/Δ cells, while being absent in differentiated cells, such as MEFs (Fig. 2B). As mentioned previously, the DCRΔ/Δ cells also expressed the ES cell-specific pre-miRNAs, miR-292as and miR-293, albeit not in their mature, processed form (Fig. 1E).
Figure 2.
Dicer-deficient ES cells retain ES cell characteristics but fail to differentiate. (A) Phase-contrast micrographs of heterozygous (neo/+) and homozygous (Δ/Δ)ES cell colonies (indicated by arrows) grown on a feeder layer of MEFs. (B, top panel) RT–PCR analysis of the ES cell-specific α6-integrin isoform (306 bp). (Bottom panel) Quantitative RT–PCR analysis of oct-4 transcripts normalized to hprt transcripts. The averages and standard deviations of three experiments are depicted in the graph. (C) Size of teratomas following injection of the indicated ES cells into nude mice is plotted as a function of time (weeks post-implantation). The averages from five injected mice are shown. (D) Light micrographs of paraffin sections from day 12 EBs stained with hematoxylin-eosin. EBs derived from heterozygous (DCRneo/+), homozygous (DCRneo/neo), and Dicer-deficient ES cells (DCRΔ/Δ) are shown in the top panel (10×). In the lower panel, higher magnification images (40×) of a portion of the same EBs are displayed. (E) Semiquantitative RT–PCR analyses of mesoderm- and ectoderm-specific differentiation markers. RNA was extracted and reverse-transcribed from EBs generated from DCRneo/+, DCRneo/neo, and two different DCRΔ/Δ clones at day 3, 5, and 8 of differentiation and analyzed for expression of differentiation markers bmp4, hnf4, gata1, and brachyury. Hprt transcripts were amplified as a loading control. (F) Quantitative real-time PCR analysis of oct-4 transcripts normalized to hprt transcripts in EBs at days 3, 5, 8, 10, and12 of differentiation. The averages and standard deviations of three experiments are depicted in the graph.
However, DCRΔ/Δ cells did not contribute to the generation of chimeric mice when injected into blastocysts, unlike DCRneo/+ and DCRneo/neo cells, which gave rise to high-percentage chimeras (data not shown). Furthermore, the Dicer-deficient cells failed to generate detectable teratomas upon subcutaneous injection into nude mice (Fig. 2C). Post-mortem evaluation of the mice 3 wk after injection revealed no identifiable aggregates of cells or tumors in mice implanted with DCRΔ/Δ cells.
To further investigate whether Dicer ablation leads to a specific defect in differentiation, several mutant and control ES cell clones were tested for their ability to form embryoid bodies (EBs) in vitro. When ES cells are cultured in suspension in the absence of leukemia inhibitory factor (LIF), they form cell aggregates known as EBs, in which cells differentiate into the three germ layers (Martin and Evans 1975). The differentiation of these cells into primitive endoderm, mesoderm, and ectoderm is a highly regulated process and correlates with expression of a panel of specific markers, which can be used to analyze the differentiation process at the molecular level (Keller et al. 1993; Di Cristofano et al. 1998). All clones analyzed formed EB-like structures that grew comparably well until day 8–10 of differentiation, at which point DCRΔ/Δ clones ceased growing. Paraffin sections of EBs harvested on day 12 were stained with hematoxylin-eosin (H&E), and differentiation into the three germ layers was visualized by microscopy. DCRneo/+ and DCRneo/neo EBs were composed of distinct cell types and clearly defined endoderm-, ectoderm-, and mesoderm-like structures (Fig. 2D). In contrast, the DCRΔ/Δ cells only formed aggregates of cells with little morphological evidence of differentiation. Accordingly, when EBs were analyzed for expression of genes characteristic of endodermal (hnf4) (Duncan et al. 1994) and mesodermal (brachyury, bmp4, gata1) (Elefanty et al. 1997) differentiation, no expression of any of these differentiation markers was detected in DCRΔ/Δ EBs, while DCRneo/+ and DCRneo/neo EBs exhibited normal expression profiles for these genes (Fig. 2E). Interestingly, oct-4 expression, which progressively decreased in the control EBs, only partially decreased in mutant EBs after day 5 of differentiation (Fig. 2F).
DCRΔ/Δ ES cells express high levels of centromeric repeat sequence RNA
RNAi-mediated heterochromatic silencing has been demonstrated at S. pombe centromeres. If RNAi-guided heterochromatin formation occurs in mammals, mouse centromeres are potential targets. These units are composed of tandem repeats of specific DNA sequences (minor and major satellite repeats) that are rich in methylated DNA residues and heterochromatin-associated histone modifications, and appear to be transcriptionally repressed (Lehnertz et al. 2003).
As a first step to investigate the effect of Dicer ablation on these heterochromatic structures, the abundance of transcripts encoded by centromeric and pericentric repeats was measured in DCRΔ/Δ and heterozygous control cells. Real-time RT–PCR analysis for pericentric (major satellite) repeats and semiquantitative RT–PCR for the centromeric (minor satellite) repeats revealed that transcripts derived from both of these elements were more abundant in DCRΔ/Δ cells than in heterozygous controls (Fig. 3A,B). These results were also confirmed by Northern analysis (Fig. 3E; Supplementary Figs. S3, S6). Trichostatin A (TSA) treatment of the cells led to increased levels of major satellite transcripts, in both DCRneo/+ and DCRΔ/Δ cells (Fig. 3A). Transcripts derived from other repetitive sequences, such as L1 and intracisternal A particle (IAP) retrotransposons, were also more readily detected in the absence of Dicer (Supplementary Fig. S2).
Figure 3.
Analyses of satellite repeat transcripts in ES cells in the presence or absence of Dicer. (A) Quantitative real-time RT–PCR was performed to determine the relative abundance of transcripts from pericentric major satellite repeats in control (Dcrneo/+, gray bars) and mutant (DcrΔ/Δ, black bars) ES cells. Total RNA samples were treated with DNase I prior to reverse transcription. Plotted are values (in arbitrary units) for abundance of major satellite transcripts normalized to levels of hprt transcripts, a housekeeping gene. ES cells were either untreated or treated with increasing concentrations of the histone deactylase inhibitor, TSA. (B) Semiquantitative RT–PCR was performed to determine the relative abundance of transcripts from centromeric minor satellite repeats in control (neo/+ and neo/neo) and mutant (Δ/Δ) ES cells. Digital photographs (negative image) of agarose gel analyses of the RT–PCR products for minor satellite and β-actin transcripts are shown. Total RNA samples were treated with DNase I prior to reverse transcription. Equivalent amounts of reverse-transcribed RNA were used for each PCR reaction as determined by the abundance of hprt transcripts, which was obtained by quantitative real-time RT–PCR (data not shown). (C) Total RNA from wild-type ES cells was either untreated (–) or treated (+) with RNase ONE and resolved in a denaturing 15% acrylamide gel. Shown in the left panel is the gel stained with ethidium bromide (EtBr). In the right panel, the gel was blotted and probed with a radiolabeled probe specific for minor satellite repeats (pMR150). A radiolabeled Decade marker (M) consisting of 20-, 30-, 40-, 50-, 60-, 70-, 80-, 90-, 100-, and 150-nt RNA species is included. (D) Total RNA was size fractionated using mirVana glass fiber filters and equivalent amounts of the small RNA fraction (<200 nt) were resolved in a denaturing 15% acrylamide gel. Shown in the left panel is the gel stained with EtBr loaded with short RNAs from wild-type (neo/+) or mutant (Δ/Δ) ES cells. In the right panel, the gel was used for a Northern blot hybridized with a probe specific for minor satellite repeats (pMR150). A radiolabeled RNA Decade ladder (M) is also included. (E) Total RNA was size fractionated using mirVana glass fiber filters, and equivalent amounts of the long RNA fraction (>200 nt) were resolved in a denaturing 10% acrylamide gel and analyzed by Northern blot as in C and D. The EtBr-stained gel is shown below.
Earlier studies have shown that noncoding poly-adenylated RNA transcripts arise from both strands of centromeric satellite repeats in mouse (Rudert et al. 1995). Conceivably, these RNA transcripts form double-stranded structures that can become subject to cleavage by Dicer. As in the case of S. pombe, the resulting siRNAs could mediate TGS of homologous DNA sequences. To ascertain whether the RNA transcripts from major and minor satellites can, indeed, form dsRNA species, total RNA from DCRneo/+ cells was treated with RNase ONE, which digests single-stranded RNA while leaving double-stranded RNA intact. After RNase ONE digestion, total RNA was largely degraded as visualized by ethidium bromide staining (Fig. 3C). Northern blot analysis of the same gel, however, using a probe specific for the minor satellite repeat, revealed that most of the specific signal was still present after the RNase ONE treatment. Similar results were obtained using major satellite-specific oligonucleotide probes (Supplementary Fig. S3). Interestingly, RNA species of diverse sizes were identified by Northern blot (Fig. 3C,D), with the predominant signals detected at ∼150 nt and 25–30 nt. The presence of the 25–30-nt species suggests that these RNA molecules could be generated by Dicer processing and, if so, might be analogous to the small heterochromatic siRNAs cloned from S. pombe (Reinhart and Bartel 2002). Indeed, small RNA species (<200 nt) homologous to minor and major satellite repeats were greatly reduced in DCRΔ/Δ cells, and no signal could be detected in the 25–30-nt range (Fig. 3D; Supplementary Fig. S3). In contrast, and consistent with the RT–PCR data, the fraction of large RNAs (>200 nt) isolated from the same cells was greatly enriched in minor and major satellite transcripts (Fig. 3E; Supplementary Fig. S3).
In summary, it appears that transcripts from different heterochromatic regions are more abundant in DCRΔ/Δ cells than in control cells. A significant fraction of these transcripts are double-stranded and, in the absence of Dicer, double-stranded centromeric RNA transcripts cannot be processed into siRNA-like species.
Defects in TGS, DNA methylation, and histone modifications
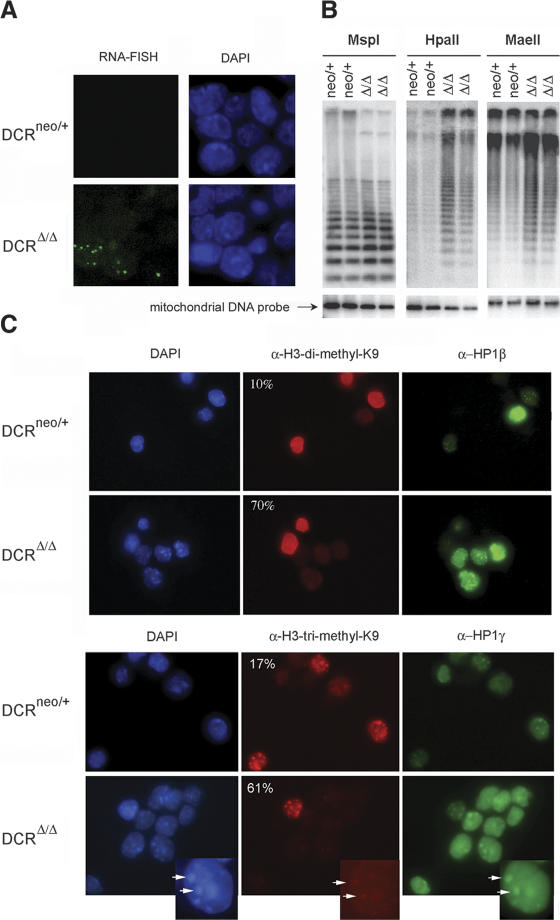
To further investigate whether the accumulation of minor satellite repeat transcripts in DCRΔ/Δ cells could be due to increased transcription, fluorescent in situ hybridization (FISH) was performed in an attempt to detect nascent centromeric transcripts. RNA FISH can detect newly synthesized RNA that is still attached to chromosomes as it is being transcribed. If centromeric repeat sequences are not transcriptionally silenced in the absence of Dicer, one would expect RNA FISH signals associated with centromeres to be enhanced in DCRΔ/Δ cells. Indeed, ∼20% of DCRΔ/Δ cells displayed strong, punctate nuclear FISH signals as opposed to heterozygous control cells, in which no signal could be detected (Fig. 4A). RNase treatment of the cells prior to the hybridization abolished the signal (Supplementary Fig. S4).
Figure 4.
Evidence for transcriptional derepression of centromeric repeats. (A) RNA FISH using a centromeric DNA probe was performed on DCRneo/+ and DCRΔ/Δ ES cells. The respective DAPI nuclear staining is shown next to each panel (63×). (B) DNA methylation analysis of genomic DNA derived from two DCRneo/+ and two DCRΔ/Δ clones. Equivalent amounts of DNA were digested with MspI, HpaII, or MaeII. (Lower panel) EtBr staining of the gel (data not shown) and rehybridization with a mitochondrial DNA probe demonstrate equivalent loading. MspI cleaves irrespective of whether the CpG dinucleotide within the cognate restriction site is methylated, whereas HpaII or MaeII will not. Gels were blotted on a nitrocellulose membrane and hybridized with a probe specific for the minor satellite repeat. (C) Histone modifications and HP1 protein localization are altered in Dicer-deficient cells. DCRneo/+ and DCRΔ/Δ cells were stained either with a rabbit polyclonal antibody specific for dimethyl-H3K9 and a monoclonal antibody against HP1β (upper panels) or a polyclonal antibody specific for trimethyl-H3K9 and a monoclonal antibody against HP1γ (lower panels). DAPI staining of nuclei for each panel is shown on the left (63× magnification). Percentages of cells unstained or dim for variously methylated H3K9 species are indicated on the top left corner of each panel. At least 200 cells from each clone were counted. Inset figures point to an example of the colocalization of HP-1γ, trimethyl H3K9 staining, and centromeric heterochromatin as reflected by the existence of DAPI dense spots.
Since we cannot exclude that the observed FISH signal may reflect lack of post-transcriptional processing of satellite transcripts by Dicer, we analyzed the DNA methylation status of centromeric repeats and associated histone modifications.
For the DNA methylation analysis, equivalent amounts of DNA were digested either with the methylation-sensitive enzyme HpaII, or its methylation-insensitive isoschizomer MspI. MaeII, another methylation-sensitive restriction enzyme that recognizes a different consensus sequence from MspI/HpaII, was also used in this analysis. Digested DNA was resolved by gel electrophoresis, and gels were blotted and hybridized with a minor satellite-specific probe. Genomic DNA from DCRΔ/Δ cells was more readily digested by both HpaII and MaeII, suggesting that it is significantly less methylated than DNA isolated from the DCRneo/+ controls. In contrast, all genomic DNA samples were equally digested by MspI, showing that equivalent amounts of DNA were digested in this experiment (Fig. 4B). Reprobing of the membranes with a mitochondrial DNA probe was performed to verify equal loading (Lei et al. 1996).
DNA methylation in mammals is linked to specific histone modifications (Lehnertz et al. 2003), and these modifications are impaired in RNAi mutants of S. pombe and Drosophila. Therefore, we asked whether heterochromatic histone modifications were also impaired in Dicer-deficient cells. Specifically, dimethylation and trimethylation of histone H3 at Lys 9 (di-H3mK9 and tri-H3mK9, respectively) are enriched at centromeric repeats (Lehnertz et al. 2003; Peters et al. 2003). Immunofluorescence staining for di-H3mK9 and tri-H3mK9 revealed that a large percentage of DCRΔ/Δ cells (∼70% and 62%, respectively) had significantly reduced staining for both modified histone H3 species, compared to the DCRneo/+ control cells (Fig. 4C). Mono-methyl H3-K9 staining was identical between DCRneo/+ and DCRΔ/Δ cells (data not shown). Interestingly, there was an inverse correlation between H3-mK9 levels and the staining of certain HP1 proteins; the staining for HP1β and γ appeared brighter and more diffuse in cells with low H3mK9 staining. Total HP1γ protein levels, however, were not significantly different between DCRneo/+ and DCRΔ/Δ cells as assessed by Western blot analysis (data not shown).
One would expect that the described centromeric heterochromatin defect would lead to an increased percentage of cells with aneuploidy. However, no significant aberrations in chromosome numbers or structure (as assessed by spectral karyotyping analysis) were seen in DCRΔ/Δ cells, compared to heterozygous and neo/neo controls (Supplementary Fig. S5).
These data demonstrate that chromatin modifications and the DNA methylation of centromeric repeats are altered in the absence of a functional RNAi machinery, suggesting that, as in S. pombe and Arabidopsis, mammalian Dicer is required for proper heterochromatin formation in certain regions of the mammalian genome.
Dicer-deficient ES cells can be rescued by Dicer alone
In order to determine whether the phenotypes we observed in the DCRΔ/Δ ES cells were solely the result of Dicer-deficiency and not an inadvertent secondary mutation that could have occurred during the generation of these cells, we reintroduced Dicer into these cells by homologous recombination (DCR rec. cells). Using the same conditional gene-targeting vector previously used for generating the DCRneo allele, we were able to correct one allele in DCRΔ/Δ ES cells such that their genotype became DCRneo/Δ. Several Dicer-reconstituted clones were isolated, and proper gene targeting was confirmed by Southern blot analysis (Supplementary Fig. S6).
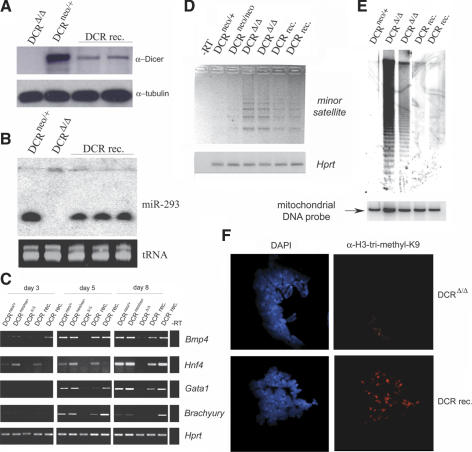
Dicer protein expression in the reconstituted clones was verified by Western blot analysis. As expected, lower levels of Dicer protein were detected in these cells, compared to the DCRneo/+ control, since only one dcr-1 allele was restored (Fig. 5A; Supplementary Fig. S6A). To determine whether this level of Dicer expression was functional, we analyzed the ability of Dicer-reconstituted cells to produce mature miRNAs. Northern blot analysis using a probe specific for miR-293 showed that miRNAs were fully processed in the Dicer-reconstituted clones (Fig. 5B).
Figure 5.
Reconstitution of Dicer expression and function in DCRΔ/Δ ES cells. (A) Western blot of extracts prepared from a homozygous (Δ/Δ) clone, a heterozygous (neo/+) ES cell clone, and two independent Dicer-reconstituted clones (DCR rec.). (Bottom) The Western blot was probed for Dicer protein using antisera (ab1414) and then stripped and reprobed for tubulin. (B) Northern analysis of miR-293 expression in total RNA from a control ES cell clone (neo/+), a Dcr-null clone (Δ/Δ), and Dicer-reconstituted clones performed as in Figure 1E. (Bottom) To demonstrate equal RNA loading, a segment of the EtBr-stained gel is shown. (C) Semiquantitative RT–PCR analyses of mesoderm- and ectoderm-specific differentiation markers. RNA was extracted and reverse-transcribed from EBs generated from DCRneo/+, DCRneo/neo, and two different DCRΔ/Δ clones at day 3, 5, and 8 of differentiation and analyzed for expression of differentiation markers bmp4, hnf4, gata1, and brachyury. Hprt transcripts were amplified as a loading control. (D) Semiquantitative RT–PCR of transcripts from centromeric minor satellite repeats in control (DCRneo/+ and DCRneo/neo), two mutants (DCRΔ/Δ), and Dicer-reconstituted (DCR rec.) ES cells. Digital photographs (negative image) of agarose gel analyses of the RT–PCR products for minor satellite and hprt transcripts are shown. Total RNA samples were treated with DNase I prior to reverse transcription. (E) DNA methylation analysis of genomic DNA derived from DCRneo/+, two DCRΔ/Δ, and two Dicer-reconstituted (DCR rec.) clones. Equivalent amounts of DNA were digested with HpaII and resolved in a 1% agarose gel. A Southern blot was performed with a probe specific for the minor satellite repeat (pMR150). The same blot was stripped and rehybridized with a mitochondrial DNA probe to demonstrate equal DNA loading. (F, right panels) Metaphase chromosomes from DCRΔ/Δ and Dicer-reconstituted cells were stained with a rabbit polyclonal antibody specific for trimethyl-H3K9. DAPI staining of chromosomes for each panel is shown on the left (100× magnification).
In order to investigate whether Dicer reconstitution also rescued the defect in differentiation, reconstituted ES cells were differentiated into EBs, as previously described. RT–PCR was performed to detect expression of differentiation markers in EBs at different time points (Fig. 5C). These markers could be detected in EBs generated from Dicer-reconstituted clones at levels comparable to those in DCRneo/+ and DCRneo/neo controls. Differentiation upon Dicer reconstitution was also evidenced by H&E staining of histological preparations (Supplementary Fig. S6B).
Dicer reconstitution also appeared to reinstate gene silencing of centromeric DNA, as indicated by the results of semiquantitative RT–PCR of minor satellite repeat transcripts (Fig. 5D). Although silencing of minor satellite repeat expression was not complete, lower levels of transcripts were detected in Dicer-reconstituted cells. This was further confirmed by Northern blot analysis for minor satellite transcripts (Supplementary Fig. S6C). To determine whether centromeric DNA methylation was similarly restored, equivalent amounts of genomic DNA were digested with the methylation-sensitive enzyme HpaII and analyzed by Southern blot as previously described (Fig. 5E). The same Southern blot was stripped and reprobed for mitochondrial DNA to test for equal loading. Methylation of minor satellite repeats also appears to be restored in Dicer-reconstituted clones.
Finally, trimethyl-H3K9 localization on metaphase chromosomes was determined using immunofluorescence microscopy (Fig. 5F). A large percentage (60%, 31/51 metaphases) of the chromosomes from DCRΔ/Δ cells displayed significantly reduced or diffuse staining for trimethyl-H3K9. In contrast, the majority of chromosomes (90%, 59/66 metaphases) from Dicer-reconstituted cells stained for trimethyl-H3K9 and the staining colocalized with DAPI-dense centromeres (Fig. 5F). Spreads from both clones stained comparably well for HP1γ (Supplementary Fig. S7). In summary, these results indicate that the phenotypes observed in DCRΔ/Δ ES cells are, indeed, specific to Dicer-deficiency since they could be rescued by Dicer re-expression.
Discussion
Murine Dicer is essential for development and stem cell maintenance in the embryo (Bernstein et al. 2003). Using a conditional gene-targeting approach, Dicer-deficient ES cells were generated. The targeting approach used yielded a functional Dicer knockout, since no Dicer protein or activity could be detected in DCRΔ/Δ cells. Analysis of these cells revealed interesting aspects of RNAi-mediated control of cellular differentiation and epigenetic silencing in mammals. The relevant phenotypes in the knockout cells were specific to Dicer-deficiency, since they were rescued upon Dicer re-expression.
Expression of the mature form of ES cell-specific miRNAs was abolished in DCRΔ/Δ cells, but was restored in Dicer-reconstituted clones. In addition, DCRΔ/Δ cells were unable to perform shRNA-mediated RNAi of a reporter gene. These observations are comparable to the phenotype of Dicer inactivation in other organisms (Grishok et al. 2001; Ketting et al. 2001), as well as to the results of functional analysis of Dicer activity in mammalian cells by knock-down experiments (Hutvagner et al. 2001; Paddison et al. 2002b; Lee et al. 2003).
PTGS mediated by synthetic siRNAs, however, was not affected in the absence of Dicer. Similarly, immunodepletion of Dicer from mammalian cell extracts did not compromise RISC activity in vitro (Martinez et al. 2002). In keeping with these findings, we present in vivo evidence, showing that, contrary to what was observed in Drosophila (Lee et al. 2004; Pham et al. 2004), there is only one mammalian Dicer and this protein is not required for RISC function.
Surprisingly, after Dicer ablation, ES cells retained their traditional colony-forming morphology and expressed ES cell-specific genes. At first glance, these findings appear inconsistent with published work in which Dicer-deficient ES cells could not be obtained (Bernstein et al. 2003). However, this discrepancy may only reflect differences in experimental procedures. Using a conditional gene-targeting approach, we generated a DCRneo allele that is functionally intact. Thus, we were able to apply stringent selection pressure (high G418 concentration) to promote the generation and survival of homozygous DCRneo/neo (Dicer-proficient) cells, from which Dicer-deficient cells could be efficiently generated in a second Cre-mediated step. In contrast, Bernstein et al. (2003) attempted to select for rare Dicer-deficient mutants among an overwhelming majority of Dicer-proficient cells and may not have recovered the former because of their competitive growth disadvantage. Indeed, DCRΔ/Δ cells proliferated more slowly than either DCRneo/+ or DCRneo/neo cells. Clearly, our experiments do not address whether Dicer is required for ES cell maintenance in vivo or for derivation of ES cells from the embryonic epiblast (Bernstein et al. 2003). Instead, the data show that Dicer is not required for ES cell viability ex vivo.
In vivo, Dicer is required for proper development of a mouse embryo (Bernstein et al. 2003). Consistent with this finding, the Dicer-deficient ES cells that were generated failed to differentiate in multiple assays. In particular, DicerΔ/Δ ES cells formed EB-like structures but showed no molecular or histological evidence of differentiation. This differentiation defect was rescued in Dicer-reconstituted clones.
Curiously, oct-4 transcript levels remained elevated in DCRΔ/Δ EBs. It is possible that the differentiation defects observed in Dicer-null cells reflect inability of these cells to down-regulate the expression of this, and possibly other, related stem cell-specific transcription factors. Transcription factors and Hox genes are prime targets for miRNA regulation (Lewis et al. 2003; Yekta et al. 2004). During ES cell differentiation, the miRNA expression profile changes (Houbaviy et al. 2003), and one or more newly expressed miRNAs could be involved in the regulation of transcription factors critical for cellular differentiation. In addition, one cannot exclude that the differentiation defect of DCRΔ/Δ cells reflects an inability of these cells to perform proper methylation and heterochromatinization of key regulatory genes. DNA methyltransferases (Dnmts) are important for ES cell differentiation (Jackson et al. 2004). During development, a wave of de novo methylation is critical for proper differentiation (for review, see Li 2002) and, as further discussed below, we present evidence that related processes are impaired in DCRΔ/Δ cells.
Heterochromatin formation depends on the RNAi machinery in S. pombe, Tetrahymena, and Drosophila, and links between DNA methylation and RNAi have been reported in plants (for review, see Schramke and Allshire 2004). However, the original premise was that the RNAi pathway is mainly involved in PTGS. When Fire et al. (1998) initially reported on the mechanism of RNAi in C. elegans, they noted that promoter and intronic sequences were ineffective in dsRNA-mediated gene silencing, thus reasoning that this process occurred post-transcriptionally. While there are examples of RNA-guided chromatin modifications in various organisms, they have, to date, been considered an oddity of the relevant host organism and not a general mechanism of epigenetic control. In fact, a recent report demonstrated that DNA methylation in Neurospora is independent of the RNAi machinery (Freitag et al. 2004). Here we present evidence of a link between the RNAi pathway and DNA methylation of an endogenous sequence in animals, raising the possibility that RNA-based TGS is a general event in higher eukaryotic gene regulation.
Specifically, the data suggest that ablation of Dicer, a central molecule in the RNAi pathway, leads to derepression of normally silenced genetic elements, such as transposons and centromeric heterochromatin. An RNA component seems to be involved in heterochromatin formation in mammalian cells, but the specific nature of this RNA component and its possible link to the RNAi machinery have not been established (Maison et al. 2002). The present experiments directly address this issue. Centromeric repeat sequence-derived transcripts were up-regulated in DCRΔ/Δ cells, suggesting that these regions escape transcriptional gene silencing in the absence of Dicer. However, Northern blot analyses showed that Dicer extensively processes the largely double-stranded RNA derived from centromeric repeats into smaller RNAs, ranging from 25 to 150 nt. While the apparent lack of centromeric region silencing might reflect failure of DCRΔ/Δ cells to efficiently convert primary transcripts into smaller dsRNA species, the loss of heterochromatin-related modifications in the centromeric regions of the mutants suggests another possibility. Conceivably, the 25–30-nt species, which are absent in the Dicer-deficient cells, could be analogous to the small, centromeric heterochromatin-encoded RNAs cloned from S. pombe (Reinhart and Bartel 2002). If so, one might hypothesize that these RNAs are incorporated into a mammalian RITS complex and function as guides for TGS of homologous genomic sequences.
One potential concern is that these RNAs do not correspond to the canonical 21-nt-long RNAs that are usually generated by Dicer cleavage, nor have any centromeric repeat small RNAs been cloned from mice (Lagos-Quintana et al. 2002). The abundance of these centromeric siRNAs may be very low, since in wild-type cells these regions are normally silenced. Furthermore, since our probe should, in principle, detect all small RNAs generated by Dicer cleavage of long dsRNA from centromeric transcripts, a more diffuse signal compared to the expected 21-nt-long RNA is not surprising. The precise sequence and length of these siRNAs remain to be determined by cloning.
Dicer-deficiency and inefficient processing of centromeric repeat transcripts correlate with defects in methylation of centromeric DNA and histone H3. Typically, centromeric satellite repeats are highly methylated and enriched in di- and trimethylated histone H3-K9. These modifications are dependent on Dnmts and Suv39h methyltransferases (DNA and histone methyltransferases, respectively) (Lei et al. 1996; Okano et al. 1999; Lehnertz et al. 2003; Peters et al. 2003). We have presented evidence that they also require an intact RNAi pathway: (1) In DCRΔ/Δ cells, DNA methylation was partially abolished at the minor satellite repeats, similar to dnmt3a–/–3b–/– ES cells (Chen et al. 2003) and (2) histone di- and trimethylation were reduced in a large percentage of the cells. Similarly, when metaphase chromosomes from DCRΔ/Δ cells were stained for trimethyl H3K9, most of them displayed a reduced signal compared to their Dicer-reconstituted counterparts. Interestingly, some cells appeared to retain proper di- and trimethyl H3K9 localization in the absence of Dicer, suggesting that loss of the methyl marks occurs progressively over time. Treatment of cells with histone deacetylase inhibitors, such as TSA, might result in a more pronounced or faster-appearing phenotype. Unfortunately, DCRΔ/Δ cells were more sensitive to TSA treatment than DCRneo/+ cells, and it was not possible to culture these cells in TSA for prolonged periods of time (data not shown).
An inverse correlation of HP1β and HP1γ staining with dimethyl- and trimethyl-H3K9, respectively, was noted. Mutation of components of the RNAi pathway in Drosophila led to loss of H3mK9 methylation and delocalization of the HP proteins (Pal-Bhadra et al. 2004) as did Dicer inactivation in DT40 cells (Fukagawa et al. 2004). The loss of proper H3K9 methylation at centromeric repeats due to Dicer-deficiency might result in delocalization of the HP1 proteins from centromeres.
The defective centromeric chromatin structure of Suv39h dn mutant mice resulted in increased genomic instability and aneuploidy in MEFs and lymphoid cells (Peters et al. 2001). However, Suv39h dn mutant ES cells were not similarly affected (Peters et al. 2001; Garcia-Cao et al. 2004). DCRΔ/Δ cells also showed no evidence of aneuploidy or genomic instability, suggesting that ES cells use different mechanisms for maintaining genomic stability than differentiated cells (Burdon et al. 2002). This possibility might also explain the fact that Dicerdeficient ES cells are viable, while DT40 Dicer mutants displayed severe growth arrest (Fukagawa et al. 2004). Therefore, the role of mammalian Dicer and the RNAi pathway in genome integrity control needs to be investigated in differentiated cells.
Thus, we have presented evidence indicating that the mammalian RNAi pathway is essential for differentiation and chromatin structure maintenance. A single dicer gene, on mouse Chromosome 12, was maintained during mammalian evolution to mediate post-transcriptional, translational, and transcriptional gene silencing. We have observed one evolutionarily conserved function of mouse Dicer: silencing of centromeric heterochromatin. Dicer-dependent regulation of other aspects of mammalian epigenetic control, such as X-chromosome inactivation, imprinting, and transposon silencing are attractive topics for further investigation.
Materials and methods
Gene targeting and cell culture
Bruce-4 ES cells derived from C57BL/6 mice were cultured, transfected with the gene-targeting vector depicted in Figure 1A, and homologous recombinants were isolated using protocols from our laboratory (Casola 2004). In order to obtain homozygous DCRneo/neo ES cells, heterozygous DCRneo/+ clones were cultured in increasing concentrations of the neomycin analog G418, as previously described (Mortensen et al. 1992). Deletion of the DCRneo allele was achieved by infection with an adenovirus encoding Cre recombinase (Harvard Gene Therapy Initiative), and deletion was verified by Southern blotting. To reconstitute Dicer expression, DCRΔ/Δ ES were transfected with the gene-targeting vector depicted in Figure 1A, and selected with G418 as previously described.
The lentiviral shRNA expression vector FSIPPW was created from the FUGW self-inactivating lentiviral vector (Lois et al. 2002) by replacing the ubiquitin C promoter and eGFP cassette with an optimized human H1 promoter to drive shRNA expression in tandem with the PGK promoter driving expression of the puromycin acetyltransferase gene. The shRNA expression vector targeting eGFP (FSIPPW-eGFP) targets the sequence 5′-AAGAACGGCATCAAGGTGAACTT-3′. Lentiviruses were packaged and pseudotyped with the vesicular stomatitis virus glycoprotein (VSV-G) as previously described (Lois et al. 2002). ES cells transduced with lentivirus were selected with 1 μg/mL of puromycin (Sigma) for 4 d prior to FACS analysis. GFP-specific siRNA was purchased from Dharmacon, and ES cell transfection was performed using Lipofectamine 2000 reagent (Invitrogen).
EB differentiation and teratoma formation
ES cells were trypsinized and resuspended in differentiation medium without LIF (DMEM, 15% FBS, penicillin, streptomycin, nonessential amino acid). Feeders were removed by selective adherence to plastic dishes for 2 h. ES cell numbers were adjusted to a final concentration of 105 cells/mL and plated into low adherence dishes (Costar, Ultra Low Cluster). EBs were fixed in Bouin's fixative (Sigma) for histology (Harvard Histology Core Facility), or lysed in Trizol (Invitrogen), and RNA was prepared according to the manufacturer's instructions. For the teratoma formation assay ∼5 × 106 cells were injected subcutaneously into the back of nude mice (five mice per ES clone). Teratomas were measured weekly. Animals were sacrificed and tumors were harvested on week 3 post-injection.
Western analysis
Antisera from rabbits immunized with KLH-coupled peptides (Dana-Farber Molecular Biology Core Facilities) corresponding to residues 1385–1405 (ab1416) or 385–405 (ab1414) of mouse Dicer were used for Western blot analyses. Cytoplasmic lysate from ES cells cultured on gelatinized plates without MEFs was prepared by lysis in hypotonic buffer (containing protease inhibitors), and 10 μg of each lysate was loaded per lane. One microliter of recombinant Dicer protein (Stratagene) was used as a control.
RNA and DNA analyses
RNA from ES cells and EBs was isolated using Trizol reagent (Invitrogen), and miRNA Northern blots were performed according to standard procedures. In order to enrich for RNA smaller than 200 nt, a mirVana RNA isolation kit (Ambion) was used according to the manufacturer's instructions. Primers for major and minor satellite repeat RT–PCR were previously described (Lehnertz et al. 2003), while the sequence of the actin primers was 5′-CCTAAGGCCAACCGTGAAAAG-3′ and 5′-TCTTCATGGTGCTAGGAGCCA-3′. For quantitative RT–PCR, we used the following primers to amplify hprt: 5′-TGAAGAGC TACTGTAATGATCAGTCAAC-3′ and 5′-AGCAAGCTTGC AACCTTAACCA-3′. A PstI fragment from pMR150 was used as probe for Southern and Northern blots of the minor satellite repeats (Chapman et al. 1984). The following oligonucleotides were used to probe Northern blots for major satellite repeats: 5′-AATGTCCACTGTAGGACGTGGAATATGGCA-3′ and 5′-CACGGAAAATGAGAAATACACACTTTAGGATGTG-3′. oct-4-and α6-integrin-specific primers as well as the primers for amplification of EB differentiation markers and hprt have been described elsewhere (Di Cristofano et al. 1998; Houbaviy et al. 2003).
Immunofluorescence and RNA FISH
Indirect immunofluorescence was performed as previously described (Lehnertz et al. 2003). Briefly, ES cells were allowed to adhere on positively charged slides, fixed in paraformaldehyde, and stained with different antibodies. The mono-, di-, and tri-H3mK9 antibodies were prepared in T. Jenuwein's laboratory. Monoclonal anti-HP1β and γ antibodies were purchased from Chemicon. RNA FISH was performed using directly labeled FITC pan-centromeric probe (Cambio) as previously described (Ganesan et al. 2002). Metaphase spreads for IF staining were prepared as previously described (Guenatri et al. 2004) with the exception that cells were fixed in paraformaldehyde after centrifugation.
Karyotypic analysis
Karyotypes were analyzed in the CBR Institute for Biomedical Research SKY Core Facility as previously described (Mills et al. 2004). For assessment of chromosome numbers and centromere structure, 50–100 metaphase spreads from each clone were analyzed.
Acknowledgments
We thank S. Casola and S. Willms for the plasmid pEASY-FLIRTneo, P. Oberdoerffer and Y. Sasaki for pCAGGS-GFP, D. Baltimore for pFUGW, C. Beard and R. Jaenisch for pMR150, T. Chen and E. Li for the mitochondrial DNA probe, and W. Filipowicz for generously sharing antibody reagents. We thank K. Mills and F. Alt for performing SKY analyses and help with karyotypic analysis, C. Marrese for help with the RNA FISH, R. Wright for the teratoma injections, and R. Bronson for the EB histology. We thank S. Casola, S. Frietz, D. Li, R. Mostoslavsky, P. Oberdoerffer, V. Rebel, Y. Sasaki, M. Schlissel, M. Schmidt-Supprian, and T. Tuschl for advice and/or critical reading of the manuscript. S.A.M. is supported by a Ruth L. Kirschstein NRSA post-doctoral fellowship. This work was supported by grants from the National Cancer Institute and National Institutes of Health.
Supplemental material is available at http://www.genesdev.org.
Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.1248505.
Corresponding authors.
References
- Aravin A.A., Naumova, N.M., Tulin, A.V., Vagin, V.V., Rozovsky, Y.M., and Gvozdev, V.A. 2001. Double-stranded RNA-mediated silencing of genomic tandem repeats and transposable elements in the D. melanogaster germline. Curr. Biol. 11: 1017–1027. [DOI] [PubMed] [Google Scholar]
- Aravin A.A., Lagos-Quintana, M., Yalcin, A., Zavolan, M., Marks, D., Snyder, B., Gaasterland, T., Meyer, J., and Tuschl, T. 2003. The small RNA profile during Drosophila melanogaster development. Dev. Cell 5: 337–350. [DOI] [PubMed] [Google Scholar]
- Bernstein E., Caudy, A.A., Hammond, S.M., and Hannon, G.J. 2001. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409: 363–366. [DOI] [PubMed] [Google Scholar]
- Bernstein E., Kim, S.Y., Carmell, M.A., Murchison, E.P., Alcorn, H., Li, M.Z., Mills, A.A., Elledge, S.J., Anderson, K.V., and Hannon, G.J. 2003. Dicer is essential for mouse development. Nat. Genet. 35: 215–217. [DOI] [PubMed] [Google Scholar]
- Burdon T., Smith, A., and Savatier, P. 2002. Signalling, cell cycle and pluripotency in embryonic stem cells. Trends Cell Biol. 12: 432–438. [DOI] [PubMed] [Google Scholar]
- Carmell M.A. and Hannon, G.J. 2004. RNase III enzymes and the initiation of gene silencing. Nat. Struct. Mol. Biol. 11: 214–218. [DOI] [PubMed] [Google Scholar]
- Casola S. 2004. Conditional gene mutagenesis in B-lineage cells. Methods Mol. Biol. 271: 91–109. [DOI] [PubMed] [Google Scholar]
- Catalanotto C., Pallotta, M., ReFalo, P., Sachs, M.S., Vayssie, L., Macino, G., and Cogoni, C. 2004. Redundancy of the two dicer genes in transgene-induced posttranscriptional gene silencing in Neurospora crassa. Mol. Cell. Biol. 24: 2536–2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan S.W., Zilberman, D., Xie, Z., Johansen, L.K., Carrington, J.C., and Jacobsen, S.E. 2004. RNA silencing genes control de novo DNA methylation. Science 303: 1336. [DOI] [PubMed] [Google Scholar]
- Chapman V., Forrester, L., Sanford, J., Hastie, N., and Rossant, J. 1984. Cell lineage-specific undermethylation of mouse repetitive DNA. Nature 307: 284–286. [DOI] [PubMed] [Google Scholar]
- Chen T., Ueda, Y., Dodge, J.E., Wang, Z., and Li, E. 2003. Establishment and maintenance of genomic methylation patterns in mouse embryonic stem cells by Dnmt3a and Dnmt3b. Mol. Cell. Biol. 23: 5594–5605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Cristofano A., Pesce, B., Cordon-Cardo, C., and Pandolfi, P.P. 1998. Pten is essential for embryonic development and tumour suppression. Nat. Genet. 19: 348–355. [DOI] [PubMed] [Google Scholar]
- Duncan S.A., Manova, K., Chen, W.S., Hoodless, P., Weinstein, D.C., Bachvarova, R.F., and Darnell Jr., J.E. 1994. Expression of transcription factor HNF-4 in the extraembryonic endoderm, gut, and nephrogenic tissue of the developing mouse embryo: HNF-4 is a marker for primary endoderm in the implanting blastocyst. Proc. Natl. Acad. Sci. 91: 7598–7602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elefanty A.G., Robb, L., Birner, R., and Begley, C.G. 1997. Hematopoietic-specific genes are not induced during in vitro differentiation of scl-null embryonic stem cells. Blood 90: 1435–1447. [PubMed] [Google Scholar]
- Finnegan E.J., Margis, R., and Waterhouse, P.M. 2003. Posttranscriptional gene silencing is not compromised in the Arabidopsis CARPEL FACTORY (DICER-LIKE1) mutant, a homolog of Dicer-1 from Drosophila. Curr. Biol. 13: 236–240. [DOI] [PubMed] [Google Scholar]
- Fire A., Xu, S., Montgomery, M.K., Kostas, S.A., Driver, S.E., and Mello, C.C. 1998. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391: 806–811. [DOI] [PubMed] [Google Scholar]
- Freitag M., Lee, D.W., Kothe, G.O., Pratt, R.J., Aramayo, R., and Selker, E.U. 2004. DNA methylation is independent of RNA interference in Neurospora. Science 304: 1939. [DOI] [PubMed] [Google Scholar]
- Fukagawa T., Nogami, M., Yoshikawa, M., Ikeno, M., Okazaki, T., Takami, Y., Nakayama, T., and Oshimura, M. 2004. Dicer is essential for formation of the heterochromatin structure in vertebrate cells. Nat. Cell Biol. 6: 784–791. [DOI] [PubMed] [Google Scholar]
- Ganesan S., Silver, D.P., Greenberg, R.A., Avni, D., Drapkin, R., Miron, A., Mok, S.C., Randrianarison, V., Brodie, S., Salstrom, J., et al. 2002. BRCA1 supports XIST RNA concentration on the inactive X chromosome. Cell 111: 393–405. [DOI] [PubMed] [Google Scholar]
- Garcia-Cao M., O'Sullivan, R., Peters, A.H., Jenuwein, T., and Blasco, M.A. 2004. Epigenetic regulation of telomere length in mammalian cells by the Suv39h1 and Suv39h2 histone methyltransferases. Nat. Genet. 36: 94–99. [DOI] [PubMed] [Google Scholar]
- Grishok A., Pasquinelli, A.E., Conte, D., Li, N., Parrish, S., Ha, I., Baillie, D.L., Fire, A., Ruvkun, G., and Mello, C.C. 2001. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell 106: 23–34. [DOI] [PubMed] [Google Scholar]
- Guenatri M., Bailly, D., Maison, C., and Almouzni, G. 2004. Mouse centric and pericentric satellite repeats form distinct functional heterochromatin. J. Cell Biol. 166: 493–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall I.M., Shankaranarayana, G.D., Noma, K., Ayoub, N., Cohen, A., and Grewal, S.I. 2002. Establishment and maintenance of a heterochromatin domain. Science 297: 2232–2237. [DOI] [PubMed] [Google Scholar]
- Hamilton A., Voinnet, O., Chappell, L., and Baulcombe, D. 2002. Two classes of short interfering RNA in RNA silencing. EMBO J. 21: 4671–4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houbaviy H.B., Murray, M.F., and Sharp, P.A. 2003. Embryonic stem cell-specific microRNAs. Dev. Cell 5: 351–358. [DOI] [PubMed] [Google Scholar]
- Hutvagner G., McLachlan, J., Pasquinelli, A.E., Balint, E., Tuschl, T., and Zamore, P.D. 2001. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science 293: 834–838. [DOI] [PubMed] [Google Scholar]
- Jackson J.P., Lindroth, A.M., Cao, X., and Jacobsen, S.E. 2002. Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature 416: 556–560. [DOI] [PubMed] [Google Scholar]
- Jackson M., Krassowska, A., Gilbert, N., Chevassut, T., Forrester, L., Ansell, J., and Ramsahoye, B. 2004. Severe global DNA hypomethylation blocks differentiation and induces histone hyperacetylation in embryonic stem cells. Mol. Cell. Biol. 24: 8862–8871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones L., Hamilton, A.J., Voinnet, O., Thomas, C.L., Maule, A.J., and Baulcombe, D.C. 1999. RNA–DNA interactions and DNA methylation in post-transcriptional gene silencing. Plant Cell 11: 2291–2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki H. and Taira, K. 2004. Induction of DNA methylation and gene silencing by short interfering RNAs in human cells. Nature 431: 211–217. [DOI] [PubMed] [Google Scholar]
- Keller G., Kennedy, M., Papayannopoulou, T., and Wiles, M.V. 1993. Hematopoietic commitment during embryonic stem cell differentiation in culture. Mol. Cell. Biol. 13: 473–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketting R.F., Fischer, S.E., Bernstein, E., Sijen, T., Hannon, G.J., and Plasterk, R.H. 2001. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes & Dev. 15: 2654–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight S.W. and Bass, B.L. 2001. A role for the RNase III enzyme DCR-1 in RNA interference and germ line development in Caenorhabditis elegans. Science 293: 2269–2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogan G.L., Tulin, A.V., Aravin, A.A., Abramov, Y.A., Kalmykova, A.I., Maisonhaute, C., and Gvozdev, V.A. 2003. The GATE retrotransposon in Drosophila melanogaster: Mobility in heterochromatin and aspects of its expression in germline tissues. Mol. Genet. Genomics 269: 234–242. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M., Rauhut, R., Yalcin, A., Meyer, J., Lendeckel, W., and Tuschl, T. 2002. Identification of tissue-specific microRNAs from mouse. Curr. Biol. 12: 735–739. [DOI] [PubMed] [Google Scholar]
- Lee R.C. and Ambros, V. 2001. An extensive class of small RNAs in Caenorhabditis elegans. Science 294: 862–864. [DOI] [PubMed] [Google Scholar]
- Lee R.C., Feinbaum, R.L., and Ambros, V. 1993. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75: 843–854. [DOI] [PubMed] [Google Scholar]
- Lee Y., Ahn, C., Han, J., Choi, H., Kim, J., Yim, J., Lee, J., Provost, P., Radmark, O., Kim, S., et al. 2003. The nuclear RNase III Drosha initiates microRNA processing. Nature 425: 415–419. [DOI] [PubMed] [Google Scholar]
- Lee Y.S., Nakahara, K., Pham, J.W., Kim, K., He, Z., Sontheimer, E.J., and Carthew, R.W. 2004. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell 117: 69–81. [DOI] [PubMed] [Google Scholar]
- Lehnertz B., Ueda, Y., Derijck, A.A., Braunschweig, U., Perez-Burgos, L., Kubicek, S., Chen, T., Li, E., Jenuwein, T., and Peters, A.H. 2003. Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr. Biol. 13: 1192–1200. [DOI] [PubMed] [Google Scholar]
- Lei H., Oh, S.P., Okano, M., Juttermann, R., Goss, K.A., Jaenisch, R., and Li, E. 1996. De novo DNA cytosine methyltransferase activities in mouse embryonic stem cells. Development 122: 3195–3205. [DOI] [PubMed] [Google Scholar]
- Lewis B.P., Shih, I.H., Jones-Rhoades, M.W., Bartel, D.P., and Burge, C.B. 2003. Prediction of mammalian microRNA targets. Cell 115: 787–798. [DOI] [PubMed] [Google Scholar]
- Li E. 2002. Chromatin modification and epigenetic reprogramming in mammalian development. Nat. Rev. Genet. 3: 662–673. [DOI] [PubMed] [Google Scholar]
- Lindroth A.M., Cao, X., Jackson, J.P., Zilberman, D., McCallum, C.M., Henikoff, S., and Jacobsen, S.E. 2001. Requirement of CHROMOMETHYLASE3 for maintenance of CpXpG methylation. Science 292: 2077–2080. [DOI] [PubMed] [Google Scholar]
- Llave C., Kasschau, K.D., Rector, M.A., and Carrington, J.C. 2002. Endogenous and silencing-associated small RNAs in plants. Plant Cell 14: 1605–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lois C., Hong, E.J., Pease, S., Brown, E.J., and Baltimore, D. 2002. Germline transmission and tissue-specific expression of transgenes delivered by lentiviral vectors. Science 295: 868–872. [DOI] [PubMed] [Google Scholar]
- Maison C., Bailly, D., Peters, A.H., Quivy, J.P., Roche, D., Taddei, A., Lachner, M., Jenuwein, T., and Almouzni, G. 2002. Higher-order structure in pericentric heterochromatin involves a distinct pattern of histone modification and an RNA component. Nat. Genet. 30: 329–334. [DOI] [PubMed] [Google Scholar]
- Martin G.R. and Evans, M.J. 1975. Differentiation of clonal lines of teratocarcinoma cells: Formation of embryoid bodies in vitro. Proc. Natl. Acad. Sci. 72: 1441–1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez J., Patkaniowska, A., Urlaub, H., Luhrmann, R., and Tuschl, T. 2002. Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell 110: 563–574. [DOI] [PubMed] [Google Scholar]
- Matzke M., Aufsatz, W., Kanno, T., Daxinger, L., Papp, I., Mette, M.F., and Matzke, A.J. 2004. Genetic analysis of RNA-mediated transcriptional gene silencing. Biochim. Biophys. Acta 1677: 129–141. [DOI] [PubMed] [Google Scholar]
- Melquist S. and Bender, J. 2003. Transcription from an upstream promoter controls methylation signaling from an inverted repeat of endogenous genes in Arabidopsis. Genes & Dev. 17: 2036–2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mette M.F., Aufsatz, W., van der Winden, J., Matzke, M.A., and Matzke, A.J. 2000. Transcriptional silencing and promoter methylation triggered by double-stranded RNA. EMBO J. 19: 5194–5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills K.D., Ferguson, D.O., Essers, J., Eckersdorff, M., Kanaar, R., and Alt, F.W. 2004. Rad54 and DNA Ligase IV cooperate to maintain mammalian chromatid stability. Genes & Dev. 18: 1283–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki K., Fine, N.A., Fujisawa, T., and Gorovsky, M.A. 2002. Analysis of a piwi-related gene implicates small RNAs in genome rearrangement in Tetrahymena. Cell 110: 689–699. [DOI] [PubMed] [Google Scholar]
- Morris K.V., Chan, S.W., Jacobsen, S.E., and Looney, D.J. 2004. Small interfering RNA-induced transcriptional gene silencing in human cells. Science 305: 1289–1292. [DOI] [PubMed] [Google Scholar]
- Mortensen R.M., Conner, D.A., Chao, S., Geisterfer-Lowrance, A.A., and Seidman, J.G. 1992. Production of homozygous mutant ES cells with a single targeting construct. Mol. Cell. Biol. 12: 2391–2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss E.G., Lee, R.C., and Ambros, V. 1997. The cold shock domain protein LIN-28 controls developmental timing in C. elegans and is regulated by the lin-4 RNA. Cell 88: 637–646. [DOI] [PubMed] [Google Scholar]
- Okano M., Bell, D.W., Haber, D.A., and Li, E. 1999. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99: 247–257. [DOI] [PubMed] [Google Scholar]
- Paddison P.J., Caudy, A.A., Bernstein, E., Hannon, G.J., and Conklin, D.S. 2002a. Short hairpin RNAs (shRNAs) induce sequence-specific silencing in mammalian cells. Genes & Dev. 16: 948–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paddison P.J., Caudy, A.A., and Hannon, G.J. 2002b. Stable suppression of gene expression by RNAi in mammalian cells. Proc. Natl. Acad. Sci. 99: 1443–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal-Bhadra M., Bhadra, U., and Birchler, J.A. 1999. Cosuppression of nonhomologous transgenes in Drosophila involves mutually related endogenous sequences. Cell 99: 35–46. [DOI] [PubMed] [Google Scholar]
- ____. 2002. RNAi related mechanisms affect both transcriptional and posttranscriptional transgene silencing in Drosophila. Mol. Cell 9: 315–327. [DOI] [PubMed] [Google Scholar]
- Pal-Bhadra M., Leibovitch, B.A., Gandhi, S.G., Rao, M., Bhadra, U., Birchler, J.A., and Elgin, S.C. 2004. Heterochromatic silencing and HP1 localization in Drosophila are dependent on the RNAi machinery. Science 303: 669–672. [DOI] [PubMed] [Google Scholar]
- Papp I., Mette, M.F., Aufsatz, W., Daxinger, L., Schauer, S.E., Ray, A., van der Winden, J., Matzke, M., and Matzke, A.J. 2003. Evidence for nuclear processing of plant micro RNA and short interfering RNA precursors. Plant Physiol. 132: 1382–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park W., Li, J., Song, R., Messing, J., and Chen, X. 2002. CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr. Biol. 12: 1484–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park C.W., Chen, Z., Kren, B.T., and Steer, C.J. 2004. Double-stranded siRNA targeted to the huntingtin gene does not induce DNA methylation. Biochem. Biophys. Res. Commun. 323: 275–280. [DOI] [PubMed] [Google Scholar]
- Peters A.H., O'Carroll, D., Scherthan, H., Mechtler, K., Sauer, S., Schofer, C., Weipoltshammer, K., Pagani, M., Lachner, M., Kohlmaier, A., et al. 2001. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell 107: 323–337. [DOI] [PubMed] [Google Scholar]
- Peters A.H., Kubicek, S., Mechtler, K., O'Sullivan, R.J., Derijck, A.A., Perez-Burgos, L., Kohlmaier, A., Opravil, S., Tachibana, M., Shinkai, Y., et al. 2003. Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol. Cell 12: 1577–1589. [DOI] [PubMed] [Google Scholar]
- Pham J.W., Pellino, J.L., Lee, Y.S., Carthew, R.W., and Sontheimer, E.J. 2004. A Dicer-2-dependent 80s complex cleaves targeted mRNAs during RNAi in Drosophila. Cell 117: 83–94. [DOI] [PubMed] [Google Scholar]
- Reinhart B.J. and Bartel, D.P. 2002. Small RNAs correspond to centromere heterochromatic repeats. Science 297: 1831. [DOI] [PubMed] [Google Scholar]
- Reinhart B.J., Slack, F.J., Basson, M., Pasquinelli, A.E., Bettinger, J.C., Rougvie, A.E., Horvitz, H.R., and Ruvkun, G. 2000. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403: 901–906. [DOI] [PubMed] [Google Scholar]
- Reinhart B.J., Weinstein, E.G., Rhoades, M.W., Bartel, B., and Bartel, D.P. 2002. MicroRNAs in plants. Genes & Dev. 16: 1616–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudert F., Bronner, S., Garnier, J.M., and Dolle, P. 1995. Transcripts from opposite strands of gamma satellite DNA are differentially expressed during mouse development. Mamm. Genome 6: 76–83. [DOI] [PubMed] [Google Scholar]
- Schramke V. and Allshire, R. 2003. Hairpin RNAs and retrotransposon LTRs effect RNAi and chromatin-based gene silencing. Science 301: 1069–1074. [DOI] [PubMed] [Google Scholar]
- ____. 2004. Those interfering little RNAs! Silencing and eliminating chromatin. Curr. Opin. Genet. Dev. 14: 174–180. [DOI] [PubMed] [Google Scholar]
- Sijen T., Vijn, I., Rebocho, A., van Blokland, R., Roelofs, D., Mol, J.N., and Kooter, J.M. 2001. Transcriptional and post-transcriptional gene silencing are mechanistically related. Curr. Biol. 11: 436–440. [DOI] [PubMed] [Google Scholar]
- Slack F.J., Basson, M., Liu, Z., Ambros, V., Horvitz, H.R., and Ruvkun, G. 2000. The lin-41 RBCC gene acts in the C. elegans heterochronic pathway between the let-7 regulatory RNA and the LIN-29 transcription factor. Mol. Cell 5: 659–669. [DOI] [PubMed] [Google Scholar]
- Svoboda P., Stein, P., Filipowicz, W., and Schultz, R.M. 2004. Lack of homologous sequence-specific DNA methylation in response to stable dsRNA expression in mouse oocytes. Nucleic Acids Res. 32: 3601–3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taverna S.D., Coyne, R.S., and Allis, C.D. 2002. Methylation of histone H3 at lysine 9 targets programmed DNA elimination in Tetrahymena. Cell 110: 701–711. [DOI] [PubMed] [Google Scholar]
- Verdel A., Jia, S., Gerber, S., Sugiyama, T., Gygi, S., Grewal, S.I., and Moazed, D. 2004. RNAi-mediated targeting of heterochromatin by the RITS complex. Science 303: 672–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpe T.A., Kidner, C., Hall, I.M., Teng, G., Grewal, S.I., and Martienssen, R.A. 2002. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 297: 1833–1837. [DOI] [PubMed] [Google Scholar]
- Volpe T., Schramke, V., Hamilton, G.L., White, S.A., Teng, G., Martienssen, R.A., and Allshire, R.C. 2003. RNA interference is required for normal centromere function in fission yeast. Chromosome Res. 11: 137–146. [DOI] [PubMed] [Google Scholar]
- Wassenegger M., Heimes, S., Riedel, L., and Sanger, H.L. 1994. RNA-directed de novo methylation of genomic sequences in plants. Cell 76: 567–576. [DOI] [PubMed] [Google Scholar]
- Wienholds E., Koudijs, M.J., van Eeden, F.J., Cuppen, E., and Plasterk, R.H. 2003. The microRNA-producing enzyme Dicer1 is essential for zebrafish development. Nat. Genet. 35: 217–218. [DOI] [PubMed] [Google Scholar]
- Wightman B., Ha, I., and Ruvkun, G. 1993. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 75: 855–862. [DOI] [PubMed] [Google Scholar]
- Xie Z., Johansen, L.K., Gustafson, A.M., Kasschau, K.D., Lellis, A.D., Zilberman, D., Jacobsen, S.E., and Carrington, J.C. 2004. Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2: E104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yekta S., Shih, I.H., and Bartel, D.P. 2004. MicroRNA-directed cleavage of HOXB8 mRNA. Science 304: 594–596. [DOI] [PubMed] [Google Scholar]
- Zilberman D., Cao, X., and Jacobsen, S.E. 2003. ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science 299: 716–719. [DOI] [PubMed] [Google Scholar]