Abstract
Dachshund homolog 1 (DACH1), a key cell fate determination factor, contributes to tumorigenesis, invasion, metastasis of human breast neoplasm. However, the exact molecular mechanisms for the anti-tumor roles of DACH1 in breast carcinoma are still lack of extensive understanding. Herein, we utilized immunohistochemistry (IHC) staining and public microarray data analysis showing that DACH1 was higher in normal breast, low-grade and luminal-type cancer in comparison with breast carcinoma, high-grade and basal-like tumors respectively. Additionally, both correlation analysis of public databases of human breast carcinoma and IHC analysis of mice xenograft tumors demonstrated that DACH1 inversely related to cancer stem cells (CSCs) markers, epithelial-mesenchymal transition (EMT) inducers and basal-enriched molecules, while cluster of differentiation 44 (CD44) behaved in an opposite manner. Furthermore, mice transplanted tumor model indicated that breast cancer cells Met-1 with up-regulation of DACH1 were endowed with remarkably reduced potential of tumorigenesis. Importantly, meta-analysis of 19 Gene Expression Omnibus (GEO) databases of breast cancer implicated that patients with higher DACH1 expression had prolonged time to death, recurrence and metastasis, while CD44 was a promising biomarker predicting worse overall survival (OS) and metastasis-free survival (MFS). Collectively, our study indicated that CD44 might be a novel target of DACH1 in breast carcinoma.
Introduction
In spite of significant achievement made in early diagnosis and therapeutic strategies, breast cancer still draws great attention from the worldwide because of its high incidence rate and mortality1–3. The unsatisfactory clinical outcome is mostly due to tumor recurrence, metastasis and therapy-resistance1. Identifying novel biomarkers related to molecular subtypes, aggressive phenotypes and prognosis of breast cancer is essential for drug development, disease surveillance and precise therapy.
The retinal determination gene network (RDGN), including DACH1, EYA1 and SIX1, plays crucial roles in the development of multiple organs4. SIX1 and EYA1, two important RDGN members, exert favorable effects on tumor initiation and progression4, 5, and high expression of SIX1 and EYA1 is an adverse factor for clinical outcomes for breast cancer patients6–8. On the contrary, another key RDGN member DACH1 behaved as a tumor suppressor and reduced expression of DACH1 predicts poor survival performance of breast cancer patients9. Several lines of evidence have demonstrated that the hypermethylation of promoter region leads to the down-regulation of DACH1, which is closely associated with proliferation, invasion and metastasis of various tumors, including breast cancer10–13, lung cancer14, esophageal cancer15, renal cell carcinoma16 and hepatocellular carcinoma17. DACH1 antagonizes the transcription and translation of oncogenes and induces epithelial-mesenchymal transition (EMT) in breast cancer, resulting in the inhibition of tumor growth, invasion and migration9, 10. Recent studies prove that cancer stem cells (CSCs) possess potent self-renewal ability and are responsible for tumor relapse and metastasis and endogenous DACH1 participates in the negative regulation of CSCs18, 19.
Cluster of differentiation-44 (CD44), a ubiquitously present glycoprotein on the membrane of mammalian cells, plays essential roles in a variety of biological function such as cell division, adhesion and migration20. During the past decades, the role of CD44 in cancer development has been revealed and valued. As a well-known marker of CSCs, CD44 promotes carcinogenesis, invasion, metastasis and therapy-resistance20–23. It promotes proliferation and suppresses apoptosis by regulation of relative pathways, including Ras-Raf-Mek-Erk-Cyclin D1 pathway and phosphoinositide 3-kinase (PI3K)-Akt signaling, as well as stimulates EMT, which contributes to tumor invasion and metastasis20, 24.
Previous study has demonstrated that endogenous reduction of DACH1 was accompanied by down-regulation of CSC markers, such as SOX2, Nanog, KLF418. To further evaluate the correlation between DACH1 and CSC markers and EMT inducers in breast cancer, we performed a comprehensive analysis of immunohistochemistry (IHC) staining, publicly available microarray data, RNA profiling and western blot. Our study indicated that DACH1 was inversely correlated with CD44 and CD44 might be a novel target of DACH1 in breast cancer.
Results
DACH1 and CD44 associated with tumorigenesis and histological grade of breast cancer
In order to evaluate the expression of DACH1 and CD44 in normal breast and breast malignant tissues, we carried out IHC analysis on two TMAs (BR1502–97 and BR1502-98) with normal breast and human breast cancer tissues. DACH1 was majorly found in nucleus and CD44 was mostly detected on the membrane of breast cancer cells. Representative images of IHC staining for noncancerous and cancerous tissues were shown in Fig. 1a, showing that DACH1 decreased and CD44 increased in breast neoplasm tissues in comparison with normal breast.
Figure 1.
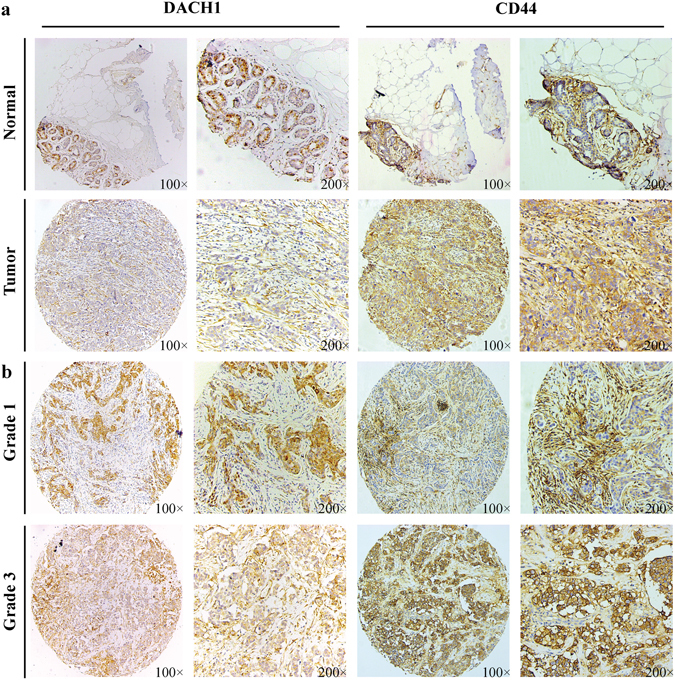
DACH1 and CD44 were correlated with tumorigenesis and histological grade of breast carcinoma. (a) Representative images of immunohistochemistry staining of DACH1 and CD44 in noncancerous and cancerous tissues were shown. (b) Representative images of immunohistochemistry staining of DACH1 and CD44 in low-grade and high-grade breast neoplasm tissues were shown.
Additionally, we also explored the correlation between the protein abundance of DACH1 and CD44 and histological grade. Representative images of IHC staining for low-grade and high-grade cancerous tissues were showed in Fig. 1b, which indicating that DACH1 was inversely correlated with tumor grade, while CD44 was positively associated with histological grade.
Expression of DACH1 and CD44 correlated with molecular subtypes of breast cancer
We also carried out IHC staining to assess the protein abundance of DACH1 and CD44 in luminal-type and basal-like breast cancer tissues. Representative images for the expression of DACH1 and CD44 in luminal and basal tissues were shown in Fig. 2a. Furthermore, expression analysis of GSE20711 including a total of 45 luminal and 22 basal-like breast tumor cases was also interrogated to evaluate the mRNA levels of DACH1 and CD44 in luminal and basal-like breast neoplasm tissues, which showed that DACH1 was enriched in luminal breast carcinoma in comparison with basal-like breast cancer (P < 0.001) (Fig. 2b), while CD44 exhibited an opposite tendency (P < 0.001) (Fig. 2c) at mRNA level. Altogether, our results indicated that luminal breast carcinoma was most likely to be DACH1high/CD44low type, and basal-like breast tumor tissues were majorly DACH1low/CD44high type.
Figure 2.
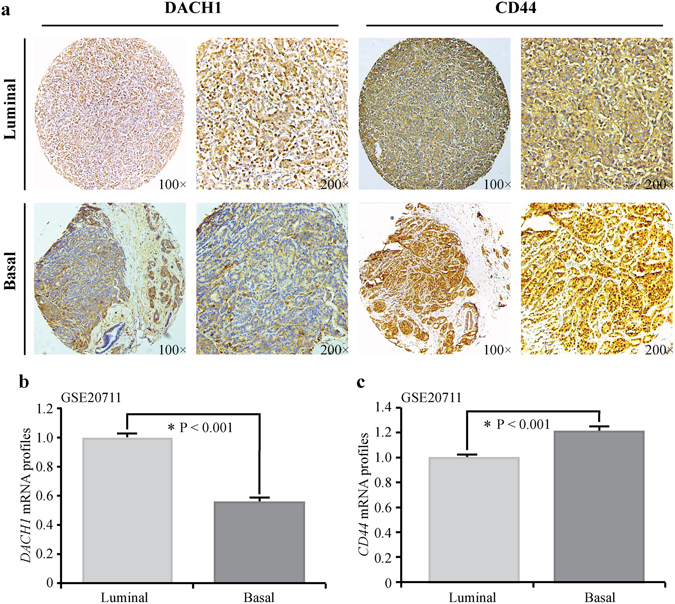
The expression of DACH1 and CD44 correlated with molecular subtypes of breast carcinoma. (a) Representative images of immunohistochemistry staining of DACH1 and CD44 in luminal-type and basal-like breast cancer tissues were shown. (b) Expression analysis of public microarray dataset GSE20711 showed that the mRNA level of DACH1 was significantly higher in luminal-type than in basal-like breast tumor. (c) Expression analysis of GSE 20711 also displayed that CD44 mRNA expression was remarkably lower in luminal-type than in basal-like breast cancer.
DACH1 down-regulated some CSC and EMT markers in vitro, as well as blocked Met-1 tumor growth in vivo
Breast cancer Met-1 cells were transducted with a DACH1 expression vector resulting in an ∼4.5-fold increase in DACH1 expression (Fig. 3a) and subsequent reduction of CSC markers CD44, KLF4 and MYC as well as EMT markers including FN1 and VIM (Fig. 3b) by mRNA analysis. Western blot also demonstrated the presence of the DACH1-tagged FLAG epitome, and the effects of DACH1 overexpression on the protein abundance of CD44, Fibronectin, Vimentin, p21 and p27, which were showed in Fig. 3c. Ectopic expression of DACH1 contributed to remarkable reduction of CD44, Fibronectin, Vimentin and significant up-regulation of p21 and p27 in Met-1 cells. Mammary tumor growth in vivo was assessed by subcutaneous implantation of Met-1 cells in nude mice (Fig. 3f). Met-1 cells with engineered expression of DACH1 were endowed with remarkably reduced potential of tumorigenesis in xenograft tumors. Up-regulation of DACH1 significantly reduced the volume of tumors by ∼90% and slowed down tumor growth (Fig. 3d). Tumor weight was also reduced by ∼90% in comparison with the control tumors (Fig. 3e). The results implicated that DACH1 suppressed the expression of some CSCs and EMT markers and serves as a potent anti-tumor factor in xenograft tumors.
Figure 3.
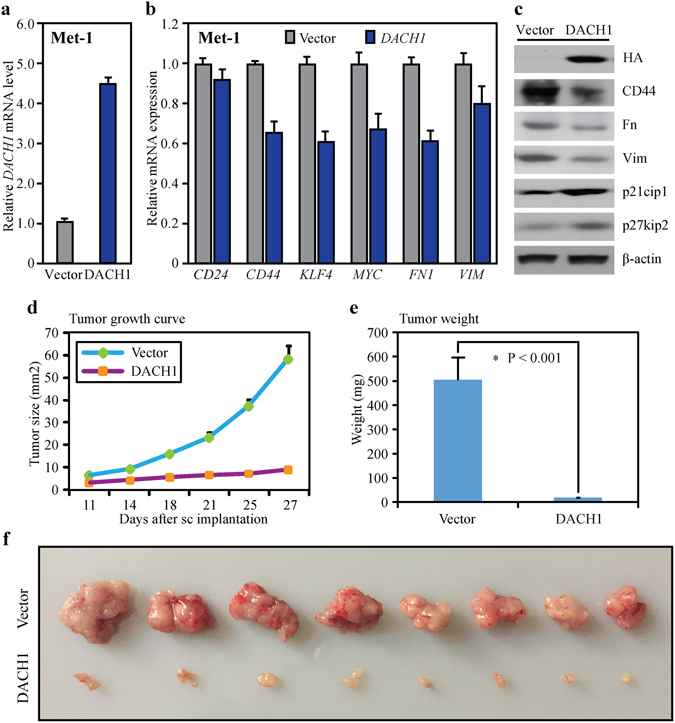
DACH1 regulated the expression of some CSCs and EMT genes in Met-1 cells and suppressed tumor growth in vivo. (a) Stable expression of DACH1 in breast cancer Met-1 cells was achieved by retrovirus infection. (b) RNA microarray and cluster analysis showed that upregulation of DACH1 reduced the mRNA levels of CD24, CD44, KLF4, MYC, FN1 and VIM in Met-1 cells. (c) Western blot indicated that upregulation of DACH1 reduced the protein abundance of CD44, Fibronectin, Vimentin, p21 and p27 in Met-1 cells. (d) DACH1 overexpression significantly reduced the volume of tumors by ∼90% and slowed down the speed of tumor growth in nude mice xenograft tumors. (e) Overexpression of DACH1 also reduced mice transplanted tumor weight by ∼90%. (f) Mammary tumor growth in vivo was evaluated by subcutaneous implantation of DACH1-overexpressing Met-1 cells and the GFP controls in nude mice.
DACH1 reduced the expression of CD44, Fibronectin, Vimentin, Myc, Sox2, EGFR, Ki-67 in vivo
Immunohistochemistry analysis was conducted to assess the protein abundance of DACH1, CD44, Myc, Sox2, Fibronectin, Vimentin, EGFR, Ki-67 in nude mice xenograft tumor tissues with overexpression of DACH1 and the GFP controls. Additionally, we also employed IHC scoring to quantize the levels of these proteins in both DACH-overexpressing and the control tumors by using semi-quantitative criteria. About six 200 magnification images of each kind of protein were selected for IHC scoring by two experienced pathologists independently. Representative images of IHC staining and scoring results for DACH1, CD44, Myc, Sox2, Fibronectin, Vimentin, EGFR and Ki-67 were shown in Fig. 4a,b,c,d,e,f,g and h, respectively. Our results displayed that over-expression of DACH1 (P < 0.001) remarkably reduced the expression of CD44 (P < 0.001), Myc (P < 0.001), Sox2 (P < 0.001), Fibronectin (P < 0.001), Vimentin (P < 0.001), EGFR (P < 0.001) and Ki-67 (P < 0.001), demonstrating that DACH1 could potently down-regulated the expression of some CSCs and EMT markers, basal-like factor EGFR and proliferative biomarker Ki-67 in vivo at protein level.
Figure 4.
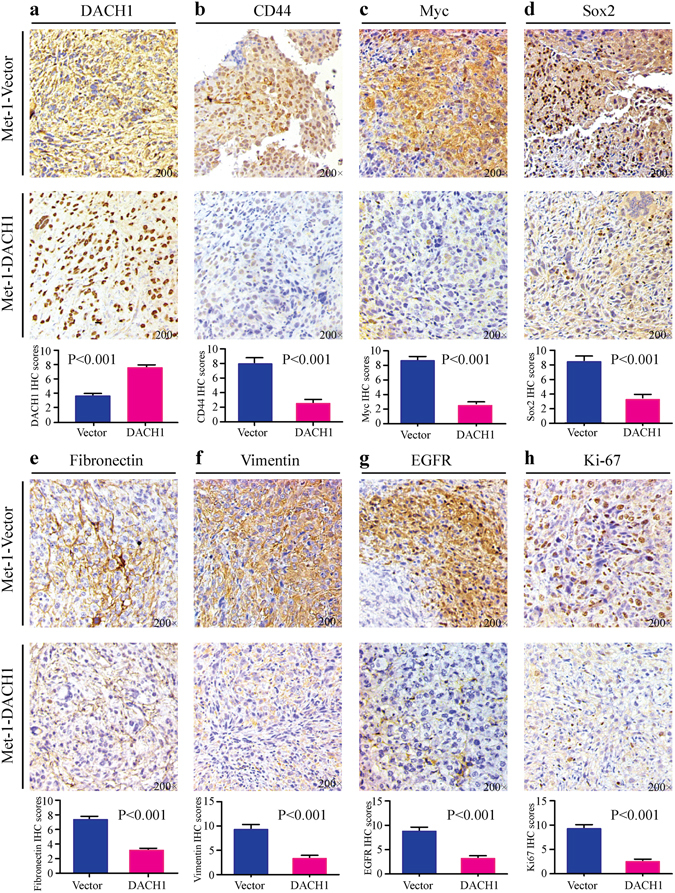
DACH1 reduced the expression of CD44, Myc, Sox2, Fibronectin, Vimentin, EGFR and Ki-67 in vivo. Both representative immunohistochemistry images and scoring results showed that overexpression of DACH1 (a) down-regulated the protein abundance of CD44 (b), Myc (c), Sox2 (d), Fibronectin (e), Vimentin (f), EGFR (g) and Ki-67 (h) in nude mice xenograft tumors.
Correlation between the expression of DACH1 and CD44 and the levels of FN1, VIM, YBX1, FOXA1, EGFR and MKI67
Previous study has implicated that DACH1 enriched in luminal A breast cancer and its expression fluctuated in direct proportion to the level of luminal-like marker FOXA113. Previously experimental study demonstrated that DACH1 participated in the inhibition of Snail-induced EMT through suppressing the activity of the Y box-binding protein (YB-1)9. Herein, public dataset GSE20685 was interrogated to assess the association between DACH1 and CD44, FN1, VIM, FOXA1, EGFR and MKI67, as well as evaluate the correlation between CD44 and the above genes. The results showed that DACH1 mRNA expression was inversely correlated with CD44 (R = −0.341, P < 0.001) (Fig. 5a), FN1 (R = −0.214, P < 0.001) (Fig. 5b), VIM (R = −0.229, P < 0.001) (Fig. 5c), EGFR (R = −0.390, P < 0.001) (Fig. 5e) and MKI67 (R = −0.376, P < 0.001) (Fig. 5f), but positively associated with the mRNA expression of FOXA1 (R = 0.608, P < 0.001) (Fig. 5d). Conversely, CD44 was found to be positively correlated with FN1 (R = 0.253, P < 0.001) (Fig. 5g), VIM (R = 0.237, P < 0.001) (Fig. 5h), YBX1 (R = 0.446, P < 0.001) (Fig. 5i), EGFR (R = 0.336, P < 0.001) (Fig. 5k) and MKI67 (R = 0.215, P < 0.001) (Fig. 5l), while there was a significantly negative association between CD44 and FOXA1 (R = −0.402, P < 0.001) (Fig. 5j).
Figure 5.
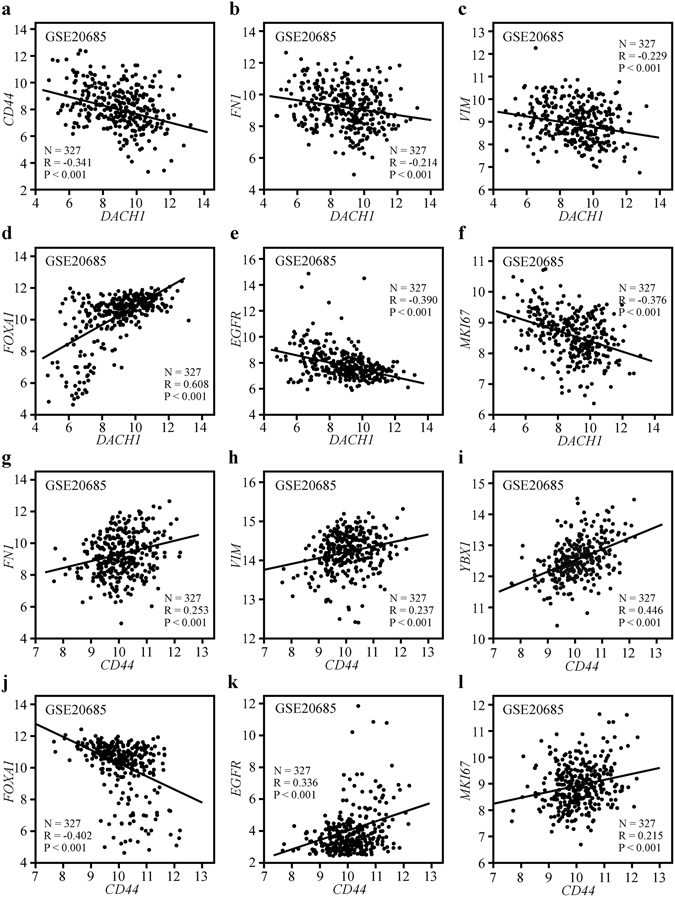
The expression of DACH1 and CD44 correlated with VIM, FN1, YBX1, FOXA1, EGFR and MKI67 in breast cancer tissues. Correlation analysis of public dataset GSE 20685 showed that DACH1 was inversely correlated with cancer stem cell marker CD44 (a), mesenchymal markers FN1 (b) and VIM (c) as well as basal-like markers EGFR (e) and MKI67 (f), while positively associated with luminal marker FOXA1 (d). CD44 was parallel with FN1 (g) and VIM (h), YBX1 (i), EGFR (k) and MKI67 (l), while negatively associated with FOXA1 (j).
Breast cancer cell line data reported by Neve RM25, including a total of 51 different breast cancer lines from luminal-type (N = 25) or basal-like (N = 26), were also employed to evaluate the correlation between the expression of DACH1 and CD44 and the levels of FN1, VIM, YBX1, FOXA1, EGFR and MKI67. The results displayed that DACH1 mRNA expression was inversely correlated with CD44 (R = −0.507, P < 0.001) (Fig. 6a), FN1 (R = −0.354, P = 0.011) (Fig. 6b), VIM (R = −0.419, P = 0.002) (Fig. 6c), EGFR (R = −0.523, P < 0.001) (Fig. 6e) and MKI67 (R = −0.336, P = 0.016) (Fig. 6f) but positively associated with the mRNA level of FOXA1 (R = 0.539, P < 0.001) (Fig. 6d), which were consistent with the results got in GSE20685. In contrast, CD44 was parallel with FN1 (R = 0.406, P = 0.003) (Fig. 6g), VIM (R = 0.614, P < 0.001) (Fig. 6h), YBX1 (R = 0.557, P < 0.001) (Fig. 6i), EGFR (R = 0.506, P < 0.001) (Fig. 6k) and MKI67 (R = 0.391, P = 0.005) (Fig. 6l) but negatively associated with FOXA1 (R = −0.689, P < 0.001) (Fig. 6j), supporting the conclusion from the correlation analysis of human breast tumor samples (GSE20685).
Figure 6.

The association between the expression of DACH1 and CD44 with VIM, FN1, YBX1, FOXA1, EGFR and MKI67 in breast cancer cell lines. Correlation analysis of data with a total of 51 breast cancer cell lines showed that DACH1 was inversely correlated with CD44 (a), FN1 (b), VIM (c), EGFR (e) and MKI67 (f), while positively associated with FOXA1 (d). In contrast, CD44 was parallel with FN1 (g), VIM (h), YBX1 (i), EGFR (k) and MKI67 (l), but negatively related to FOXA1 (j).
The opposite roles of DACH1 and CD44 in clinical outcomes of breast cancer patients
In order to assess the prognostic value of DACH1 and CD44 in breast cancer, a meta-analysis enrolling a total of 19 published Gene Expression Omnibus (GEO) databases and including 3574 breast cancer patients was performed26–44. The characteristics of these 19 GSE databases were displayed in Table 1. The results indicated that patients with higher mRNA expression of DACH1 tended to enjoy longer time to death (HR: 0.72 (0.57–0.92), I2 = 22.9%, P = 0.232) (Fig. 7a), relapse (HR: 0.73 (0.56–0.97), I2 = 64.1%, P = 0.003) (Fig. 7c) and metastasis (HR: 0.73 (0.56–0.95), I2 = 27.2%, P = 0.202) (Fig. 7e). On the contrary, higher mRNA expression of CD44 was directly related to worse OS (HR: 1.27 (1.03–1.57), I2 = 3.0%, P = 0.412) (Fig. 7b) and MFS (HR: 1.40 (1.02–1.93), I2 = 48.4%, P = 0.050) (Fig. 7f), but did not insignificantly contribute to worse RFS (HR: 1.19 (0.94–1.51), I2 = 45.7%, P = 0.056) (Fig. 7d). Our results demonstrated that DACH1 was a promising biomarker predictive of better clinical outcomes, while CD44 was an adverse factor for the survival performance of breast cancer patients.
Table 1.
Characteristics of the included public microarray datasets in the meta-analysis.
| First Author | GSE accession | Year | Duration (Months) | Patient Number | Detection | Platform |
|---|---|---|---|---|---|---|
| Desmedt C26 | GSE7390 | 2007 | 163.2 | 198 | Microarray | GPL96 |
| Pawitan Y27 | GSE1456 | 2005 | 102 | 159 | Microarray | GPL96 |
| Hennessy BT28 | GSE10885 | 2009 | 106 | 89 | Microarray | GPL887 |
| Dedeurwaerder S29 | GSE20711 | 2011 | 169 | 88 | Microarray | GPL570 |
| Kao KJ30 | GSE20685 | 2011 | 156 | 327 | Microarray | GPL570 |
| Clarke C31 | GSE42568 | 2013 | 100.9 | 104 | Microarray | GPL570 |
| Desmedt C32 | GSE16446 | 2011 | 60 | 120 | Microarray | GPL570 |
| Loi S33 | GSE6532 | 2010 | 176.8 | 327 | Microarray | GPL96 |
| Bild AH34 | GSE3143 | 2006 | 156 | 158 | Microarray | GPL8300 |
| Terunuma A35 | GSE39004 | 2014 | 120 | 61 | Microarray | GPL6244 |
| Heikkinen T36 | GSE24450 | 2011 | 120 | 183 | Microarray | GPL6947 |
| Hatzis C37 | GSE25066 | 2011 | 120 | 508 | Microarray | GPL96 |
| Symmans WF38 | GSE17705 | 2010 | 196 | 298 | Microarray | GPL96 |
| Wang Y39 | GSE2034 | 2005 | 180 | 286 | Microarray | GPL96 |
| Tofigh A40 | GSE58644 | 2014 | 145 | 321 | Microarray | GPL6244 |
| Minn AJ41 | GSE2603 | 2005 | 130 | 99 | Microarray | GPL96 |
| Minn AJ42 | GSE5327 | 2007 | 156 | 58 | Microarray | GPL96 |
| Sircoulomb F43 | GSE17907 | 2010 | 112 | 51 | Microarray | GPL570 |
| Nagalla S44 | GSE45255 | 2013 | 127.4 | 139 | Microarray | GPL96 |
Figure 7.
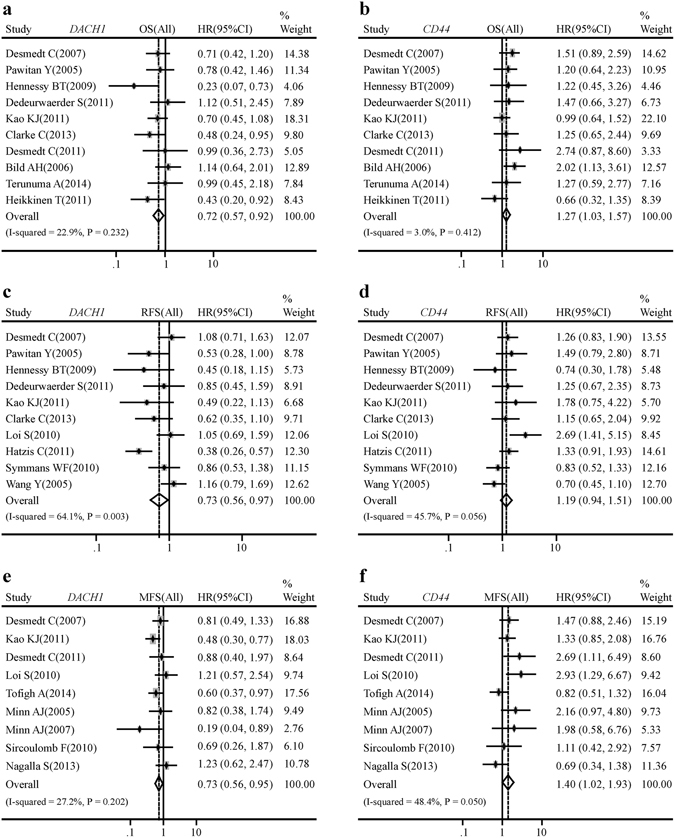
DACH1 and CD44 were related to clinical outcomes of breast cancer patients. Meta-analysis of a total of 19 public databases showed that breast cancer patients with higher DACH1 mRNA level tended to have better overall survival (a), relapse-free survival (c), metastasis-free survival (e). On the contrary, patients with higher CD44 mRNA expression had shorter time to death (b) and metastasis (f), but did not acquire statistically significant worse relapse-free survival in comparison to patients with comparatively lower CD44 expression (d).
Discussion
DACH1, as an important member of RDGN, is widely expressed in epithelial cells and plays critical roles in normal organ development45, 46. However, its absent or lower expression contributes to the initiation and progression of various tumor types, including lung adenocarcinoma14, pancreatic cancer47, breast cancer10 and gastric cancer48. This study provided results that expression types of DACH1 and CD44 were opposite in breast cancer. Overexpression of DACH1 reduced the levels of CSC and EMT markers in breast cancer cell line and potently inhibited the ability of tumorgenesis in xenograft model. Besides, correlation analysis exhibited that DACH1 and CD44 behaved completely differently in the correlations with FN1, VIM, YBX1, FOXA1, EGFR and MKI67. Importantly, DACH1 serves as a protective factor, while CD44 is an unfavorable element for the prognosis of breast cancer patients. Thus, we concluded that DACH1 might exert inhibitory effects on the development of breast cancer partly by suppression of EMT inducers and CSCs markers, especially CD44.
Carcinogenesis may derive from the acquisition of a plethora of oncogenic mutations, sequential suppression of endogenous growth inhibitors49 and the loss control over pivotal cellular functions50. DACH1 accounts for the carcinogenesis of various tumor types, including human breast cancer51. Previous studies have implicated that DACH1 negatively regulated cellular proliferation and tumor growth by repressing cell cycle protein cyclin D1 in both breast cancer11 and renal clear cell cancer16. Restoration of DACH1 suppressed clone formation of renal clear cell cancer cells in vitro as well as tumor growth in vivo through the inhibition of cyclin D1 transcription via associating with AP-1 protein16. Furthermore, hypermethylation of DACH1 promoter region itself led to carcinogenesis and it also exerted inhibitory effects on tumor initiation through activating transforming growth factor-beta (TGF-beta) signaling15. Besides, DACH1 inhibited cellular growth in an NAD and p53-dependent manner by direct protein-protein association in breast cancer52. On the contrary, CD44, a family of transmembrane glycoproteins, exerted promoting roles in tumorigenesis of various cancer types20. Up-regulation of the cleaved intracellular domain of CD44 (CD44ICD) enriched the mammosphere formation, while the blockage of CD44ICD reversed the effects53. Immunohistochemistry analysis on human breast cancer tissues and expression analysis of GSE42568 indicated that CD44 remarkably increased in breast carcinoma in comparison with normal breast tissues22. CD44 functions in carcinogenesis through binding to extracellular matrix components and messenger molecules in tumor environment54.
Several lines of evidence indicated that DACH1 expression correlated with tumor differentiation. Immunohistochemistry analysis of clear cell renal cancer tissues displayed that DACH1 was inversely correlated with tumor grade16. About 60% of cells in low-grade tumors expressed DACH1, and less than 20% cells in grade III tumors expressed DACH116. Similar phenomenon was also found in breast cancer13. DACH1 was remarkably enriched in low-grade breast tumors13. In contrast, CD44 positively correlated with breast tumor grade21, 22. Our previous study displayed a positive association between CD44 expression and breast tumor grade at both mRNA and protein levels21, 22. This paper further supported this opposite tendency of DACH1 and CD44 in low-grade and high-grade breast carcinoma, respectively.
Majorly according to the status of ER, PR and Her2, breast cancer is grouped into four distinct subtypes including luminal, Her2-positive, basal-type and normal-like. Among these, luminal tumor patients accounted for the most part of breast carcinoma population and have relatively good clinical outcomes, while patients with basal-like tumor endowed with malignancy features have significantly poor prognosis. Previous study has demonstrated that nuclear DACH1 is a biomarker of luminal breast cancer13. Breast tumors with positive ER–alpha and co-expressing PR-alpha were most likely to highly express DACH1, and nuclear DACH1 expression was positively correlated with luminal marker FOXA1 and inversely associated with basal-like markers EGFR13. In contrast, our previous meta-analysis showed that CD44 mRNA was remarkably enriched in basal-like breast cancer compared with luminal-type breast tumor21. CD44 was also positively correlated with basal markers EGFR, KRT5 and KRT17, and inversely associated with luminal marker FOXA1 22. Our results supported that luminal breast neoplasm tended to be with high DACH1 expression and low CD44 level, while basal-like tumors were most likely to be the inverse type. The correlation analysis further indicated that DACH1 was significantly inversely associated with CD44.
CSCs were composed of a subset of tumor cells with the expression of stem cell-associated markers and enhanced capacity for tumorigenesis, metastasis and therapy-resistance55. It has been implicated that endogenous DACH1 participated in the negative regulation of CSCs18. DACH1 suppressed the expression of stem cell markers SOX2, Nanog and KLF4 through binding to the promoters of these genes18. On the contrary, CD44, as a well-known CSCs marker, positively monitored the levels of CSCs markers20. Nuclear location of cleaved CD44 intracellular domain transcriptionally activated stemness factors Nanog, Sox2 and Oct453.
EMT is a complex and highly conserved process which enhances cellular invasiveness, being critical for metastasis of various solid tumors and considered to be promising therapeutic targets56. EMT was dysregulated by a complex network during tumor development. Knock-down of DACH1 in breast cancer cells MCF-7 and T47D promoted the morphology change from epithelial phenotype to mesenchymal pattern and interfered with cell–cell contact, accompanied by down-regulation of epithelial marker E-cadherin, resulting in cell migration and invasion10. DACH1 also transcriptionally suppressed the activity of Snail, leading to the activation of E-cadherin in breast cancer cells10, but the complex of DACH1 and Snail could bind to the E-box of E-cadherin promoter then contributing to the reduction of E-cadherin10. In addition, DACH1 reduced the expression of the mesenchymal marker Snail through suppressing the activity of the Y box-binding protein, an important EMT inducer9. Besides, previous study has revealed that DACH1 decreased both in breast cancer cell lines and tissues accompanied by relatively high proportion of CSCs18. Inversely, CD44 not only promoted EMT, but also was upregulated by some mesenchymal markers20. Previous studies have demonstrated that mesenchymal genes including ZEB1, TWIST1, SNAI1 and SLUG were positively correlated with CD44 expression20. The switch from CD44 variant isoforms to its standard isoform is essential to undergo EMT for normal epithelial cells57.
DACH1 played important roles in invasion and migration of various neoplasms, such as lung adenocarcinoma14, gastric cancer48, pancreatic cancer47 and breast cancer12. Several lines of evidence showed that DACH1 is absent or suppressed in poor prognosis breast cancer58. Breast cancer patients with reduced DACH1 expression had three years shorter time to death in comparison to those with normal DACH1 levels11. Immunohistochemistry analysis showed that higher nuclear level of DACH1 was predictive of longer disease-free interval, cancer relapse-free survival and distant metastasis-free survival over 5 years post diagnosis13. In consistence, our analysis of GSE databases showed that higher mRNA level of DACH1 was parallel with better OS, RFS and MFS. The protected effect of DACH1 in the prognosis of breast cancer patients could be explained partly by negatively interplaying CSCs and EMT10, 59. Besides, DACH1 inhibited breast cancer migration and invasion also via suppressing oncogene function through targeting interleukin-812. Altogether, these studies suggested that DACH1 could be a valuable molecular marker for prognosis, thereby detection of DACH1 level is useful for therapeutic stratification of breast cancer patients. In contrast, CD44 contributed to tumor formation and progression predictive of poor clinical outcome20. CD44 functioned as an oncoprotein and knockdown of CD44 remarkably attenuated the migration and invasion of breast cancer cells MDA-MB-231 and Hs578T by modulating c-Src transcription60. According to the analysis of 448 primary breast tumors, CD44 was parallel with enhancive distant recurrence and decreased disease-free survival61.
In this study, 19 public datasets were enrolled to assess the correlation between the mRNA levels of DACH1 and CD44 and the survival performance of breast cancer patients. However, there are still some limitations: 1) the overall sample size is limited. Some data were not available when the meta-analysis was conducted; 2) the platforms used to evaluate the mRNA expression of DACH1 and CD44 are different; 3) the results at the mRNA levels might not be consistent with those at protein abundance because there is a complex regulation network from mRNA to protein.
In conclusion, this study confirmed that DACH1 and CD44 inversely related in breast cancer, different grade tumors and different subtypes. DACH1 was negatively associated with CD44 in vitro and vivo. DACH1 served as a good prognostic marker, and CD44 was an unfavorable element of breast cancer patients. Thus, we concluded that CD44 might be a novel target of DACH1.
Materials and Methods
Immunohistochemical staining
To evaluate the expression of DACH1 and CD44 in normal breast versus breast tumor tissues, grade 1–2 versus grade 3 tissues as well as luminal-type and basal-like human breast carcinoma tissues, two commercially available tissue microarray (TMA) slides (BR1502–97 and BR1502-98, US Biomax, Inc, Rockville, MD) containing histologically confirmed tissues were purchased for immunohistochemistry (IHC) analysis. In addition, IHC was also employed to compare the expression of CD44, Fibronectin, Vimentin, Sox2, Myc, EGFR and Ki-67 in Met-1 nude mice xenograft tumors with overexpression of DACH1 and the controls. The Specific primary antibodies against DACH1 (10914-1-AP, ProteinTech Group), CD44 (15675-1-AP, ProteinTech Group), Fibronectin (sc-8422, Santa Cruze), Vimentin (5741, Cell Signaling Technology), Sox2 (AB5603, Millipore), Myc (sc-40, Santa Cruze), EGFR (sc-03, Santa Cruze) and Ki-67 (ab15680, Abcam) were utilized for IHC with a 2-step protocal62.
Analysis and quantification of staining
For quantification, a total of six 200× magnifications of each kind of protein were selected for IHC scoring. Two experienced pathologists assessed the immunohistochemical score independently. Scores were calculated on intensity and proportion of positive staining tumor cells in the whole tissue stains according to the Fromowitz Standard as described above63. The staining intensity was scored as 0 (no staining), 1 (weak staining, light yellow), 2 (moderate staining, yellow brown) and 3 (strong staining, brown). The proportions of stained tumor cells were classified as 1 (0–25% positive cells), 2 (26–50% positive cells), 2 (51–75% positive cells) and 3 (76–100% positive cells). The multiplication for intensity and proportion was utilized to represent the protein levels of DACH1, CD44, Myc, Sox2, Fibronectin, Vimentin, EGFR and Ki-67.
Cell culture
The breast cancer cell line Met-1, was cultured in high-glucose Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS, Life Technologies, Inc.). All cells were grown 37 °C in a humidified incubator with 5% CO2.
Western blot
Cells were washed with cold PBS, scraped into RIPA buffer and centrifuged. The cell lysates were subjected to 10% SDS-PAGE and transferred to a polyvinylidene fluoride (PVDF) hybridization transfer membrane. The primary antibodies used were as follows: HA (Sc-7392, Santa Cruze), CD44 (15675-1-AP, ProteinTech group), Fibronectin (sc-8422, Santa Cruze), Vimentin (5741, Cell Signaling Technology), p21cip1 (sc-6246, Santa Cruze), p27kip2 (sc-1641, Santa Cruze) and β-actin (Sc-47778, Santa Cruze). Secondary staining and detection were carried out in accordance with standard protocols16, 64.
RNA profiling by microarray
DNA-free total RNA isolated from Met-1 cells expressing GFP or DACH1 were used to probe Affymetrix Gene 1.0 arrays (Affymetrix, Santa Clara, CA). RNA quality was determined by gel electrophoresis. Analysis of the arrays was performed using GeneSpring. Arrays were normalized using robust multi-array analysis, and the p value of 0.05 was applied as a statistical criterion for differentially expressed genes.
Meta-analysis for DACH1 on published Gene Expression Omnibus (GEO) databases
We carried out a comprehensive search of relevant GEO databases for mRNA expression of DACH1 and CD44 through ArrayExpress and Oncomine. The datasets meeting the following criteria were included: 1) the datasets were about human breast cancer; 2) the mRNA expression of DACH1 and CD44 was measured in these databases; 3) clinical outcomes of patients were showed in these databases; 4) the sample capacity was more than 50. Only the latest and most complete datasets were included when several databases shared common patients. At last, a total of 19 independent human breast cancer microarray databases with the mRNA expression of DACH1 and CD44 and required the survival information of breast cancer patients were enrolled in this systematic analysis.
Cutoff value for DACH1 and CD44 was median expression. OS, RFS and MFS were evaluated by Cox proportional hazard ratio (HR) and 95% confidence interval (95% CI). HRs was employed to assess the survival outcome of breast cancer patients with high mRNA expression of DACH1 and CD44 and HR >1 indicated that high expression of DACH1 and CD44 predicted worse survival of patients. The random-effect model was employed when heterogeneity was present, and the fixed-effect model was used when homogeneity was demonstrated. Heterogeneity of publication was evaluated by means of the Chi-square-based-Q statistic and inconsistency index (I2) statistic. Statistical analysis was performed based on the guidelines of Meta-Analysis of Observational Studies. The STATA software package (version 12.0) (Stata Corp LP, College Station, TX, USA) was employed to perform the meta-analysis.
Analysis of gene expression data
GSE20711, available through GEO databases and containing 45 luminal-type and 22 basal-like breast carcinoma cases, was analyzed to evaluate the mRNA expression of DACH1 and CD44 in basal-like carcinoma in comparison with that in luminal-type tumors.
GSE20685, containing a total of 327 patients of breast cancer, was interrogated to assess the correlation between DACH1 and CD44, FN1, VIM, FOXA1, EGFR, MKI67 as well as the correlation between CD44 and FN1, VIM, YBX1, FOXA1, EGFR and MKI67 in breast cancer tissues.
Breast cancer cell line data of 51 breast cancer cell lines of different molecular subtypes published by Neve RM, et al.25 were also employed to evaluate whether the correlation in breast cancer cells was consistent with that in breast tumor tissues.
Nude Mice Study
The animal protocols were approved by the ethics committee of the Tongji Hospital of Huazhong University of Science and Technology. The methods used in this section were in accordance with the relevant guidelines and regulations. 1 × 105 Met-1 cells expressing GFP control or DACH1 were implanted subcutaneously into 4–6-week-old athymic female nude mice purchased from Wuhan Hamilton Biological Polytron Technologies Inc. The tumor growth was measured twice weekly for 3 weeks by using a digital caliper. Tumor weight was measured when mice were sacrificed on day 27 after cells implantation.
Statistical analysis
Correlation analyses were performed using SPSS 20 statistical software (SPSS Inc., Chicago, IL, USA). The Student’s t-test was applied to evaluate the differences in groups as appropriate and the significance level was set at 0.05. The correlation analysis was evaluated by a χ2- test. A two-tailed p value < 0.05 was considered statistically significant.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (NSFC) No. 81572608 and 81172422 (KW), 81502296 (JX), and was also supported by NIH grant R01CA132115 (RGP).
Author Contributions
H.X.X. collected and analyzed the data, did experiments and wrote the manuscript. S.N.Y., X.Y., J.X., and D.K. carried out immunohistochemical analysis. K.M.W. and R.G.P. designed the study and revised the manuscript. All authors edited and approved the final manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J. Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.Xu H, et al. Recent advances of highly selective CDK4/6 inhibitors in breast cancer. J. Hematol. Oncol. 2017;10:97. doi: 10.1186/s13045-017-0467-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hu X, Huang W, Fan M. Emerging therapies for breast cancer. J. Hematol. Oncol. 2017;10:98. doi: 10.1186/s13045-017-0466-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kong D, et al. The retinal determination gene network: from developmental regulator to cancer therapeutic target. Oncotarget. 2016;7:50755–50765. doi: 10.18632/oncotarget.9394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu Y, et al. The DACH/EYA/SIX gene network and its role in tumor initiation and progression. Int. J. Cancer. 2016;138:1067–1075. doi: 10.1002/ijc.29560. [DOI] [PubMed] [Google Scholar]
- 6.Xu HX, et al. Expression profile of SIX family members correlates with clinic-pathological features and prognosis of breast cancer: A systematic review and meta-analysis. Medicine (Baltimore) 2016;95:e4085. doi: 10.1097/MD.0000000000004085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu K, et al. EYA1 phosphatase function is essential to drive breast cancer cell proliferation through cyclin D1. Cancer Res. 2013;73:4488–4499. doi: 10.1158/0008-5472.CAN-12-4078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu Q, et al. The expression profile and clinic significance of the SIX family in non-small cell lung cancer. J. Hematol. Oncol. 2016;9:119. doi: 10.1186/s13045-016-0339-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu K, et al. Cell fate factor DACH1 represses YB-1-mediated oncogenic transcription and translation. Cancer Res. 2014;74:829–839. doi: 10.1158/0008-5472.CAN-13-2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao F, et al. DACH1 inhibits SNAI1-mediated epithelial-mesenchymal transition and represses breast carcinoma metastasis. Oncogenesis. 2015;4:e143. doi: 10.1038/oncsis.2015.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu K, et al. DACH1 is a cell fate determination factor that inhibits cyclin D1 and breast tumor growth. Mol. Cell Biol. 2006;26:7116–7129. doi: 10.1128/MCB.00268-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu K, et al. Dachshund inhibits oncogene-induced breast cancer cellular migration and invasion through suppression of interleukin-8. Proc. Natl. Acad. Sci. USA. 2008;105:6924–6929. doi: 10.1073/pnas.0802085105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Powe DG, et al. DACH1: its role as a classifier of long term good prognosis in luminal breast cancer. PLoS One. 2014;9:e84428. doi: 10.1371/journal.pone.0084428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Han N, et al. DACH1 inhibits lung adenocarcinoma invasion and tumor growth by repressing CXCL5 signaling. Oncotarget. 2015;6:5877–5888. doi: 10.18632/oncotarget.3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu L, et al. Silencing DACH1 promotes esophageal cancer growth by inhibiting TGF-beta signaling. PLoS One. 2014;9:e95509. doi: 10.1371/journal.pone.0095509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chu Q, et al. DACH1 inhibits cyclin D1 expression, cellular proliferation and tumor growth of renal cancer cells. J. Hematol. Oncol. 2014;7:73. doi: 10.1186/s13045-014-0073-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu Y, et al. DACH1 is a novel predictive and prognostic biomarker in hepatocellular carcinoma as a negative regulator of Wnt/beta-catenin signaling. Oncotarget. 2015;6:8621–8634. doi: 10.18632/oncotarget.3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu K, et al. Cell fate determination factor Dachshund reprograms breast cancer stem cell function. J. Biol. Chem. 2011;286:2132–2142. doi: 10.1074/jbc.M110.148395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schulenburg A, et al. Cancer stem cells in basic science and in translational oncology: can we translate into clinical application? J. Hematol. Oncol. 2015;8:16. doi: 10.1186/s13045-015-0113-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu H, et al. The role of CD44 in epithelial-mesenchymal transition and cancer development. Onco. Targets Ther. 2015;8:3783–3792. doi: 10.2147/OTT.S95470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu H, et al. Enrichment of CD44 in basal-type breast cancer correlates with EMT, cancer stem cell gene profile, and prognosis. Onco. Targets Ther. 2016;9:431–444. doi: 10.2147/OTT.S97192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu H, et al. CD44 correlates with clinicopathological characteristics and is upregulated by EGFR in breast cancer. Int. J. Oncol. 2016;49:1343–1350. doi: 10.3892/ijo.2016.3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Orian-Rousseau V. CD44 Acts as a Signaling Platform Controlling Tumor Progression and Metastasis. Front Immunol. 2015;6:154. doi: 10.3389/fimmu.2015.00154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zoller M. CD44: can a cancer-initiating cell profit from an abundantly expressed molecule? Nat. Rev. Cancer. 2011;11:254–267. doi: 10.1038/nrc3023. [DOI] [PubMed] [Google Scholar]
- 25.Neve RM, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515–527. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Desmedt C, et al. Strong time dependence of the 76-gene prognostic signature for node-negative breast cancer patients in the TRANSBIG multicenter independent validation series. Clin. Cancer Res. 2007;13:3207–3214. doi: 10.1158/1078-0432.CCR-06-2765. [DOI] [PubMed] [Google Scholar]
- 27.Pawitan Y, et al. Gene expression profiling spares early breast cancer patients from adjuvant therapy: derived and validated in two population-based cohorts. Breast Cancer Res. 2005;7:R953–964. doi: 10.1186/bcr1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hennessy BT, et al. Characterization of a naturally occurring breast cancer subset enriched in epithelial-to-mesenchymal transition and stem cell characteristics. Cancer Res. 2009;69:4116–4124. doi: 10.1158/0008-5472.CAN-08-3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dedeurwaerder S, et al. DNA methylation profiling reveals a predominant immune component in breast cancers. EMBO Mol. Med. 2011;3:726–741. doi: 10.1002/emmm.201100801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kao KJ, Chang KM, Hsu HC, Huang AT. Correlation of microarray-based breast cancer molecular subtypes and clinical outcomes: implications for treatment optimization. BMC Cancer. 2011;11:143. doi: 10.1186/1471-2407-11-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Clarke C, et al. Correlating transcriptional networks to breast cancer survival: a large-scale coexpression analysis. Carcinogenesis. 2013;34:2300–2308. doi: 10.1093/carcin/bgt208. [DOI] [PubMed] [Google Scholar]
- 32.Desmedt C, et al. Multifactorial approach to predicting resistance to anthracyclines. J. Clin. Oncol. 2011;29:1578–1586. doi: 10.1200/JCO.2010.31.2231. [DOI] [PubMed] [Google Scholar]
- 33.Loi S, et al. PIK3CA mutations associated with gene signature of low mTORC1 signaling and better outcomes in estrogen receptor-positive breast cancer. Proc. Natl. Acad. Sci. USA. 2010;107:10208–10213. doi: 10.1073/pnas.0907011107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bild AH, et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 2006;439:353–357. doi: 10.1038/nature04296. [DOI] [PubMed] [Google Scholar]
- 35.Terunuma A, et al. MYC-driven accumulation of 2-hydroxyglutarate is associated with breast cancer prognosis. J. Clin. Invest. 2014;124:398–412. doi: 10.1172/JCI71180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Heikkinen T, et al. Variants on the promoter region of PTEN affect breast cancer progression and patient survival. Breast Cancer Res. 2011;13:R130. doi: 10.1186/bcr3076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hatzis C, et al. A genomic predictor of response and survival following taxane-anthracycline chemotherapy for invasive breast cancer. Jama. 2011;305:1873–1881. doi: 10.1001/jama.2011.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Symmans WF, et al. Genomic index of sensitivity to endocrine therapy for breast cancer. J. Clin. Oncol. 2010;28:4111–4119. doi: 10.1200/JCO.2010.28.4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang Y, et al. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet. 2005;365:671–679. doi: 10.1016/S0140-6736(05)70933-8. [DOI] [PubMed] [Google Scholar]
- 40.Tofigh A, et al. The prognostic ease and difficulty of invasive breast carcinoma. Cell Rep. 2014;9:129–142. doi: 10.1016/j.celrep.2014.08.073. [DOI] [PubMed] [Google Scholar]
- 41.Minn AJ, et al. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436:518–524. doi: 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Minn AJ, et al. Lung metastasis genes couple breast tumor size and metastatic spread. Proc. Natl. Acad. Sci. USA. 2007;104:6740–6745. doi: 10.1073/pnas.0701138104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sircoulomb F, et al. Genome profiling of ERBB2-amplified breast cancers. BMC Cancer. 2010;10:539. doi: 10.1186/1471-2407-10-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nagalla S, et al. Interactions between immunity, proliferation and molecular subtype in breast cancer prognosis. Genome Biol. 2013;14:R34. doi: 10.1186/gb-2013-14-4-r34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Davis RJ, et al. Dach1 mutant mice bear no gross abnormalities in eye, limb, and brain development and exhibit postnatal lethality. Mol. Cell Biol. 2001;21:1484–1490. doi: 10.1128/MCB.21.5.1484-1490.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Heanue TA, et al. Dach1, a vertebrate homologue of Drosophila dachshund, is expressed in the developing eye and ear of both chick and mouse and is regulated independently of Pax and Eya genes. Mech. Dev. 2002;111:75–87. doi: 10.1016/S0925-4773(01)00611-6. [DOI] [PubMed] [Google Scholar]
- 47.Bu XN, Qiu C, Wang C, Jiang Z. Inhibition of DACH1 activity by short hairpin RNA represses cell proliferation and tumor invasion in pancreatic cancer. Oncol. Rep. 2016;36:745–754. doi: 10.3892/or.2016.4843. [DOI] [PubMed] [Google Scholar]
- 48.Yan W, et al. Epigenetic silencing of DACH1 induces the invasion and metastasis of gastric cancer by activating TGF-beta signalling. J. Cell Mol. Med. 2014;18:2499–2511. doi: 10.1111/jcmm.12325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/S0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 50.Chen Y, et al. Identifying potential cancer driver genes by genomic data integration. Sci. Rep. 2013;3:3538. doi: 10.1038/srep03538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Popov VM, et al. The Dachshund gene in development and hormone-responsive tumorigenesis. Trends Endocrinol. Metab. 2010;21:41–49. doi: 10.1016/j.tem.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen K, et al. Acetylation of the cell-fate factor dachshund determines p53 binding and signaling modules in breast cancer. Oncotarget. 2013;4:923–935. doi: 10.18632/oncotarget.1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cho Y, et al. Cleaved CD44 intracellular domain supports activation of stemness factors and promotes tumorigenesis of breast cancer. Oncotarget. 2015;6:8709–8721. doi: 10.18632/oncotarget.3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Morath I, Hartmann TN, Orian-Rousseau V. CD44: More than a mere stem cell marker. Int. J. Biochem. Cell Biol. 2016;81:166–173. doi: 10.1016/j.biocel.2016.09.009. [DOI] [PubMed] [Google Scholar]
- 55.Velasco-Velazquez MA, Homsi N, De La Fuente M, Pestell RG. Breast cancer stem cells. Int. J. Biochem. Cell Biol. 2012;44:573–577. doi: 10.1016/j.biocel.2011.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jayachandran A, Dhungel B, Steel JC. Epithelial-to-mesenchymal plasticity of cancer stem cells: therapeutic targets in hepatocellular carcinoma. J. Hematol. Oncol. 2016;9:74. doi: 10.1186/s13045-016-0307-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Klingbeil P, Isacke CM. The ‘alternative’ EMT switch. Breast Cancer Res. 2011;13:313. doi: 10.1186/bcr2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu K, et al. Cell fate determination factor DACH1 inhibits c-Jun-induced contact-independent growth. Mol. Biol. Cell. 2007;18:755–767. doi: 10.1091/mbc.E06-09-0793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wu K, et al. DACH1 inhibits transforming growth factor-beta signaling through binding Smad4. J. Biol. Chem. 2003;278:51673–51684. doi: 10.1074/jbc.M310021200. [DOI] [PubMed] [Google Scholar]
- 60.Nam K, Oh S, Lee KM, Yoo SA, Shin I. CD44 regulates cell proliferation, migration, and invasion via modulation of c-Src transcription in human breast cancer cells. Cell. Signal. 2015;27:1882–1894. doi: 10.1016/j.cellsig.2015.05.002. [DOI] [PubMed] [Google Scholar]
- 61.McFarlane S, et al. CD44 increases the efficiency of distant metastasis of breast cancer. Oncotarget. 2015;6:11465–11476. doi: 10.18632/oncotarget.3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu, Q. Q. et al. Decreased DACH1 expression in glomerulopathy is associated with disease progression and severity. Oncotarget (2016). [DOI] [PMC free article] [PubMed]
- 63.Fromowitz FB, et al. ras p21 expression in the progression of breast cancer. Hum. Pathol. 1987;18:1268–1275. doi: 10.1016/S0046-8177(87)80412-4. [DOI] [PubMed] [Google Scholar]
- 64.Tian Y, et al. Modification of platinum sensitivity by KEAP1/NRF2 signals in non-small cell lung cancer. J. Hematol. Oncol. 2016;9:83. doi: 10.1186/s13045-016-0311-0. [DOI] [PMC free article] [PubMed] [Google Scholar]


