Abstract
Development of miniaturized devices that enable rapid and direct analysis of the specific binding of small molecules to proteins could be of substantial importance to the discovery of and screening for new drug molecules. Here, we report highly sensitive and label-free direct electrical detection of small-molecule inhibitors of ATP binding to Abl by using silicon nanowire field-effect transistor devices. Abl, which is a protein tyrosine kinase whose constitutive activity is responsible for chronic myelogenous leukemia, was covalently linked to the surfaces of silicon nanowires within microfluidic channels to create active electrical devices. Concentration-dependent binding of ATP and concentration-dependent inhibition of ATP binding by the competitive small-molecule antagonist STI-571 (Gleevec) were assessed by monitoring the nanowire conductance. In addition, concentration-dependent inhibition of ATP binding was examined for four additional small molecules, including reported and previously unreported inhibitors. These studies demonstrate that the silicon nanowire devices can readily and rapidly distinguish the affinities of distinct small-molecule inhibitors and, thus, could serve as a technology platform for drug discovery.
Keywords: inhibitors, kinase, Gleevec, chronic myclogenous leukemia, drug discovery
Identification of organic molecules that bind specifically to proteins is central to the discovery and development of new pharmaceuticals and to chemical genetic approaches for elucidating complex pathways in biological systems (1–4). Broadly representative of the importance of this concept for developing drugs to treat disease have been efforts focused on identifying inhibitors to protein tyrosine kinases (1, 5). Tyrosine kinases represent especially attractive targets because they are central elements in the networks that mediate signal transduction in mammalian cells. The regulatory function of tyrosine kinases occurs through phosphorylation of a tyrosine residue of a substrate protein by using ATP as a phosphate source (Fig. 1A) and subsequent transmission of this event through signal transduction cascade. Deregulation of phosphorylation through, for example, mutation or overexpression of protein tyrosine kinases has been linked to a number of diseases, including cancer (1, 5, 6).
Fig. 1.
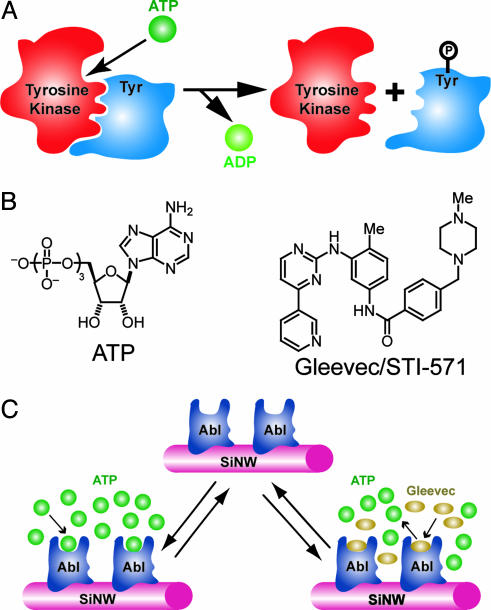
Detection of small-molecule–protein interactions for tyrosine kinases. (A) Scheme illustrating basic activity of a tyrosine kinase: ATP binds to the tyrosine kinase active site, and then the γ-phosphate group is transferred to the tyrosine (Tyr) residue of the substrate protein. (B) Structures of ATP and the small-molecule inhibitor of ATP binding to the Abl tyrosine kinase, Gleevec, or STI-571. (C) Detection of ATP binding and small-molecule inhibition of binding by using a SiNW sensor device. The tyrosine kinase Abl is covalently linked to the surface of a SiNW, and then the conductance of the nanowire device is monitored to detect ATP binding and the competitive inhibition of ATP binding by Gleevec.
The identification of inhibitors to ATP or substrate protein binding thus can serve as a means of treating diseases linked to a tyrosine kinase. A successful example of this strategy has been the introduction of the small molecule STI-571, or Gleevec (Fig. 1B), which competitively inhibits ATP binding to the tyrosine kinase Abl and is a highly effective treatment for chronic myelogenous leukemia (1, 5, 7, 8). This success and the recognition that Gleevec may be unable to cure late-stage chronic myelogenous leukemia because of mutations in the kinase (5, 8–10) suggest that the development of approaches that enable rapid, flexible, and quantitative comparison of small-molecule inhibitors of ATP or substrate protein binding to tyrosine kinases, including those with mutations, could substantially improve drug discovery and development.
Here, we report a highly sensitive detection scheme for identifying small-molecule inhibitors that does not require labeling of the protein, ATP, or small molecule and can be carried out in real-time by using silicon nanowire (SiNW) field-effect transistor (FET) devices. Previously, we have shown that SiNW FETs could be used to detect binding and unbinding of proteins to their corresponding ligands linked to nanowire surfaces in aqueous solutions (11), and, more recently, proteins and nucleic acids also have been detected by using carbon nanotubes (12–14) and SiNW (15) FETs, respectively. To demonstrate the potential for screening small-molecule inhibitors to tyrosine kinases, we linked the Abl kinase to the surface of SiNW FETs and investigated the binding of ATP and competitive inhibition of ATP binding with organic molecules as shown schematically in Fig. 1C. In this configuration, binding or inhibition of binding of the negatively charged ATP to Abl linked at the SiNW surface is detected simply as an increase or decrease in the conductance of the device resulting from the gating effect of surface charge (11).
Materials and Methods
SiNW FETs. SiNWs were synthesized by chemical vapor deposition using 20-nm gold nanoclusters as catalysts, silane as reactant, and diborane as p-type dopant with a B:Si ratio of 1:4,000 (16). The as-grown SiNWs were dispersed from ethanol suspension onto silicon substrates with 600-nm-thick oxide layer (11, 17, 18), and then source/drain metal contacts to the ends of the SiNWs were defined by electron beam lithography followed by electron beam evaporation of Ti (60 nm) and Au (40 nm). The SiNW devices exhibited transconductance values (18) of 1,200–1,600 nS/V in buffer solution following the modification procedure described below.
Surface Modification. Abl was covalently linked to the surfaces of SiNW devices by using a three-step procedure. First, SiNW devices were cleaned in an oxygen plasma [0.3 torr (1 torr = 133 Pa), 25 W for 60 s] to remove contaminants and leave a uniform silicon oxide surface. Second, devices were reacted with a 2% ethanol solution of 3-(trimethoxysilyl)propyl aldehyde (United Chemical Technologies, Bristol, PA) containing 4% water and 0.1% acetic acid for 1 hr. After reaction, the device chip was rinsed with absolute ethanol and heated at 120°C for 10 min in the N2 atmosphere. Third, Abl tyrosine kinase (New England Biolabs, Beverly, MA) was coupled to the SiNW surfaces by flowing through a microfluidic channel (11) at a concentration of 5 μg/ml with 4 mM sodium cyanoborohydride at a flow rate of 0.15 ml/hr; unreacted aldehyde groups were quenched by a flow of 15 mM Tris buffer (pH 7.5) for 5–10 min. The Abl protein was purified by dialysis against 15 mM Hepes buffer (pH 7.5) containing 0.1 mM MgCl2 and 0.1 mM EGTA before reaction with the SiNW surfaces.
Competitive Inhibitors. Extraction of Gleevec. A capsule of Gleevec was broken, and the powder was dissolved in ethyl acetate, followed by washing twice with 1.0 M sodium carbonate aqueous solution and once with saturated sodium chloride solution. The organic layer was dried over anhydrous sodium sulfate, filtered, and concentrated to provide pure Gleevec as white powder. 1H NMR (600 MHz, CD3OD): δ 9.27 (d, J = 1.8 Hz, 1H), 8.62 (dd, J = 4.8, 1.8 Hz, 1H), 8.59 (dt, J = 8.4, 1.8 Hz, 1H), 8.45 (d, J = 5.4 Hz, 1H), 8.19 (s, 1H), 7.90 (d, J = 7.8 Hz, 2H), 7.54 (dd, J = 7.8, 4.8 Hz, 1H), 7.48 (d, J = 8.4 Hz, 2H), 7.40 (dd, J = 8.4, 1.8 Hz, 1H), 7.35 (d, J = 4.8 Hz, 1H), 7.25 (d, J = 7.8 Hz, 1H), 3.61 (s, 3H), 2.51 (br), 2.31 (s, 3H), 2.28 (s, 3H); MS(APCI+) C29H32N7O (M+H) 493.8.
A2, A3, and biotin. A2 [N-(3-nitro-6-methylphenyl)-4-(3′-pyridyl)-2-pyrimidineamine] was obtained from Rintech (Gaithersburg, MD). A3 [methyl 4-(2,5-dihydroxybenzylamino)benzoate] was obtained from Calbiochem. Biotin was obtained from Sigma–Aldrich and used without further purification.
Synthesis of A1. To a solution of N-(3-nitro-6-methylphenyl)-4-(3′-pyridyl)-2-pyrimidineamine in ethanol was added tin(II) chloride. After stirring at 65°C overnight, the reaction was cooled to room temperature, diluted by methylene chloride, and washed three times with saturated sodium chloride solution. The organic layer was dried over anhydrous sodium sulfate, filtered, concentrated, and purified with silica gel chromatography to provide N-(3-amino-6-methylphenyl)-4-(3′-pyridyl)-2-pyrimidineamine (A1) as a pale yellow oil. 1H NMR (600 MHz, CDCl3): δ 9.27 (s, 1H), 8.72 (d, J = 4.2 Hz, 1H), 8.50 (d, J = 5.4 Hz, 1H), 8.35 (d, J = 7.8 Hz, 1H), 7.62 (d, J = 1.8 Hz, 1H), 7.44 (dd, J = 8.1, 4.5 Hz, 1H), 7.15 (d, J = 5.4 Hz, 1H), 7.01 (d, J = 8.4 Hz, 1H), 6.96 (s, 1H), 6.42 (dd, J = 7.8, 2.4 Hz, 1H), 2.26 (s, 3H); MS(ES+) C16H16N5 (M+H) 277.7.
Solution Sensing Measurements. The sensing measurements were carried out in similar manner to previous studies (11). In short, microfluidic channels (200 μm height and width) made with poly(dimethylsiloxane) mold, which was passivated with polyethylene glycol (molecular weight 5,000; Shearwater Polymers, Huntsville, AL) to reduce unspecific adsorption. Before experiments, buffer (1.5 μM Hepes buffer at pH 7.5 containing 1 μM MgCl2 and 1 μM EGTA) was flowed through the sensor to establish a baseline. This same buffer was used in all experiments to assess ATP binding and competitive inhibition of ATP binding. A constant flow of 0.2 ml/hr was used in all of the experiments. The time-dependent conductance of SiNW FET devices was recorded by using a lock-in technique at a frequency of 31 Hz and an amplitude of 30 mV; the dc source-drain bias was zero in all of the measurements. Change of conductance (ΔG) values were obtained from average values of the conductance baseline (≈150 data points) before solution change and signal conductance (200–300 data points), with error bars corresponding to ± 1 SD; signal spikes immediately after solution changes were not included in the averages. In all of the ATP binding/inhibition experiments, the ionic strength of the buffer solutions was 10–1,000 times greater than the ATP or small-molecule inhibitor concentrations.
Results
ATP Sensing. Typical time-dependent data recorded from an Abl-modified SiNW device (Fig. 2A) exhibited reversible, concentration-dependent increases in conductance upon introducing buffer solutions containing ATP. The reversibility of these concentration-dependent increases was evident from the corresponding decreases in conductance to baseline value upon introducing buffer solution without ATP. The observed increases in conductance are consistent with the binding of negatively charged ATP to Abl. Specifically, a p-type SiNW FET will exhibit an increase (decrease) in conductance when the gate-voltage is negative (positive) because of the accumulation (depletion) of carriers. Binding of negatively charged ATP to the Abl kinase increases the negative surface-charge density and increases conductance similar to a negative gate voltage (11). The conductance changes also can exhibit variations vs. time after switching between buffer and ATP/buffer solutions, for example, between sets of arrows in Fig. 2 A. These variations are believed to be due to several factors, including (i) electronic noise, which is produced when connecting solution reservoirs at different potentials to the microfluidic cell, at short time scales, and (ii) the sampling of protein-binding sites with different accessibility at longer time scales. Notably, control experiments carried out with devices prepared in the same way except that Abl protein was not coupled to the surfaces showed little or no change in conductance upon addition of the same concentration ATP solutions. These experiments demonstrate that the conductance changes observed for the Abl-modified SiNW devices correspond to specific binding of ATP to the Abl tyrosine kinase.
Fig. 2.
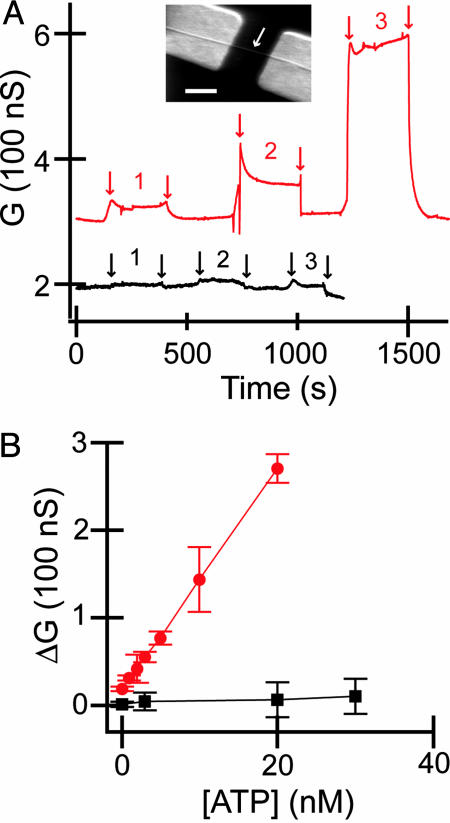
Detection of ATP binding. (A) Conductance (G) vs. ATP concentration for SiNWs modified with Abl (red curve) and a device prepared in an identical fashion, except Abl was not coupled to the surface (black curve). Regions 1, 2, and 3 correspond to 0.1, 3, and 20 nM ATP, respectively. Arrows indicate the points where solution is changed. (Inset) Scanning electron micrograph of a typical SiNW FET device. The nanowire is highlighted by a white arrow and is contacted on either end with Ti/Au metal electrodes. (Scale bar: 500 nm.) (B) Change in conductance (ΔG) vs. ATP concentration for Abl-modified SiNW (red) and SiNW without Abl (black).
In addition, the data demonstrate that ATP binding to Abl can be distinguished above background at concentrations at least as low as 100 pM. Plots summarizing the concentration-dependent ATP binding to Abl monitored by the SiNW devices exhibit characteristic linear response at low concentrations (Fig. 2B) and saturation at higher concentrations (see below). Devices without Abl linked to the surface showed essentially no concentration-dependent response. The ability to rapidly quantify ATP binding without specific labels suggests that this approach could serve as a simple and potentially quantitative screen for ATP binding to proteins.
Competitive Inhibition of ATP Binding by Gleevec. Central to the focus of this work is the use of the SiNW devices to monitor directly competitive inhibition of ATP binding by small molecules. We investigated competitive inhibition by recording the conductance of Abl-modified SiNW FETs in the presence of buffer solutions containing ATP and ATP/Gleevec. We found that increases in the Gleevec concentration at fixed ATP concentration yielded systematic and reproducible decreases in the conductance associated with ATP binding (Fig. 3A); that is, Gleevec, which is not negatively charged, competes with ATP for the binding site in Abl. Measurements of the conductance changes as a function of ATP concentration for different fixed concentrations of Gleevec (Fig. 3B) demonstrate several key points. First, the ATP binding curves shifted systematically to the right (higher ATP concentration) as Gleevec was increased from 1 to 3 nM, although the saturation conductance changes at high ATP concentrations are very similar. These results are consistent with reversible competitive inhibition of an agonist (ATP) with an antagonist (Gleevec). The presence of Gleevec reduces the total number of available binding sites at relatively low ATP concentrations, and this reduction effectively translates into lower sensor response at a fixed ATP concentration. However, sufficiently high ATP concentrations overwhelm the influence of Gleevec, and a saturation response due to total receptor occupancy is ultimately observed.
Fig. 3.
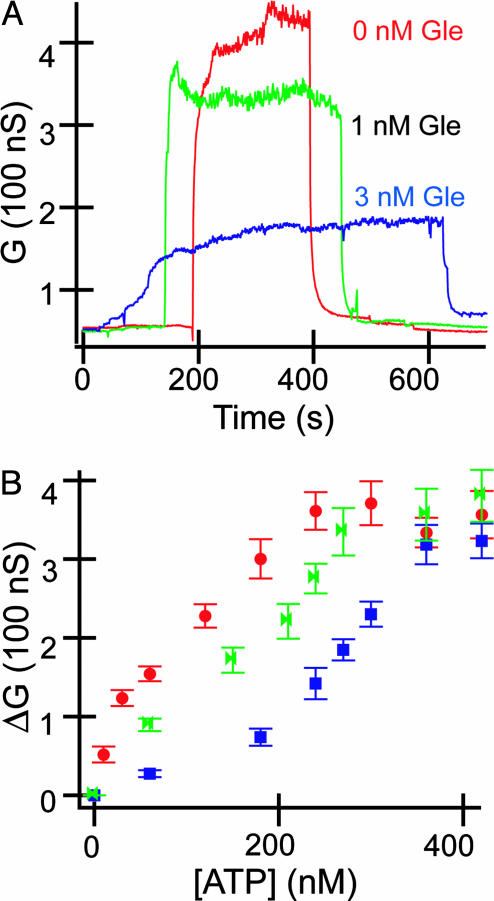
Inhibition of ATP binding by Gleevec. (A) Conductance vs. time data for ATP binding in the presence of different concentrations of Gleevec (Gle). The ATP concentration was fixed at 240 nM in the three experiments; the Gleevec concentrations in the solutions are indicated. (B) Change in conductance (ΔG) vs. ATP concentration for Abl-modified SiNW in the presence of different base concentrations of Gleevec: red, green, and blue correspond to 0, 1, and 3 nM Gleevec, respectively.
Second, these data can be analyzed to provide a measure of the ATP dissociation constant and Gleevec or other small-molecule inhibition constant. The ATP binding constant estimated from the linear response region (11) of the data is ≈50 nM. This value is in good agreement with the constant, 65 nM, determined from the analysis of concentration-dependent kinetic data for substrate phosphorylation by using radioactively labeled ATP (8). The shift in the ATP binding curves in Fig. 3B can be analyzed (19) by using C′/C = 1 + ([I]/KI), where C and C′ are the concentrations of ATP required to produce a conductance response in the absence and presence, respectively, of inhibitor at [I], and KI is the inhibition constant. Analysis of our data yields a KI of ≈2 nM, which is smaller than a value, 25 nM, obtained from kinetic assays (8), although we note that values reported from different studies have varied by more than an order of magnitude (8, 9).
Competitive Inhibition of ATP Binding by Additional Small Molecules. In addition, we have investigated inhibition of ATP binding by four additional small molecules, two of which are known inhibitors for Abl. Molecules A1, A2, and A3 shown in Fig. 4A have structural homology with Gleevec (Fig. 1B), whereas the fourth molecule tested, biotin, was chosen as a control. Plots of the normalized conductance vs. time recorded from Abl-modified SiNW devices (Fig. 4B) exhibit reversible decreases in conductance due to competitive inhibition of ATP binding by small molecules. These data are displayed as normalized conductance to compare devices with different absolute responses. Notably, the conductance decrease at constant small-molecule concentration, which is indicative of the degree of inhibition, depends strongly on molecular structure with Gleevec > A1 > A2 > A3; the control biotin shows essentially no change above background as expected. The ordering for Gleevec, A1, and A3 is in agreement with reported inhibition constants of 25 nM, 1.5 μM, and 9 μM, respectively (8, 20, 21). Interestingly, A2, whose KI value was not found in the published literature, shows clear inhibition with a magnitude less than A1 but greater than A3.
Fig. 4.
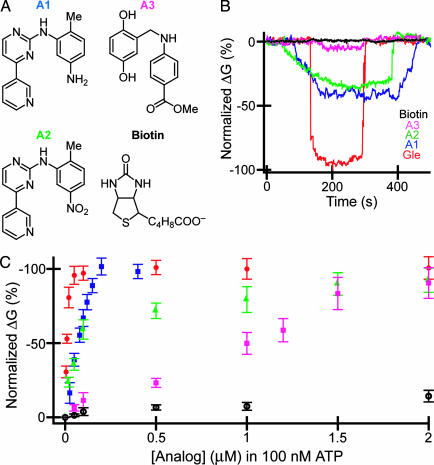
Screening of small-molecule inhibitors. (A) Structures of small molecules investigated for inhibition of ATP binding to Abl. (B) Normalized conductance vs. time data recorded from Abl-modified SiNW devices by using solutions containing 100 nM ATP and 50 nM small molecules for Gleevec (red), A1 (blue), A2 (green), A3 (pink), and biotin (black). The ATP and small molecules were dissolved in the same buffer as described in Materials and Methods. (C) Normalized change in conductance (ΔG) vs. small-molecule concentration in fixed 100 nM of ATP. The red, blue, green, pink, and black data points correspond to Gleevec, A1, A2, A3, and biotin, respectively. To correct for different absolute device sensitivity, the data were plotted as the normalized ΔG(ΔG, specific concentration)/(saturation ΔG) × 100%, where ΔG is the difference between the measured and baseline conductance.
To further characterize the small-molecule binding and inhibition, we recorded data as a function of the concentration of small molecule at a fixed ATP concentration of 100 nM (Fig. 4C). The results for Gleevec, A1, A2, and A3 exhibit a linear increase in the inhibition at low concentrations, followed by saturation at higher values, whereas biotin shows almost no concentration dependence. The data for the inhibitors also shifted systematically to right (higher inhibitor concentration), which is indicative of reduced inhibition for Gleevec > A1 > A2 > A3. The interaction of A2 with Abl has not been characterized previously by x-ray crystallography; however, based on the structure of Gleevec bound to Abl (7), we speculate that A2 and A1 should have similar interactions, except for potential loss of one hydrogen bond due to replacement of the amino group in A1 with a nitro group in A2 (Fig. 4A). Confirmation of this suggestion will require direct structural measurements, although we believe the detected binding of A2 highlights the potential of our SiNW detectors for direct and rapid discovery of molecular inhibitors.
Discussion
These studies demonstrate label-free and highly sensitive detection of ATP binding and small-molecule inhibition of ATP binding to Abl by using SiNW FET devices. Label-free detection is of considerable interest because it represents the most general approach for screening and assay development and often can facilitate analysis of key thermodynamic and kinetic parameters (22). In this regard, it is important to compare our nanowire-based approach to other label-free methods used to study small-molecule–protein binding, including surface plasmon resonance (SPR) and calorimetry. In recent SPR studies of small-molecule binding to kinases and other proteins (23–26), the protein has been immobilized on the SPR sensor chip, and then the optical response was recorded as a function of time under different experimental conditions. These studies have shown that SPR can provide association and dissociation rate constants and correspondingly equilibrium dissociation constants for small-molecule–protein binding, information that can be valuable to understanding origin of relative binding behavior. We have not achieved this level of quantitative analysis in our initial studies with SiNW FETs but note that implementation of background subtraction techniques used in SPR (23, 24, 26), which could be readily implemented with small arrays (27), should enable similar parameter analysis with our approach. We also believe that there are possible advantages to the SiNW devices compared with SPR, including (i) higher sensitivity, (ii) smaller quantity of protein required to make active sensor chips, and (iii) the potential for very large integrated arrays.
In addition, calorimetry is a general technique for determining thermodynamic properties of small-molecule–protein and other molecular interactions (28, 29). The use of calorimetry has, however, been restricted due to the requirement of relatively large protein concentration of the order of milligrams per milliliter (23). In many systems, such as new proteins being screened for kinase activity and/or inhibition of ATP binding, only small or trace quantities of protein are available, thus precluding the use of calorimetry. Very recently, a potential solution to this size scale in calorimetry has been described with the fabrication of enthalpy arrays (30). Ref. 30 demonstrates that the general nature of calorimetry can be on orders-of-magnitude smaller scale, although the quantities of protein required for an experiment are still much larger and the sensitivity lower than we report for the SiNW devices.
In summary, we have described a highly sensitive, direct electrical detection methodology for investigating ATP binding and small-molecule inhibition of ATP binding to the tyrosine kinase, Abl. We have shown that concentration-dependent binding of ATP and concentration-dependent inhibition of ATP binding by Gleevec can be characterized by monitoring the conductance of SiNW FET devices with Abl linked to the SiNW surface and that this data can be used to determine binding and inhibition constants. In addition, concentration-dependent inhibition of ATP binding by four other small molecules was characterized and shown to provide a rapid assay of relative binding/inhibition affinities, which suggests that this approach could serve as a technology platform for drug discovery. Our method is also attractive from the standpoint of requiring very little protein to make active devices, which could make studies of systems produced at low expression levels possible, and has the potential to be extended to large, integrated arrays by using recently reported assembly methods (27). Lastly, these results suggest that it should be straightforward to use our SiNW detection method to probe small-molecule-mediated inhibition of protein–protein interactions, and thus we believe these SiNW nanosensors could impact broadly drug discovery and chemical genetics.
Acknowledgments
We thank T. Mitchison (Harvard University) for Gleevec and helpful discussion. C.M.L. was supported by the National Cancer Institute, Ellison Medical Foundation, and Defense Advanced Research Projects Agency.
Author contributions: C.M.L. designed research; W.U.W. and C.C. performed research; K.-h.L. and Y.F. contributed new reagents/analytic tools; W.U.W. and C.M.L. analyzed data; and W.U.W. and C.M.L. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: FET, field-effect transistor; SiNW, silicon nanowire; SPR, surface plasmon resonance; A1, N-(3-amino-6-methylphenyl)-4-(3′-pyridyl)-2-pyrimidineamine; A2, N-(3-nitro-6-methylphenyl)-4-(3′-pyridyl)-2-pyrimidineamine; A3, methyl 4-(2,5-dihydroxybenzylamino)benzoate.
References
- 1.Grosios, K. & Traxler, P. (2003) Drugs Future 28, 670-697. [Google Scholar]
- 2.Strausberg, R. L. & Schreiber, S. L. (2003) Science 300, 294-295. [DOI] [PubMed] [Google Scholar]
- 3.Stockwell, B. R. (2000) Trends Biotechnol. 18, 449-455. [DOI] [PubMed] [Google Scholar]
- 4.MacBeath, G. (2001) Genome Biol. 2, 2005.1-2005.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Becker, J. (2004) Nat. Biotechnol. 22, 15-18. [DOI] [PubMed] [Google Scholar]
- 6.Blume-Jensen, P. & Hunter, T. (2001) Nature 411, 355-365. [DOI] [PubMed] [Google Scholar]
- 7.Schindler, T., Bornmann, W., Pellicena, P., Miller, W. T., Clarkson, B. & Kuriyan, J. (2000) Science 289, 1938-1942. [DOI] [PubMed] [Google Scholar]
- 8.Corbin, A. S., Buchdunger, E., Pascal, F. & Druker, B. J. (2002) J. Biol. Chem. 277, 32214-32219. [DOI] [PubMed] [Google Scholar]
- 9.Azam, M., Latek, R. R. & Daley, G. Q. (2003) Cell 112, 831-843. [DOI] [PubMed] [Google Scholar]
- 10.Shah, N. P., Tran, C., Lee, F. Y., Chen, P., Norris, D. & Sawyers, C. L. (2004) Science 305, 399-401. [DOI] [PubMed] [Google Scholar]
- 11.Cui, Y., Wei, Q., Park, H. & Lieber, C. M. (2001) Science 293, 1289-1292. [DOI] [PubMed] [Google Scholar]
- 12.Chen, R. J., Bangsaruntip, S., Drouvalakis, K. A., Kam, N. W. S., Shim, M., Li, Y., Kim, W., Utz, P. J. & Dai, H. (2003) Proc. Natl. Acad. Sci. USA 100, 4984-4989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Besteman, K., Lee, J. O., Wiertz, F. G. M., Heering, H. A. & Dekker, C. (2003) Nano Lett. 3, 727-730. [Google Scholar]
- 14.Chen, R. J., Choi, H. C., Bangsaruntip, S., Yenilmez, E., Tang, X. W., Wang, Q., Chang, Y. L. & Dai, H. J. (2004) J. Am. Chem. Soc. 126, 1563-1568. [DOI] [PubMed] [Google Scholar]
- 15.Hahm, J. & Lieber, C. M. (2004) Nano Lett. 4, 51-54. [Google Scholar]
- 16.Cui, Y. Lauhon, L. J., Gudiksen, M. S., Wang, J. & Lieber, C. M. (2001) Appl. Phys. Lett. 78, 2214-2216. [Google Scholar]
- 17.Cui, Y. & Lieber, C. M. (2001) Science 291, 851-853. [DOI] [PubMed] [Google Scholar]
- 18.Cui, Y., Zhong, Z., Wang, D., Wang, W. U. & Lieber, C. M. (2003) Nano Lett. 3, 149-152. [Google Scholar]
- 19.Bourne, H. R. & Zastrow, M. V. (2001) in Basic and Clinical Pharmacology, ed. Katzung, B. G. (McGraw–Hill, New York), 8th ed., pp. 9-34.
- 20.Zimmermann, J., Buchdunger, E., Mett, H., Meyer, T. & Lydon, N. B. (1997) Bioorg. Med. Chem. Lett. 7, 187-192. [Google Scholar]
- 21.Anafi, M., Gazit, A., Gilon, C., Ben-Neriah, Y. & Levitzki, A. (1992) J. Biol. Chem. 267, 4518-4523. [PubMed] [Google Scholar]
- 22.Cooper, M. A. (2003) Anal. Bioanal. Chem. 377, 834-842. [DOI] [PubMed] [Google Scholar]
- 23.Casper, D., Bukhtiyarova, M. & Springman, E. B. (2004) Anal. Biochem. 325, 126-136. [DOI] [PubMed] [Google Scholar]
- 24.Myszka, D. G. (2004) Anal. Biochem. 329, 316-323. [DOI] [PubMed] [Google Scholar]
- 25.Day, Y. S. N., Baird, C. L., Rich, R. L. & Myszka, D. G. (2002) Protein Sci. 11, 1017-1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rich, R. L. & Myszka, D. G. (2000) Curr. Opin. Biotechnol. 11, 54-61. [DOI] [PubMed] [Google Scholar]
- 27.Jin, S., Whang, D. M., McAlpine, M. C., Friedman, R. S., Wu, Y. & Lieber, C. M. (2004) Nano Lett. 4, 915-919. [Google Scholar]
- 28.Leavitt, S. & Freire, E. (2001) Curr. Opin. Struc. Biol. 11, 560-566. [DOI] [PubMed] [Google Scholar]
- 29.Jelesarov, I. & Bosshard, H. R. (1999) J. Mol. Recognit. 12, 3-18. [DOI] [PubMed] [Google Scholar]
- 30.Torres, F. E., Kuhn, P., De Bruyker, D., Bell, A. G., Wolkin, M. V., Peeters, E., Williamson, J. R., Anderson, G. B., Schmitz, G. P., Recht, M. I., et al. (2004) Proc. Natl. Acad. Sci. USA 101, 9517-9522. [DOI] [PMC free article] [PubMed] [Google Scholar]