Abstract
Background
Cultivated ginseng is often introduced as a substitute and adulterant of Russian wild ginseng due to its lower cost or misidentification caused by similarity in appearance with wild ginseng. The aim of this study is to develop a simple and reliable method to differentiate Russian wild ginseng from cultivated ginseng.
Methods
The mitochondrial NADH dehydrogenase subunit 7 (nad7) intron 3 regions of Russian wild ginseng and Chinese cultivated ginseng were analyzed. Based on the multiple sequence alignment result, a specific primer for Russian wild ginseng was designed by introducing additional mismatch and allele-specific polymerase chain reaction (PCR) was performed for identification of wild ginseng. Real-time allele-specific PCR with endpoint analysis was used for validation of the developed Russian wild ginseng single nucleotide polymorphism (SNP) marker.
Results
An SNP site specific to Russian wild ginseng was exploited by multiple alignments of mitochondrial nad7 intron 3 regions of different ginseng samples. With the SNP-based specific primer, Russian wild ginseng was successfully discriminated from Chinese and Korean cultivated ginseng samples by allele-specific PCR. The reliability and specificity of the SNP marker was validated by checking 20 individuals of Russian wild ginseng samples with real-time allele-specific PCR assay.
Conclusion
An effective DNA method for molecular discrimination of Russian wild ginseng from Chinese and Korean cultivated ginseng was developed. The established real-time allele-specific PCR was simple and reliable, and the present method should be a crucial complement of chemical analysis for authentication of Russian wild ginseng.
Keywords: allele-specific polymerase chain reaction, cultivated ginseng, nad7, single nucleotide polymorphism, wild ginseng
1. Introduction
Panax ginseng Meyer is a valuable medicinal plant that has been widely used in Asian countries for thousands of years. Modern medical science has verified that P. ginseng is effective in improving blood circulation and brain function [1], enhancing immune function [2], preventing diabetes [3], and improving sexual performance [4], as well as having anticancer and antiaging properties [5], [6]. The active constituents responsible for these pharmacological effects are ginsenosides and nonsaponin components such as polysaccharides, peptides, polyacetylene compounds, and essential fatty acids [7]. There are two different types of ginseng on the market: wild and cultivated ginseng. Wild ginseng grows in nature without artificial intervention. Cultivated ginseng is grown in forests and mountains, and the growth conditions are humanly controlled. Due to their different genotypes and growth environments, wild ginseng and cultivated ginseng have different ages of maturity. Generally, cultivated ginseng is harvested after 5–6 yr cultivation but wild ginseng takes more than 30 yr to mature. Wild ginseng has traditionally been known to be more effective than cultivated ginseng and a host of studies have demonstrated their differences in composition of active compounds [8], [9]. Therefore, wild ginseng is much more valuable and expensive than cultivated ginseng.
As a result of the increasing demand in the world market and excessive commercial collections, wild ginseng is in danger of extinction and thus cultivated ginseng accounts for most of the ginseng in the market to meet the demand of wild ginseng. However, wild ginseng originating from the Russian Sikhote-Alin Mountains comprises a significant proportion in the market due to its effective conservation and high yield. Accordingly, cultivated ginseng is often introduced as substitute and adulterant of Russian wild ginseng intentionally, mainly because of its lower cost or misidentification caused by similarity in appearance with wild ginseng. Morphological identification of Russian wild ginseng is subjective and error-prone because its accuracy depends heavily on the examiner’s experience and morphological differences are susceptible to environmental and developmental stages. Although previous studies have revealed the differences between cultivated and wild ginseng in composition of active compounds, contents of chemical constituents are easily affected by physiological conditions and stages of development [10]. The aim of this study is to develop a simple and reliable DNA method for differentiation of Russian wild ginseng from cultivated ginseng, in order to ensure therapeutic effects as well as to protect consumers’ rights.
2. Materials and methods
2.1. Plant materials and DNA isolation
Cultivated ginseng landraces, Damaya, Ermaya, Biantiao, and Huangguo were provided by the Chinese institute of Jilin ginseng. Wild ginseng was collected in the Russian Sikhote-Alin Mountains, and Korean ginseng cultivars were provided by Korean Ginseng Center for Most Valuable Products & Ginseng Genetic Resource Bank (Table 1). All the voucher specimens were morphologically identified by Professor Woo-Saeng Kwon and deposited in Korean Ginseng Center for Most Valuable Products & Ginseng Genetic Resource Bank. The fresh ginseng roots were frozen in liquid nitrogen and ground into fine powders. Genomic DNA was isolated using a Plant DNA extraction kit (TransGen Biotech, Beijing, China), according to the manufacturer's instructions.
Table 1.
Ginseng plant materials used in this study
| Ginseng sample | Voucher | Location | Number of samples | Accession number of nad7 intron3 |
|---|---|---|---|---|
| Damaya | Da01 | Jilin, China | 10 | KU239110 |
| Ermaya | Er01 | Jilin, China | 10 | KU239110 |
| Biantiao | Bi01 | Jilin, China | 10 | KU239110 |
| Huangguo | Hu01 | Jilin, China | 10 | KU239110 |
| Chunpoong | GB001 | Kochang, Korea | 10 | HQ241271 |
| Yunpoong | GB002 | Kochang, Korea | 10 | HQ241265 |
| Gopoong | GB003 | Chuncheon, Korea | 10 | HQ241266 |
| Sunpoong | GB004 | Kochang, Korea | 10 | HQ241267 |
| Sunweon | GBD043 | Daejeon, Korea | 10 | HQ241268 |
| Sunwon | GBD048 | Daejeon, Korea | 10 | HQ241270 |
| Russian wild ginseng | RWG041 | Sikhote-Alin, Russia | 20 | KU239111 |
2.2. Polymerase chain reaction amplification and Gel electrophoresis
The third intron of mitochondrial nad7 gene was amplified using primers nad73F (5′- CAA CAA CGG TTC TGC CTG AC-3′) and nad73R (5′-GCC CAC CAC TTA ACT TTC AC-3′). The 20 μL polymerase chain reaction (PCR) mixture contained 0.5μM of each primer, 20 ng of template DNA, and 10 μL of 2× EasyTaq PCR SuperMix (TransGen Biotech). PCR amplifications were carried out using one cycle of 4 min at 94°C, 35 cycles of 30 s at 94°C, 30 s at 60°C, and 1 min at 72°C. The final extension was at 72°C for 7 min. PCR products were analyzed via 1.0% agarose gel electrophoresis and visualized by ethidium bromide staining under UV.
2.3. Sequencing and analysis
The PCR products were purified with a PCR purification kit (TransGen Biotech) according to the manufacturer’s instructions. DNA sequencing of both forward and reverse directions was then conducted on an automatic DNA sequencer (ABI PRISM 3700; Applied Biostystems, Waltham, MA, USA). The sequences were assembled and analyzed using SeqMan software. Multiple sequence alignments were conducted using the Clustal Omega program [11].
2.4. Allele-specific PCR
According to the multiple sequence alignment result, primer nadwR was designed specific to Russian wild ginseng from its mutation site. The substitution of T for G at the third base from the 3′ terminus was an additional mismatch introduced to ensure its absolute allele specificity. Molecular authentication of Russian wild ginseng was performed by using multiplex PCR with primers nad73F, nadwR, and nad73R. The combination of primers nad73F and nad73R worked as a positive control to show that the reagents and PCR process was not problematic. The constitution of 20 μL PCR reaction mixture was identical with described above, except that 0.05μM of specific primer nadwR was added. Allele-specific PCR cycling parameters used were 1 cycle of 4 min at 94°C, 35 cycles of 30 s at 94°C, 30 s at 54°C, and 1 min at 72°C, with a final extension of 72°C for 7 min.
2.5. Cloning and sequencing of specific fragment
The Russian wild ginseng specific fragments were recycled with a EasyPure Quick Gel Extraction kit (TransGen Biotech), and then ligated into the pGEM-T Easy vector (Promega, Fitchburg, WI, USA) and transformed into competent Escherichia coli cells. White clones were selected and cultured in LB liquid medium at 37°C overnight with shaking. Plasmid DNA was isolated with a EasyPure Plasmid DNA miniPrep kit (TransGen Biotech) and sequenced by the same method as 2.3.
2.6. Real-time PCR assay for validation of Russian wild ginseng marker
To check the reliability and specificity of the SNP marker, 20 individuals of Russian wild ginseng were collected and identified. Primer nadwF (5′-AGG TTA GGT GCT ATT GAT GGA-3′) was designed to be compatible with primer nadwR for real-time PCR identification of Russian wild ginseng. Real-time allele-specific PCR assays were performed on a Rotor-Gene 6000 machine (Corbett Life Science, Mortlake, NSW, Australia). The 10-μL reaction mixture consisted of 5–10 ng DNA, 5μM of each primer, and 5 μL 2× SYBR Green I Mastermix (SensiMixPlus SYBR; Bioline, London, UK). The cycling profile was 10 min of activation at 95°C, followed by 45 cycles of a three-step thermal profile involving 10 s at 95°C for denaturation, 15 s at 54°C, and 20 s at 72°C for extension. The melting analysis condition was performed with a ramp from 85°C to 98°C, rising by 1°C at each step. Endpoint analysis was used for molecular authentication of different ginseng individuals.
3. Results and discussion
The P. ginseng genome is tetraploid and about 3.2 Gb in size [12]. The high complexity of P. ginseng genome makes it difficult to obtain a complete genomic sequence, and the low level of intraspecific polymorphism has limited the application of molecular markers for wild ginseng authentication. However, the conserved exon sequences and variable occurrence of introns of mitochondrial DNA provide an attractive reservoir for phylogenetic studies [13]. The nad7 gene, encoding the subunit 7 of NADH dehydrogenase complex I, carries three or four introns in flowering plants [14]. In this study, the third intron of mitochondrial nad7 gene was proved to be effective for SNP exploitation of Russian wild ginseng.
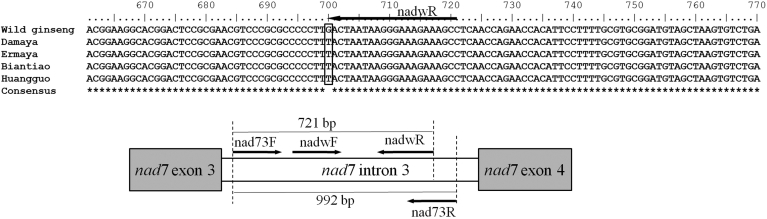
The amplified PCR products of nad7 intron 3 domain were determined to be 992 bp. The nad7 intron 3 regions of Russian wild ginseng and Chinese cultivated ginseng were registered in GenBank with accession numbers KU239110 and KU239111. Multiple sequence alignment result showed that nad7 intron 3 domains of Russian wild ginseng, Korean cultivated ginseng, and Chinese cultivated ginseng are almost identical, except that a SNP site specific to Russian wild ginseng was detected. As shown in Fig. 1, at the 700th nucleotide position, Russian wild ginseng contains G, but Chinese cultivated ginseng landraces were replaced with T at the same location. Based on this mutation site, primer nadwR (5′-GGC TTT CTT TGC CTT ATT ATT C-3′) was designed for specific authentication of Russian wild ginseng. The substitution of T for G at the third base from the 3′ terminus was introduced intentionally to ensure its absolute specificity and reliable discrimination between the alleles.
Fig. 1.
A diagram of positions of primers used in this study.
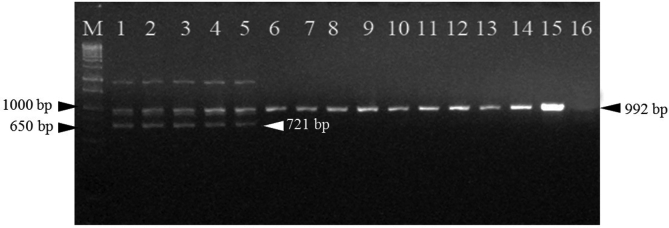
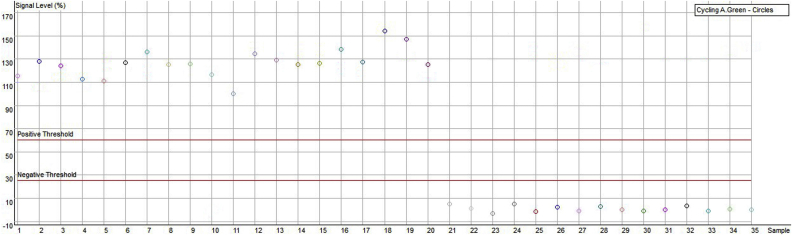
Molecular authentication of Russian wild ginseng was performed via allele-specific PCR with three primers: nad73F, nad73R, and the specific primer nadwR. As shown in Fig. 2, all the ginseng samples yielded amplicons of 992 bp with universal primer pairs. However, only Russian wild ginseng generated the band of 721 bp by the combination of nad73F and nadwR, which represents its G allele. The specific 721 bp PCR fragment of Russian wild ginseng was recycled and sequenced after cloning, and the sequence was indeed partial of the nad7 intron 3 region. Besides, there is a nonspecific fragment (about 1800 bp) produced in Russian wild ginseng by the three-primer combination. This amplicon does not affect the identification and further can be an additional marker of wild ginseng. The fragment patterns of different ginseng samples indicated that primer nadwR showed required specificity and the present marker could be used for authentication of Russian wild ginseng. To validate the reliability of this authentication method, 20 individual Russian wild ginseng samples were collected and analyzed using real-time allele-specific PCR assay. As shown in Fig. 3, the signal levels of the Korean and Chinese ginseng samples are lower than the negative threshold. Therefore, the 20 Russian wild ginseng samples could be easily discriminated from Chinese ginseng landraces and Korean ginseng cultivars by endpoint analysis method, and the present method showed 100% accuracy. Therefore, the established real-time allele-specific PCR assay is effective for molecular authentication of Russian wild ginseng from other Chinese and Korean cultivated ginseng samples.
Fig. 2.
Agarose gel image of allele-specific polymerase chain reaction. Lane M: 1,000 bp DNA ladder; lanes 1–5: Russian wild ginseng; lanes 6–11: Chunpoong, Yunpoong, Gopoong, Sunpoong, Sunweon, Sunwon, respectively; lanes 12–15: Damaya, Ermaya, Biantiao, Huangguo, respectively; lane 16: negative control.
Fig. 3.
Real-time polymerase chain reaction assay of different ginseng samples using endpoint analysis results. 1–20: Russian wild ginseng individuals; 21–26: ginseng cultivated in Korea; 27–34: ginseng cultivated in China; 35: no template control.
Nowadays, chemical analysis is the principle method for identification of wild ginseng. Ginsenosides and amino acids are the main marker compounds for chemical authentication. According to the differences in composition or contents of active compounds, HPLC/MS and UPLC can be used for authentication of wild ginseng [15], [16]. However, wild ginseng is expensive and purification of certain marker compounds may consume a certain quantity of sample. Furthermore, the ultimate chemical profiles may be affected by extrinsic factors such as harvesting, drying, and storage conditions.
In comparison, DNA markers are reliable for informative polymorphisms as the genetic composition is unique for each species and is not affected by age, physiological conditions as well as environmental factors. Various DNA markers have been developed for Panax species [17], [18], [19], but these markers are based on interspecific polymorphisms and few intraspecific polymorphisms were reported in P. ginseng. However, mitochondrial DNA noncoding regions show their superiority at intraspecific diversity studies [20], [21]. Here, we developed a simple and reliable method for identification of Russian wild ginseng, by exploiting its specific SNP in mitochondrial nad7 intron 3. Because mitochondria are relatively intact during processing and mitochondrial genes appear in multiple copies [22], the present method is degradation-tolerant and should be a crucial complement of chemical analysis for authentication of Russian wild ginseng.
Conflicts of interest
All contributing authors declare no conflicts of interest.
Acknowledgments
This study was supported by National Natural Science Foundation of China (81503178).
Contributor Information
Hongtao Wang, Email: wht1211@gmail.com.
Deok-Chun Yang, Email: dcyang@khu.ac.kr.
References
- 1.Kennedy D.O., Scholey A.B. Ginseng: potential for the enhancement of cognitive performance and mood. Pharmacol Biochem Behav. 2003;75:687–700. doi: 10.1016/s0091-3057(03)00126-6. [DOI] [PubMed] [Google Scholar]
- 2.Rivera E., Pettersson F.E., Inganäs M., Paulie S., Grönvik K.O. The Rb1 fraction of ginseng elicits a balanced Th1 and Th2 immune response. Vaccine. 2005;23:5411–5419. doi: 10.1016/j.vaccine.2005.04.007. [DOI] [PubMed] [Google Scholar]
- 3.Lai D.M., Tu Y.K., Liu I.M., Chen P.F., Cheng J.T. Mediation of beta-endorphin by ginsenoside Rh2 to lower plasma glucose in streptozotocin-induced diabetic rats. Planta Med. 2006;72:9–13. doi: 10.1055/s-2005-916177. [DOI] [PubMed] [Google Scholar]
- 4.de Andrade E., de Mesquita A.A., Claro Jde A., de Andrade P.M., Ortiz V., Paranhos M., Srougi M. Study of the efficacy of Korean Red Ginseng in the treatment of erectile dysfunction. Asian J Androl. 2007;9:241–244. doi: 10.1111/j.1745-7262.2007.00210.x. [DOI] [PubMed] [Google Scholar]
- 5.Keum Y.S., Park K.K., Lee J.M., Chun K.S., Park J.H., Lee S.K., Kwon H., Surh Y.J. Antioxidant and anti-tumor promoting activities of the methanol extract of heat-processed ginseng. Cancer Lett. 2000;150:41–48. doi: 10.1016/s0304-3835(99)00369-9. [DOI] [PubMed] [Google Scholar]
- 6.Chen Z.K., Fan C.X., Ye Y.H., Yang L., Jiang Q., Xing Q.Y. Isolation and characterization of a group of oligopeptides related to oxidized glutathione from the root of Panax ginseng. J Pept Res. 1998;52:137–142. doi: 10.1111/j.1399-3011.1998.tb01368.x. [DOI] [PubMed] [Google Scholar]
- 7.Choi K.T. Botanical characteristics, pharmacological effects and medicinal components of Korean Panax ginseng C A Meyer. Acta Pharmacol Sin. 2008;29:1109–1118. doi: 10.1111/j.1745-7254.2008.00869.x. [DOI] [PubMed] [Google Scholar]
- 8.Jang A.Y., Song E.J., Shin S.H., Hwang P.H., Kim S.Y., Jin Y.W., Lee E.K., Lim M.J., Oh I.S., Ahn J.Y. Potentiation of natural killer (NK) cell activity by methanol extract of cultured cambial meristematic cells of wild ginseng and its mechanism. Life Sci. 2015;135:138–146. doi: 10.1016/j.lfs.2015.06.018. [DOI] [PubMed] [Google Scholar]
- 9.Sun H., Liu F., Sun L., Liu J., Wang M., Chen X., Xu X., Ma R., Feng K., Jiang R. Proteomic analysis of amino acid metabolism differences between wild and cultivated Panax ginseng. J Ginseng Res. 2016;40:113–120. doi: 10.1016/j.jgr.2015.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang W.T., Thissen U., Ehlert K.A., Koek M.M., Jellema R.H., Hankemeier T., van der Greef J., Wang M. Effects of growth conditions and processing on Rehmannia glutinosa using fingerprint strategy. Planta Med. 2006;72:458–467. doi: 10.1055/s-2005-916241. [DOI] [PubMed] [Google Scholar]
- 11.Sievers F., Higgins D.G. Clustal Omega, accurate alignment of very large numbers of sequences. Methods Mol Biol. 2014;1079:105–116. doi: 10.1007/978-1-62703-646-7_6. [DOI] [PubMed] [Google Scholar]
- 12.Li C., Zhu Y., Guo X., Sun C., Luo H., Song J., Li Y., Wang L., Qian J., Chen S. Transcriptome analysis reveals ginsenosides biosynthetic genes, microRNAs and simple sequence repeats in Panax ginseng C.A. Meyer. BMC Genomics. 2013;14:245. doi: 10.1186/1471-2164-14-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Knoop V. The mitochondrial DNA of land plants: peculiarities in phylogenetic perspective. Curr Genet. 2004;46:123–139. doi: 10.1007/s00294-004-0522-8. [DOI] [PubMed] [Google Scholar]
- 14.Bonen L., Williams K., Bird S., Wood C. The NADH dehydrogenase subunit 7 gene is interrupted by four group II introns in the wheat mitochondrial genome. Mol Gen Genet. 1994;244:81–89. doi: 10.1007/BF00280190. [DOI] [PubMed] [Google Scholar]
- 15.Sun H., Zhang Y.Y., Wang Y.P., Zheng P.H. Ginsenoside analysis on the wild ginseng and the ginseng cultivated in forest by HPLC/MS. Chin J Spec Wild Econ Anim Plant Res. 2011;3:52–54. [Google Scholar]
- 16.Zhang L., Gao W., Zhou S., Cai N. Application of UPLC to separation and analysis of ginsenosides from cultivated ginseng and forest-grown wild ginseng. Lat Am J Pharm. 2011;30:991–995. [Google Scholar]
- 17.Um J.Y., Chung H.S., Kim M.S., Na H.J., Kwon H.J., Kim J.J., Lee K.M., Lee S.J., Lim J.P., Do K.R. Molecular authentication of Panax ginseng species by RAPD analysis and PCR-RFLP. Biol Pharm Bull. 2001;24:872–875. doi: 10.1248/bpb.24.872. [DOI] [PubMed] [Google Scholar]
- 18.Lee J.W., Bang K.H., Kim Y.C., Seo A.Y., Jo I.H., Lee J.H., Kim O.T., Hyun D.Y., Cha S.W., Cho J.H. CAPS markers using mitochondrial consensus primers for molecular identification of Panax species and Korean ginseng cultivars (Panax ginseng C. A. Meyer) Mol Biol Rep. 2012;39:729–736. doi: 10.1007/s11033-011-0792-4. [DOI] [PubMed] [Google Scholar]
- 19.Wang H., Kim M.K., Kwon W.S., Jin H., Liang Z., Yang D.C. Molecular authentication of Panax ginseng and ginseng products using robust SNP markers in ribosomal external transcribed spacer region. J Pharm Biomed Anal. 2011;55:972–976. doi: 10.1016/j.jpba.2011.03.037. [DOI] [PubMed] [Google Scholar]
- 20.Wang H., Sun H., Kwon W.S., Jin H., Yang D.C. A PCR-based SNP marker for specific authentication of Korean ginseng (Panax ginseng) cultivar “Chunpoong”. Mol Biol Rep. 2010;37:1053–1057. doi: 10.1007/s11033-009-9827-5. [DOI] [PubMed] [Google Scholar]
- 21.Wang H., Sun H., Kwon W.S., Jin H., Yang D.C. Molecular identification of the Korean ginseng cultivar “Chunpoong” using the mitochondrial nad7 intron 4 region. Mitochondrial DNA. 2009;20:41–45. doi: 10.1080/19401730902856738. [DOI] [PubMed] [Google Scholar]
- 22.Foran D.R. Relative degradation of nuclear and mitochondrial DNA: an experimental approach. J Forensic Sci. 2006;51:766–770. doi: 10.1111/j.1556-4029.2006.00176.x. [DOI] [PubMed] [Google Scholar]