Abstract
Progression of mammalian cells through the G1 and S phases of the cell cycle is driven by D-type and E-type cyclins. According to the current models, at least one of these cyclin families must be present to allow cell proliferation. Here, we show that several cell types can proliferate in the absence of all G1 cyclins. However, upon ablation of G1 cyclins, embryonic stem (ES) cells attenuated their pluripotent characteristics, with majority of cells acquiring the trophectodermal cell fate. We established that G1 cyclins, together with their associated cyclin-dependent kinases (CDKs) phosphorylate and stabilize core pluripotency factors Nanog, Sox2 and Oct4. Treatment of murine ES cells, patient-derived glioblastoma tumor-initiating cells, or triple-negative breast cancer cells with a CDK-inhibitor strongly decreased Sox2 and Oct4 levels. Our findings suggest that CDK-inhibition might represent an attractive therapeutic strategy by targeting glioblastoma tumor-initiating cells, which depend on Sox2 to maintain their tumorigenic potential.
Progression of cells through the G1 phase as well as entry into and passage through the DNA-synthesis (S phase) of the cell cycle is driven by a class of proteins called G1 cyclins. Two families of G1 cyclins operate in mammalian cells, D-type (D1, D2 and D3), which activate the cyclin-dependent kinases CDK4 and CDK6 and E-type (E1 and E2), which activate CDK2 (ref. 1).
Analyses of mice lacking all three D-type cyclins (D1−/−D2−/−D3−/−) revealed that these proteins are essential for proliferation only in a few selected compartments, such as hematopoietic cells. Importantly, the overwhelming majority of cells in cyclin D-null embryos proliferated normally, revealing that normal cell cycle progression can take place in the absence of D-cyclins2.
Furthermore, studies of embryos lacking cyclins E1 and E2 (E1−/−E2−/−) revealed a requirement for these proteins only in specific compartments, such as placenta and heart. Again, the majority of cell types proliferated normally, revealing that E-cyclins are dispensable for proliferation of most cell types3,4. Collectively, these findings led to the current model that G1 cyclins can perform overlapping functions, and that at least one class of G1 cyclins (D-type or E-type) must be present to allow mammalian cell proliferation.
In this study we decided to test this notion by generating embryonic stem (ES) cells, tissues and chimeric embryos lacking all five G1 cyclins. These studies revealed that, contrary to the prevailing view, G1 cyclins are not uniformly required for cell proliferation, but they play essential, direct roles in maintaining cell stemness and in regulating cell fate specification.
RESULTS
Generation of G1 cyclin-deficient ES cells
We interbred cyclin D1−/−, D2−/−, D3−/−, E1F/F (conditional cyclin E1 knockout) and E2−/− mice and generated cyclin D1+/−D2+/−D3+/−E1F/FE2+/− animals. We then intercrossed these mice, harvested blastocysts and cultured them in vitro to derive pluripotent ES cells (Fig. 1a). We succeeded in generating one cell line of the desired D1−/−D2−/−D3−/−E1F/FE2−/− genotype (expected ratio: 1:256), and one additional independent ES cell line heterozygous at the cyclin D2 locus (D1−/−D2+/−D3−/−E1F/FE2−/−). The latter cell line was then converted to the D1−/−D2−/−D3−/−E1F/FE2−/− genotype by re-targeting the remaining cyclin D2 allele (Supplementary Fig. 1a, b, Table 1).
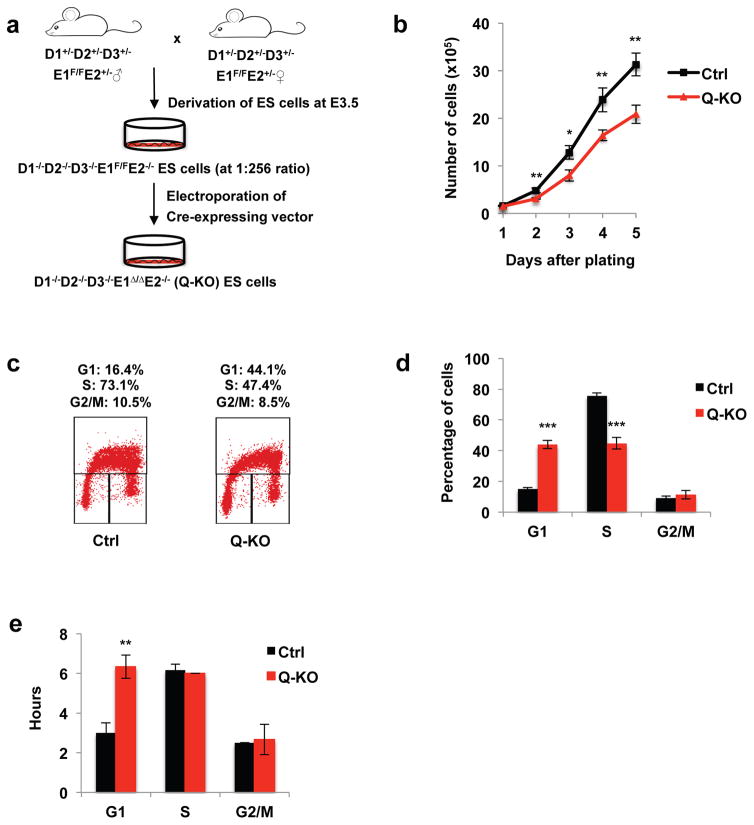
Figure 1. Generation and cell cycle analyses of ES cells lacking all G1 cyclins.
a, A strategy to generate cyclin D1−/−D2−/−D3−/−E1Δ/Δ E2−/− (Q-KO) ES cells. b, Growth curves of control (Ctrl) and Q-KO ES cells during in vitro culture, mean ± standard deviation (s.d.) of n=3 independent experiments. P=0.274, 0.004, 0.01, 0.009, 0.004. c, Cell cycle distribution of control (Ctrl) and Q-KO ES cells. Cells were pulsed with BrdU, stained with an anti-BrdU antibody and propidium iodide and analyzed by flow cytometry. The percentages of cells in the indicated cell cycle phases are shown, representative of n=4 independent experiments. d, Quantification of flow cytometric analyses performed as in c, mean ± s.d. of n=4 independent experiments. P<0.001, <0.001, =0.265. e, The mean length of G1, S and G2/M phases of the cell cycle in Ctrl and Q-KO ES cells. Cells were pulsed with BrdU, and progression of BrdU+ cells through the cell cycle was monitored over time by propidium iodide staining of BrdU+ cells at different time-points. Shown are mean ± s.d. of n=3 independent experiments. P=0.002, 0.374, 0.725. Two-tailed t-tests were used (*, p < 0.05; **, p < 0.01; ***, p < 0.001). Source data for b, d and e can be found in Supplementary Table 5.
ES cells proliferate in the absence of G1 cyclins
We introduced Cre recombinase into cyclin D1−/−D2−/−D3−/−E1F/FE2−/− ES cells, thereby acutely deleting cyclin E1 and rendering cells devoid of all G1 cyclins (D1−/−D2−/−D3−/−E1 Δ/ΔE2−/−). Very unexpectedly, these quintuple-knockout (hereafter termed Q-KO) ES cells continued to proliferate, albeit at a modestly reduced rate (Fig. 1b). Cell cycle analyses revealed an increased fraction of cells in the G1 phase and lengthening of the G1 phase by 3 hours in Q-KO ES cells (Fig. 1c–e, Supplementary Fig. 1d, e). By contrast, the length of S and G2/M phases was unaltered in Q-KO cells and spanned 6 and 3 hours, respectively (Fig. 1e). Hence, the total cell division cycle was slightly prolonged in Q-KO ES cells (15 hr vs. 12 hr in control cells), explaining mildly reduced proliferation rates of the mutant cells (Fig. 1b). Analyses of the expression levels of the remaining cyclins and CDKs revealed that the levels of these proteins were either unchanged or lower in Q-KO cells (Supplementary Fig. 1b). We concluded that ES cells can proliferate in the absence of all five G1 cyclins, and without any obvious upregulation of the remaining cell cycle proteins.
Ablation of G1 cyclins in mouse embryonic fibroblasts
These observations were very surprising, given the prevailing view that at least one class of G1 cyclins is required for cell proliferation. However, most of current cell cycle models are based on analyses of cultured mouse embryonic fibroblasts (MEFs). To extend our observations to this cell type, we utilized tetraploid blastocyst complementation method, which allows one to generate embryos derived entirely from mutant ES cells3 (Fig. 2a).
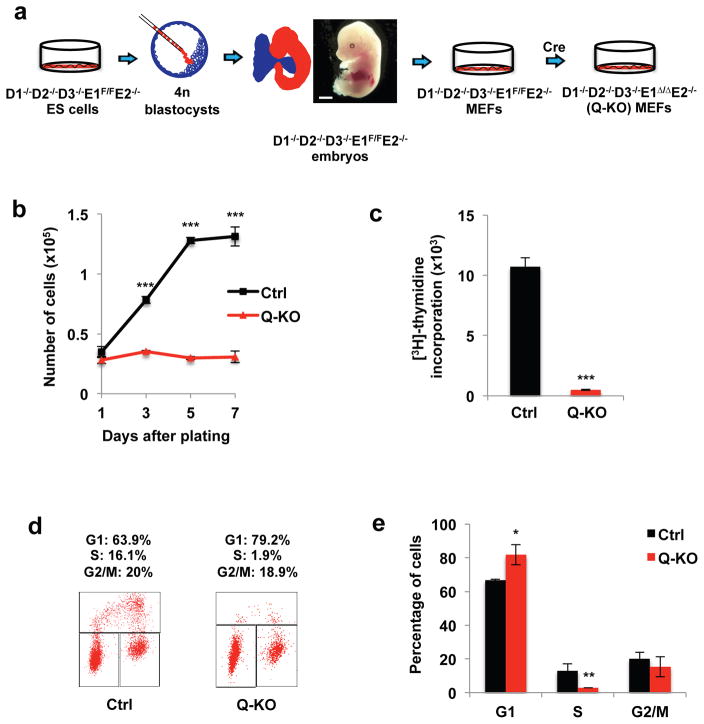
Figure 2. Generation and analyses of Q-KO MEFs.
a, Strategy to obtain Q-KO mouse embryonic fibroblasts (MEFs). Note normal appearance of a cyclin D1−/−D2−/−D3−/−E1F/FE2−/− embryo (representative of n=15 embryos), harvested at day 13.5 of gestation. Scale bar, 500 μm. b, Growth curves of Ctrl and Q-KO MEFs during in vitro culture, mean ± s.d. of n=3 independent experiments. P=0.093, <0.001, <0.001, <0.001. c, Ctrl and Q-KO MEFs were cultured in the presence of [3H]-thymidine, and incorporation into cells was determined by scintillation counting, mean ± s.d. of n=3 independent experiments. P<0.001. d, Cell cycle distribution of control (Ctrl) and Q-KO MEFs. Cells were pulsed with BrdU, stained with an anti-BrdU antibody and propidium iodide and analyzed by flow cytometry. The percentages of cells in the indicated cell cycle phases are shown representative of 3 independent experiments. e, Quantification of flow cytometric analyses performed as in d, mean ± s.d. of n=3 independent experiments. P=0.027, 0.016, 0.614. Two-tailed t-tests were used (*, p < 0.05; **, p < 0.01; ***, p < 0.001). Source data for b, c and e can be found in Supplementary Table 5.
We injected D1−/−D2−/−D3−/−E1F/FE2−/− ES cells into tetraploid blastocysts, and collected embryos at day 13.5 of gestation. Mutant embryos were viable and displayed normal appearance (Fig. 2a), indicating that a single G1 cyclin (cyclin E1) is sufficient for normal cell proliferation and development up to this stage. We then derived MEFs from the mutant embryos and transduced cells with Cre recombinase, thereby acutely shutting off cyclin E1 expression and rendering MEFs devoid of all G1 cyclins. Strikingly, ablation of all G1 cyclins completely blocked proliferation of MEFs (Fig. 2b–e, Supplementary Fig. 1c). We concluded that while ES cells can proliferate in the absence of G1 cyclins, in MEFs at least one G1 cyclin protein must be expressed to allow cell cycle progression.
Attenuation of stem cell pluripotency upon shutdown of G1 cyclins
Staining of in vitro cultured ES cells with alkaline phosphatase (AP), a marker of undifferentiated pluripotent stem cells5, revealed that ablation of G1 cyclins resulted in a strong decrease in AP staining. Whereas in control ES cells ~90% of colonies were uniformly AP-positive, this fraction was reduced to ~30% following ablation of all G1 cyclins (Fig. 3a, b).
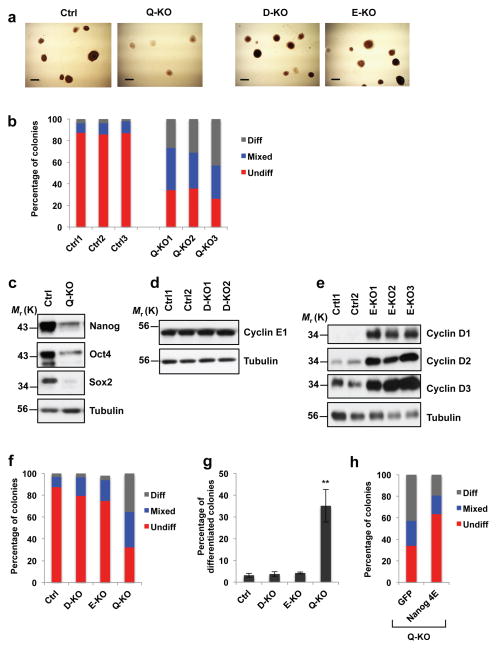
Figure 3. Attenuation of ES cell pluripotency upon ablation of G1 cyclins.
a, Alkaline phosphatase (AP) staining of Ctrl, D1−/−D2−/−D3−/−E1Δ/Δ E2−/− (Q-KO), D1−/−D2−/−D3−/− (D-KO) and E1Δ/Δ E2−/− (E-KO) ES cell colonies. Cells were analyzed 5 days after plating, representative of 5 independent experiments Scale bar, 250 μm. b, Bars show mean percentages of differentiated (Diff) AP-negative, mixed (some cells AP-positive, some -negative) and undifferentiated (Undiff) uniformly AP-positive ES cell colonies, 3 independent lines. c, Immunoblot analysis of Nanog, Oct4 and Sox2 protein levels in Ctrl and Q-KO ES cells, representative of 4 independent experiments. d, Immunoblot analysis of cyclin E1 protein levels in two independent lines of control (Ctrl1 and Ctrl2) and cyclin D-KO (D-KO1 and D-KO2) ES cells, representative of 3 independent experiments. e, Immunoblot analysis of the levels of D-type cyclins in independent lines of control (Ctrl1 and Ctrl2) and cyclin E-KO ES cells (E-KO1, E-KO2, and E-KO3), representative of 3 independent experiments. In c–e tubulin served as a loading control. f, Same analysis as in b, showing mean percentages of differentiated, mixed and undifferentiated colonies, 3 independent lines. g, Mean percentages of differentiated (AP-negative) colonies in ES cells of the indicated genotypes, mean ± s.d. of n=3 independent experiments. P=0.388, 0.056, 0.002. Two-tailed t-tests were used (**, p < 0.01). h, Q-KO ES cells were transduced with lentiviruses encoding GFP, or encoding a Nanog mutant containing phospho-mimicking glutamic acid substitutions in all four cyclin E-CDK2-dependent phosphoresidues (Nanog 4E), and colonies were stained with alkaline phosphatase. Bars show mean percentages of differentiated (Diff), mixed, and undifferentiated (Undiff) colonies (as in b), 3 independent lines. Source data for b, f, g and h can be found in Supplementary Table 5. Unprocessed original scans of blots are shown in Supplementary Fig. 9.
To probe this phenomenon at the molecular level, we analyzed protein levels of core pluripotency factors Nanog, Oct4 and Sox2 in Q-KO ES cells. These proteins are essential to maintain cell stemness, and their depletion causes the loss of the pluripotent state6–9. We found that ablation of G1 cyclins resulted in a downregulation of all three pluripotency factors (Fig. 3c). Importantly, the levels of transcripts encoding Nanog, Oct4 and Sox2 were unchanged in Q-KO ES cells (Fig. 4a, Supplementary Fig. 2a), indicating that G1 cyclins maintain the levels of these proteins at the posttranscriptional level.
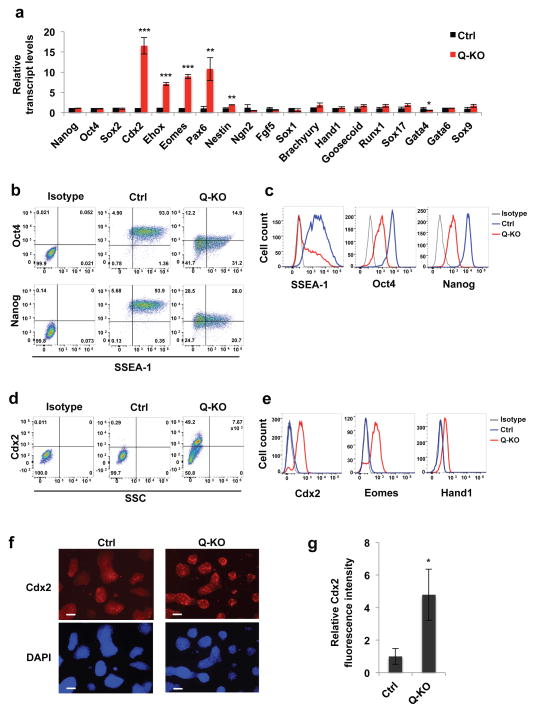
Figure 4. Analyses of differentiation markers in Q-KO ES cells.
a, Relative expression levels of the indicated transcripts in Ctrl and Q-KO ES cells, determined by reverse-transcription quantitative PCR. Shown are mean values ± s.d. of n=3 independent experiments. P=0.485, 0.612, 0.56, <0.001, <0.001, <0.001, =0.004, 0.001, 0.22, 0.091, 0.113, 0.06, 0.138, 0.061, 0.058, 0.145, 0.013, 0.142, 0.057. Mean expression levels observed in Ctrl ES cells were set as 1. b, c, Ctrl and Q-KO ES cells were permeabilized, stained with antibodies against SSEA-1, Oct4 or Nanog, and analyzed by flow cytometry. Panel c shows distribution of staining intensities. Isotype, isotype-specific IgG control. The mean percentages of SSEA-1+ cells for Ctrl and Q-KO cells were 95.4 ± 2.7 and 49.4 ± 6.5 (± s.d.), respectively, p < 0.001; for Oct4+ cells 94.7 ± 1.2 and 35.7 ± 9.7, respectively, p = 0.0025; and for Nanog+ cells 96.1 ± 2.8 and 44.3 ± 11.0, respectively, p < 0.001 (two-tailed t-test). d, e, Cells were permeabilized, stained with antibodies against Cdx2, Eomes or Hand1, and analyzed by flow cytometry. SSC, side scatter. Isotype, isotype-specific IgG control. Panel e shows distribution of staining intensities. The mean percentages of Cdx2+ cells were 1.3 ± 1.0 for control cells and 47.2 ± 14.1 for Q-KO cells (± s.d.), p < 0.0016 (two-tailed t-test). f, Immunostaining of Ctrl and Q-KO ES cell colonies for Cdx2 protein. Lower panels represent DAPI staining (to visualize cells). Scale bar, 100 μm. g, Quantification of immunofluorescence staining of Ctrl and Q-KO ES cell colonies for Cdx2, mean ± s.d. of n=3 independent experiments. P=0.016. Two-tailed t-tests were used (*, p < 0.05; **, p < 0.01; ***, p < 0.001). Shown in b–f are representatives of n=3 independent experiments Source data for a and g can be found in Supplementary Table 5.
To gauge the contribution of the D-type and E-type cyclins to maintenance of the pluripotent state, we compared wild-type, cyclin D-null (D1−/−D2−/−D3−/−), E-null (E1Δ/Δ E2−/−) and Q-KO ES cells. We observed that wild-type ES cells expressed low levels of D-type cyclins, whereas cyclin E was strongly expressed, as reported before10 (Fig. 3d, e). Ablation of D-type cyclins did not affect the levels of cyclin E (Fig. 3d), and resulted in only a slight decrease in the percentage of undifferentiated AP-positive ES cell colonies (see D-KO in Fig. 3a, f, g), and modest reduction of Nanog, Oct4 and Sox2 levels (Supplementary Fig. 2b). Cyclin E-null (E1Δ/Δ E2−/−) ES cells displayed very strong compensatory upregulation of all three D-type cyclins (Fig. 3e), a modestly reduced fraction of AP-positive colonies (see E-KO in Fig. 3a, f, g), and modestly reduced levels of Nanog, Oct4 and Sox2 proteins (Supplementary Fig. 2b). We concluded that G1 cyclins cooperate to maintain the levels of Nanog, Oct4 and Sox2 proteins in ES cells via a posttranscriptional mechanism, with E-cyclins playing the primary role and D-cyclins representing a likely ‘backup’ mechanism.
To better characterize the population of Q-KO ES cells, we stained control and Q-KO cells with antibodies against SSEA-1 (a marker of undifferentiated mouse stem cells), as well as Oct4 and Nanog, and analyzed cells by flow cytometry. As expected, nearly all control ES cells were positive for SSEA-1. In contrast only 45–50% of Q-KO ES cells were SSEA-1-positive, with majority of Q-KO cells being SSEA-1-negative (Fig. 4b, c). As expected, 95% of control ES cells expressed high Nanog and Oct4 levels. In contrast, approximately 40% of Q-KO ES cells were Oct4low and Nanoglow, and the rest was Oct4/Nanog-negative (Fig. 4b, c). Hence, ablation of G1 cyclins led to an attenuation of markers that characterize undifferentiated pluripotent stem cell state.
Ablation of G1 cyclins promotes trophectodermal differentiation
In addition to maintaining pluripotency, Oct4 and Sox2 proteins actively repress the trophectodermal cell fate. Oct4 was shown to block transcriptional upregulation of a caudal-related homeobox protein Cdx2, a key regulator of the trophectoderm lineage that confers the trophectodermal specification11–13. Consequently, reduced Oct4 (or Sox2) expression in ES cells triggers differentiation of ES cells into trophectoderm, by transcriptionally upregulating Cdx2 and another regulator of trophectodermal differentiation, Eomesodermin (Eomes)6,9,11,14,15. Given strongly reduced levels of Oct4 and Sox2 proteins upon ablation of G1 cyclins, we tested expression of trophectodermal markers in Q-KO ES cells.
We stained ES cells with antibodies against Cdx2, Eomes and another trophectodermal cell marker, Hand1, and analyzed by flow cytometry. As expected, control ES cells were negative for each of these markers. In contrast, 50–60% of Q-KO ES cells were Cdx2- and Eomes-positive, and expressed low levels of Hand1 (Fig. 4d, e). Immunostaining of intact ES cell colonies for Cdx2 confirmed increased Cdx2 protein levels in Q-KO cells (Fig. 4f, g). As expected, upregulation of the trophectodermal markers (Cdx2, Eomes, Ehox) in Q-KO cells occurred at the transcriptional level (Fig. 4a, Supplementary Fig. 2a). Importantly, markers of mesodermal, endodermal and ectodermal lineages remained unchanged upon ablation of G1 cyclins, except for an upregulation of a neuroectodermal marker, Pax6 (Fig. 4a, Supplementary Fig. 2a, and Table 2).
To further extend these findings, we used RNA-sequencing to compare transcript abundance, in an unbiased fashion, between Q-KO and control ES cells. We found that 66 transcripts were strongly (>4 fold) upregulated and 16 downregulated upon ablation of G1 cyclins (Supplementary Fig. 2c, d, Table 3). Several of these upregulated transcripts corresponded to trophectodermal/trophoblast genes, in addition to strongly increased levels of Cdx2 and Eomes (Supplementary Table 3). Collectively, these results indicate that ablation of G1 cyclins leads to the attenuation of pluripotency, and promotes trophectodermal cell fate.
Contribution of Q-KO ES cells to different lineages in vivo
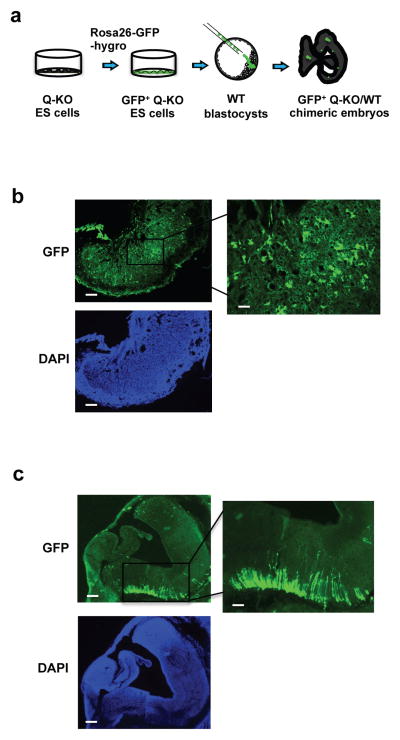
To gauge the ability of Q-KO ES cells to contribute to different lineages in vivo, we knocked-in a cDNA encoding green fluorescent protein (GFP) into the ubiquitously expressed Rosa26 locus16 in Q-KO ES cells. We then injected GFP+ Q-KO ES cells into wild-type blastocysts and implanted the chimeric embryos for further development (Fig. 5a). Consistent with our observation that ablation of G1 cyclins triggered an upregulation of trophectodermal markers (Fig. 4a, d–g, Supplementary Fig. 2a and Table 3), which specify the lineage that forms the placenta, we found that Q-KO ES cells preferentially contributed to embryonic placentas in vivo (Fig. 5b). On the other hand, Q-KO ES cells contributed poorly to the embryo proper, however we detected a clear contribution of GFP+ cells to neural cells (Fig. 5c).
Figure 5. Contribution of Q-KO ES cells to the developing embryo.
a, Strategy used to gauge the contribution of Q-KO (GFP+) ES cells to different compartments of the developing embryos. b, Strong contribution of Q-KO (GFP+) ES cells to the embryonic part of placenta. Shown is a section of a placenta from a chimeric embryo, stained for GFP. Scale bar, 100 μm. Right panel shows higher magnification of the boxed area, scale bar, 400 μm. Lower panel represents DAPI staining (to visualize cells). c, A section of embryonic brain from a chimeric embryo stained for GFP. Scale bar, 100 μm. Right panel shows higher magnification of the boxed area, scale bar, 400 μm. Lower panel represents DAPI staining. Three Q-KO and one Ctrl GFP+ ES cell lines were injected, 10 Q-KO and 3 Ctrl embryos were analyzed. The imagines are representative of 10 Q-KO chimeric embryos.
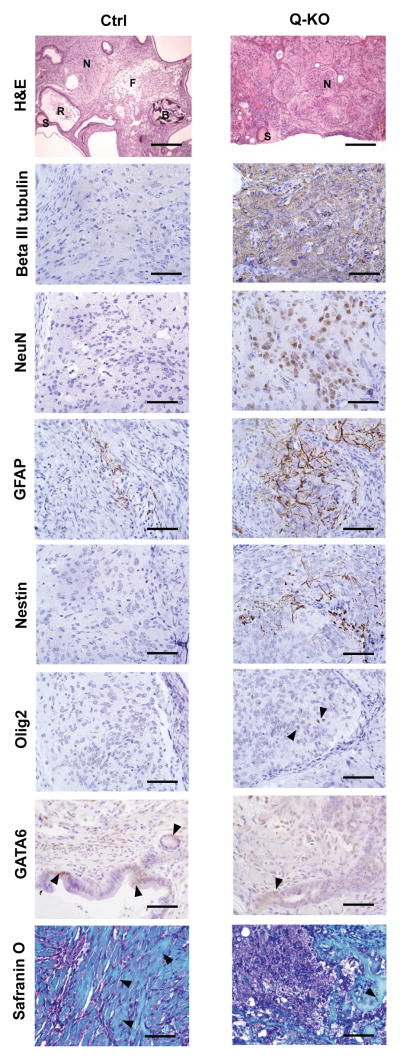
We extended these analyses by subcutaneously injecting Q-KO ES cells into immunocompromised nu/nu mice. Under these conditions ES cells proliferate, differentiate and form teratomas composed of tissues derived from ectodermal, endodermal and mesodermal lineages17. As expected, teratomas derived from control ES cells displayed roughly equal proportion of these three lineages. In contrast, tumors derived from Q-KO ES cells showed a predominance of the neural tissue (Fig. 6, upper panel). Immunostaining of teratoma sections revealed the Q-KO ES cells give rise to neural stem/progenitor cells (Nestin+), immature postmitotic neuronal precursors (beta III tubulin+), mature neurons (NeuN+), astrocytes (GFAP+), oligodendrocytes (Olig2+), as well as endodermal (GATA6+) and mesodermal lineages (skeletal muscle, cartilage and bone) (Fig. 6 and Supplementary Fig. 3). Collectively, these analyses reveal that Q-KO ES cells retain the ability to give rise to several different lineages in vivo.
Figure 6. Analyses of teratomas.
Analyses of teratomas. Ctrl and Q-KO ES cells were injected subcutaneously into nude mice. Teratomas were collected and sections were stained with haematoxylin and eosin (H&E), or analysed by immunohistochemistry with antibodies against beta III tubulin (a marker of immature postmitotic neuronal precursors), NeuN (a marker of mature neurons), GFAP (astrocytes), nestin (neural stem cells), Olig2 (oligodendrocytes), GATA6 (endoderm), or stained with Safranin O (to highlight mesoderm-derived skeletal muscle, cartilage and bone). Arrowheads point to Olig2- and GATA6- positive cells, and in the Safranin O panel to mesoderm-derived muscle (Ctrl) and bone (Q-KO). S, squamous epithelium; R, respiratory epithelium; N, neural tissue; F, fat; B, bone. Scale bars, 250 μM in (H&E panel) and 50 μM (all other panels). 12 Ctrl and 12 Q-KO teratomas (derived from 3 independent ES cell lines per genotype) were analysed by histology; sections from 2 teratomas per genotype were used for immunohistochemical confirmation of the identity of the lineages.
Decreased protein stability and increased polyubiquitination of pluripotency factors in Q-KO ES cells
To elucidate the exact role of G1 cyclins in maintaining Nanog, Oct4 and Sox2 protein levels in ES cells, we blocked new protein synthesis by treating wild-type and Q-KO ES cells with cycloheximide, and followed the decay of these proteins over time. We found that ablation of all G1 cyclins decreased the half-life and strongly accelerated degradation of Nanog, Oct4 and Sox2 proteins (Supplementary Fig. 4a–d). Importantly, treatment of Q-KO cells with a proteasomal inhibitor MG132 partially restored normal Nanog, Oct4 and Sox2 protein levels, indicating that these proteins undergo accelerated proteasomal degradation in the absence of G1 cyclins (Supplementary Fig. 4e).
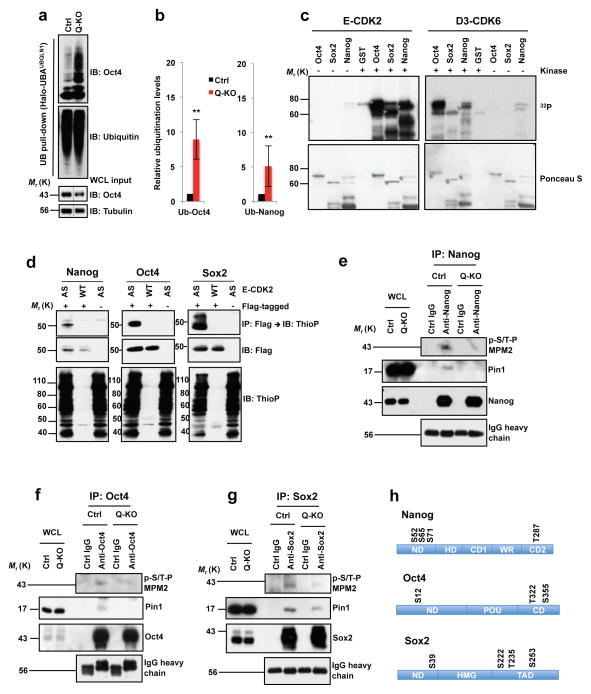
Since degradation of Nanog, Oct4 and Sox2 proteins is triggered by polyubiquitination18–20, we analyzed polyubiquitination status of the endogenous Nanog and Oct4 proteins in Q-KO cells. We observed that ablation of G1 cyclins strongly increased polyubiquitination of both proteins (Fig. 7a, b). Collectively these analyses indicate that G1 cyclins promote the stability of core pluripotency proteins by inhibiting their polyubiquitination, and by protecting them from proteasomal degradation.
Figure 7. Regulation of Nanog, Oct4 and Sox2 levels by G1 cyclins.
a, Polyubiquitin affinity purification (UB pull-down), immunoblotted with Oct4 and ubiquitin antibodies. WCL input: Oct4 levels in whole cell lysates (WCL), prior to purification (same as lanes 2, 4 in Supplementary Fig. 4e; middle portion spliced out (dashed lines). b, Left panel: quantification of a, mean ± s.d. of n=4 independent experiments. P=0.001. Band intensities corresponding to polyubiquitinated Oct4 were normalized against Oct4 total levels in Ctrl or Q-KO lysates prior to purification. Ctrl ES cells value is set at 1. Right panel: similar analysis of Nanog polyubiquitination, mean ± s.d. of n=4 independent experiments. P=0.001. Two-tailed t-tests (**, p < 0.01). c, In vitro kinase reactions. Recombinant GST-Nanog, GST-Oct4, GST-Sox2, or GST incubated in presence (+) or absence (−) of E1-CDK2 or D3-CDK6 with 32PγATP. Lower panel: Ponceau S staining of membranes. Green stars mark full-length GST-Nanog, -Oct4 and -Sox2. d, In-cell kinase reactions. 293T cells transfected with cyclin E1 and analog-sensitive CDK2 (AS), or wild-type CDK2 (WT), and Flag-tagged Nanog, Oct4 or Sox2 (+Flag-tagged) or empty vectors (−), and incubated with N6-PhEt-ATPγS. Upper panels: Flag-tagged Nanog, Oct4 or Sox2 proteins were immunoprecipitated (IP) with anti-Flag, immunoblots probed (IB) with anti-thiophosphate ester (ThioP). Middle panels: anti-Flag. Lower panels: lysates immunoblotted with anti-ThioP. Control lanes 3 and 6 are the same. e–g, ES cells were treated with MG132 to equalize Nanog, Oct4 and Sox2 levels in Ctrl and Q-KO cells. Endogenous Nanog, Oct4 or Sox2 proteins were immunoprecipitated (control: lysates treated with IgG), and immunoblotted with antibody against phosphorylated Ser/Thr-Pro (p-S/T-P MPM2, upper panel). Middle and third panels: anti-Pin1 and anti-Nanog/Oct4/Sox2. Bottom panel: IgG heavy chain control. Proteins levels in WCLs were analyzed. h, Cyclin E-CDK2-dependent phosphoresidues, mass spectrometric analysis. ND, N-terminal domain; HD, homeodomain; CD1, C-terminal domain 1; WR, tryptophan repeat; CD2, C-terminal domain 2; POU, POU-specific DNA-binding domain; CD, C-terminal domain; HMG, High mobility group DNA-binding domain; TAD, transactivation domain. Results representative of 3 (c–g) or 4 (a) independent experiments. Source data for c–e in Supplementary Table 5. Unprocessed blots in Supplementary Fig. 9.
G1 cyclins-CDKs directly phosphorylate and stabilize Nanog, Oct4 and Sox2 proteins
We next asked how at the molecular level G1 cyclins regulate the stability of Nanog, Oct4 and Sox2. The best documented function of G1 cyclins and their associated cyclin-dependent kinases is to phosphorylate target proteins1. Therefore, we used in vitro kinase reactions with purified recombinant proteins to test the ability of cyclin E-CDK2 and D3-CDK6 kinases to phosphorylate Nanog, Oct4 and Sox2. We found that all three pluripotency factors were readily phosphorylated by these kinases (Fig. 7c). We also verified the ability of cyclin E-CDK2 to phosphorylate Nanog, Oct4 and Sox2 proteins in vivo using cells expressing analog-sensitive CDK2. Analog-sensitive kinases, unlike wild-type kinases, can use ‘bulky’ N6-substituted ATP-analogs such as N6-phenylethyl-ATP (6-PhEt-ATP) to phosphorylate their substrates. Therefore, by supplementing cells expressing analog-sensitive kinases with thio-containing 6-PhEt-ATP (6-PhEt-ATPγS), one can specifically label their substrates with thiophosphate moieties21,22. Immunoprecipitation of Nanog, Oct4 or Sox2, followed by immunoblotting with an anti-thiophosphate antibody revealed that cyclin E-CDK2 kinase indeed readily phosphorylates all three proteins in vivo (Fig. 7d). In support for these findings, we established that an acute ablation of G1 cyclins strongly reduced phosphorylation of the endogenous Nanog, Oct4 and Sox2 proteins in Q-KO ES cells (Fig. 7e–g, upper panels).
Using mass spectrometry we identified cyclin E-CDK2-dependent phosphoresidues on the three pluripotency factors. We found that cyclin E-CDK2 kinase phosphorylates Nanog on S52, S65, S71 and T287 residues, Oct4 on S12, T322 and S355, and Sox2 on S39, S222, T235 and S253 (Fig. 7h, Supplementary Table 4). Intriguingly, a previous report established that phosphorylation of Nanog on these four residues stabilizes Nanog by promoting a physical interaction between Nanog and Pin1 (ref. 23). Pin1 is peptidyl-prolyl cis/trans isomerase that specifically binds to phosphorylated S/T-P motifs to catalytically regulate the conformation of its substrates24. Consistent with these findings, we found that ablation of G1 cyclins diminished association of the endogenous Nanog and Pin1 proteins (Fig. 7e, second panel). Moreover, a Nanog mutant containing inactivating alanine substitutions within cyclin E-CDK2-dependent phosphoresidues displayed reduced half-life (Supplementary Fig. 5a, b) and increased polyubiquitination (Supplementary Fig. 5g), in agreement with reduced protein levels (Fig. 3c, Supplementary Fig. 2b) and increased polyubiquitination of the endogenous Nanog in Q-KO ES cells (Fig. 7b). Conversely, phospho-mimicking glutamic acid substitutions within cyclin E-CDK2-dependent residues of Nanog stabilized the protein by extending its half-life (Supplementary Fig. 5a, b).
In case of Oct4, one of the E-CDK2-dependent residues identified in our mass spectrometric analyses was S12 (Fig. 7h, Supplementary Table 4). Phosphorylation of S12 was shown to stabilize Oct4, by triggering phosphorylation-dependent interaction between Oct4 with Pin1 (ref. 25). Consistent with this observation, we found that ablation of G1 cyclins diminished interaction between the endogenous Oct4 and Pin1 proteins in Q-KO ES cells (Fig. 7f, second panel). Moreover, inactivating alanine substitutions within cyclin E-CDK2-dependent phosphoresidues of Oct4 strongly decreased protein half-life (Supplementary Fig. 5c, d) and led to an increased polyubiquitination (Supplementary Fig. 5h). Conversely, phospho-mimicking substitutions within cyclin E-CDK2-dependent residues of Oct4 stabilized the protein by extending its half-life (Supplementary Fig. 5c, d).
Ablation of G1 cyclins also decreased interaction between the endogenous Sox2 and Pin1 proteins in Q-KO ES cells (Fig. 7g, second panel), suggesting a similar molecular mechanism of phosphorylation-dependent stabilization by Pin1. As it was the case for Nanog and Oct4, alanine substitutions within cyclin E-CDK2-dependent phosphoresidues of Sox2 decreased protein half-life (Supplementary Fig. 5e, f) and led to an increased polyubiquitination (Supplementary Fig. 5i), while phospho-mimicking substitutions extended the half-life (Supplementary Fig. 5e, f).
To further establish the causative link between hypophosphorylation of core pluripotency factors in Q-KO cells and the loss of pluripotency, we transduced Q-KO ES cells with a virus encoding Nanog protein containing phospho-mimicking glutamic acid substitutions within all four cyclin E-CDK2-dependent phosphoresidues. Strikingly, this partially corrected the phenotype of G1 cyclin-deficiency, and reduced the fraction of differentiated (AP-negative) colonies while increasing the fraction of undifferentiated AP+ colonies (Fig. 3h, Supplementary Fig. 2e).
Collectively these findings indicate that cyclin E-CDK2 and D3-CDK6 kinases phosphorylate Nanog, Oct4 and Sox2, thereby promoting their interaction with Pin1, and protecting these proteins from polyubiquitination and proteasomal degradation. Consistent with this model, ablation of G1 cyclins in ES cells decreased phosphorylation of the endogenous Nanog, Oct4 and Sox2 proteins, decreased their interaction with Pin1, leading to accelerated polyubiquitination, increased proteasomal degradation and strongly decreased half-life of these proteins.
Effect of CDK kinase inhibition on Nanog, Oct4 and Sox2 levels in ES cells
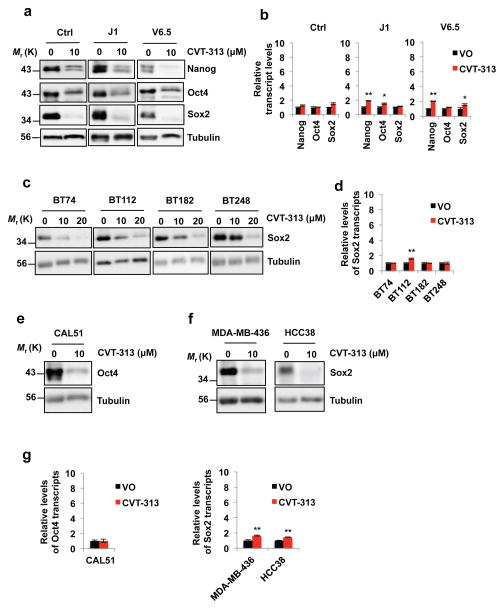
As described above, E-type cyclins are highly expressed in ES cells, whereas D-cyclins are present at low levels (Fig. 3d, e). However, ablation of E-cyclins in ES cells caused only a modest downregulation of Nanog, Oct4 and Sox2 protein levels (Supplementary Fig. 2b), most likely due to a strong compensatory upregulation of the D-type cyclins (Fig. 3e). We hypothesized that a chemical inhibition of cyclin E-associated kinase might not trigger such a compensatory mechanism, and hence might suffice to reduce the levels of pluripotency factors. Since cyclin E activates predominantly CDK2 (and to a lesser extent CDK1), we treated three different types of wild-type ES cells with the most selective inhibitor of CDK2 available, CVT-313, which inhibits CDK2 (IC50 = 0.5 μM) as well as CDK1, albeit with a lower potency (IC50 = 4.2 μM)26. We observed that reatment with CVT-313 did not cause upregulation of the D-type cyclins, and it triggered a strong decrease of Nanog, Oct4 and Sox2 protein levels (Fig. 8a, Supplementary Fig. 6). Importantly, the levels of Nanog, Oct4 and Sox2 transcripts were unchanged upon CVT-313 treatment of ES cells (Fig. 8b), similar to what we observed upon G1 cyclin ablation (Fig. 4a). These findings are consistent with our model that G1 cyclin-CDK kinases regulate Nanog, Oct4 and Sox2 at the posttranslational level, via direct phosphorylation. Consequently, combined ablation of E-type and D-type cyclins, or inhibition of CDK kinase, lead to destabilization of Nanog, Oct4 and Sox2 proteins in ES cells.
Figure 8. Decreased levels of Nanog, Oct4 and Sox2 proteins upon treatment of ES cells and human cancer cells with a CDK Inhibitor CVT-313.
a, In vitro cultured control ES cells derived from a mouse blastocyst of the mixed genetic background C57BL/6 × 129Sv (Ctrl), or wild-type J1 ES cells (from 129S4/SvJae strain), or wild-type V6.5 ES cells (from C57BL/6 × 129/Sv cross) were treated with vehicle only - DMSO (0) or with 10 μM CVT-313. The levels of Nanog, Oct4 and Sox2 proteins were assessed by immunoblotting. b, Reverse transcription-quantitative PCR (RT-qPCR) analysis of Nanog, Oct4 and Sox2 transcript levels, from ES cells treated with vehicle only (VO) or CVT-313, as in a, mean ± s.d. of n=3 independent experiments. P=0.053, 0.565, 0.550, 0.001, 0.020, 0.064, 0.002, 0.053, 0.016. c, Patient-derived glioblastoma cell lines were cultured as tumor-initiating cells in serum-free neural stem cell medium in the form of neurospheres. Cells were treated with the indicated concentrations of CVT-313 or vehicle only (0), and the levels of Sox2 protein were determined by immunoblotting. d, RT-qPCR analysis of Sox2 transcript levels, from glioblastoma cells treated with vehicle only (VO) or CVT-313, as in c, mean ± s.d. of n=3 independent experiments. P=0.627, 0.001, 0.962, 0.98. e and f, In vitro cultured human triple-negative breast cancer cell lines CAL51, MDA-MB-436 and HCC38 were treated with 10 μM CVT-313 or with vehicle only (0), and the levels of Oct4 (e), or Sox2 proteins (f) were determined by immunoblotting. g, RT-qPCR analysis of Oct4 or Sox2 transcript levels, from breast cancer cell lines treated with vehicle only (VO) or CVT-313, as in e and f, mean ± s.d. of n=3 independent experiments. P=0.928, 0.002, 0.001. In a, c, e, f, tubulin served as a loading control. Results are representative of n=3 independent experiments. Two-tailed t-tests were used (*, p < 0.05; **, p < 0.01). Source data for b, d and g can be found in Supplementary Table 5. Unprocessed original scans of blots are shown in Supplementary Fig. 9.
Strong reduction of Sox2 and Oct4 levels upon CDK inhibition in human cancer cells
In addition to regulating pluripotency of ES cells, Sox2 and Oct4 are also expressed and play critical roles in maintaining cell stemness and the tumorigenic potential of cancer ‘stem cell-like’ cells27,28. This was best documented in case of glioblastoma, the most frequent and most aggressive malignant brain tumor29. The tumor contains a high proportion of tumor-initiating ‘stem cell-like’ cells that uniformly exhibit high Sox2 levels29–33. Importantly, Sox2 was shown to represent a master regulator of glioblastoma cell stemness33,34. Depletion of Sox2 in tumor-initiating cells resulted in the loss of their ‘stem cell-like’ phenotype and markers and abrogated their tumorigenic potential in vivo35–37. For this reason, we asked whether inhibition of CDK activity might reduce the levels of Sox2 in human tumor cells. To do so, we utilized primary patient-derived glioblastoma tumor-initiating cells that are propagated as neurospheres in neural stem cell media to maintain their ‘stem cell-like’ properties. These cells are highly tumorigenic in vivo, and form aggressive glioblastomas upon intracranial injection into recipient mice38. We treated cultures of glioblastoma tumor-initiating cell lines derived from four independent patients with CVT-313, and monitored Sox2 levels. As it was the case in murine ES cells, inhibition of CDK kinase in human tumor-initiating cells led to a strong reduction of Sox2 protein levels, without having any impact on the levels of Sox2 transcripts (Fig. 8c, d, Supplementary Fig. 7).
We extended these analyses to human triple-negative basal breast cancers. This cancer type displays gene expression profile that resembles that of ES cells, and was shown to express high levels of Nanog, Oct4 and Sox2 transcriptional targets39. We screened thirteen human triple-negative breast cancer cell lines and identified two that express high Sox2 levels (MDA-MB-436 and HCC38) and one with high expression of Oct4 (CAL51). Again, treatment of these three cell lines with CVT-313 led to a strong downregulation of Sox2 and Oct4 protein levels (Fig. 8e–g, Supplementary Fig. 7).
To confirm that G1 cyclin-CDK kinases regulate Sox2 and Oct4 levels in human cancer cells via direct phosphorylation, we ectopically expressed in cancer cells mutant Sox2 or Oct4 proteins containing phospho-mimicking glutamic acid substitutions within all cyclin E-CDK2-dependent phosphoresidues, and treated cells with CVT-313. Consistent with our earlier findings, the levels of endogenous Sox2 and Oct4 proteins dropped precipitously upon CDK inhibition. In contrast, CVT-313 treatment had no impact on the levels phospho-mimicking Sox2 and Oct4 mutants (Supplementary Fig. 8a–c). Hence, mutation of CDK2-dependent phosphoresidues of Sox2 or Oct4 renders the levels of these proteins insensitive to CDK inhibition in cancer cells, consistent with our model that CDK2 stabilizes Sox2 and Oct4 via direct phosphorylation at the residues identified in this study. Collectively, these analyses reveal that – like in murine ES cells – also in human cancer cells cyclin-CDK kinases regulate Oct4 and Sox2 at the posttranscriptional level through protein phosphorylation.
DISCUSSION
According to the current cell division models, G1 cyclins are essential for cell proliferation. In the work described above we tested this model by generating ES cells, chimeric embryos, teratomas and MEFs lacking all G1 cyclins. We report that – contrary to the prevailing view – several cell types can proliferate without G1 cyclins. ES cells lacking all G1 cyclins proliferated nearly normally in vitro (Fig. 1b). Upon injection of only fifteen Q-KO ES cells into mouse blastocysts, these cells expanded in vivo and contributed to the developing placentas (Fig. 5b). Moreover, upon injection into nude mice, Q-KO ES cells formed teratomas composed of all lineages with a preponderance of neural cells (Fig. 6). In contrast, and in agreement with the current models, we found that an acute shutdown of G1 cyclins in MEFs completely halted their proliferation (Fig. 2b–e). Hence, G1 cyclins are required for proliferation of some, but not all cellular compartments.
Unexpectedly, we found that the major, rate-limiting function of G1 cyclins in ES cells is the maintenance of stem cell pluripotency, as ablation of G1 cyclins led to a rapid attenuation of the pluripotent state (Fig. 3a–c, f, g and 4b). Several previous studies pointed to a link between the cell cycle and stem cell pluripotency. It has been well established that murine embryonic stem cells display an elevated and constitutive CDK2 activity throughout the entire cell cycle. This phenomenon was linked to elevated and constitutive expression of cyclins E and A, leading to constitutive inactivation of the retinoblastoma protein and activation of E2F target genes10,40,41. In addition, murine ES cells do not express CDK inhibitors, which further contributes to the elevated CDK2 activity10. The exit from pluripotency and onset of differentiation is accompanied by downregulation of global CDK activity and lengthening of the G1 phase10. Several lines of evidence suggest that this restructuring of the cell cycle may play a causative role in the loss of pluripotency. Thus, in primed human embryonic stem cells, inhibition of CDK2 led to decrease in Oct4 levels42. Others reported that expression of protein CDK inhibitors, or knock-down of CDK2 in human stem cells resulted in the loss of the pluripotent state43,44. CDK2 was reported to directly phosphorylate Sox2 (ref. 45). The authors suggested that this phosphorylation has no impact on maintenance of ES cell pluripotency, but it affects efficiency of reprogramming45. Another study postulated that CDKs regulate pluripotency indirectly, by preventing endodermal differentiation through phosphorylation of Smad2/3 transcription factors46.
In the current study we provide a direct genetic evidence that G1 cyclins directly regulate the pluripotent state by driving phosphorylation of Nanog, Oct4 and Sox2. We show that an acute genetic ablation of all G1 cyclins leads to a strongly decreased phosphorylation of the endogenous Nanog, Oct4 and Sox2 proteins. This disables the ability of these proteins to interact with Pin1, resulting in their increased polyubiquitination and increased proteasomal degradation. This, in turn triggers attenuation of the pluripotent state. We interpret these findings as an indication that under normal physiological conditions, elevated CDK activity contributes to maintenance of the pluripotent state by directly phosphorylating and thereby stabilizing core pluripotency factors. Attenuation of CDK2 activity and upregulation of CDK inhibitors, seen during restructuring of the cell cycle from the stem cell to somatic cell mode10,40,41, leads to decreased levels of Nanog, Oct4 and Sox2, thereby contributing to the exit from the pluripotent state.
Another unexpected finding of our study is that expression of G1 cyclins prevents ES cells from acquiring the trophectodermal fate. Mouse ES cells are derived from blastocyst inner cell mass (which normally does not contribute to trophectoderm), and hence they have limited ability to give rise to this lineage47. In contrast, reduction of Oct4 (or Sox2) expression induces preferential differentiation of ES cells into trophectoderm, by transcriptionally upregulating a caudal-related homeobox 2 (Cdx2) and Eomesodermin (Eomes) proteins6,9,11,14,15. Consistent with strongly reduced levels of Oct4 and Sox2 upon ablation of G1 cyclins, Q-KO cells upregulated expression of Cdx2 and Eomes transcripts and proteins (Fig. 4a, d–g, Supplementary Fig. 2a and Table 3), and preferentially contributed to placentas in chimeric mice (Fig. 5b). These results indicate that G1 cyclins regulate lineage specification through stabilization of the Cdx2 repressors Oct4 and Sox2. Of note, Oct4 was also shown to repress specification of the neuroectodermal cell fate, a process that requires the presence of Sox248. We hypothesize that increased propensity of Q-KO ES cells to give rise to neural cells in teratoma assays (Fig. 6), might be linked to the loss of Oct4 expression in Sox2low cells, i.e., cells expressing Sox2 above the threshold required for the neuroectodermal differentiation.
Sox2 and Oct4 proteins were also shown to play important roles in maintaining ‘stem cell-like’ properties of tumor-initiating cells. This has been best documented in case of Sox2 and glioblastoma31–37,49. We found that inhibition of CDK in glioblastoma tumor-initiating cells caused a strong downregulation of Sox2 levels (Fig. 8c). Likewise, inhibition of CDK kinase strongly downregulated Sox2 and Oct4 protein levels in triple-negative breast cancer cells (Fig. 8e, f), another tumor type displaying a high proportion of ‘stem cell-like’ (CD44+) cells50. Collectively, these results suggest that inhibition of CDK might represent a very efficacious therapeutic strategy for glioblastoma (and possibly breast cancer) patients, by ablating stem cell properties of tumor-initiating cells.
METHODS
Derivation and culture of quintuple knockout (Q-KO) mouse embryonic stem (ES) cells
Cyclin D1+/− (ref. 51), D2+/− (ref. 52), and D3+/− mice53 were intercrossed with cyclin E1F/FE2−/− mice54 giving rise to D1+/−D2+/−D3+/−E1F/FE2+/− animals. These were further intercrossed, females sacrificed at day 3.5 post coitum and ES cell cultures were derived from blastocyst-stage embryos as described3. We succeeded in generating one cell line of the D1−/−D2−/−D3−/−E1F/FE2−/− genotype, and one additional cell line heterozygous at the cyclin D2 locus (D1−/−D2+/−D3−/−E1F/FE2−/−). The heterozygous D1−/−D2+/−D3−/−E1F/FE2−/− ES cells were electroporated with a modified cyclin D2 gene-targeting construct52 containing hygromycin resistance cassette in place of the Neo gene. This way we obtained several additional clones of D1−/−D2−/−D3−/−E1F/FE2−/− ES cells. Both the original D1−/−D2−/−D3−/−E1F/FE2−/− cell line, and several independent clones of the second cell line (derived from D1−/−D2+/−D3−/−E1F/FE2−/− cells) were used for all subsequent analyses, with identical results. To delete the cyclin E1 gene, D1−/−D2−/−D3−/−E1F/FE2−/− cells were electroporated with pOG231-Cre vector, giving rise to D1−/−D2−/−D3−/−E1Δ/Δ E2−/− (Q-KO) ES cells. Cyclin D1−/−D2−/−D3−/− (D-KO) and E1F/FE2−/− ES cells were derived in the same way; the latter were electroporated with pOG231-Cre yielding E1Δ/Δ E2−/− (E-KO) ES cells. ES cells were expanded on feeder layers of mitotically inactivated embryonic fibroblasts in Knockout DMEM (Invitrogen) with 15% fetal calf serum (HyClone) and 100 U/ml LIF (Millipore). Subsequently, ES cells were kept in ground pluripotent state by culturing them in the presence of MEK inhibitor PD184352 (Cayman Chemical, 1 μM), GSK3 inhibitor CHIR99021 (Cayman Chemical, 3 μM) and 100 U/ml LIF, as described55,56. ES cell genotyping and deletion analysis (shown in Supplementary Fig. 1a) were performed with PCR primers (Supplementary Table 2, Integrated DNA Technologies): 94°C 5 min, followed by 33 cycles of: 94°C 30 sec, 60°C 30 sec, 72°C 30 sec, and a final extension step at 72°C for 10 min. All mouse procedures were carried out according to a protocol approved by the Institutional Animal Care and Use Committee of the Dana-Farber Cancer Institute.
Generation of Q-KO mouse embryonic fibroblasts (MEFs)
Cyclin D1−/−D2−/−D3−/−E1F/FE2−/− or wild-type ES cells were injected into tetraploid blastocysts as described3. MEFs were derived from embryos at day 13.5 of gestation as before3 and used for transduction and analyses at passage 1 or 2. D1−/−D2−/−D3−/−E1F/FE2−/− or wild-type MEFs were transduced with Ad5CMVCre-eGFP (MOI100, Gene Transfer Vector Core) to delete the cyclin E1 gene and to generate D1−/−D2−/−D3−/−E1 Δ/ΔE2−/− (Q-KO) MEFs. For control, D1−/−D2−/−D3−/−E1F/FE2−/− MEFs were transduced with Ad5CMVeGFP (MOI100, Gene Transfer Vector Core).
Cell cycle analyses
For cell proliferation analyses, equal numbers of cells were plated at the beginning of experiment. Cells were counted at the indicated time points. For cell cycle distribution, cells were pulsed with bromodeoxyuridine (BrdU) for 1 hr (ES cells), 2hr (cancer cells) or 4 hr (MEFs), then stained with an anti-BrdU antibody (Supplementary Table 3) and with propidium iodide followed by flow cytometry. MEFs were also pulsed with [3H]-thymidine for 1 hr and analyzed as described3.
Pulse-chase experiments were used to measure the length of each cell cycle phase in ES cells (Fig. 1e). S phase cells were labeled by addition of 75 μM BrdU to the culture medium for 1 hr. Subsequently, cells were returned to BrdU-free medium, harvested at 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11 and 12 hr after the end of BrdU labeling, and stained with propidium iodide. We gated on BrdU-positive cells and analyzed their DNA content by flow cytometry. The length of the S phase was defined as the time period during which BrdU-positive cells exit the S-phase (i.e. the time of disappearance of BrdU-positive cells with DNA content between 2N and 4N DNA). The length of G2/M: time period between the end of BrdU labeling and the time when BrdU positive cells start accumulating in G1 phase (2N DNA content). The length of G1: time period between appearance of BrdU-positive G1 cells (2N) and the time when BrdU positive cells start entering the next S phase (>2N DNA content).
Alkaline phosphatase staining of ES cells
Alkaline Phosphatase Detection Kit (Millipore) was used. ES cells were cultured for five days (500 cells in one well of 12-well plates), fixed with 4% paraformaldehyde in PBS for 1–2 min and rinsed with TBST (20 mM Tris-HCl, pH 7.4, 0.15 M NaCl, 0.05% Tween-20). Fast Red Violet/Naphthol solution (Fast Red Violet mixed with Naphthol AS-BI phosphate solution and water at 2:1:1 ratio) was added (1 ml per well). Cells were incubated in dark at room temperature (RT) for 15 min, rinsed with TBST and covered with 1x PBS. Subsequently, undifferentiated alkaline phosphatase-positive (red) colonies, differentiated (colorless) and mixed ES cell colonies were counted under the scope.
Flow cytometry analysis of intracellular proteins in ES cells
Cells were fixed and permeabilized with Fixation/Permeabilization Solution Kit (BD Biosciences), then stained with conjugated (SSEA-1, Oct4, Nanog and Hand1, Supplementary Table 1) or non-conjugated antibodies or with IgG isotype control. For staining with non-conjugated antibodies, cells were blocked with 2% BSA in PBS for 30 min at RT, and incubated with anti-Cdx2 or Eomes antibodies or rabbit IgG control for 60 min at RT (Supplementary Table 1). Cells were then washed three times with PBS and incubated for 40 min at RT in the dark using a Dylight 488-conjugated goat anti-rabbit IgG (H+L) secondary antibody (Thermo Fisher), and then re-suspended in PBS for flow cytometry. Cells were analyzed using BD LSRFortessa analyzer (BD Biosciences) and FlowJo software.
Cdx2 staining of ES cell colonies
ES cells were fixed with 4% paraformaldehyde in PBS for 10 min, permeabilized with 0.1% Triton X-100 in PBS for 10–15 min, blocked with 5% BSA in PBS at RT for 30 min, and incubated with an anti-Cdx2 antibody in 5% BSA in PBS at 4°C overnight in a humidified chamber. Cells were then washed with PBS-Tween 20 and incubated with Alexa Fluor®488-conjugated goat anti-rabbit IgG antibody and 1 μg/ml DAPI for 1 hr at RT in the dark (Supplementary Table 1).
Annexin V staining
FITC Annexin V Apoptosis Detection Kit I (BD Biosciences) was used. Cells were washed twice with PBS and resuspended in 1X Binding Buffer at 1 × 106 cells/ml. 100 μl of cell suspension were mixed with 5 μl of FITC Annexin V and 5 μl propidium iodide, incubated for 15 min at RT in the dark, diluted in 400 μl of 1X Binding Buffer and analyzed by flow cytometry.
Lentiviral transduction of Q-KO ES cells and breast cancer cells
The coding sequences of Nanog, Oct4 or Sox2 mutants containing glutamic acid phospho-mimicking substitutions in all cyclin E-CDK2-dependent phosphoresidues (Nanog-4E, Oct4-3E or Sox2-4E) were subcloned into pDEST-hUbc-3xFLAG-AttR-IRES-Hygro (for Nanog-4E) or pDEST-hUbc-3xFLAG-AttR-IRES-Neo (for Oct4-3E or Sox2-4E) (modified FUGW plasmid, a gift from Dr. David Baltimore, Addgene plasmid # 14883), giving rise to 3 x Flag-mutant (Nanog-4E, Oct4-3E or Sox2-4E) transfer plasmids. On day 0, 3 x Flag-mutant plasmids were co-transfected with the lentiviral envelope (VSVG) and packaging (Δ8.9) plasmids into 293FT cells (Invitrogen). Medium with virus particles was collected on day 2 and passed through a 0.45 μm syringe filter followed by virus concentration on Amicon Ultra-15 100 kDa centrifugal columns. Concentrated virus was added in 1 ml of cell growth medium and 10 μg/ml Polybrene (Millipore) to 25 × 103 Q-KO ES cells or breast cancer cells. On day 3, the medium was replaced with the concentrated virus from the second harvest from the original 293FT cells. On day 4, cells were fed with fresh medium and subjected to a 6-days-long selection with hygromycin (100 μg/ml for ES cells) or neomycin (1000 μg/ml for MDA-MB-436 and CAL51, 500 μg/ml for HCC38 cells).
Generation and analyses of GFP+ Q-KO chimeric embryos
GFP cDNA was knocked-in into the Rosa26 locus of Q-KO ES cells by electroporating Q-KO ES cells with Rosa26-GFP-Hygro vector (see Plasmids and Table 2 for PCR genotyping primers). To obtain chimeric embryos, C57BL/6J mice (Taconics) were mated and blastocysts collected at day 3.5 post coitum. GFP+ Q-KO ES cells were injected into blastocysts (~15 ES cells per blastocyst), and the blastocysts were transferred to the uteri of pseudopregnant Swiss Webster females (Taconic). Embryos were collected at embryonic day 12.5, fixed and frozen. Immunofluorescence staining was performed with anti-GFP and Alexa Fluor®488-conjugated goat anti-rabbit IgG polyclonal antibodies (Supplementary Table 1) and analyzed using a fluorescent microscope (Zeiss, Ax10, Germany).
Teratoma formation
1×106 of wild-type or Q-KO ES cells were injected under the skin of female NCr nude mice (10 weeks old, Taconic). Approximately 2–3 weeks after the injection, when tumors were visible under the skin, teratomas were dissected, fixed in 4% paraformaldehyde in PBS overnight and embedded in paraffin. Sections were stained with hematoxylin and eosin, or processed for immunohistochemistry.
Immunohistochemistry and histological staining of teratomas
Immunohistochemistry was performed using the Dako Envision+ system. Sections were subjected to heat-mediated antigen retrieval with 10 mM sodium citrate buffer (pH 6.0, 20 min) and blocked with endogenous peroxidase blocking reagent (S2003, Dako). Slides were incubated overnight at 4°C with primary antibodies (Supplementary Table 1), then for 1–2 hr at RT with secondary antibodies (Supplementary Table 1), and chromogenic detection was achieved using 3,3′-diaminobenzidine (DAB) (K3468, Dako). Counterstaining was performed with Mayer’s hematoxylin. Safranin O histological staining was performed by the Dana-Farber/Harvard Cancer Center Specialized Histopathology Core to highlight skeletal muscle, cartilage and bone.
Western blotting and immunoprecipitation
Whole cell extracts were obtained and proteins solubilized for immunoblotting as described3. Antibodies are listed in Supplementary Table 1. For immunoprecipitation of endogenous Nanog, Sox2 or Oct4 (Fig. 7e–g), 2 mg of cell extracts from control or Q-KO ES cells were incubated with 30 μl slurry of agarose conjugated with anti-Nanog, Oct4 or Sox2 antibodies (Supplementary Table 1) overnight at 4°C. Immunocomplexes were resolved on SDS–PAGE, and immunoblotted with the indicated antibodies.
Plasmids
pOG231-Cre plasmid was used to express Cre recombinase in ES cells. Gene-targeting Rosa26-GFP construct57 was kindly provided by Dr. Konrad Hochedlinger (Massachusetts General Hospital, Boston, MA). The construct was modified by inserting the hygromycin resistance gene into the XhoI site, giving rise to Rosa26-GFP-Hygro vector, which was used for gene-targeting. Plasmids expressing HA-Nanog, HA-Oct4 and HA-Sox2 were constructed by subcloning mouse cDNAs into pcDNA3. All Nanog, Oct4 and Sox2 mutants were generated with the QuikChange XL Site-Directed Mutagenesis kit (Stratagene).
Quantitative PCR with reverse transcription
RNA was extracted using the RNeasy Mini Kit (Qiagen), and the reverse transcription (RT) reaction was performed using High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). After mixing the resultant template with primers (Supplementary Table 2, Integrated DNA Technologies) and with SYBR green PCR master mix (Applied Biosystems), PCR was performed with 7300 Real Time PCR System (Applied Biosystems).
RNA-Seq library preparation, sequencing and data analysis
Libraries were prepared from 500 ng of purified total RNA using Illumina TruSeq Stranded mRNA sample preparation kit. The dsDNA libraries were quantified by Qubit fluorometer, Agilent TapeStation 2200, and RT-qPCR using the Kapa Biosystems library quantification kit. Uniquely indexed libraries were pooled in equimolar ratios and sequenced on an Illumina NextSeq500 with single-end 75bp reads by the Dana-Farber Cancer Institute Molecular Biology Core.
RNA-Seq analysis was performed using Visualization Pipeline (VIPER: https://bitbucket.org/cfce/viper/). Briefly, reads were aligned to mm9 reference genome assembly and gene expression quantified with STAR aligner (v2.5.1b). DESeq2 was used to test for differential gene expression.
Cycloheximide and MG132 treatment
ES cells were treated with 100 μg/ml cycloheximide (Sigma) for the indicated times before harvesting. Whole cell extracts were subjected to immunoblot analysis with the indicated antibodies. Nanog, Oct4 and Sox2 band intensities were quantified, normalized with tubulin, and plotted.
HeLa cells were co-transfected with HA-tagged wild-type Nanog, Sox2 or Oct4, or with mutant versions containing alanine or glutamic acid substitutions within cyclin E-CDK2-dependent residues together with CMV-GFP plasmid, and treated with 100 μg/ml cycloheximide for the indicated times before harvesting. Whole cell extracts were subjected to immunoblot analysis with anti-HA, anti-GFP (control of transfection efficiency) and anti-tubulin (loading control) antibodies. HA band intensities were quantified, normalized with GFP, and plotted.
In experiments shown in Fig. 7a, b, e–g and Supplementary Fig. 4e, ES cells were incubated in the presence of 10 μM MG132 (EMD Chemicals) for 2 hr.
Capture of polyubiquitinated proteins from cell extracts
ES cells were treated with 10 μM MG132 for 2 hr prior to lysis. Cells were lysed in lysis buffer (50 mM Tris/HCl pH 7.5, 1 mM EDTA, 1 mM EGTA, 50 mM NaF, 5 mM sodium pyrophosphate, 10 mM sodium 2-glycerol 1-phosphate, 1mM sodium orthovanadate, 1% (v/v) NP-40, 1 μg/ml aprotinin and leupeptin, 1 mM benzamidine, 1mM AEBSF, 10 μM PR-619 and 100 mM chloroacatemide), to produce whole cell lysates. Polyubiquitinated proteins were then purified from whole cell lysates using Halo-4×UBAUBQLN1 as described58. Briefly, whole-cell lysates (1.5 mg) were incubated at 4°C for 6 hr with 50 μl of Halo-4×UBAUBQLN1 beads (pack volume). Following four washes with lysis buffer containing 0.5 M NaCl and one wash in 10 mM Tris (pH 8.0), proteins were released from Halo-4×UBAUBQLN1 beads using 1x LDS Novex sample buffer. Captured proteins were resolved using SDS-PAGE and immunoblotted with antibodies against ubiquitin (in 5% skimmed milk in TBST, 1 hr at RT), Nanog or Oct4 (both overnight at 4°C) (Supplementary Table 1).
In-cell polyubiquitination detection
293T cells were transfected with HA-Nanog, HA-Oct4 or HA-Sox2, and His–ubiquitin expression plasmids. Twenty four hr after transfection, cells were treated with 15 μM MG132 overnight before protein extraction. Cell extracts were incubated with Ni-NTA matrices at RT for 3 hr in the presence of 6 M guanidine-HCl, pH 8.0. Immobilized proteins were resolved by SDS–PAGE, and immunoblotted with an anti-HA antibody (Supplementary Table 1).
In-cell kinase reactions with analog-sensitive CDKs
Six hundred thousand 293T cells per well were seeded in a 6-well plate one day before co-transfection with plasmids encoding 3xFlag-Oct4, 3xFlag-Sox2 or 3xFlag-Nanog (0.25 μg) and analog sensitive CDK2 (F80G) or wild-type CDK2 (0.8 μg) together with cyclin E1, or analog sensitive CDK1 (M32V, F80G) or wild-type CDK1 together with cyclin B1. Next day, cells were incubated for 20 min at 30°C with 200 μl of kinase reaction buffer (20 mM HEPES pH 7.5, 100 mM KOAc, 5 mM NaOAc, 2 mM MgOAc2, 1 mM EGTA, 10 mM MgCl2, 0.5 mM DTT, 30 μg/ml digitonin, 5 mM GTP, 0.1 mM ATP, 0.1 mM N6-(furfuryl) ATPγS (Biolog), 1X HALT phosphatase and protease inhibitors cocktail (Thermo) as described21. Cells were lysed by adding 200 μl of 2x RIPA buffer (100 mM Tris pH 8.0, 300 mM NaCl, 2% NP-40, 0.2% SDS, 20 mM EDTA). Flag-tagged proteins were immunoprecipitated for 2 hr at 4°C using anti-Flag M2 antibody-coupled resin (Sigma). Beads were resuspended in 20 μl of 2.5 mM p-nitrobenzyl mesylate (PNBM, Abcam) and incubated for 30 min at RT. Proteins were resolved by SDS-PAGE and immunoblotted with an anti-thiophosphate ester antibody (Abcam).
In vitro kinase assays
GST-Nanog, GST-Oct4, and GST-Sox2 were expressed from pGEX-5x-3 (GE Life Sciences) in BL21-Gold (Agilent) and purified on glutathione Sepharose (GE Life Sciences). One microgram of each fusion protein was subjected to kinase activity of 20 ng cyclin D3-CDK6 or E1-CDK2 (ProQinase) in the presence of 10 μCi 32PγATP, 0.1 mM ATP, 10 mM MgCl2, 1 μM DTT, for 30 min at 30°C. Reactions were terminated by adding SDS sample loading buffer, resolved by SDS-PAGE, transferred to a nitrocellulose membrane and visualized by autoradiography and by Ponceau S staining of membranes.
For identification of phosphorylated residues, recombinant proteins were incubated with cyclin E1-CDK2 kinase for 2 hr at 30°C in the presence of 5 mM ATP, 10 mM MgCl2, 1 μM DTT followed by SDS-PAGE and visualization with Coomassie Blue. Bands containing GST-Nanog, -Oct4, and -Sox2 were excised, digested with trypsin or Lys C and analyzed by mass spectrometry.
Detection of Nanog, Oct4 and Sox2 phosphorylation sites
Nanog, Oct4 or Sox2 proteins were digested from SDS-PAGE gels using Trypsin, Asp-N, Glu-C or chymotrypsin in a 50 mM Tris buffer (pH 8.8) at 37°C. Peptides were extracted from the gel using 50% acetonitrile and 1% formic acid, which was dried using a vacuum centrifuge. Samples were then desalted using Stage tips and dried by vacuum centrifugation.
For the Sox2 phosphorylation site, S222, synthetic peptides (SQTYMNGSPTYSMSYSK, SQTYMNGAPTYSMSYSK, AQAYMNGSPAYAMAYAK) were designed to determine whether cyclin E1-CDK2 could phosphorylate these peptides in vitro. These peptides were designed to contain the native Sox2 peptide sequence, or S222A mutation, or all serines and threonines mutated to alanines except S222. In vitro kinase reactions were performed the same way as for recombinant proteins and the reaction was quenched with acetic acid. Samples were then desalted using Stage tips.
For in vitro analysis of phosphorylation sites on Nanog, Oct4 or Sox2, mass spectrometry was performed using an LTQ-Orbitrap Elite hybrid or an Orbitrap Fusion Tribrid mass spectrometer (MS) (Thermo Fisher, San Jose, CA). LTQ-Orbitrap Elite instrument was equipped with a Famos autosampler (LC Packings, Sunnyvale, CA) and an Accela 600 pump (Thermo Fisher). The Orbitrap Fusion Tribrid mass spectrometer was equipped with a Proxeon ultra high pressure liquid chromoatography (LC). The LC separation and MS methods for Orbitrap Elite experiments was performed as described59. For the Orbitrap Fusion experiments, LC separation and MS methods was performed as described60.
An in-house suite of software tools was used to convert mass spectrometric data from raw file to mzmxl format. Erroneous peptide ion charge state and monoisotopic m/z was corrected as described61. MS/MS spectra assignments were made using the Sequest algorithm62 using the entire mouse Ensembl database (version 3.6). Sequest searches were performed using a target-decoy strategy63 with the mouse Ensembl database in correct orientation (forward database) and the same database but with all sequences in reverse orientation (reverse database). The mouse Ensembl database contained 109,972 entries. The data was searched using a precursor ion tolerance of 20 ppm, fragment ions tolerance of 0.8 Da, ASP-N, Trypsin, Glu-C or chymotrypsin specificity and allowing two missed cleavages. A dynamic modification on methionine (+15.99492 Da) and phosphorylation (79.96633 Da) was considered in each search. A peptide level false discovery (FDR) rate of less than 1% was used as a threshold for peptide identifications using the target decoy strategy. Additional filtering was achieved using a linear discriminant analysis, which combined several parameters into a single probability for each peptide and these probabilities were used to achieve a less than 1% FDR. The parameters used for linear discriminant analysis were Xcorr, DCn, peptide mass accuracy and charge state, and peptide length.
CVT-313 treatment
Cells were cultured in the presence of 10 or 20 μM CVT-313 (Santa Cruz) for 48 hr.
Cell lines
Patient-derived glioblastoma multiforme cell lines BT74, BT112, BT182, BT248 were established from GBM resections38,64. Cells were cultured as neurospheres in neural stem cell media: DMEM/F12 (Invitrogen) with N2 and B27 Supplements (Gibco Technologies), 20 ng/ml human recombinant bFGF and EGF (Millipore), and 50 μg/mL penicillin/streptomycin. Glioblastoma cell lines were not authenticated.
MDA-MB-436 and HCC38 cells were cultured in RPMI (Invitrogen) with 10% fetal bovine serum (Sigma), CAL51 cells in DMEM (Invitrogen) with 20% fetal bovine serum. These cell lines were authenticated by STR profiling at the Dana-Farber Cancer Institute Molecular Biology Core.
Human trophoblast cell lines HTR-8/SVneo (CRL-3271), BeWo (CCL-98), JEG-3 (HTB-36) were from ATCC. Mouse neural stem/progenitor cells NE-4C (CRL-2925) and NE-GFP-4C (CRL-2926) were from ATCC, rat fetal neural stem/progenitor cells N7744100 from Thermo Fisher, and an additional murine neural stem/progenitor cell line from Dr. C. Stiles (Dana-Farber Cancer Institute, Boston, MA).
All cell lines were routinely tested for mycoplasma contamination.
Statistics and reproducibility
ImageJ software was used to quantify the intensity of immunoblotting images. Non-saturating exposure of immunoblot films was used for quantification with the appropriate loading controls as standards. For mouse experiments, no statistical methods were used to predetermine sample size. The experiments were not randomized and the investigators were not blinded to allocation during experiments and outcome assessment. No animals or samples were excluded from the analysis. Statistical data were analyzed using two-tailed unpaired t-tests. Error bars represent s.d. and significance is denoted. Sample sizes are shown in corresponding figure legends. Raw data can be found in the Statistics Source Data (Supplementary Table 5). Representative figures are shown in Fig. 1c, 2a, d, 3a, c–e, 4b–f, 5b, c, 6, 7a, c, d–g, 8a, c, e, f, Supplementary Fig. 1a–d, 2b, c, 3, 4a, e, 5, 6b, c, 8. Each experiment was repeated independently: 2 repeats (Fig. 6), 3 repeats (Fig. 1d, 1e, 2c–e, 3b, 3h, 4, 7b–g, 8, Supplementary Fig. 1a–c, 1e, 2a–b, 2e, 3, 4a–d, 5, 6a, 6c, 7, 8), 4 repeats (Fig. 1b, 1c, 2b, 7a, Supplementary Fig. 1d, 4e), 5 repeats (Fig. 3a, 3f–g, Supplementary Fig. 6b), 10 repeats (Fig. 5b–c), 15 repeats (Fig. 2a).
Data availability
The RNA-Seq data have been deposited in the Gene Expression Omnibus (GEO) database under the accession code GSE90091. Source data for Fig. 1b, 1d, 1e, 2b, 2c, 2e, 3b, 3f–h, 4a, 4g, 7b, 8b, 8d, 8g and Supplementary Fig. 1e, 2a, 2b, 4b–d, 5b, 5d, 5f, 6a, 7, 8 have been provided as Supplementary Table 5. All other data supporting the findings of this study are available from the corresponding author upon reasonable request.
Supplementary Material
Acknowledgments
We thank Drs Hye-Jung Kim, Myles Brown, Henry Long, Prakash Rao, Zach Herbert, Quifu Ma, and John Alberta, Jing Zhou and Charles D. Stiles for helpful discussions and help with experiments. This work was supported by NIH grants R01 CA202634, R01 CA132740, and P01 CA080111 (to P.S.) and AG011085 (to J.W.H). A.O. was supported by an Edward R. and Anne G. Lefler Center Postdoctoral Fellowship.
Footnotes
Author Contributions L.L. and P.S. conceived the study. L.L. designed and performed the experiments with help from collaborators. W.M. contributed in vitro and in vivo kinase assays, viral transduction and conceptually helped with design of experiments. H.I., K.S., N.T.N., and W.W. contributed analyses of endogenous Nanog/Oct4/Sox2 phosphorylation, interaction with Pin1, and analyses of mutant proteins. J.M.C. and S.P.G. contributed mass spectrometric analyses. N.L. initiated the study and bred compound mutant animals, Y.G. performed cultures of human cancer cells, A.Y.M. and K.L.L. provided immunostaining analyses of teratomas, K.L.L. provided human patient-derived primary glioblastoma cells, A.O. and J.W.H. contributed analyses of polyubiquitination of endogenous proteins, A.K. helped with in-cell kinase assays, R.T.B. analyzed and interpreted all histological specimens, K.P. provided human breast cancer cell lines and helped with the design of breast cancer analyses. L.L. and P.S. wrote the paper. P.S. directed the study.
P.S. is a consultant and a recipient of a research grant from Novartis.
References
- 1.Malumbres M. Cyclin-dependent kinases. Genome Biol. 2014;15:122. doi: 10.1186/gb4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kozar K, et al. Mouse development and cell proliferation in the absence of D-cyclins. Cell. 2004;118:477–491. doi: 10.1016/j.cell.2004.07.025. [DOI] [PubMed] [Google Scholar]
- 3.Geng Y, et al. Cyclin E ablation in the mouse. Cell. 2003;114:431–443. doi: 10.1016/s0092-8674(03)00645-7. [DOI] [PubMed] [Google Scholar]
- 4.Parisi T, et al. Cyclins E1 and E2 are required for endoreplication in placental trophoblast giant cells. Embo J. 2003;22:4794–4803. doi: 10.1093/emboj/cdg482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martello G, et al. Esrrb is a pivotal target of the Gsk3/Tcf3 axis regulating embryonic stem cell self-renewal. Cell Stem Cell. 2012;11:491–504. doi: 10.1016/j.stem.2012.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Avilion AA, et al. Multipotent cell lineages in early mouse development depend on SOX2 function. Genes & development. 2003;17:126–140. doi: 10.1101/gad.224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ivanova N, et al. Dissecting self-renewal in stem cells with RNA interference. Nature. 2006;442:533–538. doi: 10.1038/nature04915. [DOI] [PubMed] [Google Scholar]
- 8.Mitsui K, et al. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell. 2003;113:631–642. doi: 10.1016/s0092-8674(03)00393-3. [DOI] [PubMed] [Google Scholar]
- 9.Nichols J, et al. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell. 1998;95:379–391. doi: 10.1016/s0092-8674(00)81769-9. [DOI] [PubMed] [Google Scholar]
- 10.Boward B, Wu T, Dalton S. Concise review: Control of cell fate through cell cycle and pluripotency networks. Stem Cells. 2016;34:1427–1436. doi: 10.1002/stem.2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Niwa H, et al. Interaction between Oct3/4 and Cdx2 determines trophectoderm differentiation. Cell. 2005;123:917–929. doi: 10.1016/j.cell.2005.08.040. [DOI] [PubMed] [Google Scholar]
- 12.Ralston A, Rossant J. Cdx2 acts downstream of cell polarization to cell-autonomously promote trophectoderm fate in the early mouse embryo. Dev Biol. 2008;313:614–629. doi: 10.1016/j.ydbio.2007.10.054. [DOI] [PubMed] [Google Scholar]
- 13.Rossant J, Tam PP. Blastocyst lineage formation, early embryonic asymmetries and axis patterning in the mouse. Development. 2009;136:701–713. doi: 10.1242/dev.017178. [DOI] [PubMed] [Google Scholar]
- 14.Masui S, et al. Pluripotency governed by Sox2 via regulation of Oct3/4 expression in mouse embryonic stem cells. Nat Cell Biol. 2007;9:625–635. doi: 10.1038/ncb1589. [DOI] [PubMed] [Google Scholar]
- 15.Niwa H, Miyazaki J, Smith AG. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat Genet. 2000;24:372–376. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- 16.Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- 17.Chung Y, et al. Embryonic and extraembryonic stem cell lines derived from single mouse blastomeres. Nature. 2006;439:216–219. doi: 10.1038/nature04277. [DOI] [PubMed] [Google Scholar]
- 18.Baltus GA, et al. Acetylation of sox2 induces its nuclear export in embryonic stem cells. Stem Cells. 2009;27:2175–2184. doi: 10.1002/stem.168. [DOI] [PubMed] [Google Scholar]
- 19.Ramakrishna S, et al. PEST motif sequence regulating human NANOG for proteasomal degradation. Stem Cells Dev. 2011;20:1511–1519. doi: 10.1089/scd.2010.0410. [DOI] [PubMed] [Google Scholar]
- 20.Xu H, et al. WWP2 promotes degradation of transcription factor OCT4 in human embryonic stem cells. Cell Res. 2009;19:561–573. doi: 10.1038/cr.2009.31. [DOI] [PubMed] [Google Scholar]
- 21.Banko MR, et al. Chemical genetic screen for AMPKalpha2 substrates uncovers a network of proteins involved in mitosis. Mol Cell. 2011;44:878–892. doi: 10.1016/j.molcel.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hertz NT, et al. Chemical Genetic Approach for Kinase-Substrate Mapping by Covalent Capture of Thiophosphopeptides and Analysis by Mass Spectrometry. Curr Prot Chem Bio. 2010:15–36. doi: 10.1002/9780470559277.ch090201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moretto-Zita M, et al. Phosphorylation stabilizes Nanog by promoting its interaction with Pin1. Proc Natl Acad Sci U S A. 2010;107:13312–13317. doi: 10.1073/pnas.1005847107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shen M, Stukenberg PT, Kirschner MW, Lu KP. The essential mitotic peptidyl-prolyl isomerase Pin1 binds and regulates mitosis-specific phosphoproteins. Genes & development. 1998;12:706–720. doi: 10.1101/gad.12.5.706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nishi M, et al. A distinct role for Pin1 in the induction and maintenance of pluripotency. The Journal of biological chemistry. 2011;286:11593–11603. doi: 10.1074/jbc.M110.187989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brooks EE, et al. CVT-313, a specific and potent inhibitor of CDK2 that prevents neointimal proliferation. The Journal of biological chemistry. 1997;272:29207–29211. doi: 10.1074/jbc.272.46.29207. [DOI] [PubMed] [Google Scholar]
- 27.Vanner RJ, et al. Quiescent sox2(+) cells drive hierarchical growth and relapse in sonic hedgehog subgroup medulloblastoma. Cancer cell. 2014;26:33–47. doi: 10.1016/j.ccr.2014.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boumahdi S, et al. SOX2 controls tumour initiation and cancer stem-cell functions in squamous-cell carcinoma. Nature. 2014;511:246–250. doi: 10.1038/nature13305. [DOI] [PubMed] [Google Scholar]
- 29.Sturm D, et al. Paediatric and adult glioblastoma: multiform (epi)genomic culprits emerge. Nature reviews. Cancer. 2014;14:92–107. doi: 10.1038/nrc3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Singh SK, et al. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 31.Lopez-Bertoni H, et al. DNMT-dependent suppression of microRNA regulates the induction of GBM tumor-propagating phenotype by Oct4 and Sox2. Oncogene. 2015;34:3994–4004. doi: 10.1038/onc.2014.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Holmberg J, et al. Activation of neural and pluripotent stem cell signatures correlates with increased malignancy in human glioma. PloS one. 2011:6. doi: 10.1371/journal.pone.0018454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.de la Rocha AM, Sampron N, Alonso MM, Matheu A. Role of SOX family of transcription factors in central nervous system tumors. American journal of cancer research. 2014;4:312–324. [PMC free article] [PubMed] [Google Scholar]
- 34.Liu K, et al. The multiple roles for Sox2 in stem cell maintenance and tumorigenesis. Cell Signal. 2013;25:1264–1271. doi: 10.1016/j.cellsig.2013.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gangemi RM, et al. SOX2 silencing in glioblastoma tumor-initiating cells causes stop of proliferation and loss of tumorigenicity. Stem Cells. 2009;27:40–48. doi: 10.1634/stemcells.2008-0493. [DOI] [PubMed] [Google Scholar]
- 36.Alonso MM, et al. Genetic and epigenetic modifications of Sox2 contribute to the invasive phenotype of malignant gliomas. PloS one. 2011;6:e26740. doi: 10.1371/journal.pone.0026740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hagerstrand D, et al. Identification of a SOX2-dependent subset of tumor- and sphere-forming glioblastoma cells with a distinct tyrosine kinase inhibitor sensitivity profile. Neuro Oncol. 2011;13:1178–1191. doi: 10.1093/neuonc/nor113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Holmberg Olausson K, et al. Prominin-1 (CD133) defines both stem and non-stem cell populations in CNS development and gliomas. PloS one. 2014;9:e106694. doi: 10.1371/journal.pone.0106694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ben-Porath I, et al. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet. 2008;40:499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stead E, et al. Pluripotent cell division cycles are driven by ectopic Cdk2, cyclin A/E and E2F activities. Oncogene. 2002;28:8320–8333. doi: 10.1038/sj.onc.1206015. [DOI] [PubMed] [Google Scholar]
- 41.White J, Stead E, Faast R, Conn SCP, Dalton S. Developmental activation of the Rb-E2F pathway and establishment of cell cycle-regulated cyclin-dependent kinase activity during embryonic stem cell differentiation. Molecular biology of the cell. 2005;16:2018–2027. doi: 10.1091/mbc.E04-12-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Filipczyk AA, Laslett AL, Mummery C, Pera MF. Differentiation is coupled to changes in the cell cycle regulatory apparatus of human embryonic stem cells. Stem Cell Res. 2007;1:45–60. doi: 10.1016/j.scr.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 43.Neganova I, Zhang X, Atkinson S, Lako M. Expression and functional analysis of G1 to S regulatory components reveals an important role for CDK2 in cell cycle regulation in human embryonic stem cells. Oncogene. 2009;28:20–30. doi: 10.1038/onc.2008.358. [DOI] [PubMed] [Google Scholar]
- 44.Raiz S, et al. A high proliferation rate is required for cell reprogramming and maintenance of human embryonic stem cell identity. Curr Biol. 2011;21:45–52. doi: 10.1016/j.cub.2010.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ouyang J, et al. Cyclin-dependent kinase-mediated Sox2 phosphorylation enhances the ability of Sox2 to establish the pluripotent state. The Journal of biological chemistry. 2015;290:22782–22794. doi: 10.1074/jbc.M115.658195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pauklin S, Vallier L. The cell-cycle state of stem cells determines cell fate propensity. Cell. 2013;155:135–147. doi: 10.1016/j.cell.2013.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Beddington RS, Robertson EJ. An assessment of the developmental potential of embryonic stem cells in the midgestation mouse embryo. Development. 1989;105:733–737. doi: 10.1242/dev.105.4.733. [DOI] [PubMed] [Google Scholar]
- 48.Thomson M, et al. Pluripotency factors in embryonic stem cells regulate differentiation into germ layers. Cell cycle. 2011;145:875–889. doi: 10.1016/j.cell.2011.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Suva ML, et al. Reconstructing and reprogramming the tumor-propagating potential of glioblastoma stem-like cells. Cell. 2014;157:580–594. doi: 10.1016/j.cell.2014.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Marotta LL, et al. The JAK2/STAT3 signaling pathway is required for growth of CD44(+)CD24(−) stem cell-like breast cancer cells in human tumors. J Clin Invest. 2011;121:2723–2735. doi: 10.1172/JCI44745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sicinski P, et al. Cyclin D1 provides a link between development and oncogenesis in the retina and breast. Cell. 1995;82:621–630. doi: 10.1016/0092-8674(95)90034-9. [DOI] [PubMed] [Google Scholar]
- 52.Sicinski P, et al. Cyclin D2 is an FSH-responsive gene involved in gonadal cell proliferation and oncogenesis. Nature. 1996;384:470–474. doi: 10.1038/384470a0. [DOI] [PubMed] [Google Scholar]
- 53.Sicinska E, et al. Requirement for cyclin D3 in lymphocyte development and T cell leukemias. Cancer cell. 2003;4:451–461. doi: 10.1016/s1535-6108(03)00301-5. [DOI] [PubMed] [Google Scholar]
- 54.Odajima J, et al. Cyclin E constrains Cdk5 activity to regulate synaptic plasticity and memory formation. Dev Cell. 2011;21:655–668. doi: 10.1016/j.devcel.2011.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ying QL, et al. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dunn SJ, Martello G, Yordanov B, Emmott S, Smith AG. Defining an essential transcription factor program for naive pluripotency. Science. 2014;344:1156–1160. doi: 10.1126/science.1248882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Blelloch RH, et al. Nuclear cloning of embryonal carcinoma cells. Proc Natl Acad Sci U S A. 2004;101:13985–13990. doi: 10.1073/pnas.0405015101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ordureau A, et al. Quantitative proteomics reveal a feedforward mechanism for mitochondrial PARKIN translocation and ubiquitin chain synthesis. Mol Cell. 2014;56:360–375. doi: 10.1016/j.molcel.2014.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li N, et al. Cyclin C is a haploinsufficient tumour suppressor. Nat Cell Biol. 2014;16:1080–1091. doi: 10.1038/ncb3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chick JM, et al. Defining the consequences of genetic variation on a proteome-wide scale. Nature. 2016;534:500–505. doi: 10.1038/nature18270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Huttlin EL, et al. A tissue-specific atlas of mouse protein phosphorylation and expression. Cell. 2010;143:1174–1189. doi: 10.1016/j.cell.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Eng JK, McCormack AL, Yates JR. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 63.Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods. 2007;4:207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- 64.Pandita A, Aldape KD, Zadeh G, Guha A, James CD. Contrasting in vivo and in vitro fates of glioblastoma cell subpopulations with amplified EGFR. Genes Chromosomes Cancer. 2004;39:29–36. doi: 10.1002/gcc.10300. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-Seq data have been deposited in the Gene Expression Omnibus (GEO) database under the accession code GSE90091. Source data for Fig. 1b, 1d, 1e, 2b, 2c, 2e, 3b, 3f–h, 4a, 4g, 7b, 8b, 8d, 8g and Supplementary Fig. 1e, 2a, 2b, 4b–d, 5b, 5d, 5f, 6a, 7, 8 have been provided as Supplementary Table 5. All other data supporting the findings of this study are available from the corresponding author upon reasonable request.