Abstract
Background
Ginsenoside Rh2 (G-Rh2) is a ginseng saponin that is widely investigated because of its remarkable antitumor activity. However, the molecular mechanism by which (20S) G-Rh2 triggers its functions and how target animals avoid its cytotoxic action remains largely unknown.
Methods
Phage display was used to screen the human targets of (20S) G-Rh2. Fluorescence spectroscopy and UV-visible absorption spectroscopy were used to confirm the interaction of candidate target proteins and (20S) G-Rh2. Molecular docking was utilized to calculate the estimated free energy of binding and to structurally visualize their interactions. MTT assay and immunoblotting were used to assess whether human serum albumin (HSA), bovine serum albumin (BSA), and bovine serum can reduce the cytotoxic activity of (20S) G-Rh2 in HepG2 cells.
Results
In phage display, (20S) G-Rh2-beads and (20R) G-Rh2-beads were combined with numerous kinds of phages, and a total of 111 different human complementary DNAs (cDNA) were identified, including HSA which had the highest rate. The binding constant and number of binding site in the interaction between (20S)-Rh2 and HSA were 3.5 × 105 M−1 and 1, and those in the interaction between (20S) G-Rh2 and BSA were 1.4 × 105 M−1 and 1. The quenching mechanism is static quenching. HSA, BSA and bovine serum significantly reduced the proapoptotic effect of (20S) G-Rh2.
Conclusion
HSA and BSA interact with (20S) G-Rh2. Serum inhibited the activity of (20S) G-Rh2 mainly due to the interaction between (20S) G-Rh2 and serum albumin (SA). This study proposes that HSA may enhance (20S) G-Rh2 water solubility, and thus might be used as nanoparticles in the (20S) G-Rh2 delivery process.
Keywords: cytotoxic effect, fluorescence spectroscopy, G-Rh2 human target, phage display, serum albumin
1. Introduction
Ginseng has long been used as a tonic, prophylactic and restorative agent for thousands of years in ancient China and Korea [1]. Ginsenoside, a metabolite of ginseng and a class of triterpenoid saponins, is the main active constituent of ginseng [2]. Over 150 natural ginsenosides have been identified, which are classified along with different aglycones into the two major types of dammarane and leonine [2]. With increased empirical observations, many pharmacological effects have been proven [1], for example, ginsenosides could increase learning ability, protect cerebral damnification, and improve comprehension efficiency [3], [4], [5]. Saponins from Korean Red Ginseng exhibited a potent anti-inflammatory activity under in vitro and in vivo inflammatory conditions [6], [7] and promoted breast cancer cell apoptosis [8]. The anticancer activity of ginseng saponins is correlated with the number and position of hydroxyl groups on the dammarane skeleton, number of sugar molecules, and stereoselectivity [9].
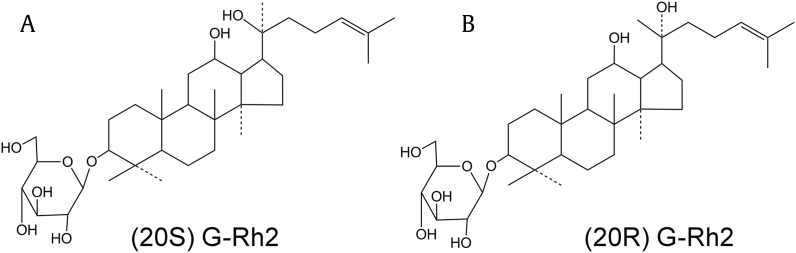
Recent extensive pharmacokinetic studies have shown that ginsenosides possess very poor oral bioavailability (< 5%) because of their poor oral absorption [10]. Rb1, Rb2, Rd, Rc, Re and Rg1, constitute more than 90% of total ginsenosides in normal ginseng extract [11]. Rb, the most abundant ginsenoside, can be easily converted into more pharmaceutically potent minor ginsenosides, such as Rh2 (Fig. 1), compound K, and PPDol, through the hydrolysis of its sugar moieties by gastric acid or microorganisms in the stomach or enteric canal [11], [12].
Fig. 1.
Chemical structures of two ginsenosides. (A) (20S) G-Rh2. (B) (20R) G-Rh2.
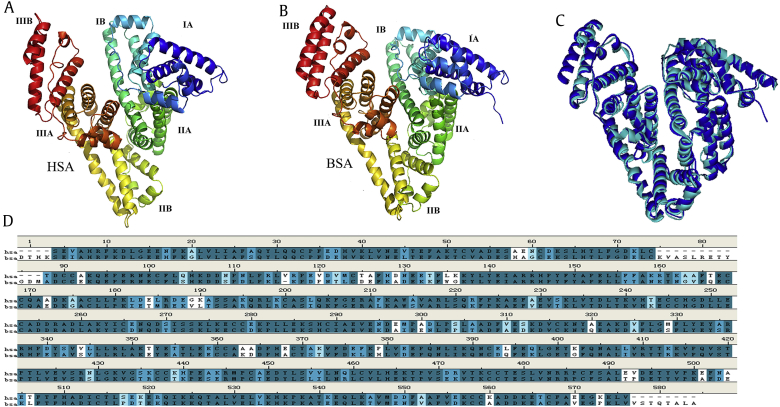
Human serum albumin (HSA), a product of the liver, is the main compound of blood. HSA contributes to many important physiological functions, such as colloidal oncotic blood pressure and the maintenance of blood pH. HSA is also a transport protein with vitamins, nutrients, toxic substances and hydronium. HSA has a long half-life of 18 days in the circulatory and lymphatic systems. Structurally, HSA is a 585 amino-acid protein, a product of three series-wound gene duplications. These three homologous domains fuse to form a predominantly alpha-helical heart-shaped molecule (Fig. 2A) that is highly cross-linked by 17 disulfides. Each domain consists of two subdomains, A and B. Several small-molecule binding sites exist in HSA, especially sites I and II (Fig. 2A) [13], [14], [15], [16]. HSA also binds a wide variety of drug molecules, and albumin–drug binding is important in our understanding of the pharmacokinetics and pharmacological effects of drugs [15], [17]. Bovine serum albumin (BSA), the counterpart of HSA, has a similar sequence, structure, and almost the same physiological functions in bovine blood (Figs. 2B–2D) [18]. Dozens of HSA-delivering drugs are currently being used in clinical trials or are being developed [19]. HSA nanoparticles are a new drug-delivery system with a number of important benefits, such as preferential uptake in tumor or inflamed tissue, high stability, high biodegradability, ready availability, and lack of toxicity and immunogenicity [20], [21], [22].
Fig. 2.
Crystal structures and sequence. (A) Indicating the subdivisions of HSA into domains (I-III) and subdomains (A, B). (B) Indicating the subdivisions of BSA into domains (I-III) and subdomains (A, B). (C) Structure aligned between HSA and BSA. (D) Sequence aligned between HSA and BSA. Atomic coordinates were obtained from the PDB (4IW1 and 4OR0). The illustration was made with Discovery Studio (version 4, BIOVIA, USA). The picture was drown according to reference [13]. BSA, bovine serum albumin; HSA, human serum albumin.
About 20 kinds of ginsenosides are absorbed through the oral consumption of ginseng, and these ginsenosides are transformed into several cytotoxic compounds such as G-Rh2, G-Rg3, compound K, and PPDol in the stomach and intestinal tract. However, little damage has been found in people who have taken ginseng. In the current study we screened cellular targets of G-Rh2 with the human liver cancer cell cDNA library by phage display technology. HSA was one of the target proteins screened out with the highest binding rate. We also experimentally demonstrated the interaction of G-Rh2 with both HSA and BSA by fluorescence spectroscopy and showed that interaction of HSA or BSA with G-Rh2 remarkably reduced its cytotoxic effect.
2. Materials and methods
2.1. Materials
(20S)-Rh2 and (20R)-Rh2 were obtained from Abcam (Cambridge, MA, USA). Amino polyethyleneglycol-polyacrylamide copolymer (PEGA) resin and T7 Select Human Liver Tumor cDNA phage library were purchased from Millipore (MA, USA). 4-Nitrophenyl bromoacetate was purchased from Alfa Aesar (MA, USA). Dimethyl sulfoxide (DMSO), 3-(4,5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide (MTT), and BSA were purchased from Sigma–Aldrich (St. Louis, MO, USA). Recombinant HSA was purchased from Healthgen Biotechnology Ltd. (Wuhan, China). Primer synthesis and sequencing were provided by Sangon Biotech (Shanghai, China). Newborn calf serum and Dulbecco modified Eagle's medium (DMEM) were purchased from Gibco (Life Technologies, Grand Island, NY, USA). Antibody against poly (adenosine diphosphateribose) polymerase (PARP) was purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). The electrogenerated chemiluminescence revelation system used was from TransGen Biotech (Beijing, China).
2.2. Methods
2.2.1. Preparation of Rh2-PEGA beads
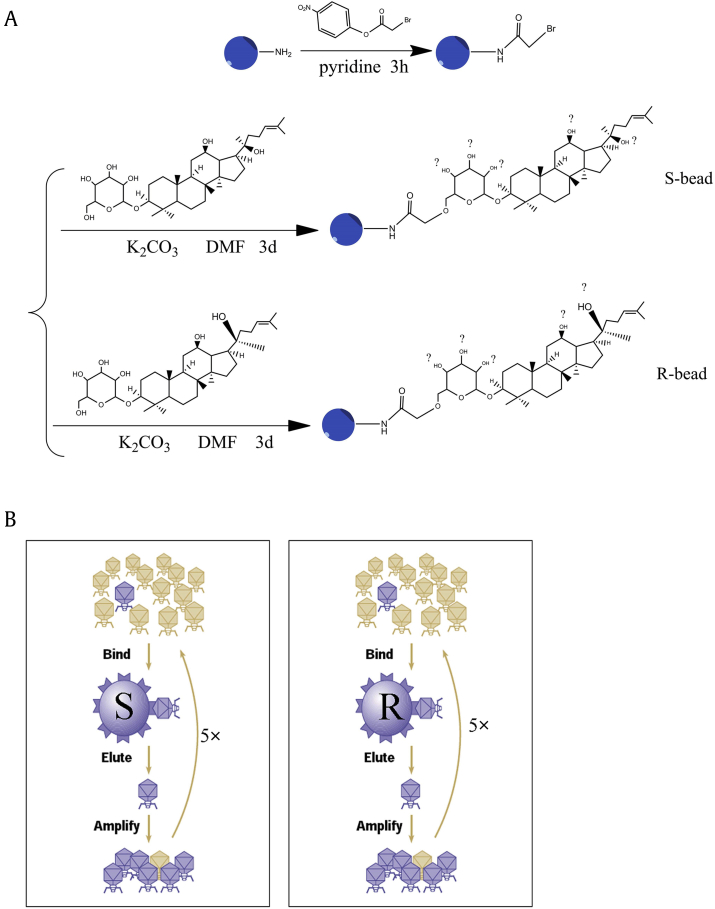
G-Rh2 was immobilized onto amino PEGA beads (Millipore) [23]. In a typical procedure, 750 mg of PEGA beads were swelled, washed three times with pyridine, and mixed with 0.33 mmol equivalents (to the amino group loaded on the beads, 0.4 mmol/g) of 4-nitrophenyl bromoacetate (Alfa Aesar). The beads were stirred at room temperature for 3 h, filtered, and washed three times with ∼10 mL each of dichloromethane (CH2Cl2), MeOH, and N, N-dimethylformamide (DMF). Then, the brominated PEGA beads were mixed with 0.3 mmol equivalents of (20S) G-Rh2 or (20R) G-Rh2, and 0.3 mmol K2CO3 in DMF. The mixture was stirred at room temperature for 3 days, filtered, and washed three times with ∼10 mL each of CH2Cl2 and MeOH. The Rh2-immobilized PEGA beads were stored in MeOH, and before use, the beads were centrifuged and washed three times with an appropriate buffer.
2.2.2. Phage display screening
According to the manufacturer's instructions and published article [23], phage display screening was performed using a T7 Select Human Liver Tumor cDNA phage library (Millipore). The original library was first amplified by infecting host cells. The amplified library was precleared by incubating 1 mL of T7 phage (∼1011 pfu/mL) with 100 μL of native PEGA-beads at 4°C overnight. The amplified precleared phage suspension (800 μL) was incubated with 200 μL of S-beads or R-beads at 4°C overnight and washed with 1 mL of TBST buffer (10 mM Tris-HCl, pH 8.0, 150 mM NaCl, and 0.1% TWEEN-20) 10 times. The beads were incubated with 200 μL of elution buffer (1% sodium dodecyl sulfate) for 30 min at room temperature by shaking gently. Eluted fractions (5 μL) were inoculated into 5 mL of Escherichia coli BLT5615 (Millipore) host bacteria cells and incubated for about 3 h at 37°C. Phage-infected cells were mixed with NaCl to a final concentration of 0.5 M and then centrifuged at 800g for 5 min. Supernatants containing phage particles were used for the subsequent biopanning step. Phage titers for each biopanning step were evaluated by counting pfu/mL according to the manufacturer's protocol (Millipore).
2.2.3. Sequence analysis of selected phage recombinants
After the final biopanning step, phages were diluted to obtain an individual plaque. About 100 μL of lysis buffer (10 mM EDTA, pH 8.0) was placed in a tube, which was heated at 65°C for 10 min, and then briefly vortexed. The solution was cooled to room temperature and centrifuged at 12,000g at 4°C for 3 min to clarify. The lysate served as the template to perform polymerase chain reaction (PCR) with the primers 5′-GGAGCTGTCGTATTCCAGTC-3′ and 5′-AACCCCTCAAGACCCGTTTA-3′. The thermal cycler program was 1 cycle at 95°C for 5 min, followed by 40 cycles at 95°C for 30 s, 58°C for 90 s, 72°C for 100 s and final extension at 72°C for 10 min. After sequencing, data were aligned to the National Center for Biotechnology Information to identify DNA and protein.
2.2.4. Cell culture
HepG2 cells were grown in DMEM supplemented with 10% (V/V) heat-inactivated newborn calf serum, 100 μg/mL of streptomycin, and 100 U/mL of penicillin at 37°C in a humidified atmosphere with 5% CO2 in saturated humidity.
2.2.5. MTT assay
Exponentially growing HepG2 cells were seeded into a 96-well plate at 1 × 104 cells/well in triplicate. HSA and BSA were diluted to a concentration of 33.6 mg/mL with DMEM medium to be a substitute for newborn calf serum (containing 33.6 mg/mL BSA). After incubation for 20 h, cells were treated with 7.5 μg/mL (20S) G-Rh2 mixed with different concentrations of albumin supplied by HSA, BSA and newborn calf serum for 48 h. At 44 h post-treatment, 20 μL of MTT (5 mg/mL; Sigma) was added to each well, and the plate was incubated for 4 h. Then, 150 μL of dimethyl sulfoxide was added to each well to solubilize the formazan crystals formed by viable cells, and color intensity was measured at 550 nm with a microplate reader (TECAN, Männedorf, Switzerland).
2.2.6. Immunoblotting analysis
The cells were washed with ice-cold phosphate buffered-saline and solubilized in a lysis buffer containing 20 mM Tris with a pH of 7.4, 1% NP-40, 150 mM NaCl, 2 mM MgCl2, 1 mM dithioreitol, 0.5% Triton X-100, 1 mM EGTA, 25 mM NaF, 1 mM Na3VO4, 50 mM β-glycerol phosphate, 2 mg/mL leupeptin, 2 mg/mL pepstatin A, 2 mg/mL antipain, and 1mM PMSF for 1 h on ice. After centrifugation at 12,000g for 15 min, 50 μg of soluble protein from each sample was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, followed by electrotransfer onto a polyvinylidene difluoride membrane (Millipore). The membrane was blocked with 5% nonfat milk in phosphate-buffered saline with 0.1% Tween 20 and probed with antibodies. The membrane was washed, incubated with horseradish peroxidase coupled with antimouse immunoglobulin G (IgG) (Pierce, Rockford, IL, USA), and detected with an electrogenerated chemiluminescence revelation system (TransGen Biotech).
2.2.7. Fluorescence spectroscopy
To confirm the interactions of HSA and BSA with G-Rh2, fluorescence spectroscopy was performed. HSA and BSA concentrations were fixed at 1 μM, transferred into a quartz cell with 1 cm length path, and titrated manually through successive additions of 1 mM G-Rh2 at 10 min time intervals. Spectra were recorded within the wavelength range of 290–450 nm, with excitation set at 280 nm, excitation bandwidth set at 5 nm, and emission bandwidth set at 5 nm at 293 K, 298 K and 303 K. Each spectrum was the mean of at least three scans.
2.2.8. UV-Visible absorption spectroscopy
UV-Visible (UV-vis) measurements in the presence and absence of G-Rh2 were conducted in the range of 250–450 nm at 298 K on UV-2550 spectrophotometer equipped with 1.0 cm quartz cells. HSA and BSA concentrations were fixed at 25 μM, while the G-Rh2 concentration was fixed at 50 μM. Following addition of HSA or BSA and G-Rh2, the solution was equilibrated for 5 min, and absorbance values were recorded thereafter.
2.2.9. Virtual screening and docking
Virtual screening was conducted using the software AutoDock (version 4.2.6) based on Lamarckian Genetic Algorithm (Scripps Research Institute, La Jolla, CA, USA) according to the manual handbook and other publications. This software uses a sophisticated gradient optimization method in its local optimization procedure. The target used in our study was the crystal structures of HSA and BSA from the Research Collaboratory for Structural Bioinformatics Protein Data Bank (http://www.rcsb.org/pdb). The predicted complexes were optimized and ranked according to the empirical scoring function, Screen Score, which estimates the binding free energy of the ligand–receptor complex. The most stable distinguished conformation with the minimal binding energy was shown using Discovery Studio 4.0 Visualizer (BIOVIA, CA, USA).
2.2.10. Statistical analysis
All values were performed in triplicate and expressed as mean ± standard deviation with Microsoft Office 2013 and imaged with Sigmaplot 10 (Systat Software Inc., San Jose, CA, USA). A Student t test was used for quantitative analysis, and the significant difference is shown as **p < 0.01 and ***p < 0.001.
3. Results and discussion
3.1. Synthesis of G-Rh2 beads and target screen
For the comprehensive identification of G-Rh2 cellular targets, we coupled (20S/R) G-Rh2 to amino polyethyleneglycol-polyacrylamide copolymer beads (PEGA beads, Millipore) after activation with 4-nitrophenyl bromoacetate. This method resulted in (20S/R) G-Rh2 being coupled to the beads through either one of six hydroxyl in the two molecules, exposing different faces of G-Rh2 to proteins (Fig. 3A). The (20S)-Rh2-loaded beads (S-beads) or the (20R)-Rh2-loaded beads (R-beads) were used to screen, in parallel, the human liver cancer phage display cDNA library (Millipore). Based on the stereoselectivity and activity of each hydroxyl, we deduced that the –OH at C6 is the most likely site and that the beads may be a mixture of different C-linked PEGA with C6-O-PEGA as a main component.
Fig. 3.
Synthesis of Rh2 beads and phage display outline. (A) Scheme for the chemical synthesis of S- and R-beads (blue). The coupling of Rh2 to the beads occurred at the end of a polyethylene glycol linker (PEGA beads). Depending on the Rh2 hydroxyl group participating in the coupling to the phenyl bromoacetate group, S- and R-beads may consist of a combination of six products. (B) Schematic of the biopanning steps in the screening of a phage display cDNA library generated from human liver tumor cell mRNA. Five rounds of biopanning (5×), each including binding to the beads, washing, elution and amplification, were performed in parallel using S- or R-beads.
Five rounds of biopanning with S- and R-beads were performed in parallel. After the final selection, phages were collected (Fig. 3B), amplified, gradually diluted, and infected with E. coli BLT5615. DNA from a monocolony of phages was used to perform PCR amplification and sequencing. As shown in Table 1, from over 500 PCRs, we obtained 94 human cDNA containing phages screened by S-beads, and these phages encoded 46 different kinds of proteins. About 16/94 of the complementary DNAs (cDNA) were HSA. Meanwhile, from the R-bead screening, we obtained 136 human cDNAs containing phages encoding 65 different kinds of proteins, and 6/136 of the cDNAs were HSA.
Table 1.
Screen of phage display
| Number of clones encoding human proteins | Number of clones encoding HSA | Number of different target proteins | Rate of HSA encoding phages | |
|---|---|---|---|---|
| S-beads | 94 | 16 | 46 | 17% (16/94) |
| R-beads | 136 | 6 | 65 | 4.4% (6/136) |
HSA, human serum albumin; R-beads, (20R)-Rh2-loaded beads; S-beads, (20S)-Rh2-loaded beads.
3.2. Fluorescence quenching studies of HSA or BSA and G-Rh2
Fluorescence spectroscopy is regarded as one of the most comprehensive methods of studying protein–ligand interaction. In the present work, we used fluorescence spectroscopy to investigate the interaction between HSA or BSA and G-Rh2. Protein fluorescence absorption originates from Trp, Tyr and Phe residues, whereas intrinsic fluorescence can be mainly attributed only to the Trp residue [24].
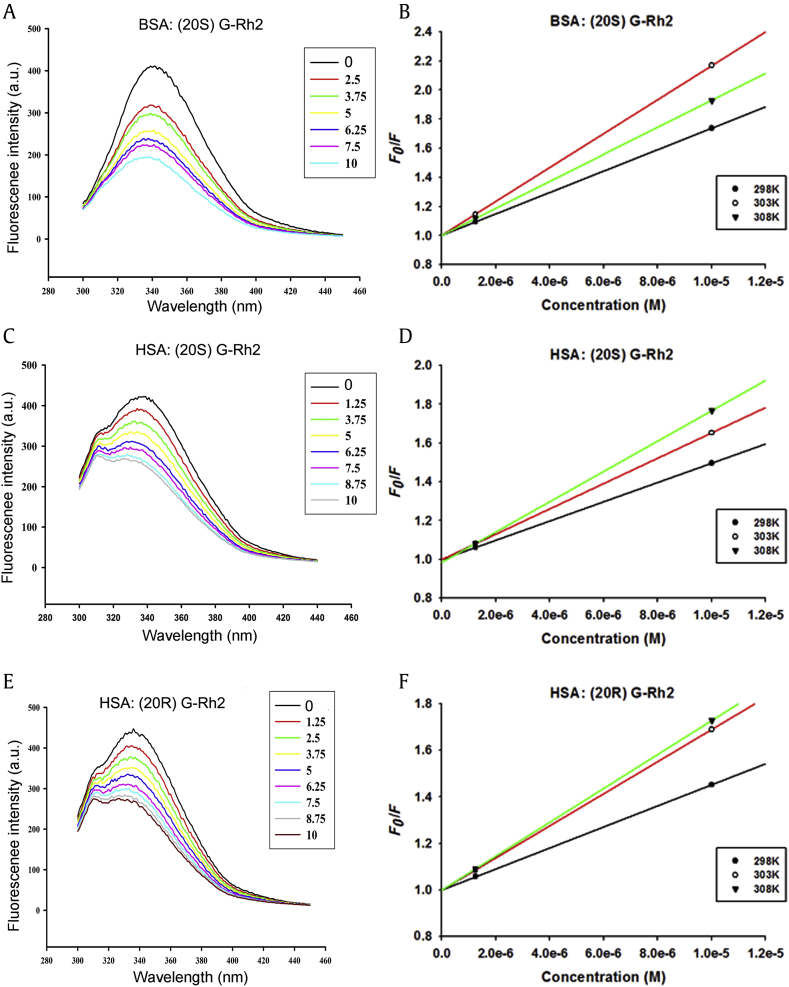
The fluorescence spectra of HSA and BSA with different concentrations of G-Rh2 were determined, as shown in Fig. 4 and Table 2. HSA and BSA fluorescence intensity decreased with increased G-Rh2 concentrations (Figs. 4A, 4C, and 4E), indicating that G-Rh2 interacted with HSA and BSA. The fluorescence intensity of G-Rh2 were not observed clearly at 290–450 nm (data not shown).
Fig. 4.
Fluorescence spectroscopy of HSA or BSA with G-Rh2 (pH 7.4). (A, B) 1 μM BSA and increasing concentrations of (20S) G-Rh2. (C, D) 1μM HSA and increasing concentrations of (20S) G-Rh2. (E, F) 1μM HSA and increasing concentrations of (20R) G-Rh2. (A, C, E) Fluorescence spectroscopy at 298 K. (B, D, F) Stern–Volmer plots for quenching of HSA and BSA fluorescence by G-Rh2 at different temperatures (298 K, 303 K and 308 K). λex = 280 nm, λem = 300–450 nm. BSA, bovine serum albumin; G, ginsenoside; HSA, human serum albumin.
Table 2.
The results of fluorescence quenching
| Temperature (K) |
KSV (104 M−1) |
kq (1012 M−1 s−1) |
K (105 M−1) |
n | |
|---|---|---|---|---|---|
| BSA: (20S) G-Rh2 | 298 | 7.3470 | 7.3470 | 3.615763 | 1.1329 |
| 303 | 11.6703 | 11.6703 | 1.425936 | 1.0192 | |
| 308 | 9.2848 | 9.2848 | 0.646398 | 0.9702 | |
| HSA: (20S) G-Rh2 | 298 | 4.9578 | 4.9578 | 5.458835 | 1.2027 |
| 303 | 6.5117 | 6.5117 | 3.50187 | 1.1449 | |
| 308 | 7.2510 | 7.2510 | 1.853958 | 1.0786 | |
| HSA: (20R) G-Rh2 | 298 | 4.5120 | 4.5120 | 2.063004 | 1.131 |
| 303 | 6.9016 | 6.9016 | 1.450775 | 1.0626 | |
| 308 | 7.2751 | 7.2751 | 0.866363 | 1.0284 |
BSA, bovine serum albumin; G, ginsenoside; HSA, human serum albumin.
Fluorescence quenching could be distinguished by collision and static quenching, whereas higher temperature could result in strong collision quenching [24]. The maximum scatter collision quenching constant reported for various kinds of quencher to a biopolymer is 2 × 1010 mol−1 s−1 [25].The Ksv and kq can be calculated as in Eq. (1) [26], below:
| (1) |
where F0 and F are the steady-state fluorescence intensities in the absence and presence of quencher, respectively; [Q] is the concentration of quencher; kq is the quenching rate constant of biomolecule; τ0 is the average lifetime of the protein without the quencher and is of the order, 10−8 s [27]; and Ksv is the Stern–Volmer dynamic quenching rate constant.
Meanwhile, in static quenching, if similar and independent binding sites in the biomolecule were assumed, the binding constant and number of binding sites can be calculated as in Eq. (2), below:
| (2) |
where F0 is the fluorescence intensity of HSA or BSA, F is the fluorescence intensity of HSA or BSA with G-Rh2, and Q is the corresponding concentration of G-Rh2.
To confirm the quenching mechanism, we detected the kq values at different temperatures. As shown in Figs. 4B, 4D, and 4F, and in Table 2, with the temperature increasing, the kq and KSV increased. This result suggested the quenching mechanism is dynamic quenching, however all the kq values were much larger than the maximum scatter collision quenching constant, 2 × 1010 mol−1 s−1. Thus, the quenching mechanism could be assumed to be due to static quenching, a new complex formation between G-Rh2 and HSA or BSA, rather than dynamic collision, if any. The number of binding sites was one in both (20S) G-Rh2 and (20R) G-Rh2. The binding constants of (20S) G-Rh2 and (20R) G-Rh2 with HSA were 3.5 × 105 M−1, and 1.5 × 105 M−1, respectively at 298K. The binding constants of (20S) G-Rh2 and BSA was 1.4 × 105 M−1 at 298K.
3.3. UV-Vis absorption spectra
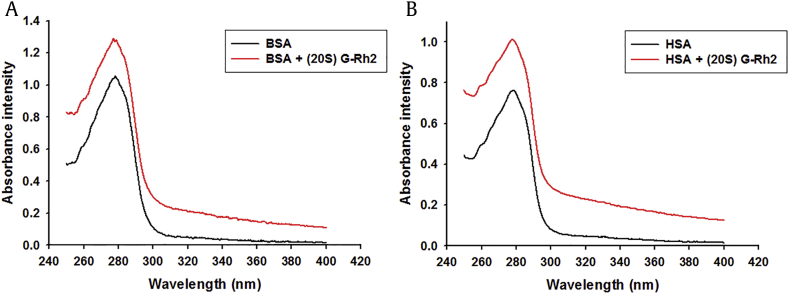
To further confirm the interaction status between BSA or HSA with G-Rh2, a UV-Vis absorption spectra was performed. UV-Vis absorption measurement is a simple and pertinent method that is used to investigate the secondary structural changes of proteins and to explore complex formations [28]. As shown in Fig. 5, the absorptions were significantly increased with a slight blue shift of the maximum absorption wavelength of BSA or HSA by adding the G-Rh2, indicating the exact binding of the G-Rh2 and BSA or HSA, and an increase of hydrophobicity for the light emitting residue.
Fig. 5.
UV-Vis absorption spectra of BSA or HSA in the presence of G-Rh2 (298K, pH7.4). Effect of (20S) G-Rh2 on the UV-Vis absorption spectra. (A) BSA. (B) HSA. CBSA = CHSA = 25 μM; CG-Rh2 = 50 μM; λ = 250–400 nm. BSA, bovine serum albumin; G, ginsenoside; HSA, human serum albumin.
In summary, the spectrophotometric analyses collectively showed that G-Rh2 binds and interacts with both BSA and HSA.
3.4. Docking analysis
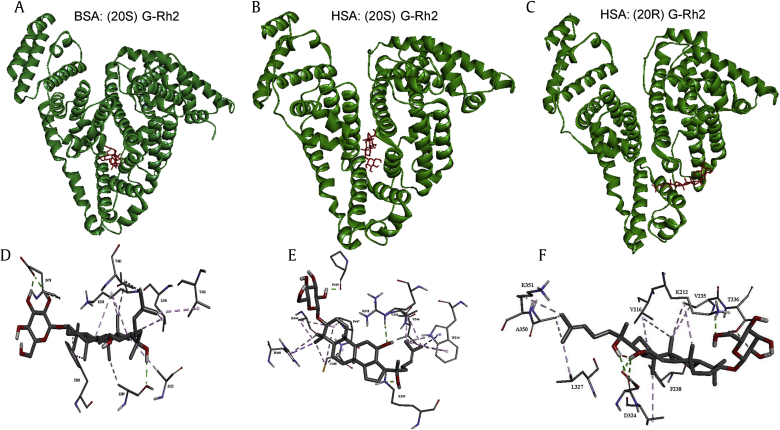
Molecular docking was used to elucidate the interaction between HSA or BSA and G-Rh2 and to ultimately elucidate the interaction sites (Fig. 6). Results obtained by AutoDock tools (version 4.2.6, Molecular Graphics Laboratory, The Scripps Research Institute, La Jolla, CA, USA) presented 10 different conformations, and we selected the conformation with the lowest estimated free energy for further analysis. This analysis showed that K195, W214, R218, P339, V343, V344, K444 and C448 contributed to the interaction between (20S) G-Rh2 and HSA. Residues F205, A209, A212, K350, V342, L346, V481 and E478 were the most vital residues present in the interaction site in (20S) G-Rh2 and BSA interaction. Residues K212, V216, V235, T236, F238, D324, L327, A350 and K351 contributed to the interaction between (20R) G-Rh2 and HSA. The possible interacting model and the main residues involved in the interaction are depicted in Figs. 6D–6F. The ligand core in contact with the protein was anchored to the binding site mainly by H-bonds (in green) and hydrophobic (in pink) interaction. The binding sites were around the chromophore in the proteins, which may explain the interaction between G-Rh2 with HSA or BSA to quench the fluorescence. The estimated free energy of HSA-G-Rh2 and BSA-G-Rh2 were all negative (Table 3), indicating their spontaneous interactions.
Fig. 6.
Docking analysis. (A–C) Whole views of binding sites. (D–F) Details of binding sites. (A, D) Visualization of BSA and (20S) G-Rh2. (B, E) Visualization of HSA and (20S) G-Rh2. (C, F) Visualization of HSA and (20R) G-Rh2. The enlargement of ligand in the binding site is shown. H-bonds are depicted by green dashed lines, and hydrophobic interactions are depicted by purple dashed lines. BSA, bovine serum albumin; G, ginsenoside; HSA, human serum albumin.
Table 3.
Docking results
| Name | Estimated free energy of binding (kcal/mol) | Estimated inhibition constant, Ki (298.15 K) |
|---|---|---|
| HSA:(20S) G-Rh2 | −6.65 | 13.29 μM |
| HSA:(20R) G-Rh2 | −6.28 | 24.81 μM |
| BSA:(20S) G-Rh2 | −5.23 | 147.2 μM |
BSA, bovine serum albumin; G, ginsenoside; HSA, human serum albumin.
3.5. Albumin inhibited the cytotoxic activity of (20S) G-Rh2
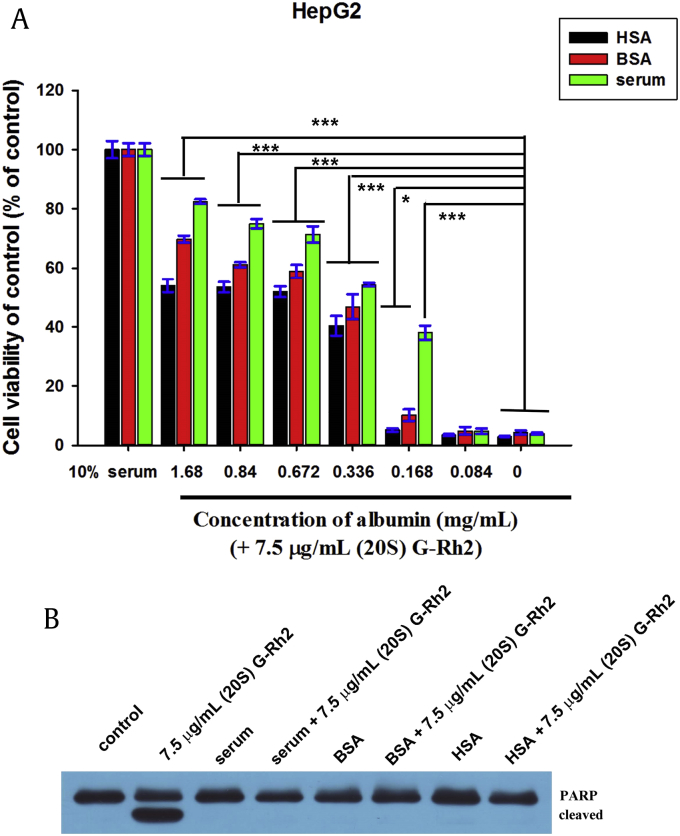
Previous studies have shown that adding serum to culture medium remarkably reduces the cytotoxic activity of (20S) G-Rh2 [29], [30]. Thus, the impaired cytotoxic effect of (20S) G-Rh2 by serum may result from the interaction between BSA and (20S) G-Rh2. To examine this possibility, MTT assay was carried out with the same concentration of serum albumin (SA) by adding free HSA, BSA and bovine serum to (20S) G-Rh2, which was contained in culture medium. As shown in Fig. 7A, treatment of 7.5 μg/mL (20S)-Rh2 without SA killed almost all cells, but with increased concentration of albumin by adding HSA, BSA or bovine serum, cell viabilities significantly increased in an albumin-dose-dependent manner. Cell viabilities as influenced by HSA and BSA were similar and slightly increased after adding bovine serum, which may be due to the growth factors and other nutrients in bovine serum that could increase cell growth and proliferation. As shown in Fig. 7B, about 7.5 μg/mL (20S) G-Rh2 without HSA, BSA or bovine serum caused the cleavage of poly-adenosine diphosphate-ribose polymerase (PARP), a substrate of active caspase-3, to yield an 85-kD fragment. However, HSA, BSA or bovine serum totally inhibited PARP cleavage, indicating that HSA, BSA or bovine serum prevented the apoptosis-inducing activity of (20S) G-Rh2. This phenomenon may explain why different IC50 values of (20S) G-Rh2 have been reported for the same cell lines [30], [31]. Humans absorb a number of cytotoxic ginsenosides including G-Rh2 by taking ginseng, but no obvious damage has been reported. This finding may be explained by the present data showing that HSA interactions with these ginsenosides including (20S) G-Rh2 downregulate their cytotoxic effect.
Fig. 7.
HSA, BSA and bovine serum significantly reduced the cytotoxic activity of 20(S)-Rh2. (A) HepG2 cells were treated with 7.5 μg/mL 20(S)-Rh2 and increasing concentrations of SA for 48 h. Cell viability was estimated by MTT assay and expressed as the percentage of viable cells in treated and controlled samples. Data are presented as the mean ± standard deviation of experiments performed in triplicate. Student t test was used for quantitative analysis, and the significant difference is shown as *p < 0.05, ***p < 0.001. (B) HepG2 cells were treated with 7.5 μg/mL (20S)-Rh2 and with or without 1.68mg/mL albumin by adding HSA, BSA or bovine serum into medium, for 4 h. The equal amounts of cell extracts were resolved by sodium dodecyl sulfate–polyacrylamide gel electrophoresis and analyzed by immunoblotting using specific antibody against PARP. BSA, bovine serum albumin; G, ginsenoside; HSA, human serum albumin; PARP, poly-adenosine diphosphate-ribose polymerase.
Like most active natural compounds, (20S) G-Rh2 has very low water solubility and is thus difficult to be used as a drug. Guang-Ji Wang et al [32] showed that the principal factors affecting the relatively low bioavailability of G-Rh2 are: (1) low solubility; (2) little membrane permeability; (3) potent efflux transport; and (4) presystemic elimination. In their studies, the micronization of Rh2 enhanced its oral absorption, Cmax, bioavailability, short Tmax, and MRT. Micronizing decreased particle size and consequently increased dissolution rate [33]. HSA, a well-known medical protein, is used to increase the solubility and transportation efficiency of clinical drugs with little toxicity. We assumed that HSA may enhance G-Rh2 solubility, bioavailability, and half time in circulation and be used as a nanoparticle to deliver these ginsenosides to tumor tissues. These possibilities should be examined in future studies.
SA is mainly produced by liver cells and secreted into the extracellular environment, however, a large amount of this protein exists in liver cells. Intracellular SA, as a G-Rh2 target protein, might mediate G-Rh2-triggered signal transductions in cells, and this possibility should be studied in future research.
In the current study, we identified HSA as a potent G-Rh2 human target by phage display technology. Both fluorescence spectroscopy and molecular docking analysis demonstrated that the interactions between HSA or BSA and G-Rh2, and the interaction of HSA or BSA with (20S) G-Rh2 resulted in the downregulation of (20S) G-Rh2 cytotoxicity.
Conflicts of interest
All authors hereby declare no conflicts of interest.
Acknowledgments
This work was supported by grants from the National Nature Science Foundation of China (Project 31240078), Grant of Talent Exploitation in 2012 from Jilin Province.
References
- 1.Lin Y., Jiang D., Li Y., Han X., Yu D., Park J.H., Jin Y.H. Effect of sun ginseng potentiation on epirubicin and paclitaxel-induced apoptosis in human cervical cancer cells. J Ginseng Res. 2015;39:22–28. doi: 10.1016/j.jgr.2014.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim Y.J., Zhang D., Yang D.C. Biosynthesis and biotechnological production of ginsenosides. Biotechnol Adv. 2015;33(6 Pt 1):717–735. doi: 10.1016/j.biotechadv.2015.03.001. [DOI] [PubMed] [Google Scholar]
- 3.Zhang J.T., Qu Z.W., Liu Y., Deng H.L. Preliminary study on antiamnestic mechanism of ginsenoside Rg1 and Rb1. Chin Med J (Engl) 1990;103:932–938. [PubMed] [Google Scholar]
- 4.Zhang G., Liu A., Zhou Y., San X., Jin T., Jin Y. Panax ginseng ginsenoside-Rg2 protects memory impairment via anti-apoptosis in a rat model with vascular dementia. J Ethnopharmacol. 2008;115:441–448. doi: 10.1016/j.jep.2007.10.026. [DOI] [PubMed] [Google Scholar]
- 5.Kennedy D.O., Scholey A.B. Ginseng: potential for the enhancement of cognitive performance and mood. Pharmacol Biochem Behav. 2003;75:687–700. doi: 10.1016/s0091-3057(03)00126-6. [DOI] [PubMed] [Google Scholar]
- 6.Baek K.S., Hong Y.D., Kim Y., Sung N.Y., Yang S., Lee K.M., Park J.Y., Park J.S., Rho H.S., Shin S.S. Anti-inflammatory activity of AP-SF, a ginsenoside-enriched fraction, from Korean ginseng. J Ginseng Res. 2015;39:155–161. doi: 10.1016/j.jgr.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang Y., Lee J., Rhee M.H., Yu T., Baek K.S., Sung N.Y., Kim Y., Yoon K., Kim J.H., Kwak Y.S. Molecular mechanism of protopanaxadiol saponin fraction-mediated anti-inflammatory actions. J Ginseng Res. 2015;39:61–68. doi: 10.1016/j.jgr.2014.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim S.J., Kim A.K. Anti-breast cancer activity of Fine black ginseng (Panax ginseng Meyer) and ginsenoside Rg5. J Ginseng Res. 2015;39:125–134. doi: 10.1016/j.jgr.2014.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nag S.A., Qin J.J., Wang W., Wang M.H., Wang H., Zhang R.W. Ginsenosides as anticancer agents: in vitro and in vivo activities, structure-activity relationships, and molecular mechanisms of action. Front Pharmacol. 2012;3 doi: 10.3389/fphar.2012.00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang Z., Gao S., Wang J.R., Yin T.J., Teng Y., Wu B.J., You M., Jiang Z.H., Hu M. Enhancement of oral bioavailability of 20(S)-Ginsenoside Rh2 through improved understanding of its absorption and efflux mechanisms. Drug Metab Disposition. 2011;39:1866–1872. doi: 10.1124/dmd.111.040006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim H., Kim J.H., Lee P.Y., Bae K.H., Cho S., Park B.C., Shin H., Park S.G. Ginsenoside Rb1 is transformed into Rd and Rh2 by Microbacterium trichothecenolyticum. J Microbiol Biotechnol. 2013;23:1802–1805. doi: 10.4014/jmb.1307.07049. [DOI] [PubMed] [Google Scholar]
- 12.Chi H., Kim D.H., Ji G.E. Transformation of ginsenosides Rb2 and Rc from Panax ginseng by food microorganisms. Biol Pharm Bull. 2005;28:2102–2105. doi: 10.1248/bpb.28.2102. [DOI] [PubMed] [Google Scholar]
- 13.Yamasaki K., Chuang V.T., Maruyama T., Otagiri M. Albumin-drug interaction and its clinical implication. Biochim Biophys Acta. 2013;1830:5435–5443. doi: 10.1016/j.bbagen.2013.05.005. [DOI] [PubMed] [Google Scholar]
- 14.Curry S. Lessons from the crystallographic analysis of small molecule binding to human serum albumin. Drug Metab Pharmacokinet. 2009;24:342–357. doi: 10.2133/dmpk.24.342. [DOI] [PubMed] [Google Scholar]
- 15.Ghuman J., Zunszain P.A., Petitpas I., Bhattacharya A.A., Otagiri M., Curry S. Structural basis of the drug-binding specificity of human serum albumin. J Mol Biol. 2005;353:38–52. doi: 10.1016/j.jmb.2005.07.075. [DOI] [PubMed] [Google Scholar]
- 16.Wang Z.M., Ho J.X., Ruble J.R., Rose J., Ruker F., Ellenburg M., Murphy R., Click J., Soistman E., Wilkerson L. Structural studies of several clinically important oncology drugs in complex with human serum albumin. Biochim Biophys Acta. 2013;1830:5356–5374. doi: 10.1016/j.bbagen.2013.06.032. [DOI] [PubMed] [Google Scholar]
- 17.Otagiri M. A molecular functional study on the interactions of drugs with plasma proteins. Drug Metab Pharmacokinet. 2005;20:309–323. doi: 10.2133/dmpk.20.309. [DOI] [PubMed] [Google Scholar]
- 18.Xu H., Yao N., Xu H., Wang T., Li G., Li Z. Characterization of the interaction between eupatorin and bovine serum albumin by spectroscopic and molecular modeling methods. Int J Mol Sci. 2013;14:14185–14203. doi: 10.3390/ijms140714185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miele E., Spinelli G.P., Miele E., Tomao F., Tomao S. Albumin-bound formulation of paclitaxel (Abraxane ABI-007) in the treatment of breast cancer. Int J Nanomedicine. 2009;4:99–105. doi: 10.2147/ijn.s3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fasano M., Curry S., Terreno E., Galliano M., Fanali G., Narciso P., Notari S., Ascenzi P. The extraordinary ligand binding properties of human serum albumin. IUBMB Life. 2005;57:787–796. doi: 10.1080/15216540500404093. [DOI] [PubMed] [Google Scholar]
- 21.Kratz F. Albumin as a drug carrier: design of prodrugs, drug conjugates and nanoparticles. J Control Release. 2008;132:171–183. doi: 10.1016/j.jconrel.2008.05.010. [DOI] [PubMed] [Google Scholar]
- 22.Bae S., Ma K., Kim T.H., Lee E.S., Oh K.T., Park E.S., Lee K.C., Youn Y.S. Doxorubicin-loaded human serum albumin nanoparticles surface-modified with TNF-related apoptosis-inducing ligand and transferrin for targeting multiple tumor types. Biomaterials. 2012;33:1536–1546. doi: 10.1016/j.biomaterials.2011.10.050. [DOI] [PubMed] [Google Scholar]
- 23.Arango D., Morohashi K., Yilmaz A., Kuramochi K., Parihar A., Brahimaj B., Grotewold E., Doseff A.I. Molecular basis for the action of a dietary flavonoid revealed by the comprehensive identification of apigenin human targets. Proc Natl Acad Sci USA. 2013;110:E2153–E2162. doi: 10.1073/pnas.1303726110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li D., Zhu M., Xu C., Chen J., Ji B. The effect of Cu2+ or Fe3+ on the noncovalent binding of rutin with bovine serum albumin by spectroscopic analysis. Spectrochim Acta A Mol Biomol Spectrosc. 2011;78:74–79. doi: 10.1016/j.saa.2010.08.069. [DOI] [PubMed] [Google Scholar]
- 25.Ware W.R. Oxygen quenching of fluorescence in solution: an experimental study of the diffusion process. J Phys Chem. 1962;66:455–458. [Google Scholar]
- 26.Li D., Mei Z., Chen X., Ji B. Characterization of the baicalein-bovine serum albumin complex without or with Cu2+ or Fe3+ by spectroscopic approaches. Eur J Med Chem. 2011;46:588–599. doi: 10.1016/j.ejmech.2010.11.038. [DOI] [PubMed] [Google Scholar]
- 27.Lakowicz J.R., Weber G. Quenching of fluorescence by oxygen. A probe for structural fluctuations in macromolecules. Biochemistry. 1973;12:4161–4170. doi: 10.1021/bi00745a020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen T., Zhu S., Cao H., Shang Y., Wang M., Jiang G., Shi Y., Lu T. Studies on the interaction of salvianolic acid B with human hemoglobin by multi-spectroscopic techniques. Spectrochim Acta A Mol Biomol Spectrosc. 2011;78:1295–1301. doi: 10.1016/j.saa.2010.12.081. [DOI] [PubMed] [Google Scholar]
- 29.Li Q., Li Y., Wang X., Fang X., He K., Guo X., Zhan Z., Sun C., Jin Y.H. Co-treatment with ginsenoside Rh2 and betulinic acid synergistically induces apoptosis in human cancer cells in association with enhanced capsase-8 activation, bax translocation, and cytochrome c release. Mol Carcinog. 2011;50:760–769. doi: 10.1002/mc.20673. [DOI] [PubMed] [Google Scholar]
- 30.Guo X.X., Li Y., Sun C., Jiang D., Lin Y.J., Jin F.X., Lee S.K., Jin Y.H. p53-dependent Fas expression is critical for Ginsenoside Rh2 triggered caspase-8 activation in HeLa cells. Protein Cell. 2014;5:224–234. doi: 10.1007/s13238-014-0027-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li B., Zhao J., Wang C.Z., Searle J., He T.C., Yuan C.S., Du W. Ginsenoside Rh2 induces apoptosis and paraptosis-like cell death in colorectal cancer cells through activation of p53. Cancer Lett. 2011;301:185–192. doi: 10.1016/j.canlet.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gu Y., Wang G.J., Sun J.G., Jia Y.W., Wang W., Xu M.J., Lv T., Zheng Y.T., Sai Y. Pharmacokinetic characterization of ginsenoside Rh2, an anticancer nutrient from ginseng, in rats and dogs. Food Chem Toxicol. 2009;47:2257–2268. doi: 10.1016/j.fct.2009.06.013. [DOI] [PubMed] [Google Scholar]
- 33.Rogers T.L., Johnston K.P., Williams R.O., 3rd Solution-based particle formation of pharmaceutical powders by supercritical or compressed fluid CO2 and cryogenic spray-freezing technologies. Drug Dev Ind Pharm. 2001;27:1003–1015. doi: 10.1081/ddc-100108363. [DOI] [PubMed] [Google Scholar]