Abstract
Development of cancer metastasis is a key contributor to mortality in patients with colorectal cancer. High expression of RING-box 2 (RBX2) in cancer cells is known to play a key role in tumor progression. However, the role of RBX2 in colorectal cancer progression is not well elucidated. In this study, we silenced RBX2 via CRISPR/Cas9 in two colorectal cancer cell lines, HCT116 and SW480. RBX2 knockout attenuated proliferation, colony formation and enhanced sensitivity of colorectal cancer cells to paclitaxel treatment. Invasive property of HCT116 and SW480 cells was also attenuated by RBX2 silencing. We confirmed that increased RBX2 correlated with higher tumor cells growth and metastasis abilities by ectopic expression of RBX2 in HCT116 and SW480 cells. In vivo studies suggested that knockout of RBX2 inhibited xenografts growth and metastasis to lung tissue, whereas ectopic expression of RBX2 promoted these cellular functions. Mechanically, RBX2 induced gastric cancer cell growth and metastasis by activating mammalian target of rapamycin/S6 kinase 1 (mTOR/S6K1). Treatment of everolimus, the specific mTOR inhibitor, significantly attenuated RBX2-mediated cell proliferation and mobility in vitro. Taken together, these results revealed a novel role of RBX2 in colorectal cancer cell growth and metastasis via the mTOR pathway and suggested RBX2 may serve as a therapeutic target in colorectal cancer.
Keywords: RBX2, colorectal cancer, metastasis, mTOR
Introduction
Colorectal cancer (CRC) is one of the most common malignancies, ranking as high as the second leading cause of cancer mortality worldwide [1]. Although surgical resection followed by adjuvant chemotherapy or radiotherapy has been considered as the standard treatment option for CRC patient, who have resectable disease, 30%-50% of CRC patients under curative resection subsequently experience local and systemic recurrence. Hence, the poor prognosis and the major cause of death in patients with CRC are attributed to relapse and distant metastasis [2]. Therefore, these is urgently need for elucidating the molecular mechanism involved CRC metastasis and identifying novel molecular biomarker, to improve the prognosis of advanced CRC patients.
RING-box 2 (RBX2), also referred to as regulator of cullins 2 (ROC2), sensitive to apoptosis gene (SAG) or rING finger protein 7 (RNF7), consists of 113 amino acids and has a molecular weight of 12.6 kDa [3]. As a highly conserved protein, RBX2 is extensively expressed in several human tissues, including skeletal muscles, testis and heart. RBX2 is a redox-inducible anti-oxidant protein, which can be induced by redox agents, and to protect cells from apoptosis process induced by oxidation. Previous study demonstrates that RBX2 is also a component of E3 ubiquitin ligases and when it combines with other components to assemble E3 ubiquitin ligases, RBX2 exhibits E3 ubiquitin ligase activity [4]. Although RNF7 is inducible by the transcription factor of activator protein-1 (AP-1), RBX2 inhibits tumor-promoting functionality of activator protein-1 (AP-1) by ubiquitinylation and degradation of c-Jun. RNF7 has been shown to mediate ubiquitinylation of various cellular proteins such as p27, p21, pro-caspase-3, IκBα, hypoxia inducible factor 1 alpha (Hif-1α) and neurofibromin 1 (NF-1) [5]. Recently, RBX2 is reported over-expressed in many human cancer s, such as lung, colon, liver and stomach. Silencing RBX2 expression by RNA interference is proved to inhibit lung cancer cells, glioblastoma cells and pancreatic carcinoma cells proliferation in vitro and reverse radiation or chemotherapeutics resistance [5]. The mechanisms underlying may include RBX2 inactivates mammalian target of rapamycin (mTOR) and nuclear factor kappa B (NF-κB) pathway and/or accumulates of tumor suppressive genes, such as NF-1, procaspase-3, p27, p21, and phosphatase and tensin homolog (PTEN).
mTOR, which is a threonine/serine kinase, is considered to be the master coordinator of extracellular signals that regulates cell proliferation and metabolism. mTOR nucleates two distinct multi-protein complexes known as mTOR complex 1 (mTORC1) and mTOR complex 2 (mTORC2), characterized by the presence of raptor and Rictor respectively. mTORC1 regulates tumor cell growth and metastasis via eIF-4E-binding protein-1 (4E-BP1) and S6 kinase 1 (S6K1), both are key regulators participated in protein biosynthesis. mTORC2 comprises mTOR, mammalian lethal with SEC13 protein 8 (mLST8), Rictor, stress-activated protein kinase (SAPK)-interacting protein (Sin1), and phosphorylates AGC kinases, including Protein Kinase B (AKT), serum/glucocorticoid-regulated kinase 1 (SGK1), and protein kinase C (PKC), all of which are associated with malignant tumor. mTORC2 phosphorylates at Ser473 site on the Ser/Thr kinase AKT as well as the stability of AKT, which is necessary for its activation. Moreover, mTORC2 is involved in a variety of processes that are regulated by extracellular regulated protein kinases 1/2 (ERK1/2), including tumor cell survival, glucose metabolism, and cellular differentiation. Previous study demonstrates that RBX2 knock-down inactivated ERK1/2 pathway and inhibits prostate cancer tumorigenesis, which suggesting RBX2 might be a potential therapeutic target for prostate cancer [6].
Accumulating evidence suggests that RBX2 is involved in carcinogenesis and tumor progression [7], nevertheless, the functional role of RBX2 in colorectal carcinoma (CRC), especially in CRC metastasis, is not yet clear is still unknown. In the present study, we determined the elevated expression of RBX2 in CRC cancer tissues compared with the matched normal tissues. We further identified the role of RBX2 in promoting CRC cancer cell growth and metastasis through RBX2 up- or down-regulation models. Finally, we found that RBX2 induced CRC cancer cell growth and metastasis were associated with mTOR/S6K1 signaling pathway. Taken together, these findings suggest that RBX2 plays a pivotal role in colorectal cancer progression and might serve as a potential therapeutic target.
Methods
Cell culture and tissue specimens
Human colon cancer cell (HT29, HCT116, SW480 and LOVO) and normal colonic epithelial cell (HCoEpiC) were purchased from the Shanghai Institute of Cell Biology, Chinese Academy of Sciences (Shanghai, China). These cells were cultured in DMEM or RPMI 1640 medium with 10% fetal bovine serum. All methods for humans were performed in accordance with the relevant guidelines and regulations. A total of 56 fresh primary cancer and 12 paired adjacent normal tissue specimens were collected from patients with colorectal cancer in Affiliated Hospital of Nanjing University of Chinese Medicine. Tumor staging was determined according to the American Joint Committee on Cancer criteria. Informed consent was obtained from each patient before study. None of these patients underwent preoperative chemotherapy and/or radiation therapy. This study was approved by the Research Ethics Committee of Nanjing University of Chinese Medicine.
Establishment of RBX2 knockout and stable expression cells
HCT116 and SW480 cells were transfected with RBX2 CRISPR/Cas9 knock out (KO) Plasmid (Santa Cruz, sc-417138) using lipofectamine 2000 (Invitrogene) according to the manufacturer’s instructions. Stable overexpression of RBX2 was carried out using the lentiviral expression system (Genecopoeia, USA) according to the manufacturer’s instructions. The generation of retrovirus supernatants and transfection of colon carcinoma cells were conducted as described previously. The expression of RBX2 was confirmed by qPCR and western blotting assays.
Real-time PCR
Total RNA was extracted from gastric cancer tissues and the matched adjacent normal tissues using Trizol reagent (Invitrogen, Carlsbad, CA, USA). 1 μg of total RNA from each sample were reverse transcribed into cDNA using PrimeScript™ RT Master Mix Kit (Takara, Dalian, China). Real-time PCR was performed using SYBR Premix Ex Taq II Kit (Takara, Dalian, China) according to the instructions of the manufacturer. The specific primers used were as follows: RBX2 sense: 5’-ACCCTGCGTCCTTTCTTCG-3’ and antisense: 5’-GGCACAGGTATCGCACTCAA-3’; β-actin sense: 5’-AAGGAGCCCCACGAGAAAAAT-3’ and antisense: 5’-ACCGAACTTGCATTGATTCCAG-3’.
Immunohistochemistry
Immunohistochemistry (IHC) analysis was performed as described previously. The paraffin-embedded sections were deparaffinized in xylene, rehydrated in descending percentages of ethanol and heated in citrate buffer (pH 6.0) for antigen retrieval. After washing steps, slides were blocked with 3.0% hydrogen peroxide and 10% goat serum and incubated with a primary antibody at 4°C overnight. The following antibodies were used: rabbit anti-RBX2 antibody (1:1000; Santa Cruz), rabbit anti-Ki67 (1:500; Cell Signaling Technology), rabbit anti-p-mTORC1 antibody (1:1000; Santa Cruz), rabbit anti- p-mTORC2 antibody (1:1000; Santa Cruz) and rabbit anti-S6K1 antibody (1:1000; Santa Cruz). After the sequential incubation with biotinylated secondary antibody, streptavidin-horseradish peroxidase complex and diaminobenzidine (DAB), the slides were counterstained with hematoxylin, dehydrated, and mounted. Finally, sections were observed and imaged under light microscope.
Western blot analysis
Western blot was performed as previously reported. Briefly, whole-cell lysates were prepared and protein concentration was estimated using a BCA Protein Assay kit (Pierce, Rockford, MA, USA). The immune-blots were probed with primary antibodies overnight at 4°C followed by peroxidase conjugated goat anti-rabbit IgG for 2 hour. The following primary antibodies were used according to the manufacturer’s protocols: rabbit anti-RBX2 antibody (1:1000; Santa Cruz), rabbit anti-Ki67 (1:500; Cell Signaling Technology), rabbit anti-p-mTORC1Ser2448 antibody (1:1000; Santa Cruz), rabbit anti-p-mTORC2Ser2481 antibody (1:1000; Santa Cruz) and rabbit anti-p-S6K1Ser371 antibody (1:1000; Santa Cruz). The blots were visualized using enhanced chemiluminescence detection kit (Thermo). The bands were scanned and quantified by densitometric analysis using Image J software.
3-(4, 5-dimethyl-2-thiazolyl)-2, 5-diphenyl-2-H-tetrazolium bromide (MTT) assay
Cell proliferation was measured by MTT assay. Briefly, cells were plated in 96-well plates at a density of 3,000 cells per well. At various time points (24, 48, 72 and 96 h), 100 μl of MTT solution was added to each well and incubated at 37°C for 2 h, and then the absorbance was measured at 490 nm. The experiment was performed independently in triplicate.
Wound healing assay
Colorectal cancer cells were seeded in 6 well plates and starved for 24 h. After scratching with scratching with pipette tip, cell debris were washed with PBS and cultured with FBS free medium. After 24 hours, matched-pair wound regions were photographed and the areas of wound closure were calculated by image J [8].
Transwell invasion assay
Cell invasion was examined using Matrigel invasion Chambers (BD Bioscience, USA). Matrigel diluted in cold PBS was added on the upper chambers and incubated for 1 hour for gel formation. Then, cells were placed into the upper chamber and complete medium were added to the lower chamber. After 6 hours, the cells invaded through the membrane were stained with 0.1% crystal violet and counted [9].
Colony formation assay
1000 colon cancer cells were seeded into 6 cm plates and incubated for 14 days. Cells were then fixed with 4% paraformaldehyde and stained with 0.1% crystal violet. Colonies were counted and photographed [10].
Flow cytometry analysis
Cells were dissociated into single cells and labeled with FITC-conjugated anti-RBX2 (BD PharMingen) at 4°C for 30 min according to the manufacturers’ recommendation. The stained colorectal cancer cells were analyzed with the FACS Calibur machine and Cell Quest software (BD Biosciences). For cell apoptosis assay, cells were seeded in six-well plates and grown to approximately 80% confluence. Then indicated cells treating with 1 μM paclitaxel for 48 h. Cells were harvested and incubated with Annexin V/PI for 15 min in the dark, followed by fluorescence-activated cell sorting (FACS) analysis. All the assays were carried out four times.
Gene expression profiling and pathway analysis
For gene expression assay in parental and RBX2 knockout HCT116 cells, total RNA was extracted by using the phenol-chloroform method (Trizma; Sigma). RNA samples were treated with DNase-free kit (Ambion; Applied Biosystems), and RNA levels were quantified with an RNA 6000 Nano Assay Kit in Agilent 2100 Bioanalyzer (Agilent Technologies). Single-strand cDNA synthesis from total RNA was performed by using TaqMan reverse transcription reagents (Applied Biosystems), as recommended by the manufacturer. All quantitative real-time PCR reactions were performed with ABI PRISM 7900HT Fast Real-Time PCR system (Applied Biosystems) [11]. The expression of the housekeeping gene, TATA-binding protein (TBP), was used to normalize for variations in input cDNA. Heat map was visualized using Java TreeView v1.1.4r3 software. Pathway analysis was carried out with DAVID (https://david.ncifcrf.gov/) and Gene Ontology gene-sets derived from KEGG. The p-value threshold was set to 0.05.
Animal studies
HCT116 or SW480 cells were infected with RBX2 or RBX2-shRNA. Parental cell and stable infected cells (3 × 106) were subcutaneously inoculated in the lower rear flank of 5-week-old Balb/c-nude mice (Shanghai Slac Laboratory Animal Co. Ltd). On day 30 after implantation, tumors were harvested and weighed. The tumor volume was calculated (volume = 0.5 × length × width2). Immunohistochemical analysis was performed as previously described to determine the expression of RBX2, Ki67, p-mTORC1, p-mTORC2 and p-S6K1. For metastasis assay in vivo, B16F10 cells were re-suspended in PBS at a concentration of 1 × 107 cells/ml. 0.1 ml cell suspension was injected into tail veins of nude mice. All of the mice were killed by CO2 14 days after inoculation [12]. All protocols were approved and supervised by the institutional animal care and use committee of Affiliated Hospital of Nanjing University of Chinese Medicine.
Statistical analysis
All statistical analyses were carried out using the SPSS 17.0 statistical software package. Numerical data were presented as mean ± standard error. Two-tailed Student’s t-test was performed. P value less than 0.05 in all cases was considered statistically significant.
Results
Overexpression of RBX2 in colorectal cancer tissues
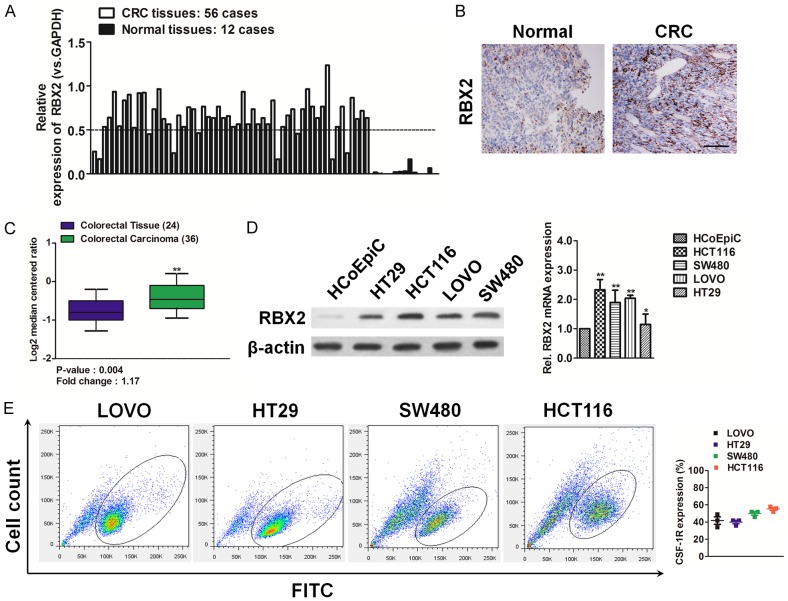
The expression of RBX2 in 56 colorectal carcinoma tissues was performed qPCR analysis and we demonstrated increased expression of RBX2 mRNA in colorectal cancer (CRC) tissues as compared to normal tissues (Figure 1A). The clinicopathologic features of the 56 patients with colorectal cancer were summarized in Supplementary Table 1. RBX2 overexpression in colorectal carcinoma was further supported by the results of immunohistochemical analysis, which showed more expression of RBX2 protein in CRC cancer tissues (Figure 1B). To investigate whether RBX2 was associated with human CRC progression, we analyzed its expression pattern in the publicly accessible Oncomine microarray database. In Skrzypczak colorectal statistics dataset containing RBX2 information [13], RBX2 was markedly increased in CRC cancer tissues, when compared with the matched normal tissues (Figure 1C). Then, we examined the protein and mRNA level of RBX2 in four CRC cell lines such as HCT116, SW480, LOVO, and HT29 by qPCR as well as immunoblotting (Figure 1D), and we further verified that RBX2 expression was relatively high in CRC cell lines and relatively low in normal colonic epithelial cell (HCoEpiC). Finally, FACS analysis after staining with anti-RBX2 antibody in CRC cell lines revealed the existence of colorectal carcinoma cells expressing RBX2 (Figure 1E).
Figure 1.
Overexpression of RBX2 in colorectal carcinoma tissues and cell lines. A. Relative expression of RBX2 mRNA in 56 CRC cancer and paired adjacent normal tissues as determined by qPCR. Increased expression of RBX2 mRNA as compared with normal tissue was observed. B. Expression of RBX2 protein was determined by IHC analysis. Representative images of IHC staining of RBX2 in CRC cancer tissues and adjacent normal tissues. Scale bar represents 100 μm. C. Box plots show increased levels of RBX2 in CRC (right) compared with normal tissues in Skrzypczak colorectal microarray data set. **P < 0.01, compared with normal colon tissues was determined by the Student’s t test. D. Western blotting analysis (left panel) of the level of RBX2 in various CRC cell lines. β-actin was used as loading controls. qPCR analysis (right panel) of RBX2 mRNA level in CRC cells. PCR values were normalized to the levels of β-actin. Data were presented as the mean ± SD from three independent measurements. E. Representative fluorescence activated cell sorter (FACS) dot plots of HCT116, SW480, LOVO, and HT29 cells stained with the anti-RBX2 antibody. Histograms reported the percentage of RBX2 positive cells as assessed by FACS. Mean ± SD of three independent experiments. Quantitative analysis demonstrates expression of RBX2 positive CRC cells range from 40%-60%.
RBX2 loss suppresses colorectal cancer growth
To dissect the functional role of RBX2 in CRC tumor growth, we generated stable RBX2 knock-out (KO) CRC cell lines by transfection of a CRISPR/Cas9 KO plasmid in HCT116 and SW480 cells. Transduction of cells with KO plasmid blocked RBX2 mRNA expression and significantly inhibited RBX2 protein expression compared to the parental cells (Figure 2A), which validated the efficiency and specific RBX2 knockout. Intriguingly, both CRC specific RBX2 KO cell lines resulted in significantly decreased cell proliferation relative to the parental cells (Figure 2B). Consistently, the frequencies of colony formation from RBX2 KO HCT116 and SW480 cells were decreased dramatically under regular medium culture condition (Figure 2C). We next compared the tumorigenic ability of parental cells vs. RBX2 KO CRC cells in Balb/c-nude mice and found that RBX2 deficient CRC cells exhibited significantly decreased growth in mice, as evaluated by tumor volume (Figure 2D). After 30 days, the xenografts were harvested and processed to immune-staining. The immunohistochemical staining of Ki67 further revealed that knockout of RBX2 inhibited CRC cancer growth in vivo (Figure 2D). Collectively, these findings identify CRC expressed RBX2 as a pro-tumorigenic mechanism.
Figure 2.
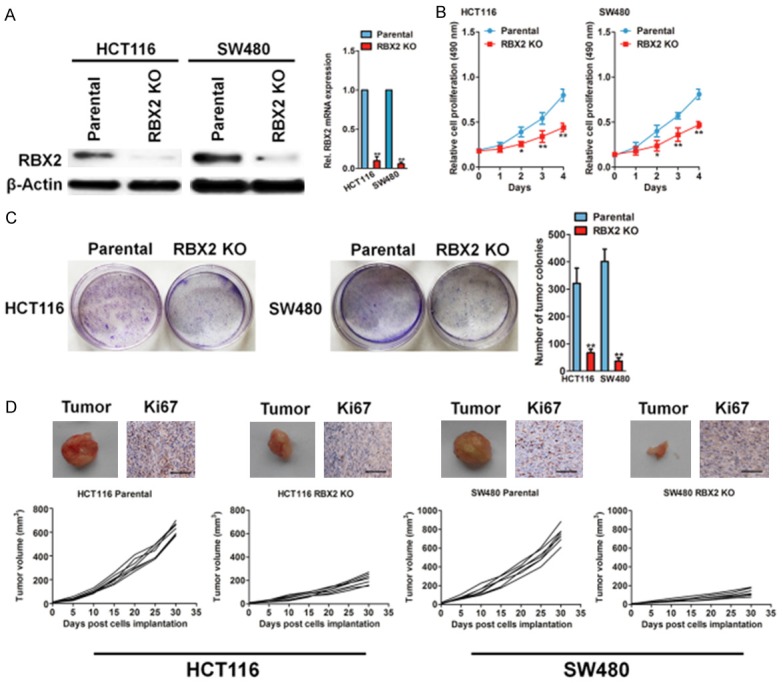
RBX2 depletion inhibits growth in colorectal cancer cells. A. Western blot to test the efficiency and specificity of RBX2 knockout plasmid transfection was shown completely loss of RBX2 relative to parental HCT116 and SW480 cells (left panel). qPCR analysis of RBX2 mRNA levels in both CRC cell lines. PCR values were normalized to the levels of β-actin. Data are presented as the mean ± SD from three independent measurements. B. Cell growth rate from parental and RBX2 knockout HCT116 cells (left) and SW480 cells (right) at indicated time point. C. Colony formation assay of parental and RBX2 knockout HCT116 and SW480 cells. 1000 cells were plated in 6-well plate in complete medium and cultured for 14 days and colonies were stained and counted. D. Representative images of xenografts. Tumor growth curve for HCT116 (left panel) and SW480 (right panel) xenogaft tumor models. Immunohistochemistry analysis of RBX2 and Ki67 in xenografts of the indicated groups. Scale bar represents 100 μm.
RBX2 loss inhibits CRC cells metastasis
Given that the poor prognosis in patients with CRC attributes to the metastatic property of cancer cells, we next addressed whether RBX2 deficient suppresses CRC cells metastasis in vitro and in vivo. Firstly, the wound healing analysis demonstrated that RBX2 knock out in two CRC cell lines resulted in a remarkably decreased cellular migration at 24 h point, when compared to the parental cells (Figure 3A). The quantitative indicated the cell migration rate was decreased from 59% to 64% in two CRC cell lines after RBX2 knockout. Consistent with the demonstration of RBX2 knockout inhibits CRC cells migration, less cells invaded into the bottom membrane when RBX2 was silenced in the Transwell assay (Figure 3B). To determine the observed CRC ells metastasis depend on RBX2, we knocked out RBX2 in B16F10 melanoma cells (Supplementary Figure 1A), which showed high invasion ability in vivo and injected the RBX2 KO B16F10 cells and their parental into nude mice via tail vein. Consistent with our in vitro findings, RBX2 KO in B16F10 cells impaired metastasis in lung tissue compared to their respective controls (Figure 3C).
Figure 3.
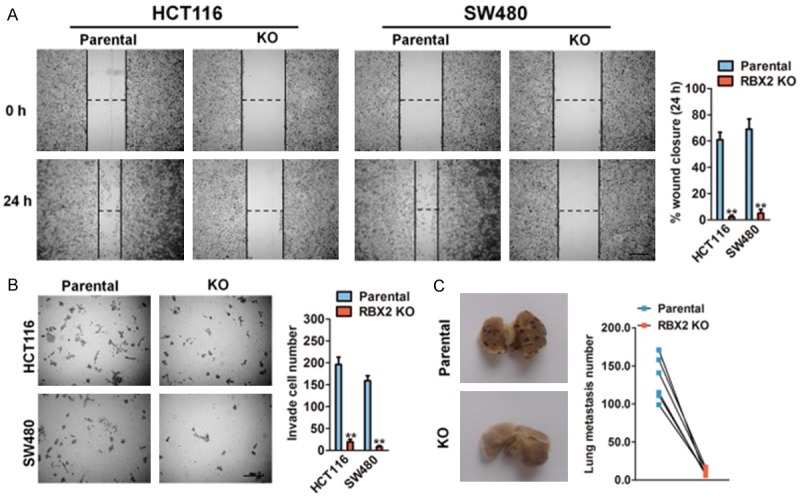
RBX2 loss suppresses cancer metastasis. A. In vitro wound healing assay with human HCT116 and SW480 cells after knock out with RBX2 expression. Image was acquired at 0, 24 h time points after scratching (left panel). Scale bar represents 100 μm. Quantification of wound closure was calculated (right panel). B. Representative staining of invasive potentials of human HCT116 and SW480 cells from Transwell assay in vitro (left panel). Quantification of invasive cells per field was analyzed (right panel). Scale bar: 100 μm. Data were compared using the two-tailed students t-test, for indicated comparisons, **P < 0.01. C. RBX2 loss significant compromised tumor metastasis in melanoma B16F10 cells lung metastasis model in vivo. Quantification of lung metastatic was calculated (right panel).
RBX2 over-expression promotes colorectal carcinoma growth
The stable overexpression of RBX2 in two CRC cell lines were retrovirally established and designated as HCT116-RBX2 and SW480-RBX2. The protein and mRNA levels of RBX2 in CRC cells were verified by immunoblotting and qPCR assay (Figure 4A), respectively. We next investigated the cell viability for four consecutive days by MTT assay and found that the induction in growth curve of cells infected with RBX2 lentvirus was more obvious on day 4, suggesting that RBX2 OE strongly promotes the proliferation of HCT116 and SW480 cells (Figure 4B). In addition, colony formation assay (Figure 4C) was performed to evaluate CRC tumor cells growth in vitro. Consistently, both the size of signal colony and total number of colonies in cells transfected that RBX2 lentvirus was remarkably increased in HCT116 and SW480 cells. To future shed light on the functional role of RBX2 in CRC cancer growth in vivo, ectopic expression of RBX2 in CRC cells from both HCT116 and SW480 exhibited significant stronger progression, as evaluated by larger tumor volume over the treatment period (Figure 4D). The immunohistochemical staining of Ki67 in tumor tissues further revealed that over-expressing of RBX2 accelerate CRC cancer growth in vivo (Figure 4D). Altogether, these results demonstrate RBX2 promote tumorigenic in colorectal carcinoma.
Figure 4.
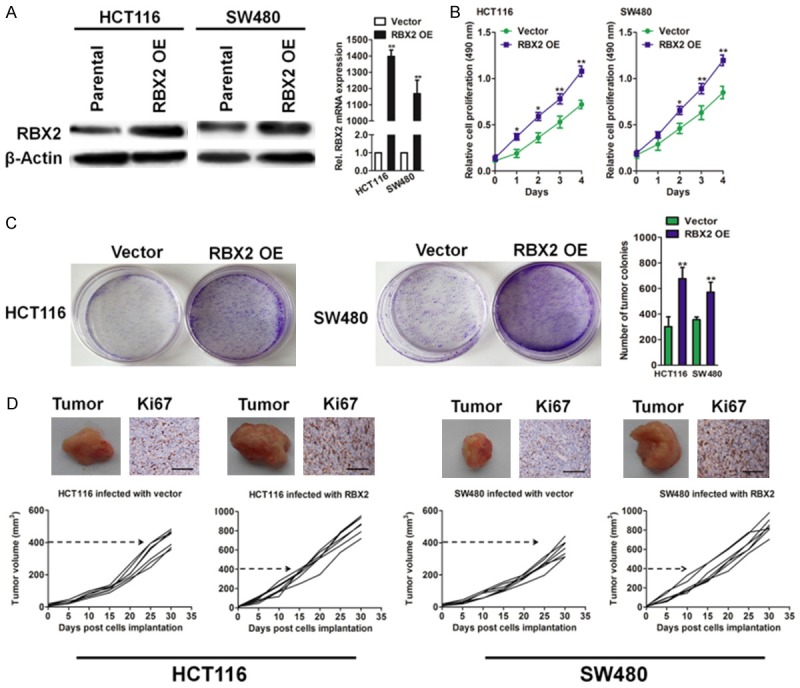
The effects of RBX2 modulation on the growth of xenografts in nude mice. A. Western blot analysis of RBX2 in HCT116 and SW480 cells transfected with vector or RBX2 (left panel). RBX2 mRNA level in HCT116 and SW480 cell lines are evaluated by qPCR analysis (right panel). PCR values were normalized to the levels of β-actin. Data are presented as the mean ± SD from three independent measurements. B. MTT analysis of CRC cancer cells infected with the indicated lentivirus. 3 × 103 cells were seeded in 96 well plates and cultured for the indicated hours. C. Colony formation assay of parental and RBX2 over-expressing CRC cells. Cells were plated in 6-well plate in complete medium and cultured for 14 days and colonies were stained and counted. D. HCT116 or SW480 cells infected with RBX2 or vector and then were implanted subcutaneously into Balb/c-nude mice to form xenografts. Representative images of tumor xenografts and immunohistochemistry analysis of Ki67 in xenografts of the indicated groups. Scale bar: 100 μm.
RBX2 promotes migration and invasion of CRC cells
Wound healing and Transwell matrigel invasion assays were performed to evaluate the effects of RBX2 on migration and invasion in HCT116-RBX2 and SW480-RBX2 cells. In wound healing assay, the wound closure ratio of cells with high RBX2 expression was obviously higher than the cells with an empty vector (Figure 5A). Matrigel assay was used to evaluate the invasive potential of cancer cells with altered RBX2 expression. The number of invaded CRC cancer cells was significantly increased by RBX2 over-expression (Figure 5B). These results indicate that RBX2 promotes the migration and invasion of CRC cancer cells in vitro. Furthermore, to detect the function of RBX2 in distant metastasis in vivo, we established B16F10-RBX2 melanoma cells (Supplementary Figure 1B) and injected the RBX2 over-expressing B16F10 cells into nude mice via tail vein. As expected, up-regulation of RBX2 in the B16F10 melanoma cells significantly promoted metastasis in lung tissue at 14 days after injection (Figure 4C), suggesting that RBX2 expression increases the number of metastatic foci in lung.
Figure 5.
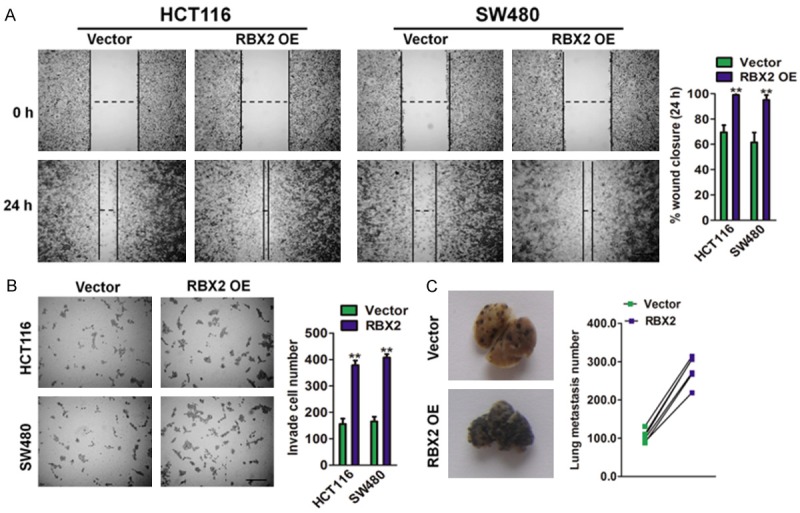
RBX2 overexpression in CRC cancer cells increase the migration and invasion. A. HCT116 and SW480 CRC cells with high RBX2 expression exhibited stronger migration abilities in wound healing assay (left panel). Scale bar represents 200 μm. Quantification of wound healing was calculated (right panel). Scale bar: 100 μm. B. HCT116 and SW480 CRC cells with high RBX2 expression exhibited stronger invasion abilities in Transwell assay. Scale bar: 200 μm. Data were compared using the two-tailed students t-test, for indicated comparisons, **P < 0.01 compared with the cells transfected with vector. Scale bar represents 100 μm. C. More metastatic foci in lung were visible in mouse injected with B16F10-RBX2 cells (left panel). Data were compared using the two-tailed Students t-test, **P < 0.01 compared with B16F10 cells transfected with vector. Quantification of lung metastasis loci was calculated (right panel).
RBX2 depletion enhances chemotherapeutic sensitivity in CRC cells
To future investigate the effect of RBX2 knock out on the sensitivity of CRC cells to chemotherapeutic drug, the parental and RBX2 stable knock-out cells were performed for proliferation and apoptosis analysis. Based on the MTT assay (Figure 6A), RBX2 depletion significantly augmented the sensitivity of HCT116 cells to paclitaxel. We next examined whether HCT116 specific RBX2 silencing affect cells colonization in vitro. As shown in Figure 6B, the colony forming ability in parental HCT116 cells were markedly compromised by paclitaxel treatment, and as inspected, RBX2 knock-out (KO) HCT116 cells showed greater sensitivity towards the chemotherapy agent. Because induced apoptosis is an important factor of the chemotherapeutic sensitivity of cancer cells, we used flow cytometry to detect the apoptosis capacity of HCT116 RBX2 knock-out/parental cells in the presence of paclitaxel. After 24 hour treatment, RBX2 KO HCT116 cells showed greater sensitivity towards paclitaxel, with more cells undergoing apoptosis after the treatment (Figure 6C). This assay indicated that RBX2 is associated with the apoptotic potential.
Figure 6.
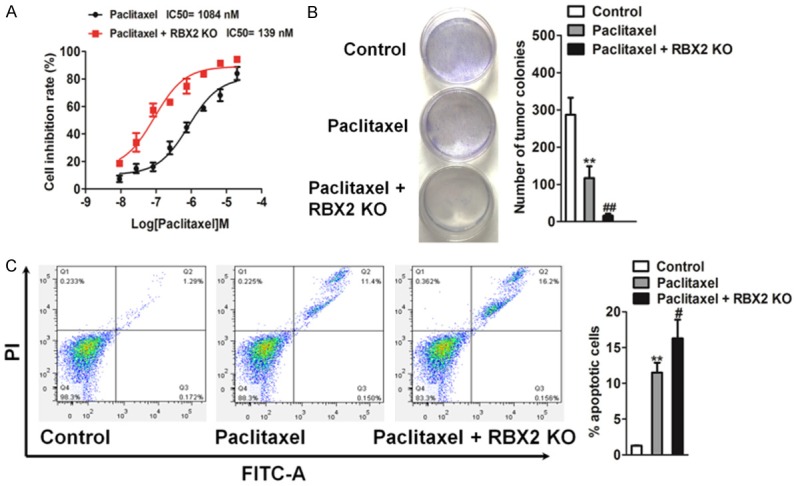
RBX2 enhances chemotherapeutic sensitivity in colon cancer cells. A. RBX2 knock-out (KO) significantly enhanced the sensitivity of HCT116 to paclitaxel and significantly reduced their IC50 based on the MTT assay. The data were presented as mean ± SD. The values were expressed as percentage of viable cells normalized to percentage of viable cells in 0.5% DMSO-treated cells. The concentration of paclitaxel resulting in 50% inhibition of control growth (IC50) was calculated by SPSS statistics software using Probit model. B. Colony formation assay was performed to detect the chemotherapeutic effects of paclitaxel on RBX2 KO HCT116 cells and control cells growth in vitro. 1000 RBX2 KO HCT116 cells or parental cells were plated in 6-well plate in complete medium and cultured with paclitaxel. After two weeks, colonies were stained and counted. C. RBX2 knock-out HCT116 cells showed greater sensitivity towards paclitaxel. Values were presented as the mean ± SD for three independent experiments. **P < 0.01 compared with control cells, ##P < 0.01 compared to control cells treated with paclitaxel.
RBX2 stimulates CRC cells proliferation via mTOR signaling pathway
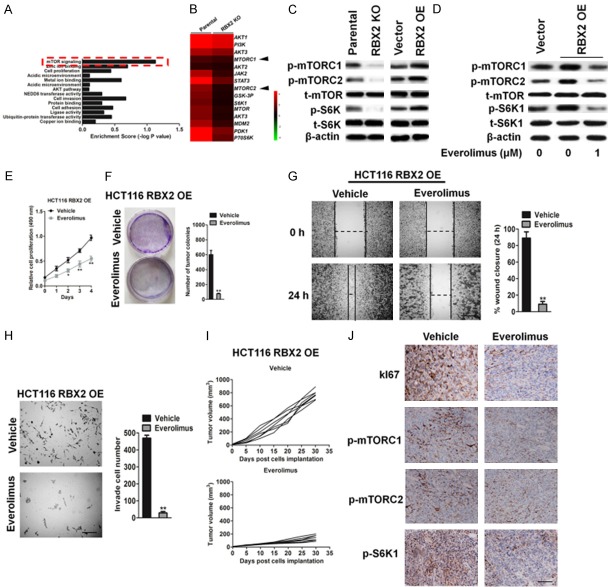
To elucidate the mechanisms in which RBX2 was engaged in the regulation of CRC cells metastasis, gene set enrichment analysis indicated that mTOR signaling was significantly enriched in RBX2 targets (Figure 7A). Furthermore, microarray assay was performed by using parental cells and RBX2 knock-out HCT116 cells. Microarray results revealed that the expression of a list of genes including mTORC1 and mTORC2 was similar between parental and HCT116 RBX2 knock out cells, which suggesting RBX2 down-regulation did not affect several kinases protein expression in CRC cells (Figure 7B). However, western blot analysis revealed that down-regulation of RBX2 significantly inhibited the phosphorylation of mTOR and its downstream molecule S6K1, while up-regulation of RBX2 increased the activity of mTOR/S6K signaling (Figure 7C). To ensure that the above approach represented a reliable readout and to explore the impact of blocking mTOR on RBX2 over-expressing cells, we studied the effects of overexpression of RBX2 on mTOR/S6K1 signaling in the presence of everolimus, a specific mTOR inhibitor. As shown in Figure 7D, everolimus significantly abrogated RBX2-induced phosphorylation of mTOR and S6K1. In line with this, RBX2-induced cell proliferation was also abolished by 1 μM everolimus as determined by MTT assay (Figure 7E). As expected, cell colony formation from everolimus-treated groups was significantly deceased compared with vehicle groups from RBX2 over-expressing HCT116 cells (Figure 7F). Wound closure from scratch assay (Figure 7G) and Transwell invasion analysis (Figure 7H) also showed that everolimus-treated group displayed significantly less metastatic potential from HCT116-RBX2 cells in vitro. In addition, to see the suppression effect of mTOR inhibitor in RBX2 knock-out cells, we treated RBX2 knockout HCT116 cells, and found treatment of mTOR inhibition in RBX2-KO HCT116 cells did not significantly reduce cell proliferation (Supplementary Figure 2A) and mobility (Supplementary Figure 2B) in vitro, indicating mTOR was one of the specific targets in RBX2 signaling. We next evaluated the tumor characterization by inhibition of mTORC1 from HCT116-RBX2 cells in vivo. As shown in Figure 7I, tumor volume was significantly decreased from in HCT116-RBX2 cells with administration of everolimus through inhibition of cell proliferation (Ki67) and mTOR pathway (Figure 7J). These results strongly suggested that RBX2 induced CRC cells growth and metastasis was mainly mediated via mTOR/S6K1 signaling pathway.
Figure 7.
Inhibition of mTOR significantly suppresses RBX2 over-expression tumor cell growth. A. Enrichment scores of signaling pathways among RBX2 targets. B. Heat-map of genes differentially expressed in parental cells and RBX2 KO HCT116 cells. C. Western blot analysis of the expression of p-mTORC1S2448, p-mTORC2S2481, t-mTOR, p-S6K1T389 and S6K1 in the HCT116 cells in response to RBX2 up-regulation. β-actin was used as loading control. D. Whole cell lysates were prepared from everolimus and vehicle treated HCT116-RBX2 cells and examined for p-mTORC1S2448, p-mTORC2S2481, t-mTOR, p-S6K1T389 and S6K1 by immunoblotting. E. Cell growth rate from everolimus and vehicle treated HCT116-RBX2. F. Representative pictures (left panel) and statistical analysis (right panel) shown colony formation from everolimus and vehicle treated HCT116-RBX2 cells. G. In vitro wound closure of everolimus and vehicle treated HCT116-RBX2 cells from 24 h after scratch assay. Scale bar: 100 μm. H. Transwell invasion assay of everolimus and vehicle treated HCT116-RBX2 cells (left panel). Quantification of invasive cells per field was analyzed (right panel). Scale bar: 100 μm. Data were compared using the two-tailed students t-test, for indicated comparisons, **P < 0.01 compared to the cells treatment with vehicle. I. Mice were injected with HCT116-RBX2 cells and then were treated with everolimus and vehicle (n = 6). Tumor volume was analyzed at indicated time point. J. Histochemistry staining of Ki67, p-mTORC1S2448, p-mTORC2S2481 and p-S6K1T389 shown decreased positive cells from everolimus treated tumors originally from injection of HCT116-RBX2 cells compare to vehicle-treated group. Scale bar: 100 μm.
Discussion
Recent studies suggest that RBX2 is frequently over-expressed in a variety of malignancies and contributes to cancer development and progression [14]. In lung, liver and stomach cancers, RBX2 were significantly overexpressed in cancerous tissues compared with normal adjacent tissues [15]. In prostate cancer, RBX2 correlates strongly with tumor cells proliferation and chemotherapy-resistant [16]. In consistence with these studies, we observed higher expression of RBX2 in colorectal cancer (CRC) relative to the matched normal tissues. Moreover, the present study identified that human CRC cells contain RBX2-expressing cancer subpopulations. Inspired by these findings, we further explored the role of RBX2 in CRC proliferation in vitro and in vivo. As expected, RBX2 interference inhibited colorectal carcinoma cells growth as well as clonogenic forming ability in vitro and tumorigenesis in vivo. Conversely, ectopic expression of RBX2 showed the opposite effects. Furthermore, RBX2 knock out increases the sensitivity of CRC HCT116 and SW480 cells target to paclitaxel treatment. These findings were consistent with the previous reports that RBX2 was a key regulator in promoting tumor cell proliferation in other tumor types.
Metastasis is one of the six initial cancer hallmarks, which is one of the main causes of CRC-associated deaths. Approximately 35% of patients with CRC have metastatic when diagnosis and more than 30% patients ultimately develop metastatic disease [17]. However, the exact molecular mechanism underlying CRC metastasis is poorly known and improving understanding the key molecules may provide novel insight for designing effective anti-CRC therapies. Since migration and invasion are the crucial events regulating CRC metastasis [18], we checked RBX2 knockout and overexpression CRC cells metastasis in vitro. Loss of RBX2 decreases the cellular migration in wound healing assay and invasion in Transwell analysis in vitro. Oppositely, RBX2 over-expressing promotes cell migration and invasion of colon cancer cells, further showing the importance of RBX2 in CRC metastasis.
Gene-annotation enrichment analysis (https://david.ncifcrf.gov/) was conducted to investigate the pathway of RBX2 in regulating and mTOR was identified as an effective mediator of RBX2 targeted. mTOR is an important upstream regulator of various signaling pathways that participates in tumor progression, including PI3K/AKT and adenosine 5’-monophosphate (AMP)-activated protein kinase (AMPK) pathway. As a protein kinase, mTOR could phosphorylate key components of the protein synthesis machinery, such as S6 kinase (S6K1). Plenty of researches show the aberrantly activated mTOR is critical for tumor cell proliferation and predicts poor prognosis in patients. Therefore, we investigated the possible role of mTOR signaling in RBX2 mediated CRC cell growth and metastasis. We found that up-regulation of RBX2 activated mTOR/S6K1 and led to increased cell proliferation. On the contrary, RBX2 knockout attenuated this pathway and resulted in suppression of CRC cells growth. To further prove these findings, we studied the effects of everolimus on vector or RBX2 over-expressed CRC cancer cells. The results suggested that treatment with everolimus dramatically abolished RBX2-induced mTOR/S6K1 activation and CRC cells proliferation as well as metastasis in vitro. These data sufficiently indicates that mTOR/S6K1 serves as downstream effecter of RBX2 in promoting colorectal carcinoma progression. Our findings were in agreement with the previous reports that mTOR pathway was an essential mediator of the RBX2-dependent cell growth in prostate cancer.
In conclusion, our in vitro and in vivo studies demonstrate that the RBX2 have profound pathogenic effects on CRC cells growth and metastasis via mTOR signaling (Figure 8), and identifies RBX2 as a potential therapeutic target for combatting colorectal carcinoma aggression.
Figure 8.
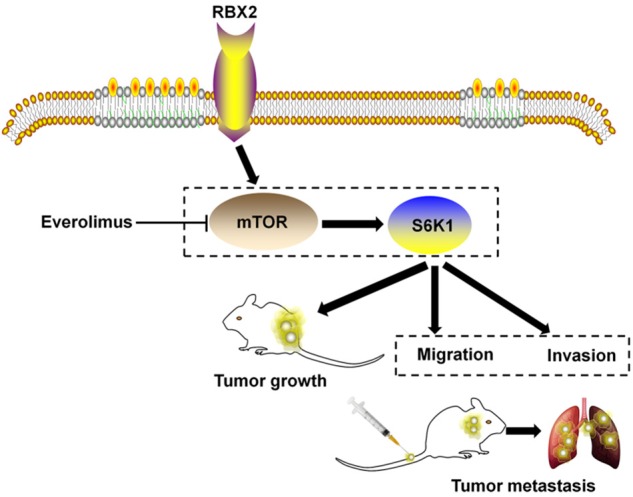
Proposed model of RBX2 promotes colon cancer metastasis via mTOR/S6K1 signaling pathway.
Acknowledgements
National science foundation of China (81373990, 81402523, 81672990 to WXY), Jiangsu Province “Six talents” high peak plan (2015-WSN-052 to WXY), Project of Jiangsu Provincial Bureau of traditional Chinese Medicine (JD201510 to WXY), the six “1” Project of Jiangsu Province (LGY2016012 to WXY).
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Zhang W, Shi X, Peng Y, Wu M, Zhang P, Xie R, Wu Y, Yan Q, Liu S, Wang J. HIF-1alpha promotes epithelial-mesenchymal transition and metastasis through direct regulation of ZEB1 in colorectal cancer. PLoS One. 2015;10:e0129603. doi: 10.1371/journal.pone.0129603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Qin Y, Tang B, Hu CJ, Xiao YF, Xie R, Yong X, Wu YY, Dong H, Yang SM. An hTERT/ZEB1 complex directly regulates E-cadherin to promote epithelial-to-mesenchymal transition (EMT) in colorectal cancer. Oncotarget. 2016;7:351–361. doi: 10.18632/oncotarget.5968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gu Q, Tan M, Sun Y. SAG/ROC2/Rbx2 is a novel activator protein-1 target that promotes c-Jun degradation and inhibits 12-O-tetradecanoylphorbol-13-acetate-induced neoplastic transformation. Cancer Res. 2007;67:3616–3625. doi: 10.1158/0008-5472.CAN-06-4020. [DOI] [PubMed] [Google Scholar]
- 4.Huang Y, Duan H, Sun Y. Elevated expression of SAG/ROC2/Rbx2/Hrt2 in human colon carcinomas: SAG does not induce neoplastic transformation, but antisense SAG transfection inhibits tumor cell growth. Mol Carcinog. 2001;30:62–70. [PubMed] [Google Scholar]
- 5.Jia L, Yang J, Hao X, Zheng M, He H, Xiong X, Xu L, Sun Y. Validation of SAG/RBX2/ROC2 E3 ubiquitin ligase as an anticancer and radiosensitizing target. Clin Cancer Res. 2010;16:814–824. doi: 10.1158/1078-0432.CCR-09-1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li H, Tan M, Jia L, Wei D, Zhao Y, Chen G, Xu J, Zhao L, Thomas D, Beer DG, Sun Y. Inactivation of SAG/RBX2 E3 ubiquitin ligase suppresses KrasG12D-driven lung tumorigenesis. J Clin Invest. 2014;124:835–846. doi: 10.1172/JCI70297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xiao Y, Jiang Y, Song H, Liang T, Li Y, Yan D, Fu Q, Li Z. RNF7 knockdown inhibits prostate cancer tumorigenesis by inactivation of ERK1/2 pathway. Sci Rep. 2017;7:43683. doi: 10.1038/srep43683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shi J, Li Q, Sheng M, Zheng M, Yu M, Zhang L. The role of TLR4 in M1 macrophage-induced epithelial-mesenchymal transition of peritoneal mesothelial cells. Cell Physiol Biochem. 2016;40:1538–1548. doi: 10.1159/000453204. [DOI] [PubMed] [Google Scholar]
- 9.Kuang J, Li L, Guo L, Su Y, Wang Y, Xu Y, Wang X, Meng S, Lei L, Xu L, Shao G. RNF8 promotes epithelial-mesenchymal transition of breast cancer cells. J Exp Clin Cancer Res. 2016;35:88. doi: 10.1186/s13046-016-0363-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu Q, Liu LZ, Qian X, Chen Q, Jiang Y, Li D, Lai L, Jiang BH. MiR-145 directly targets p70S6K1 in cancer cells to inhibit tumor growth and angiogenesis. Nucleic Acids Res. 2012;40:761–774. doi: 10.1093/nar/gkr730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang J, Zhu L, Fang J, Ge Z, Li X. LRG1 modulates epithelial-mesenchymal transition and angiogenesis in colorectal cancer via HIF-1alpha activation. J Exp Clin Cancer Res. 2016;35:29. doi: 10.1186/s13046-016-0306-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Su X, Wang J, Chen W, Li Z, Fu X, Yang A. Overexpression of TRIM14 promotes tongue squamous cell carcinoma aggressiveness by activating the NF-kappaB signaling pathway. Oncotarget. 2016;7:9939–9950. doi: 10.18632/oncotarget.6941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Skrzypczak M, Goryca K, Rubel T, Paziewska A, Mikula M, Jarosz D, Pachlewski J, Oledzki J, Ostrowski J. Modeling oncogenic signaling in colon tumors by multidirectional analyses of microarray data directed for maximization of analytical reliability. PLoS One. 2010:5. doi: 10.1371/journal.pone.0013091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun Y, Li H. Functional characterization of SAG/RBX2/ROC2/RNF7, an antioxidant protein and an E3 ubiquitin ligase. Protein Cell. 2013;4:103–116. doi: 10.1007/s13238-012-2105-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tan M, Gu Q, He H, Pamarthy D, Semenza GL, Sun Y. SAG/ROC2/RBX2 is a HIF-1 target gene that promotes HIF-1 alpha ubiquitination and degradation. Oncogene. 2008;27:1404–1411. doi: 10.1038/sj.onc.1210780. [DOI] [PubMed] [Google Scholar]
- 16.Tan M, Xu J, Siddiqui J, Feng F, Sun Y. Depletion of SAG/RBX2 E3 ubiquitin ligase suppresses prostate tumorigenesis via inactivation of the PI3K/AKT/mTOR axis. Mol Cancer. 2016;15:81. doi: 10.1186/s12943-016-0567-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xiang Y, Yao X, Chen K, Wang X, Zhou J, Gong W, Yoshimura T, Huang J, Wang R, Wu Y, Shi G, Bian X, Wang J. The G-protein coupled chemoattractant receptor FPR2 promotes malignant phenotype of human colon cancer cells. Am J Cancer Res. 2016;6:2599–2610. [PMC free article] [PubMed] [Google Scholar]
- 18.Li P, Xue WJ, Feng Y, Mao QS. MicroRNA-205 functions as a tumor suppressor in colorectal cancer by targeting cAMP responsive element binding protein 1 (CREB1) Am J Transl Res. 2015;7:2053–2059. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.