Abstract
MicroRNA-874 (miR-874), a novel cancer-associated microRNA (miRNA), has been reported to play a suppressive role in multiple malignancies. However, its function in cell migration and invasion in hepatocellular carcinoma (HCC) and the mechanisms responsible remain unclear. Here, we found miR-874 to be significantly downregulated in HCC tissues and cell lines. Moreover, this decreased expression strongly correlated with clinical stage and lymph node metastasis. Accordingly, ectopic expression of miR-874 in HCC cells markedly inhibited their migration, invasion, and epithelial-mesenchymal transition (EMT). Concerning the underlying mechanism, SRY (sex-determining region Y) -box 12 (SOX12) was identified as a direct target of miR-874, and its expression was found to be inversely correlated with that of this miRNA in HCC cells. Importantly, SOX12 knockdown had an inhibitory effect on HCC cells similar to that caused by miR-874 overexpression, whereas SOX12 overexpression in these cells negated the suppressive effects of miR-874 on migration, invasion, and EMT. Overall, these findings demonstrate that miR-874 inhibits metastasis and EMT in HCC by targeting SOX12, suggesting that this miRNA may constitute a promising therapeutic target for this disease.
Keywords: miR-874, HCC, metastasis, EMT, SOX12
Introduction
Hepatocellular carcinoma (HCC) is one of the most common malignancies and constitutes the third most frequent cause of cancer-related death [1,2]. Although many advanced therapeutic strategies, such as hepatic resection and liver transplantation, have been employed in the treatment of HCC, its prognosis remains unsatisfactory, principally due to systemic metastasis and postsurgical recurrence [3]. Therefore, elucidating the molecular mechanisms underlying HCC metastasis, which remain poorly understood, is important to identify novel therapeutic targets and improve the prognosis of this disease [3].
MicroRNAs (miRNAs) are a class of single-stranded, non-coding endogenous RNAs that inhibit gene expression at the post-transcriptional level by directly binding to the 3’-untranslated regions (3’UTRs) of messenger RNAs (mRNAs), promoting their degradation or inhibiting their translation [4]. miRNAs have been shown to be actively involved in diverse cellular processes, including proliferation, apoptosis, metastasis, differentiation, and movement [5,6]. Accumulating evidence suggests that dysregulation of miRNAs may play important roles in the pathogenesis and tumorigenicity of various human malignancies [7,8]. miRNAs have been implicated in the formation, growth, and metastasis of HCC [9] and proposed as useful diagnostic biomarkers and drug targets in this condition [10]. Thus, investigating the function of miRNAs in HCC may contribute to the identification of therapeutic targets for this malignancy.
MicroRNA-874 (miR-874), a novel cancer-associated miRNA, has been shown to be downregulated in non-small cell lung cancer [11], colorectal cancer [12,13], breast cancer [14], osteosarcoma [15], gastric cancer [16,17], and head and neck squamous cell carcinoma [18], suggesting that it plays a tumor-suppressive role in human malignancies. A recent study has revealed that expression of miR-874 is downregulated in HCC, and its overexpression inhibits HCC growth in vitro and in vivo [19]. However, the function of miR-874 in HCC cell metastasis and epithelial-mesenchymal transition (EMT) has not yet been elucidated.
In the present study, miR-874 was found to be downregulated in HCC tissues and cell lines. Moreover, restoration of miR-874 expression significantly inhibited metastasis and retarded EMT in HCC cell lines. In addition, we provide evidence that direct targeting of SOX12 by miR-874 was responsible for these effects.
Materials and methods
Tissue samples
HCC tissues (n = 48) and pair-matched noncancerous tissues were obtained from patients having undergone routine curative surgery at the First Hospital of Jilin University (Changchun, China) between July 2014 and September 2016. Specimens were collected during surgery, immediately frozen in liquid nitrogen, and stored at -80°C until use. None of the patients had received chemotherapy or radiotherapy before surgery. Written informed consent was obtained from all subjects, and this study was approved by the Ethics Committee of Jilin University.
Cell lines and cell cultures
HCC cell lines (SMMC-7721, Hep3B, HepG2, and HCCLM3) and a human immortalized normal hepatocyte line (LO2) were obtained from the Chinese Academy of Sciences (Shanghai, China). All cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Gibco, Gaithersburg, MD, USA) supplemented with 10% fetal bovine serum (FBS; HyClone, Logan, UT, USA), 100 U/ml penicillin (Sigma-Aldrich, St. Louis, MO, USA), 100 μg/ml streptomycin (Sigma-Aldrich), and 1% non-essential amino acids (Invitrogen, Carlsbad, CA, USA). Culture took place in an incubator containing a humidified atmosphere of 5% CO2 in air at 37°C.
Cell transfection
All RNA oligonucleotides were ordered from GenePharma Co., Ltd. (Shanghai, China), including an miR-874 mimic, a negative control mimic (miR-NC), siRNA against SOX12 (si-SOX12), and a scrambled negative control (si-NC). The overexpression vector pcDNA3.1-SOX12 (lacking the 3’UTR) was a kind gift from Dr. Peng Ju (Jilin University). SMMC-7721 and HepG2 cells were transiently transfected with the aforementioned oligonucleotides or the expression vector using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. Transfection efficiencies were evaluated in every experiment by qRT-PCR, performed 24 h post-transfection.
RNA isolation and qRT-PCR
Total RNA and miRNA were isolated from HCC cells with TRIzol Reagent (Invitrogen) and an miRNeasy Mini Kit (Qiagen, Hilden, Germany), respectively, according to the manufacturers’ protocols. miRNAs and mRNAs were reverse-transcribed using a TIANScript RT Kit (TiangenBiotech, Beijing, China). miR-874 and U6 snRNA levels were determined with aTaqMan MicroRNA Assay (Applied Biosystems, Carlsbad, CA, USA) using corresponding primers (Applied Biosystems). Quantification of SOX12 and GAPDH mRNA was performed using a miScript SYBR Green PCR Kit (Qiagen) and the 7900 Real-Time PCR System (Applied Biosystems), with primers that have been described previously [20]. Levels of miR-874 and SOX12 mRNA were normalized to those of U6 snRNA and GAPDH mRNA, respectively. Relative quantification was performed using the 2-∆∆Ct method.
Wound healing assay
Transfected HCC cells were seeded in 6-well plates and allowed to form a confluent monolayer. Subsequently, an artificial homogenous wound was created by scratching the monolayer with a sterile plastic micropipette tip. The wound was then photographed under a phase-contrast microscope (Olympus, Tokyo, Japan) immediately after scratching and 24 h later, and its width was measured.
Invasion assay
The invasion ability of HCC cells was evaluated using transwell chambers (BD Biosciences, Franklin Lakes, NJ, USA). Briefly, transfected cells (1×105 per well) suspended in 100 µl serum-free medium were seeded into the Matrigel-coated (BD Biosciences) upper chambers, and DMEM containing 20% FBS was added to the lower chambers. Twenty-four hours later, cells remaining on the upper surface of the membrane were removed, and those having migrated to the lower surface were fixed in 70% ethanol, stained with 0.1% crystal violet, photographed under an Axiovert 200 inverted microscope (Carl Zeiss Microscopy, Oberkochen, Germany) at 200× magnification, and counted in five randomly selected fields of view.
Luciferase reporter assay
Prediction of potential miR-874 target genes was performed using the publicly available program TargetScan7.1 (http://www.targetscan.org/). The ability of miR-874 to bind one of the possible targets identified in this manner, SOX12, was then experimentally tested with luciferase assays. The human SOX12 3’UTR, containing the putative miR-874 binding site, was subjected to PCR cloning and inserted into the pGL3 plasmid (Ambion, Austin, TX, USA), with the resulting expression vector being named WT-SOX12-3’UTR. An altered SOX12 3’UTR carrying a mutation in the miR-874 binding sequence was created using a Quik Change Site-Directed Mutagenesis Kit (Stratagene, Dallas, TX, USA) and inserted into the pGL3 vector, with the resulting construct being named MUT-SOX12-3’UTR. For the luciferase reporter assays, HCC cells were co-transfected with an oligonucleotide (miR-874 mimic or miR-NC) and one of the two reporter plasmids (WT-SOX12-3’UTR or MUT-SOX12-3’UTR) using Lipofectamine 2000. Forty-eight hours later, cells were lysed and luciferase activity was detected using a Dual-Luciferase Reporter Assay Kit (Promega, Madison, WI, USA) according to the manufacturer’s protocol. The activity of firefly luciferase was normalized to that of Renilla luciferase.
Western blotting
Total protein was extracted from cultured cells and tissues in a protein extraction buffer (Beyotime Institute of Biotechnology, Jiangsu, China) and quantified with a BCA Protein Assay Kit (Pierce Biotechnology, Rockford, IL, USA). Proteins were separated by electrophoresis on 10% SDS-polyacrylamidegels, before being transferred to polyvinylidene difluoride membranes (MerckMillipore, Billerica, MA, USA).Blots were blocked with a 5% skim milk solution and incubated overnight with an antibody against SOX12 (1:1000; Santa Cruz Biotechnology, Dallas, TX, USA), E-cadherin (1:1000; Santa Cruz Biotechnology), Twist (1:2000, Cell Signaling Technology, Danvers, MA, USA),or GAPDH (1:3000, Santa Cruz Biotechnology). Membranes were then exposed to the corresponding horseradish peroxidase-conjugated secondary antibodies for 2 h at room temperature. Protein bands were visualized using an enhanced chemiluminescence system (Thermo Fisher Scientific, Waltham, MA, USA) and X-ray film. GAPDH was used as an internal control.
Statistical analysis
All statistical analyses were performed using SPSS version 13 for Windows (SPSS Inc., Chicago, IL, USA) and GraphPad Prism5 (GraphPad Software Inc., San Diego, CA, USA). Differences among groups were evaluated for statistical significance using one-way ANOVA or two-tailed Student’s t-tests, and Spearman’s rank correlation analysis was employed to investigate correlations between pairs of factors. Data are expressed as the mean ± standard deviation (SD) from at least three independent experiments. P values <0.05 were considered statistically significant.
Results
miR-874 expression is downregulated in HCC tissues and cell lines
To examine the status of miR-874 in HCC, tissue samples from 48 cases of this disease were tested by qRT-PCR. miR-874 expression was found to be significantly lower in HCC tissues than adjacent normal tissues (P<0.01, Figure 1A). We also observed miR-874 expression to be lower in HCC tissues from patients with lymph node metastasis (P<0.01, Figure 1B) and those at an advanced tumor-node-metastasis (TNM) stage (P<0.01, Figure 1C). We next compared the miR-874 expression levels of four HCC cell lines (SMMC-7721, Hep3B, HepG2, and HCCLM3) and LO2 cells. As shown in Figure 1D, miR-874 levels in all HCC cell lines were significantly lower than those in LO2 cells (Figure 1D). These data indicate that miR-874 likely plays an suppressive role in HCC. The HCC cell lines demonstrating the lowest (SMMC-7721) and highest (HepG2) levels of miR-874 expression were used for subsequent experiments.
Figure 1.
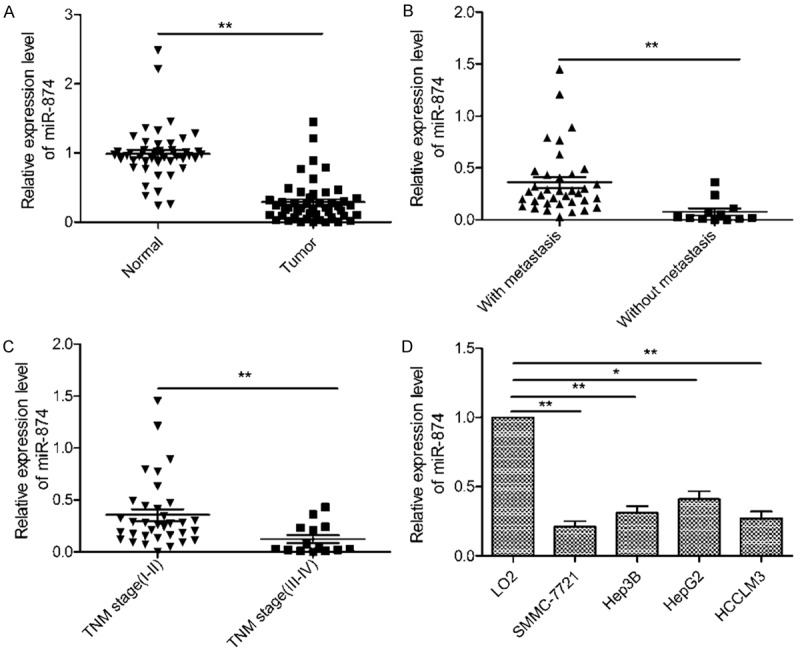
miR-874 is downregulated in human hepatocellular carcinoma (HCC) tissues and cell lines. A. Relative miR-874 levels in HCC tissues (n = 48) and matched adjacent normal tissues were determined by quantitative RT-PCR (qRT-PCR). B. qRT-PCR was used to compare miR-874 levels in HCC tissues from patients with and without lymph node metastasis. C. qRT-PCR results showing miR-874 levels in HCC tissues from patients at different tumor-node-metastasis stages. D. miR-874 expression in four HCC cell lines (SMMC-7721, Hep3B, HepG2, and HCCLM3) and a normal hepatic cell line (LO2) was analyzed using qRT-PCR. U6 was used as a loading control. *P<0.05, **P<0.01.
miR-874 suppresses HCC cell migration and invasion
As miR-874 expression was found to be lower in HCC tissues from patients with lymph node metastasis (Figure 1C), we hypothesized that this miRNA may affect metastatic activity in this disease. Therefore, to assess the effect of miR-874 on migration and invasion, we transfected SMMC-7721 and HepG2 cells with miR-874 mimic or miR-NC oligonucleotides and carried out wound healing and transwell invasion assays. qRT-PCR showed significantly increased miR-874 levels in HCC cells transfected with the miR-874 mimic, compared to those transfected with the miR-NC (Figure 2A). In addition, ectopic expression of miR-874 significantly inhibited the migration and invasion activity of both HCC cell lines (Figure 2B and 2C), indicating that miR-874 does indeed limit the metastatic potential of HCC cells.
Figure 2.
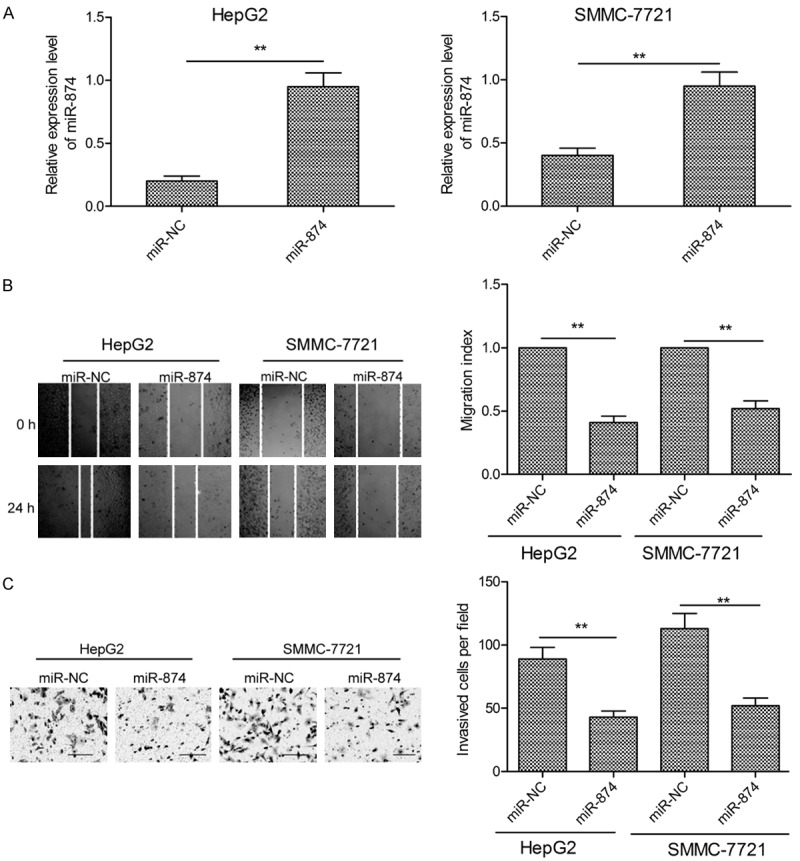
miR-874 inhibits HCC cell migration and invasion. A. Expression of miR-874 in SMMC-7721 and HepG2 cells transfected with an miR-874 mimic or miR-NC, as determined by qRT-PCR. B. Migration of SMMC-7721 and HepG2 cells was investigated using wound healing assays following transfection of an miR-874 mimic or miR-NC. C. SMMC-7721 and HepG2 cell invasion activity was determined by transwell assays following transfection of an miR-874 mimic or miR-NC. *P<0.05, **P<0.01.
SOX12 is a target of miR-874 in HCC
To understand the molecular mechanism underlying the effects of miR-874 in HCC, we searched for potential miR-874 targets using TargetScan7.1. miR-874 was predicted to be able to bind SOX12 mRNA; therefore, this gene was chosen for further study (Figure 3A). Luciferase reporter assays showed that miR-874 overexpression decreased the luciferase activity associated with the WT-SOX12-3’UTR reporter construct (Figure 3B; P<0.05), but did not affect that deriving from the MUT-SOX12-3’UTR construct (Figure 3B). To further test the association between miR-874 and SOX12, we quantified expression of endogenous SOX12 in HCC cells transfected with the miR-874 mimic or miR-NC. Transfection of the miR-874 mimic resulted in dramatically decreased expression of SOX12 at both the mRNA (Figure 3C) and protein level (Figure 3D) in both HCC cell lines.
Figure 3.
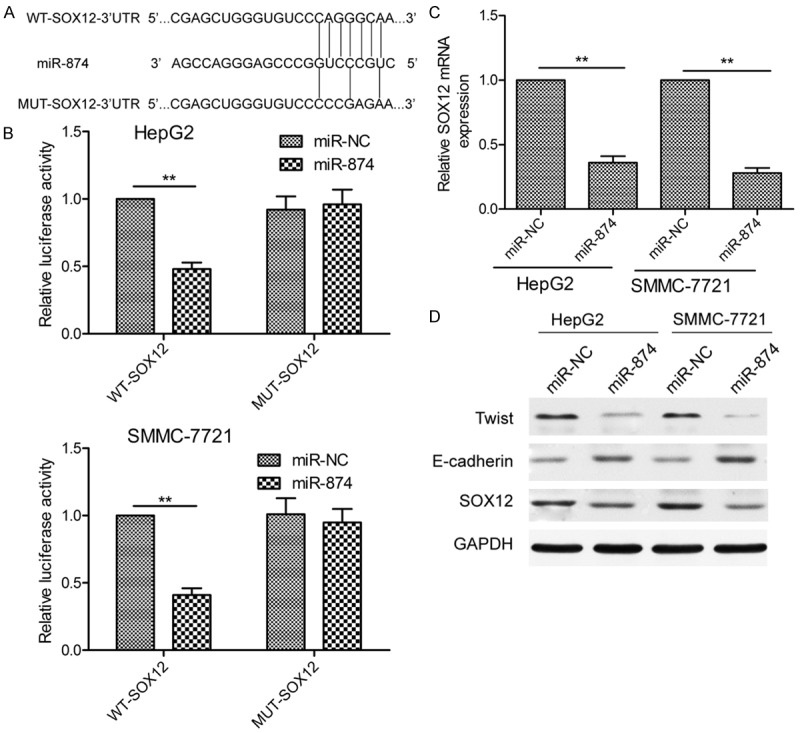
SOX12 is a direct target of miR-874 in HCC cells. A. Predicted miR-874 binding sites in the 3’-untranslated region (3’UTR) of the SOX12 mRNA sequence, and the mutations that were induced at these sites. B. Relative luciferase activity was determined using SMMC-7721 and HepG2 cells co-transfected with a wild-type or mutant SOX12 3’UTR reporter plasmid and miR-874 mimic or miR-NC RNA oligonucleotides. WT: wild-type; MUT: mutant. C. SOX12 mRNA expression in SMMC-7721 and HepG2 cells transfected with an miR-874 mimic or miR-NC, as determined by qRT-PCR. D. Levels of SOX12, E-cadherin, and Twist proteins in SMMC-7721 and HepG2 cells transfected with an miR-874 mimic or miR-NC, as determined by western blotting using GAPDH as a loading control. Data from qRT-PCR and western blot experiments were normalized to GAPDH mRNA and protein levels, respectively. *P<0.05, **P<0.01.
It has been reported that SOX12 overexpression promotes EMT by increasing the expression of Twist, which is known to induce this process [21], and decreasing that of E-cadherin [22]. Thus, we next used western blotting to examine Twist and E-cadherin expression in HCC cells transfected with the miR-874 mimic or miR-NC. Overexpression of miR-874 was found to increase expression of E-cadherin and decrease that of Twist (Figure 3D), and may therefore inhibit EMT. Taken together, these data indicate that SOX12 is indeed a target of miR-874 in HCC.
SOX12 levels are upregulated in HCC tissues and inversely correlate with miR-874 expression
Our next objective was to determine the levels of SOX12 mRNA present in the 48 HCC tumor tissue samples using qRT-PCR. SOX12 expression was significantly higher in the tumor specimens than the adjacent normal tissues (Figure 4A; P<0.01). Furthermore, two-tailed Spearman’s correlation analysis demonstrated that expression of SOX12 mRNA negatively correlated with that of miR-874 (Figure 4B; r = -0.466, P = 0.001).
Figure 4.
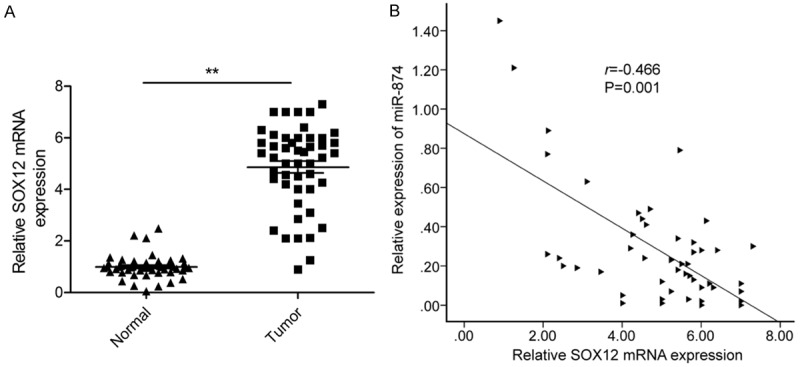
Expression of miR-874 is inversely correlated with that of SOX12 in HCC tissues. A. SOX12 mRNA expression in HCC tissues and matched normal tissues from 48 cases was determined by qRT-PCR, with GAPDH mRNA being used as an internal control. B. The negative association between SOX12 mRNA levels and miR-874 expression in HCC tissues (n = 48) was tested by Spearman’s correlation analysis. *P<0.05, **P<0.01.
SOX12 knockdown has similar effects to miR-874overexpression in HCC cells
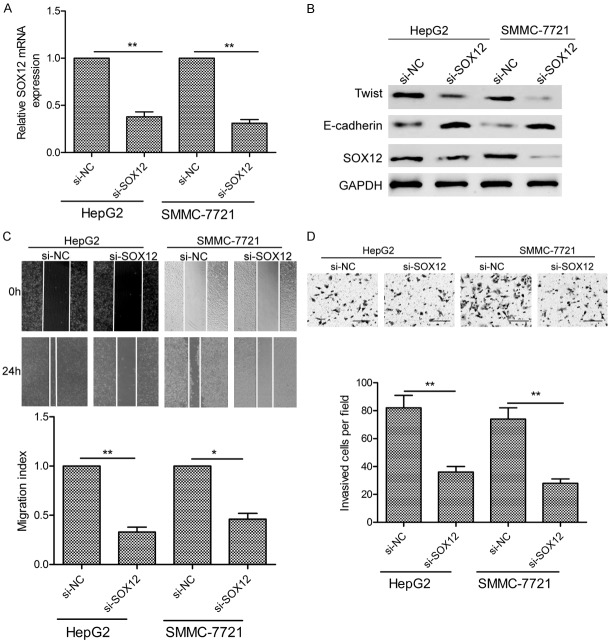
To investigate the role of SOX12 in HCC, we knocked down its expression in HCC cells using siRNA, the efficiency of which was verified by qRT-PCR and western blotting (Figure 5A and 5B). SOX12 knockdown was seen to decrease HCC cell migration and invasion in wound healing and transwell assays (Figure 5C and 5D), and respectively reduce and increase HCC cell expression of Twist and E-cadherin proteins in a western blot analysis (Figure 5B). In summary, knockdown of SOX12 replicated the effects of miR-874 overexpression in HCC cells.
Figure 5.
Downregulation of SOX12 in HCC cells replicates the effects of miR-874 overexpression. A. SOX12 mRNA levels in SMMC-7721 and HepG2 cells transfected with si-SOX12 or si-NC. GAPDH mRNA was used as an internal control. B. Western blot analysis of SOX12, E-cadherin, and Twist protein levels in SMMC-7721 and HepG2 cells transfected with si-SOX12 or si-NC. GAPDH was used as an internal control. C, D. SMMC-7721 and HepG2 cell migration and invasion were determined following transfection of si-SOX12 or si-NC. *P<0.05, **P<0.01.
miR-874 attenuates HCC cell migration, invasion, and EMT by targeting SOX12
To examine whether the inhibitory effects of miR-874 on HCC cell migration, invasion, and EMT were mediated by targeting of SOX12, we rescued SOX12 expression in HCC cells transfected with an miR-874 mimic using a SOX12 overexpression plasmid. Transfection of this vector successfully mitigated miR-874 overexpression-induced restriction of SOX12 expression at both the mRNA (Figure 6A) and protein level (Figure 6B). In addition, overexpression of SOX12 rescued the migration (Figure 6C), invasion (Figure 6D), and EMT activities (Figure 6B) of cells from suppression by miR-874 overexpression. These findings suggest that overexpression of miR-874 attenuates HCC cell metastasis and EMT by targeting SOX12, at least in part.
Figure 6.
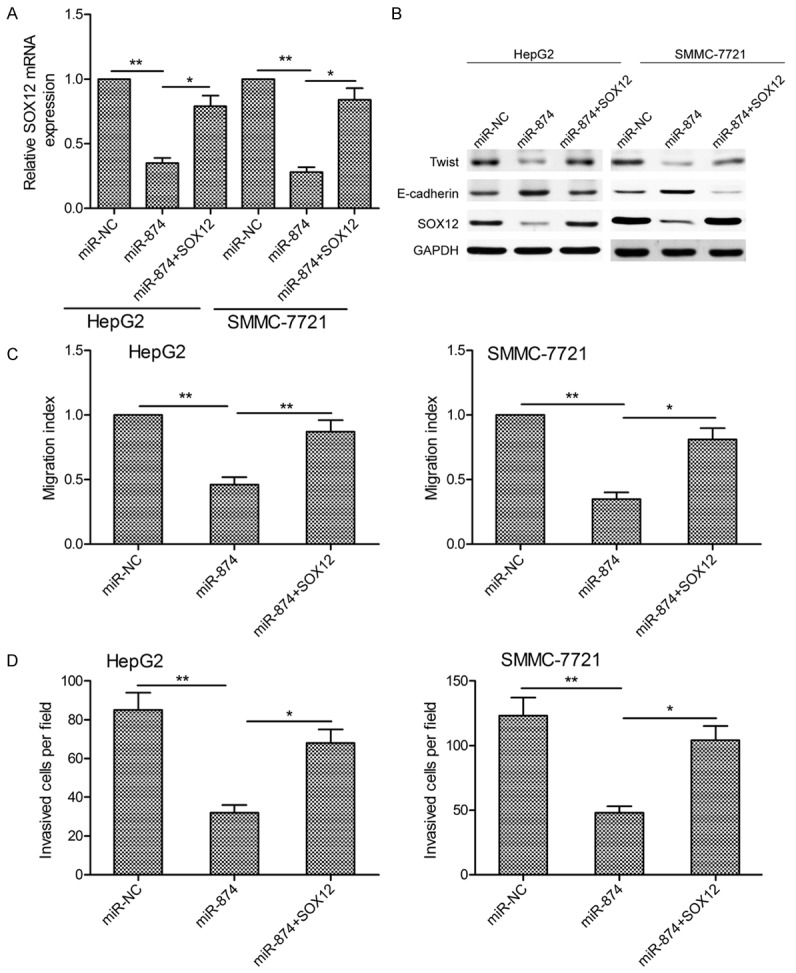
miR-874 attenuates HCC cell migration, invasion, and EMT by targeting SOX12. A. SOX12 mRNA levels in SMMC-7721 and HepG2 cells transfected with an miR-874 mimic with or without a SOX12 overexpression plasmid. GAPDH mRNA was used as an internal control. B. Western blot analysis of SOX12, E-cadherin, and Twist protein levels in SMMC-7721 and HepG2 cells transfected with an miR-874 mimic with or without a SOX12 overexpression plasmid. GAPDH was used as an internal control. C, D. SMMC-7721 and HepG2 cell migration and invasion following transfection with an miR-874 mimic with or without a SOX12 overexpression plasmid. *P<0.05, **P<0.01.
Discussion
miRNAs have been shown to be involved in the development and progression of HCC [23] and have been proposed as novel diagnostic biomarkers, effective prognostic factors, and potential therapeutic targets in this disease [24,25]. Therefore, further investigations of the functional effects of specific miRNAs in HCC and the molecular mechanisms responsible may facilitate the identification of novel markers and drug targets. In the present study, we demonstrated that miR-874 expressionin HCC tissues was significantly downregulated compared to that in adjacent non-tumor tissues, consistent with a previous study [19]. This downregulation of miR-874 was also observed in HCC cell lines. Our results, when considered together with previous data concerning the association between HCC and miR-874, strongly suggest that this miRNA plays a critical role in the progression of HCC.
Frequent intrahepatic and distant metastases are responsible for the poor overall survival of HCC patients [26]. miRNAs have been found to exert crucial effects on invasion and metastasis in various human cancers [27,28]. Previous studies have reported that miR-874can inhibit cell proliferation, migration, and invasion by targeting downstream genes [11-19]. For example, Jiang et al. revealed that ectopic miR-874 expression suppresses gastric cancer cell growth, migration, invasion, and tumorigenicity via targeting of aquaporin-3 [17]. Moreover, Dong et al. showed that this miRNA inhibits the growth and metastasis of osteosarcoma cells in part by targeting E2F transcription factor 3 [15]. Similarly, Zhang et al. demonstrated that miR-874 functions as a tumor suppressor in gastric cancer by inhibiting angiogenesis and metastasis via the STAT3/VEGF-A pathway [16]. Therefore, we attempted to establish whether miR-874 can affect the progression of HCC through its modulation of the metastatic ability of HCC cells. Our results showed that overexpression of miR-874 decreased migration and invasion by cells of HCC lines, suggesting that this miRNA can indeed suppress HCC cell metastasis.
EMT is the biological process by which cancer cells lose their epithelial polarity to assume a mesenchymal phenotype, and constitutes a crucial mechanism facilitating cancer cell metastasis [29,30]. It is characterized by decreased expression of the epithelial marker E-cadherin and increased expression of the mesenchymal markers vimentin and N-cadherin [29]. Twist proteins have been identified as important in the induction of EMT and are fundamental to the regulation of carcinoma metastasis [31]. In the present study, we found that overexpression of miR-874, by increasing expression of E-cadherin and concomitantly decreasing that of Twist, may result in inhibition of the EMT process.
To determine the route by which miR-874 exerts its anti-metastatic effect in HCC, we used bioinformatics analysis to screen for potential miR-874 target genes. One of the resulting candidates, SOX12, was selected for further investigation based on its biological functions and expression patterns. The SOX12 protein is a member of the Sox (SRY-related HMG-box) family of transcription factors and has been shown to play an essential role in embryonic development and cell fate determination [32]. Previous research has revealed SOX12 expression to be elevated in HCC tissues, with this upregulation being significantly correlated with loss of tumor encapsulation, microvascular invasion, and higher TNM stage [22]. This same study showed that overexpression of SOX12 promotes HCC invasion and metastasis by transactivating Twist1 and FGFBP1 expression [22]. Such transactivation of Twist1, along with inhibition of E-cadherin expression, by SOX12 upregulation has been shown to induce EMT [20,22]. In the current work, we confirmed SOX12 to be a functional target of miR-874 by performing luciferase reporter gene assays, qRT-PCR, and western blot analysis. Furthermore, we found that SOX12 levels were significantly higher in HCC tissues and negatively correlated with miR-874 expression. In addition, SOX12 knockdown had effects on HCC cells similar to those of miR-874 overexpression. Consistent with this, SOX12 overexpression rescued miR-874-induced inhibitory effects on HCC cell migration, invasion, and EMT. Taken together, these data indicate that miR-874 suppresses HCC cell metastasis and EMT by directly targeting SOX12.
In conclusion, the present study provided further evidence of the downregulation of miR-874 expression in HCC tissues and cell lines. Moreover, it demonstrated that miR-874 inhibits HCC cell migration, invasion, and EMT by suppressing SOX12 expression. These findings suggest that miR-874 may prove to be an attractive therapeutic target for the treatment of HCC.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (81501384).
Disclosure of conflict of interest
None.
References
- 1.Waly Raphael S, Yangde Z, Yuxiang C. Hepatocellular carcinoma: focus on different aspects of management. ISRN Oncol. 2012;2012:421673. doi: 10.5402/2012/421673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mazzola A, Costantino A, Petta S, Bartolotta TV, Raineri M, Sacco R, Brancatelli G, Camma C, Cabibbo G. Recurrence of hepatocellular carcinoma after liver transplantation: an update. Future Oncol. 2015;11:2923–2936. doi: 10.2217/fon.15.239. [DOI] [PubMed] [Google Scholar]
- 3.Forner A, Llovet JM, Bruix J. Hepatocellular carcinoma. Lancet. 2012;379:1245–1255. doi: 10.1016/S0140-6736(11)61347-0. [DOI] [PubMed] [Google Scholar]
- 4.Valinezhad Orang A, Safaralizadeh R, Kazemzadeh-Bavili M. Mechanisms of miRNAmediated gene regulation from common downregulation to mRNA-specific upregulation. Int J Genomics. 2014;2014:970607. doi: 10.1155/2014/970607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- 6.Hwang HW, Mendell JT. MicroRNAs in cell proliferation, cell death, and tumorigenesis. Br J Cancer. 2006;94:776–780. doi: 10.1038/sj.bjc.6603023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Slack FJ, Weidhaas JB. MicroRNA in cancer prognosis. N Engl J Med. 2008;359:2720–2722. doi: 10.1056/NEJMe0808667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 9.Mao B, Wang G. MicroRNAs involved with hepatocellular carcinoma (Review) Oncol Rep. 2015;34:2811–2820. doi: 10.3892/or.2015.4275. [DOI] [PubMed] [Google Scholar]
- 10.Giordano S, Columbano A. MicroRNAs: new tools for diagnosis, prognosis, and therapy in hepatocellular carcinoma? Hepatology. 2013;57:840–847. doi: 10.1002/hep.26095. [DOI] [PubMed] [Google Scholar]
- 11.Kesanakurti D, Maddirela DR, Chittivelu S, Rao JS, Chetty C. Suppression of tumor cell invasiveness and in vivo tumor growth by microRNA-874 in non-small cell lung cancer. Biochem Biophys Res Commun. 2013;434:627–633. doi: 10.1016/j.bbrc.2013.03.132. [DOI] [PubMed] [Google Scholar]
- 12.Zhao B, Dong AS. MiR-874 inhibits cell growth and induces apoptosis by targeting STAT3 in human colorectal cancer cells. Eur Rev Med Pharmacol Sci. 2016;20:269–277. [PubMed] [Google Scholar]
- 13.Han J, Liu Z, Wang N, Pan W. MicroRNA-874 inhibits growth, induces apoptosis and reverses chemoresistance in colorectal cancer by targeting X-linked inhibitor of apoptosis protein. Oncol Rep. 2016;36:542–550. doi: 10.3892/or.2016.4810. [DOI] [PubMed] [Google Scholar]
- 14.Wang L, Gao W, Hu F, Xu Z, Wang F. MicroRNA-874 inhibits cell proliferation and induces apoptosis in human breast cancer by targeting CDK9. FEBS Lett. 2014;588:4527–4535. doi: 10.1016/j.febslet.2014.09.035. [DOI] [PubMed] [Google Scholar]
- 15.Dong D, Gong Y, Zhang D, Bao H, Gu G. miR-874 suppresses the proliferation and metastasis of osteosarcoma by targeting E2F3. Tumour Biol. 2016;37:6447–6455. doi: 10.1007/s13277-015-4527-3. [DOI] [PubMed] [Google Scholar]
- 16.Zhang X, Tang J, Zhi X, Xie K, Wang W, Li Z, Zhu Y, Yang L, Xu H, Xu Z. miR-874 functions as a tumor suppressor by inhibiting angiogenesis through STAT3/VEGF-A pathway in gastric cancer. Oncotarget. 2015;6:1605–1617. doi: 10.18632/oncotarget.2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang B, Li Z, Zhang W, Wang H, Zhi X, Feng J, Chen Z, Zhu Y, Yang L, Xu H, Xu Z. miR-874 Inhibits cell proliferation, migration and invasion through targeting aquaporin-3 in gastric cancer. J Gastroenterol. 2014;49:1011–1025. doi: 10.1007/s00535-013-0851-9. [DOI] [PubMed] [Google Scholar]
- 18.Nohata N, Hanazawa T, Kinoshita T, Inamine A, Kikkawa N, Itesako T, Yoshino H, Enokida H, Nakagawa M, Okamoto Y, Seki N. Tumoursuppressive microRNA-874 contributes to cell proliferation through targeting of histone deacetylase 1 in head and neck squamous cell carcinoma. Br J Cancer. 2013;108:1648–1658. doi: 10.1038/bjc.2013.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leong KW, Cheng CW, Wong CM, Ng IO, Kwong YL, Tse E. miR-874-3p is down-regulated in hepatocellular carcinoma and negatively regulates PIN1 expression. Oncotarget. 2017;8:11343–11355. doi: 10.18632/oncotarget.14526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ding H, Quan H, Yan W, Han J. Silencing of SOX12 by shRNA suppresses migration, invasion and proliferation of breast cancer cells. Biosci Rep. 2016 doi: 10.1042/BSR20160053. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21.Yang J, Mani SA, Donaher JL, Ramaswamy S, Itzykson RA, Come C, Savagner P, Gitelman I, Richardson A, Weinberg RA. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117:927–939. doi: 10.1016/j.cell.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 22.Huang W, Chen Z, Shang X, Tian D, Wang D, Wu K, Fan D, Xia L. Sox12, a direct target of FoxQ1, promotes hepatocellular carcinoma metastasis through up-regulating Twist1 and FGFBP1. Hepatology. 2015;61:1920–1933. doi: 10.1002/hep.27756. [DOI] [PubMed] [Google Scholar]
- 23.Budhu A, Jia HL, Forgues M, Liu CG, Goldstein D, Lam A, Zanetti KA, Ye QH, Qin LX, Croce CM, Tang ZY, Wang XW. Identification of metastasis-related microRNAs in hepatocellular carcinoma. Hepatology. 2008;47:897–907. doi: 10.1002/hep.22160. [DOI] [PubMed] [Google Scholar]
- 24.Cho WC. MicroRNAs: potential biomarkers for cancer diagnosis, prognosis and targets for therapy. Int J Biochem Cell Biol. 2010;42:1273–1281. doi: 10.1016/j.biocel.2009.12.014. [DOI] [PubMed] [Google Scholar]
- 25.Giordano S, Columbano A. MicroRNAs: new tools for diagnosis, prognosis, and therapy in hepatocellular carcinoma? Hepatology. 2013;57:840–847. doi: 10.1002/hep.26095. [DOI] [PubMed] [Google Scholar]
- 26.Hussain K, El-Serag HB. Epidemiology, screening, diagnosis and treatment of hepatocellular carcinoma. Minerva Gastroenterol Dietol. 2009;55:123–138. [PubMed] [Google Scholar]
- 27.Huang S, He X. The role of microRNAs in liver cancer progression. Br J Cancer. 2011;104:235–240. doi: 10.1038/sj.bjc.6606010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dykxhoorn DM. MicroRNAs and metastasis: little RNAs go a long way. Cancer Res. 2010;70:6401–6406. doi: 10.1158/0008-5472.CAN-10-1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thiery JP. Epithelial-mesenchymal transitions in tumour progression. Nat Rev Cancer. 2002;2:442–454. doi: 10.1038/nrc822. [DOI] [PubMed] [Google Scholar]
- 30.Yang J, Weinberg RA. Epithelial-mesenchymal transition: at the crossroads of development and tumor metastasis. Dev Cell. 2008;14:818–829. doi: 10.1016/j.devcel.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 31.Rosivatz E, Becker I, Specht K, Fricke E, Luber B, Busch R, Hofler H, Becker KF. Differential expression of the epithelial-mesenchymal transition regulators snail, SIP1, and twist in gastric cancer. Am J Pathol. 2002;161:1881–1891. doi: 10.1016/S0002-9440(10)64464-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hoser M, Potzner MR, Koch JM, Bosl MR, Wegner M, Sock E. Sox12 deletion in the mouse reveals nonreciprocal redundancy with the related Sox4 and Sox11 transcription factors. Mol Cell Biol. 2008;28:4675–4687. doi: 10.1128/MCB.00338-08. [DOI] [PMC free article] [PubMed] [Google Scholar]