Abstract
Chondrocytes located in hyaline cartilage may maintain phenotype while the chondrocytes situated in calcified cartilage differentiate into hypertrophy. Chondrogenic and hypertrophic differentiation of mesenchymal stem cells (MSCs) are two subsequent processes during endochondral ossification. However, it is necessary for chondrocytes to hold homeostasis and to inhibit hypertrophic differentiation in stem cell-based regenerated cartilage. Dihydroartemisinin (DHA) is derived from artemisia apiacea which has many biological functions such as anti-malarial and anti-tumor. Whereas the effects of DHA on chondrogenic and hypertrophic differentiation are poorly understand. In this study, the cytotoxicity of DHA was determined by CCK8 assay and the cell apoptosis was analyzed by flow cytometry. Additionally, the effects of DHA on chondrogenic and hypertrophic differentiation of MSCs are explored by RT-PCR, western blotting and immunohistochemistry. The results showed that DHA inhibited expression of chondrogenic markers including Sox9 and Col2a1 by activating Nrf2 and Notch signaling. After induced to chondrogenesis, cells were treated with hypertrophic induced medium with DHA. The results revealed that hypertrophic markers including Runx2 and Col10a1 were down-regulated following DHA treatment through Pax6/HOXA2 and Gli transcription factors. These findings indicate that DHA is negative to chondrogenesis and is protective against chondrocyte hypertrophy to improve chondrocytes stability. Therefore, DHA might be not suited for chondogenesis but be potential as a new therapeutic candidate to maintain the biological function of regenerated cartilage.
Keywords: Dihydroartemisinin, chondrogenesis, chondrocyte hypertrophy, mesenchymal stem cell
Introduction
Endochondral ossification and intramembranous bone formation are the two main processes to form osseous tissues during embryogenesis [1]. Endochondral ossification, which requires a cartilage intermediate, is mainly responsible for the formation of the mammalian skeleton [2]. Endochondral bone development begins with the condensation of mesenchymal stem cells (MSCs) [3]. Next, MSCs located in the center of the condensed cells differentiate into chondrocytes, which secrete a cartilage matrix and special proteoglycans such as aggrecan [4]. This process, called chondrogenesis, is regulated by TGF-β signaling, in which Sox9 is a key transcription factor [5]. Chondrocytes in the core of the cartilage model undergo further maturation through hypertrophy [6]. The hypertrophic chondrocytes express Col10a1 and secrete matrix metalloproteinase 13 (MMP13) to degrade cartilage matrix, thus creating conditions for the entry of osteoblasts and osteoclasts [7]. Runx2, a transcription factor, plays a crucial regulatory role in chondrocyte hypertrophic differentiation [8,9]. Triiodothyronine (T3) is sufficient to support chondrocyte differentiation to hypertrophy in vitro [10]. Chondrocytes situated on the surface of the metaphysis may maintain their phenotype and not differentiate into hypertrophic cells, resulting in the formation of a thin layer of hyaline cartilageon articular surfaces that reduces the friction between adjacent bones [11].
MSCs possess multi-directional differentiation potential including osteogenesis, adipogenesis, and chondrogenesis [12-14]. Because of their extensive sources, low immunological rejection, and strong proliferation capacity, MSCs are considered promising seed cells for use in tissue engineering [15]. Studies have applied MSCs to rebuilding the damaged heart [16] and repairing bone defects [17]. Certainly, MSC-based engineered cartilage can be constructed for biologic joint replacement [18]. However, after the chondrogenesis process, chondrocytes derived from MSCs continue hypertrophic differentiation, and thus the regenerated cartilage is fibrocartilage rather than hyaline cartilage [19,20]. Therefore, prevention of chondrocyte hypertrophy is becoming increasingly crucial for the clinical application of MSCs in cartilage tissue engineering.
Dihydroartemisinin (DHA) is the main component of sweet wormwood, an herb employed in Chinese traditional medicine [21]. DHA is the most effective drug against malaria [22] and has other functions such as anticancer, antifungal, and immunomodulatory activities [23-25]. A recent study has confirmed that artemisinin confers neuroprotection against SNP-induced neuronal cell death by activating ERK phosphorylation [26]. Our group also found that DHA is protective against LPS-induced bone loss, which occurs through induction of osteoclast apoptosis [27]. However, the role of DHA in regulating chondrogenic and hypertrophic differentiation of MSCs remains unknown.
Therefore, we established cell models in vitro to simulate chondrogenic and hypertrophic differentiation of MSCs [28]. The experimental design is shown in Figure 1. Several crucial signaling pathway genes and transcription factors were also examined to explore the molecular mechanisms involved in the effect of DHA on chondrogenic and hypertrophic differentiation.
Figure 1.
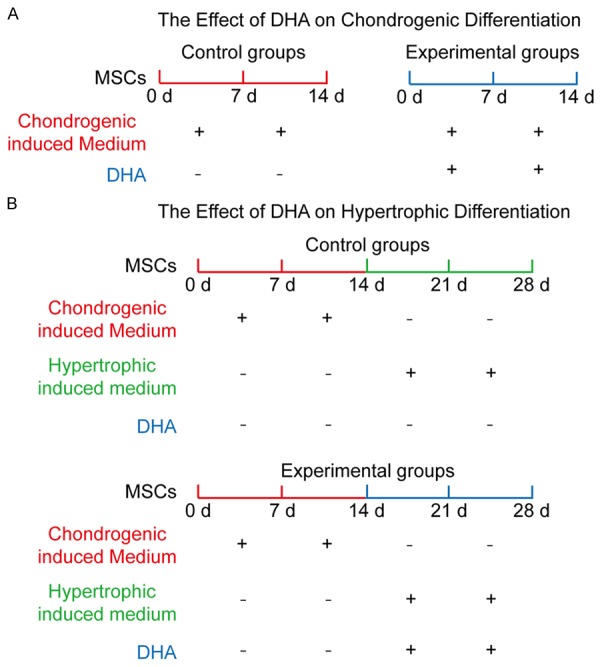
The experimental design of DHA effects on chondrogenic and hypertrophic differentiation of MSCs. A: To explore the effect of DHA on chondrogenic differentiation, MSCs were treated with chondrogenic induced medium for 7 and 14 d with DHA (experimental groups) or without DHA (control groups). B: To explore the effect of DHA on hypertrophic differentiation, MSCs were treated with chondrogenic induced medium for 14 d without DHA, and thenwere incubated by hypertrophic induced medium for another 7 and 14 d with DHA (experimental groups) or without DHA (control groups).
Materials and methods
Reagents and cell culture
DHA was purchased from Sigma Corporation of America (catalog D7439) which was dissolved in DMSO. Besides, the experimental concentration of DMSO is 0.1%. C3H10T1/2 mesenchymal stem cells were purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA). The cells were cultured in DMEM-F12 (Hyclone, Logan, Utah, USA) containing 10% fetal bovine serum (FBS) (Hyclone) and 1% Penicillin-streptomycin solution (Beyotime, Shanghai, China) at 37°C with 5% CO2. The culture medium was replaced every 2-3 d until the cells reached 90% confluence. Cells were passaged by 0.25% trypsin (Hyclone) for 2 min at room temperature. The second generation cells were used for the following experiments.
In vitro cell proliferation assay
C3H10T1/2 cells were seeded (2 × 103 per well) into 96-well plates (Corning, NY, USA) in triplicates and allowed to adhere overnight. On the following day, the medium was replaced by fresh medium with DHA. Cultures were incubated for 24, 48 and 72 hours. Then, 10 μL Cell Counting Kit-8 (CCK8, Dojindo Laboratories, Japan) reagents were added to each well and incubated for an additional 4 hours. The absorbance was read at the wavelength of 450 nm in an automated plate reader. Wells containing the CCK8 reagents without cells were used as the blank control. Cell proliferation was assessed by the absorbance values according to the manufacturer’s protocol.
Apoptosis assay
DHA induced apoptosis of MSCs was detected by flow cytometry using Annexin V-FITC Apoptosis Detection Kit (KGA, KeyGEN, Biotech) according to the manufacturer’s instructions. The apoptosis rate was assayed by using FACSCalibur Flow Cytometry (Becton Dickinson and Beckman-Coulter, San Jose, CA, USA) at 488 nm.
Chondrogenic differentiation assay
C3H10T1/2 cells were trypsinized by 0.25% trypsin and modulated at a density of 105 cells/ml. 2 ml of the suspension was placed into the center of each well on a 6-well plate (Corning). After incubation for 24 h at 37°C and 5% CO2, the medium was replaced by 2 ml chondrogenic differentiation medium (Hyclone). The chondrogenic differentiation medium composed of dexamethasone (100 nmol/L), ascorbate (50 μg/ml), ITS Supplement, proline (40 μg/ml) and TGF-β3 (10 ng/ml) was replaced every 3 d [28].
Hypertrophic differentiation assay
C3H10T1/2 cells were treated by chondrogenic differentiation medium as previously described for 14 d. Then the cells were inducted by hypertrophic differentiation medium composed of dexamethasone (1 nmol/L), ascorbate (50 μg/ml), ITS Supplement, proline (40 μg/ml) and triiodothyronine (T3, 100 ng/ml) for another 14 d. Each medium was replaced every 3 d [28].
Quantitative RT-PCR analysis
Total RNA was isolated using Trizol reagent (Life Technologies, NY, USA). Single-stranded cDNA was prepared from 1 μg of total RNA using reverse transcriptase with oligo-dT primer according the manufacturer’s instructions (Takara, Liaoning, Dalian, China). Two microlitres of each cDNA was subjected to PCR amplification using specific primers with detailed information in Table 1. The cycling conditions were set as 95°C for 30 seconds, 40 cycles of 95°C for 5 seconds, and 60°C for 30 seconds. The relative mRNA level was calculated by the normalization to that of GAPDH.
Table 1.
Primer sequences for qPCR
| Genes | Forward (5’-3’) | Reverse (5’-3’) | Tm (°C) |
|---|---|---|---|
| Gli2 | GTCACCAAGAAACAGCGTAA | GATGGCTTTGATATGTAGGC | 60 |
| Gli3 | ACAGCTCTACGGCGACTG | GCATAGTGATTGCGTTTC | 63 |
| Col2a1 | TGGTGGAGCAGCAAGAGC | TGGACAGTAGACGGAGGAAA | 61 |
| Sox9 | CCCAGCGAACGCACATCA | TGGTCAGCGTAGTCGTATT | 61 |
| HOXA2 | CCTGGAGGACTCGGACAA | AAAGGCGAAACTGGGAAA | 62 |
| Pax6 | AACAGACACCGCCCTCAC | CATAACTCCGCCCATTCA | 62 |
| Nrf2 | CAGTGCTCCTATGCGTGAA | GCGGCTTGAATGTTTGTC | 61 |
| GAPDH | GTTGTCTCCTGCGACTTCA | GGTGGTCCAGGGTTTCTTA | 62 |
| Hey1 | GAATGCCTGGCCGAAGTT | CCGCTGGGATGCGTAGTT | 62 |
| Col10a1 | CTTTCTGGGATGCCGCTTGT | GGGTCGTAATGCTGCTGCCTA | 61 |
| Runx2 | CCAACTTCCTGTGCTCCGTG | ATAACAGCGGAGGCATTTCG | 63 |
| COMP | TGGTGCTCAATCAGGGAA | TCAGTGGCGGTGTTTACAT | 61 |
Gli2, GLI family zinc finger 2; Gli3, GLI family zinc finger 3; Col2a1, collagen type II alpha 1 chain; Sox9, SRY (sex determining region Y)-box 9; HOXA2, homeobox A2; Pax6, paired box 6; Nrf2, Nf-E2 related factor 2; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; Hey1, hairy/enhancer-of-split related with YRPW motif 1; Col10a1, collagen, type X, alpha 1; Runx2, runt related transcription factor 2; COMP, cartilage oligomeric matrix protein.
Western blot analysis
The cells were extracted with lysis buffer containing 50 mM Tris (pH 7.6), 150 mM NaCl, 1% TritonX-100, 1% deoxycholate, 0.1% SDS, 1 mM PMSF and 0.2% Aprotinin (Beyotime). After we measured the protein concentration, the equal protein samples were mixed with 5 × sample buffer (Beyotime) and boiled. The samples were resolved by 10% SDS-PAGE gel and transferred on PVDF membrane (Millipore, Hong Kong, China) by using the semi-dry transfer method. After blocking in 10% nonfat dried milk in TBST for 2 h, the blots were incubated with primary antibodies including Col2a1 (rabbit polyclonal 1:500, Abcam), Sox9 (rabbit polyclonal 1:500, Abcam), Runx2 (rabbit polyclonal 1:500, Abcam), Col10a1 (rabbit polyclonal 1:500, Abcam), Gli2 (rabbit polyclonal 1:500, Bioss), Gli3 (rabbit polyclonal 1:500, Bioss), NICD (rabbit polyclonal 1:500, Bioss), Hey1 (rabbit polyclonal 1:500, Bioss), Nrf2 (rabbit polyclonal 1:500, Bioss), HOXA2 (rabbit polyclonal 1:500, Bioss), Pax6 (rabbit polyclonal 1:500, Bioss) and GAPDH (rabbit polyclonal 1:1000, Abcam) at 4°C overnight. After washing by TBST, the blots were incubated with a horseradish peroxidase-conjugated secondary antibody (diluted 1:2000, Santa Cruz) at room temperature for 1 h. Blots against GAPDH served as loading control.
Glycosaminoglycan (GAG) synthesis analysis by alcian blue staining
To demonstrate the deposition of cartilage matrix proteoglycans, representative cultures were collected at day 14, Then sulfated cartilage glycosaminoglycans (GAGs) were measured by alcian blue (Sciencell) staining. The cells were fixed in 4% Paraformaldehyde for 15 min and incubated in 3% acetic aicd for 3 min. Then the cells were stained with alcian blue for 30 min. The mean density was normalized to total cell number.
Immunohistochemistry
The cells were fixed in 4% Paraformaldehyde for 15 min. This was followed by washing in phosphate-buffered saline (PBS) and treated with H2O2 (ZSGB-BIO, Peking, China) for 10 min to inactivate endogenous peroxidase. After treatment with normal goat serum (ZSGB-BIO) at room temperature for 15 min, cells were incubated with primary antibodies including Runx2 (rabbit polyclonal 1:150, Abcam) and Col10a1 (rabbit polyclonal 1:150, Abcam) at 4°C overnight. After washing, the cells were incubated with biotinylated goat anti-rabbit (ZSGB-BIO) secondary antibodies for 30 min, followed by washing and incubation with horse radish peroxidase (HRP) (ZSGB-BIO) for 15 min. The area of the immunocomplex was visualized by chromogen 3, 3’-diaminobenzidine (DAB) for 5 min. The cells were investigated under the Olympus microscope. Image-Pro plus 6.0 software was used to analyze the IOD and area to calculate mean density of images.
Statistics
All data are representative of at least three experiments of similar results performed in triplicate unless otherwise indicated. Data are expressed as mean ± SEM. One-way ANOVA followed by Student-Newman-Keuls post hoc tests was used to determine the significance of difference between results, with P<0.05 being regarded as significant.
Results
DHA toxicity evaluation and promotive effect on apoptosis of MSCs
For toxicity evaluation, we performed CCK-8 assay to assess the effects of DHA on cell viability. MSCs were treated with basal culture medium for 24 h, 48 h and 72 h with different concentrations of DHA. The results showed that DHA above 100 μM significantly inhibited cell proliferation (P<0.05) (Figure 2B). After culturing MSCs with basic medium for 72 h, we also investigated effect of DHA on cell apoptosis by FCM (Figure 2C). In the plots, X axis represented Annexin V-FITC and Y axis represented Propidium iodide. The results revealed that DHA concentration above 100 μM affected cell apoptosis (P<0.05) (Figure 2D). According to the results, DHA concentration at 100 μM was chosen for further experiments.
Figure 2.
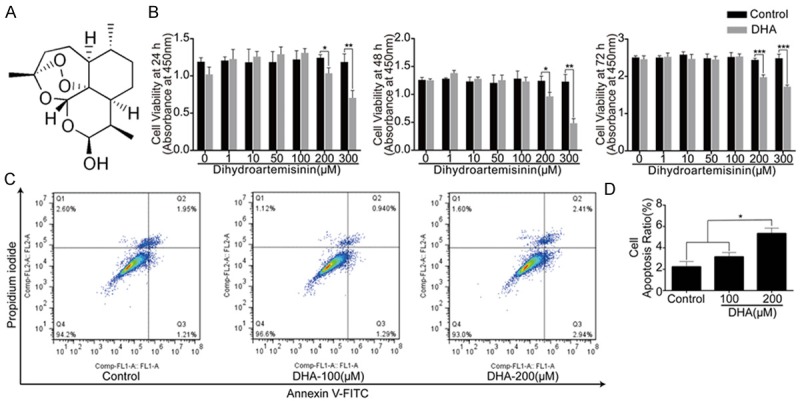
High concentrations of DHA inhibited MSCs proliferation and induced cell apoptosis of MSCs. DHA concentration of 100 μM was regarded safe and used for following experiments. A: Chemical formula of DHA. B: CCK8 analysis of cell viability of MSCs treated with different concentrations of DHA for 24 h, 48 h and 72 h. C: FCM analysis of cell apoptosis rate of MSCs treated with basic medium for 72 h with different dosages of DHA treatments. X axis represents Annexin V-FITC and Y axis represents Propidium iodide. D: Quantification analysis of cell apoptosis rate. The data in the figures represent the averages ± SD. Significant differences between the treatment and control groups are indicated in the graph with the detailed corresponding P-values from Student-Newman-Keuls post hoc tests, *(P<0.05) or **(P<0.01) or ***(P<0.001).
DHA suppressed synthesis of glycosaminoglycans and expression of chondrogenic marker genes
MSCs were incubated by chondrogenic medium containing TGF-β3 for 14 d with or without DHA, alcian blue stain was then performed to examine the effects of DHA on chondrogenesis of MSCs (Figure 3A). The results revealed that intensity of images were decreased following DHA treatment on day 14 (P<0.05) (Figure 3B). DHA had an inhibitory effect in a dose-dependent manner. On the mRNA level, the results of RT-PCR showed that chondrogenic specific markers Sox9, COMP and Col2a1 were significantly suppressed on day 3, 7 and 14 by DHA (100 Mm) treatment (P<0.01) (Figure 3C). Besides, we performed western blotting and the results showed expression of Col2a1 and Sox9 were decreased on day 7 and 14 following DHA (100 Mm) treatment (P<0.05) (Figure 3D, 3E). Our data demonstrated that DHA had inhibitory effects on chondrogenesis of MSCs at the concentration of 100 μM.
Figure 3.
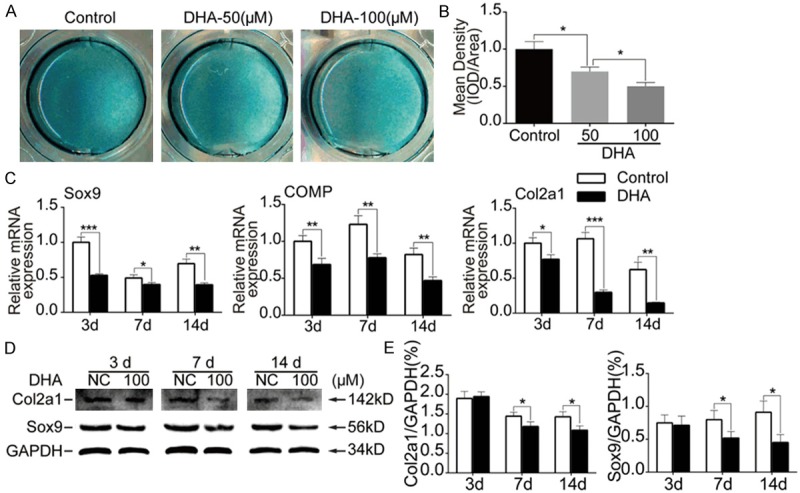
DHA inhibited chondrogenesis of MSCs. MSCs were induced by chondrogenic medium containing TGF-β3 for 3 d, 7 d and 14 d together with or without DHA. A: Representative alcian blue staining images of MSCs in different groups (day 14). B: Quantification of mean intensity of alcian blue staining. C: Relative mRNA expression levels of Sox9, COMP and Col2a1 from MSCs in different groups (day 3, 7 and 14). D: Representative western blot images of Col2a1, Sox9 and GAPDH from MSCs in different groups (day 3, 7 and 14). E: Quantification of normalized expression intensity of Col2a1 and Sox9 against GAPDH. The data in the figures represent the averages ± SD. Significant differences between the treatment and control groups are indicated in the graph with the detailed corresponding P-values from Student-Newman-Keuls post hoc tests, *(P<0.05) or **(P<0.01) or ***(P<0.001).
DHA up-regulated Nrf2 and Notch signaling during MSCs chondrogenesis
To explain the inhibitory eff-ects of DHA on chondrogenesis of MSCs, The transcriptional factors and key molecules in canonical pathways related with chondrogenesis were screened through qPCR. From the results, expressions of Nrf2 and Hey1 were decreased by the treatment of DHA (100 Mm) on day 3 but increased on day 7 and day 14 (P<0.05) (Figure 4A). Moreover we performed western blotting and the results revealed that DHA significantly promoted expressions of Nrf2, NICD and its downstream target Hey1 on day 7 and day 14 (P<0.05) (Figure 4B, 4C). These data demonstrated that DHA inhibited chondrogenesis of MSCs through up-regulating Nrf2 and Notch signaling.
Figure 4.
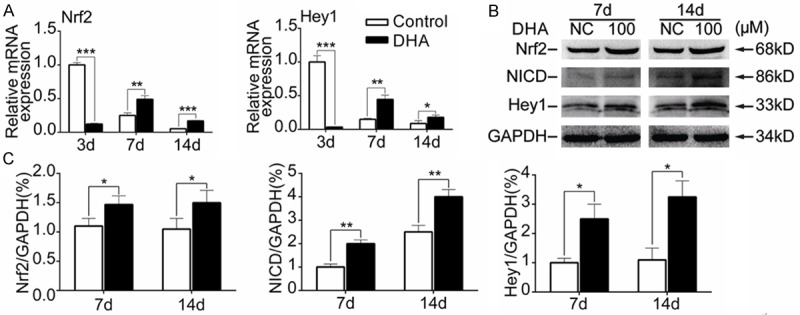
DHA activated Nrf2 and Notch signaling during MSCs chondrogenesis. MSCs were induced by chondrogenic medium containing TGF-β3 for 3 d, 7 d and 14 d together with or without DHA. A: Relative mRNA expression levels of Nrf2 and Hey1 from MSCs in different groups (day 3, 7 and 14). B: Representative western blot images of Nrf2, NICD, Hey1 and GAPDH from MSCs in different groups (day 7 and 14). C: Quantification of normalized expression intensity of Nrf2, NICD and Hey1 against GAPDH. The data in the figures represent the averages ± SD. Significant differences between the treatment and control groups are indicated in the graph with the detailed corresponding P-values from Student-Newman-Keuls post hoc tests, *(P<0.05) or **(P<0.01) or ***(P<0.001).
DHA inhibited hypertrophic differentiation of chondrocytes derived from MSCs
MSCs were incubated by chondrogenic medium containing TGF-β3 for 14 d without DHA and then were incubated by hypertrophic medium containing T3 for another 14 d with or without DHA. Through immunohistochemistry, upon DHA (100 Mm) treatment, the expressions of hypertropyic markers including Runx2 and Col10a1 were suppressed on day 21 and day 28 (P<0.05) (Figure 5A, 5B). Besides, from the results of RT-PCR, we found that the expression of Runx2 and Col10a1 were decreased on day 17, day 21 and 28 compared with control groups (P<0.05) (Figure 5C). The results of western blotting were consistent with RT-PCR. After DHA (100 Mm) treatment, Runx2 and Col10a1 expressions were decreased on day 17, day 21 and day 28 (P<0.05) (Figure 5D, 5E). Thus, these data demonstrated DHA inhibited chondrocyte hypertrophic differentiation.
Figure 5.
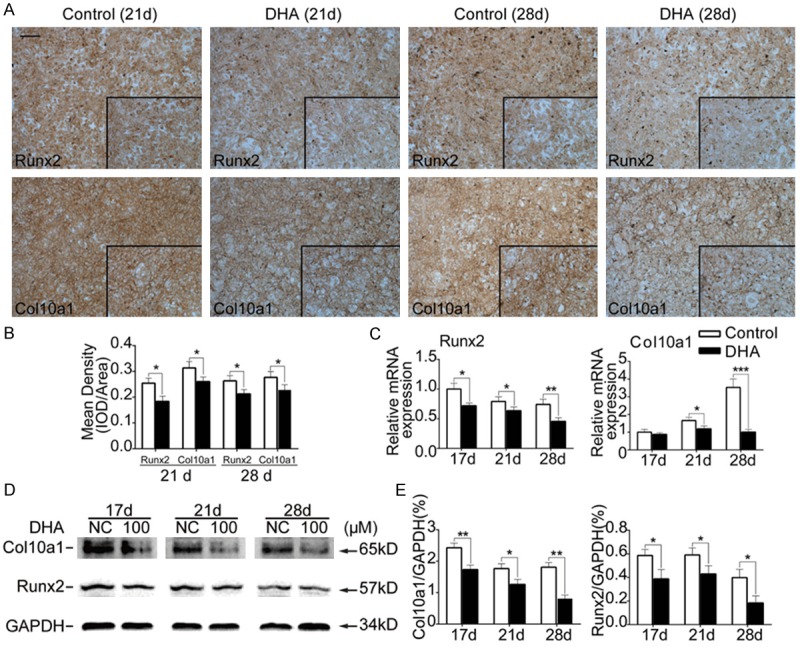
DHA inhibited expression of chondrocyte hypertrophic genes. MSCs were induced by chondrogenic medium without DHA for14 d and then treated with hypertrophic medium together with or without DHA for another 7 d and 14 d. (A) Representative immunohistochemical images of Runx2 and Col10a1 from MSCs in different groups (day 21 and 28). Scale bar represents 200 μm. (B) Quantification of mean intensity of Runx2 and Col10a1 in (A). (C) Relative mRNA expression levels of Runx2 and Col10a1 from MSCs in different groups (day 17, 21 and 28). (D) Representative western blot images of Col10a1, Runx2 and GAPDH from MSCs in different groups (day 17, 21 and 28). (E) Quantification of normalized expression intensity of Col10a1 and Runx2 against GAPDH. The data in the figures represent the averages ± SD. Significant differences between the treatment and control groups are indicated in the graph with the detailed corresponding P-values from Student-Newman-Keuls post hoc tests, *(P<0.05) or **(P<0.01) or ***(P<0.001).
DHA down-regulated Pax6/HOXA2 and Gli transcription factors in IHH signaling during hypertrophic differentiation of MSCs
We also explored the mechanism through RT-PCR to find the expressions of altered transcriptional factors and key molecules in canonical pathways related with chondrocyte hypertrophy. The results showed DHA affected the expression of IHH signaling. More specifically, DHA (100 Mm) decreased Gli2 expression and increased Gli3 expression on day 21 and day 28. Besides, following DHA treatment, expression of Pax6 was inhibited while HOXA2 expression was promoted on day 21 and day 28 (P<0.05) (Figure 6). Next we performed western blotting to verify the above results. On protein level, DHA had inhibitory roles on the expression of Gli2 and Pax6 and had promotive roles on the expression of Gli3 and HOXA2 on day 21 and day 28 (P<0.05) (Figure 7). All these results demonstrated DHA inhibited chondrocyte hypertrophy of MSCs through regulating Pax6/HOXA2 and IHH signaling.
Figure 6.
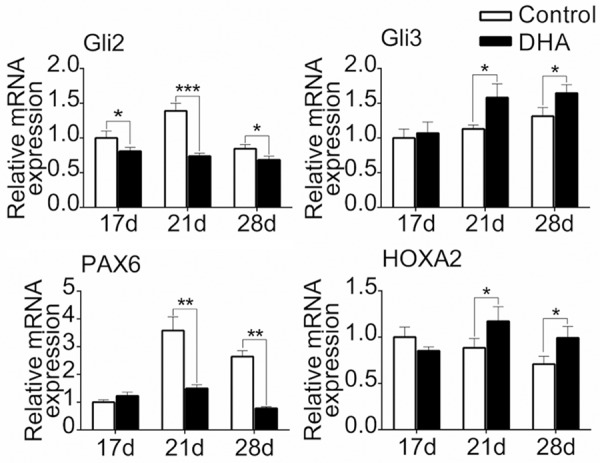
DHA down-regulated Pax6/HOXA2 and IHH signaling on mRNA level during hypertrophic differentiation of MSCs. MSCs were induced by chondrogenic medium without DHA for14 d and then treated with hypertrophic medium together with or without DHA for another 3 d, 7 d and 14 d. The mRNA expressions of Gli2, Gli3, Pax6 and HOXA2 from MSCs in different groups (day 17, 21 and 28) were detected by RT-PCR. The data in the figures represent the averages ± SD. Significant differences between the treatment and control groups are indicated in the graph with the detailed corresponding P-values from Student-Newman-Keuls post hoc tests, *(P<0.05) or **(P<0.01) or ***(P<0.001).
Figure 7.

DHA suppressed protein expression of Pax6/HOXA2 and IHH signaling during hypertrophic differentiation of MSCs. A: Representative western blot images of Gli2, Gli3, Pax6, HOXA2 and GAPDH from MSCs in different groups (day 21 and 28). B: Quantification of normalized expression intensity of Gli2, Gli3, Pax6 and HOXA2 against GAPDH. The data in the figures represent the averages ± SD. Significant differences between the treatment and control groups are indicated in the graph with the detailed corresponding P-values from Student-Newman-Keuls post hoc tests, *(P<0.05) or **(P<0.01) or ***(P<0.001).
Discussion
DHA has been considered the best drug against severe and complicated malaria [29]. It was confirmed that DHA has protective effects on the respiratory system, digestive, and nervous systems [30-32]. Because of those factors, we hypothesized that DHA might also influence chondrogenesis and chondrocyte differentiation. A recent study found that DHA is effective against glioblastoma through inhibiting cancer cell proliferation by a pharmacological effect [33]. Additionally, DHA effectively inhibits osteoclastogenesis and prevents breast cancer-induced osteolysis [34].
In this study, we demonstrate that DHA had a negative effect on the chondrogenesis of MSCs. Several chondrocyte marker genes, such as Sox9 and Col2a1, were down-regulated by DHA treatment. We then examined several crucial signaling pathway genes and transcription factors to explore the molecular mechanisms. The mRNA expression of Nrf2 and Hey1 were increased in the presence of DHA. The results of western blot analysis were consistent with RT-PCR. Overexpression of Nrf2 significantly decreased mRNA expression of several chondrocyte related genes, including Col2a1 and osteopontin, which suggested that Nrf2 negatively regulates chondrogenesis [35]. In addition, RBPjκ-dependent Notch signaling regulates multiple processes during cartilage development, including chondrogenesis and chondrocyte hypertrophy. Notch targets Hes1 and Hes5 to inhibit chondrogenesis [36]. It has been demonstrated that decreased Nrf2 expression significantly dampens Notch1 expression in non-small cell lung cancer cells [37]. In other words, Nrf2 promotes activation of Notch signaling to suppress chondrogenesis. We determined that DHA up-regulated Nrf2 and subsequently activated Notch signaling. Hence, we concluded that DHA inhibited chondrogenesis of MSCs by up-regulating Nrf2 and Notch signaling.
Differentiation of chondrocytes towards hypertrophy is a natural process in endochondral ossification. However, hypertrophy of chondrocytes in tissue-engineered cartilage should be inhibited because hypertrophic chondrocytes secrete matrix metalloproteinases to disturb chondrocyte homeostasis [38]. Interestingly, we found that the expression of Runx2 and Col10a1 in MSCs was inhibited by DHA during chondrocyte hypertrophic differentiation. We then screened several crucial signaling pathway genes and transcription factors related to chondrocyte hypertrophy, and focused on Pax6/HOXA2 and IHH signaling. We found that expression levels of Gli3 and HOXA2 were increased while those of Gli2 and Pax6 were decreased following DHA treatment. A study confirmed that persistent HOXA2 expression in chondrogenic cells results in delayed cartilage hypertrophy [39], while another study found that overexpression of Pax6 reduces HOXA2 expression [40]. Therefore, expression of Pax6 inhibits HOXA2 expression to promote chondrocyte hypertrophy. Furthermore, activated Gli transcription factors enter the nucleus to regulate expression of genes related to chondrocyte hypertrophy in IHH signaling [41]. Gli2 up-regulates hypertrophic genes while Gli3 down-regulates hypertrophic genes [1]. With DHA treatment, the expression of Pax6 was decreased. Therefore, DHA reduced the inhibitory effect of Pax6 on HOXA2, thus inhibiting chondrocyte hypertrophy. DHA also inhibited Gli2 expression and promoted Gli3 expression in IHH signaling to suppress chondrocyte hypertrophy. As a consequence, we concluded that DHA inhibited chondrocyte hypertrophy through regulation of Pax6/HOXA2 and IHH signaling (Figure 8).
Figure 8.
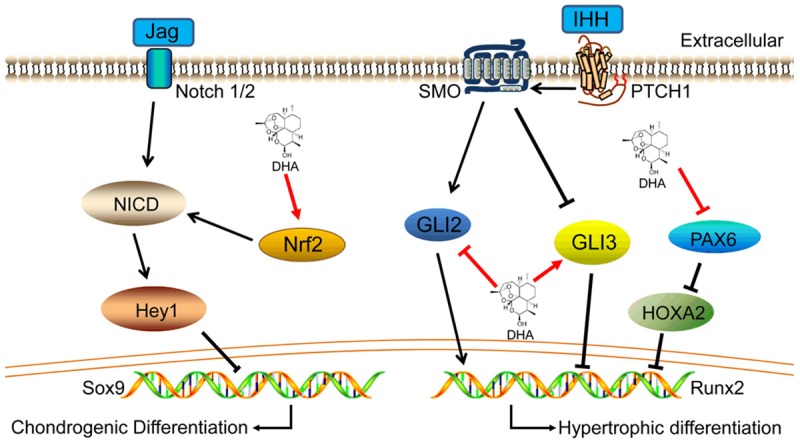
Schematic diagram of DHA function in chondrogenic and hypertrophic differentiation of MSCs. DHA promoted Nrf2 expression and then activated Notch signaling to inhibit chondrogenesis of MSCs. Besides, DHA inhibited Gli2 expression and promoted Gli3 expression in IHH signaling meanwhile DHA suppressed Pax6 expression and next reduced the inhibitory effect of Pax6 to HOXA2, thus delay chondrocyte hypertrophic differentiation.
The results of the present study demonstrate that DHA treatment inhibited chondrogenesis and chondrocyte hypertrophic differentiation of MSCs. To our knowledge, this is the only study to research the effect of DHA on MSCs chondrogenic differentiation. These data indicate that DHA should not be used during the chondrogenesis stage of endochondral ossification or in immature cartilage, due to its inhibiting effect on chondrogenesis. Because DHA might suppress degradation of cartilage matrix and potentially maintain chondrocyte homeostasis in clinical applications, it could be applied in tissue-engineered cartilage to reduce the formation of fibrocartilage.
Acknowledgements
This work was funded by grants from the Nature Science Foundation of China (81571893, 81271980), the National High-tech R&D Program of China (863 Program, 2015AA020315) and the Key project of Logistics Research Plan of PLA (BWS13C014).
Disclosure of conflict of interest
None.
References
- 1.Long F, Ornitz DM. Development of the endochondral skeleton. Cold Spring Harb Perspect Biol. 2013;5:a008334. doi: 10.1101/cshperspect.a008334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Colvin JS, Bohne BA, Harding GW, McEwen DG, Ornitz DM. Skeletal overgrowth and deafness in mice lacking fibroblast growth factor receptor 3. Nat Genet. 1996;12:390–397. doi: 10.1038/ng0496-390. [DOI] [PubMed] [Google Scholar]
- 3.Thorogood PV, Hinchliffe JR. An analysis of the condensation process during chondrogenesis in the embryonic chick hind limb. J Embryol Exp Morphol. 1975;33:581–606. [PubMed] [Google Scholar]
- 4.Kwon HJ, Lee GS, Chun H. Electrical stimulation drives chondrogenesis of mesenchymal stem cells in the absence of exogenous growth factors. Sci Rep. 2016;6:39302. doi: 10.1038/srep39302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kawakami Y, Rodriguez-Leon J, Izpisua Belmonte JC. The role of TGFbetas and Sox9 during limb chondrogenesis. Curr Opin Cell Biol. 2006;18:723–729. doi: 10.1016/j.ceb.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 6.Vega RB, Matsuda K, Oh J, Barbosa AC, Yang X, Meadows E, McAnally J, Pomajzl C, Shelton JM, Richardson JA, Karsenty G, Olson EN. Histone deacetylase 4 controls chondrocyte hypertrophy during skeletogenesis. Cell. 2004;119:555–566. doi: 10.1016/j.cell.2004.10.024. [DOI] [PubMed] [Google Scholar]
- 7.Arnold MA, Kim Y, Czubryt MP, Phan D, McAnally J, Qi X, Shelton JM, Richardson JA, Bassel-Duby R, Olson EN. MEF2C transcription factor controls chondrocyte hypertrophy and bone development. Dev Cell. 2007;12:377–389. doi: 10.1016/j.devcel.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 8.Zheng Q, Zhou G, Morello R, Chen Y, Garcia-Rojas X, Lee B. Type X collagen gene regulation by Runx2 contributes directly to its hypertrophic chondrocyte-specific expression in vivo. J Cell Biol. 2003;162:833–842. doi: 10.1083/jcb.200211089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li F, Lu Y, Ding M, Napierala D, Abbassi S, Chen Y, Duan X, Wang S, Lee B, Zheng Q. Runx2 contributes to murine Col10a1 gene regulation through direct interaction with its cis-enhancer. J Bone Miner Res. 2011;26:2899–2910. doi: 10.1002/jbmr.504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Quarto R, Campanile G, Cancedda R, Dozin B. Thyroid hormone, insulin, and glucocorticoids are sufficient to support chondrocyte differentiation to hypertrophy: a serum-free analysis. J Cell Biol. 1992;119:989–995. doi: 10.1083/jcb.119.4.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee CH, Cook JL, Mendelson A, Moioli EK, Yao H, Mao JJ. Regeneration of the articular surface of the rabbit synovial joint by cell homing: a proof of concept study. Lancet. 2010;376:440–448. doi: 10.1016/S0140-6736(10)60668-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Son HE, Kim TH, Jang WG. Curculactones A and B induced the differentiation of C3H10T1/2 and MC3T3-E1 cells to osteoblasts. Bioorg Med Chem Lett. 2017;27:1301–1303. doi: 10.1016/j.bmcl.2016.12.070. [DOI] [PubMed] [Google Scholar]
- 13.Tang Y, Qian SW, Wu MY, Wang J, Lu P, Li X, Huang HY, Guo L, Sun X, Xu CJ, Tang QQ. BMP4 mediates the interplay between adipogenesis and angiogenesis during expansion of subcutaneous white adipose tissue. J Mol Cell Biol. 2016;8:302–312. doi: 10.1093/jmcb/mjw019. [DOI] [PubMed] [Google Scholar]
- 14.Karystinou A, Roelofs AJ, Neve A, Cantatore FP, Wackerhage H, De Bari C. Yes-associated protein (YAP) is a negative regulator of chondrogenesis in mesenchymal stem cells. Arthritis Res Ther. 2015;17:147. doi: 10.1186/s13075-015-0639-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keating A. Mesenchymal stromal cells: new directions. Cell Stem Cell. 2012;10:709–716. doi: 10.1016/j.stem.2012.05.015. [DOI] [PubMed] [Google Scholar]
- 16.Golpanian S, Wolf A, Hatzistergos KE, Hare JM. Rebuilding the damaged heart: mesenchymal stem cells, cell-based therapy, and engineered heart tissue. Physiol Rev. 2016;96:1127–1168. doi: 10.1152/physrev.00019.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thompson EM, Matsiko A, Kelly DJ, Gleeson JP, O’Brien FJ. An endochondral ossificationbased approach to bone repair: chondrogenically primed mesenchymal stem cell-laden scaffolds support greater repair of criticalsized cranial defects than osteogenically stimulated constructs in vivo. Tissue Eng Part A. 2016;22:556–567. doi: 10.1089/ten.TEA.2015.0457. [DOI] [PubMed] [Google Scholar]
- 18.Saxena V, Kim M, Keah NM, Neuwirth AL, Stoeckl BD, Bickard K, Restle DJ, Salowe R, Wang MY, Steinberg DR, Mauck RL. Anatomic mesenchymal stem cell-based engineered cartilage constructs for biologic total joint replacement. Tissue Engineering Part A. 2016;22:386–395. doi: 10.1089/ten.tea.2015.0384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chang NJ, Lin CC, Shie MY, Yeh ML, Li CF, Liang PI, Lee KW, Shen PH, Chu CJ. Positive effects of cell-free porous PLGA implants and early loading exercise on hyaline cartilage regeneration in rabbits. Acta Biomater. 2015;28:128–137. doi: 10.1016/j.actbio.2015.09.026. [DOI] [PubMed] [Google Scholar]
- 20.Duan L, Liang Y, Ma B, Zhu W, Wang D. Epigenetic regulation in chondrocyte phenotype maintenance for cell-based cartilage repair. Am J Transl Res. 2015;7:2127–2140. [PMC free article] [PubMed] [Google Scholar]
- 21.Kume A, Anh DT, Shichiri M, Ishida N, Suzuki H. Probucol dramatically enhances dihydroartemisinin effect in murine malaria. Malar J. 2016;15:472. doi: 10.1186/s12936-016-1532-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Q, Weina P. Artesunate: The best drug in the treatment of severe and complicated malaria. Pharmaceuticals (Basel) 2010;3:2322–2332. doi: 10.3390/ph3072322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Y, Wang Y, Kong R, Xue D, Pan S, Chen H, Sun B. Dihydroartemisinin suppresses pancreatic cancer cells via a microRNA-mRNA regulatory network. Oncotarget. 2016;7:62460–62473. doi: 10.18632/oncotarget.11517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ho WE, Peh HY, Chan TK, Wong WS. Artemisinins: pharmacological actions beyond anti-malarial. Pharmacol Ther. 2014;142:126–139. doi: 10.1016/j.pharmthera.2013.12.001. [DOI] [PubMed] [Google Scholar]
- 25.Zhao YG, Wang Y, Guo Z, Gu AD, Dan HC, Baldwin AS, Hao W, Wan YY. Dihydroartemisinin ameliorates inflammatory disease by its reciprocal effects on Th and regulatory T cell function via modulating the mammalian target of rapamycin pathway. J Immunol. 2012;189:4417–4425. doi: 10.4049/jimmunol.1200919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zheng W, Chong CM, Wang H, Zhou X, Zhang L, Wang R, Meng Q, Lazarovici P, Fang J. Artemisinin conferred ERK mediated neuroprotection to PC12 cells and cortical neurons exposed to sodium nitroprusside-induced oxidative insult. Free Radic Biol Med. 2016;97:158–167. doi: 10.1016/j.freeradbiomed.2016.05.023. [DOI] [PubMed] [Google Scholar]
- 27.Dou C, Ding N, Xing J, Zhao C, Kang F, Hou T, Quan H, Chen Y, Dai Q, Luo F, Xu J, Dong S. Dihydroartemisinin attenuates lipopolysaccharide-induced osteoclastogenesis and bone loss via the mitochondria-dependent apoptosis pathway. Cell Death Dis. 2016;7:e2162. doi: 10.1038/cddis.2016.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mueller MB, Tuan RS. Functional characterization of hypertrophy in chondrogenesis of human mesenchymal stem cells. Arthritis Rheum. 2008;58:1377–1388. doi: 10.1002/art.23370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bahamontes-Rosa N, Gomez-Lorenzo MG, Lelievre J, Rodriguez Alejandre A, Almela MJ, Lozano S, Herreros E, Gamo FJ. A novel validated assay to support the discovery of new anti-malarial gametocytocidal agents. Malar J. 2016;15:385. doi: 10.1186/s12936-016-1429-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wei M, Xie X, Chu X, Yang X, Guan M, Wang D. Dihydroartemisinin suppresses ovalbumininduced airway inflammation in a mouse allergic asthma model. Immunopharmacol Immunotoxicol. 2013;35:382–389. doi: 10.3109/08923973.2013.785559. [DOI] [PubMed] [Google Scholar]
- 31.Sun H, Meng X, Han J, Zhang Z, Wang B, Bai X, Zhang X. Anti-cancer activity of DHA on gastric cancer--an in vitro and in vivo study. Tumour Biol. 2013;34:3791–3800. doi: 10.1007/s13277-013-0963-0. [DOI] [PubMed] [Google Scholar]
- 32.Fishwick J, Edwards G, Ward SA, McLean WG. Binding of dihydroartemisinin to differentiating neuroblastoma cells and rat cortical homogenate. Neurotoxicology. 1998;19:405–412. [PubMed] [Google Scholar]
- 33.Kim SH, Kang SH, Kang BS. Therapeutic effects of dihydroartemisinin and transferrin against glioblastoma. Nutr Res Pract. 2016;10:393–397. doi: 10.4162/nrp.2016.10.4.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Feng MX, Hong JX, Wang Q, Fan YY, Yuan CT, Lei XH, Zhu M, Qin A, Chen HX, Hong D. Dihydroartemisinin prevents breast cancer-induced osteolysis via inhibiting both breast caner cells and osteoclasts. Sci Rep. 2016;6:19074. doi: 10.1038/srep19074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hinoi E, Takarada T, Fujimori S, Wang L, Iemata M, Uno K, Yoneda Y. Nuclear factor E2 p45-related factor 2 negatively regulates chondrogenesis. Bone. 2007;40:337–344. doi: 10.1016/j.bone.2006.08.016. [DOI] [PubMed] [Google Scholar]
- 36.Rutkowski TP, Kohn A, Sharma D, Ren Y, Mirando AJ, Hilton MJ. HES factors regulate specific aspects of chondrogenesis and chondrocyte hypertrophy during cartilage development. J Cell Sci. 2016;129:2145–2155. doi: 10.1242/jcs.181271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao Q, Mao A, Yan J, Sun C, Di C, Zhou X, Li H, Guo R, Zhang H. Downregulation of Nrf2 promotes radiation-induced apoptosis through Nrf2 mediated Notch signaling in non-small cell lung cancer cells. Int J Oncol. 2016;48:765–773. doi: 10.3892/ijo.2015.3301. [DOI] [PubMed] [Google Scholar]
- 38.Gomez-Leduc T, Hervieu M, Legendre F, Bouyoucef M, Gruchy N, Poulain L, de Vienne C, Herlicoviez M, Demoor M, Galera P. Chondrogenic commitment of human umbilical cord blood-derived mesenchymal stem cells in collagen matrices for cartilage engineering. Sci Rep. 2016;6:32786. doi: 10.1038/srep32786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Massip L, Ectors F, Deprez P, Maleki M, Behets C, Lengele B, Delahaut P, Picard J, Rezsohazy R. Expression of Hoxa2 in cells entering chondrogenesis impairs overall cartilage development. Differentiation. 2007;75:256–267. doi: 10.1111/j.1432-0436.2006.00132.x. [DOI] [PubMed] [Google Scholar]
- 40.Kayam G, Kohl A, Magen Z, Peretz Y, Weisinger K, Bar A, Novikov O, Brodski C, Sela-Donenfeld D. A novel role for Pax6 in the segmental organization of the hindbrain. Development. 2013;140:2190–2202. doi: 10.1242/dev.089136. [DOI] [PubMed] [Google Scholar]
- 41.Ma RS, Zhou ZL, Luo JW, Zhang H, Hou JF. The Ihh signal is essential for regulating proliferation and hypertrophy of cultured chicken chondrocytes. Comp Biochem Physiol B Biochem Mol Biol. 2013;166:117–122. doi: 10.1016/j.cbpb.2013.07.010. [DOI] [PubMed] [Google Scholar]


